i. Summary
Planarians have gained a well-deserved reputation as an excellent model organism for research on the biology of adult stem cells and their role in regeneration. Though less widely recognized, these animals also offer many advantages for investigating mechanisms and functions of programmed cell death in self-renewing tissues. Apoptosis complements stem cell division during physiological cell turnover and constitutes a prominent feature of the tissue remodeling process that restores anatomical scale and proportion during regeneration. One technical advantage to studying apoptosis in planarians is the availability of a whole-mount TUNEL assay for visualizing dying cells throughout the animal. Here, we provide a detailed protocol for this assay that is likely to benefit researchers investigating planarian cell death in either physiological or pathological contexts.
Keywords: TUNEL, Planarian, Apoptosis, Cell Death, DNA Fragmentation
1. Introduction
First developed in 1992, the TUNEL (Terminal deoxynucleotidyl transferase-mediated dUTP Nick End Labeling) assay (1) has become one of the most widely used experimental approaches for the study of apoptotic cell death. This technique employs the terminal transferase enzyme (TdT) to catalyze template-independent addition of modified deoxynucleotides to the 3′ ends of DNA strand breaks; the incorporated nucleotides are then directly visualized (e.g., using a fluorochrome-conjugated dUTP) or indirectly detected using an antibody to the chemical modification. Because DNA fragmentation is a hallmark of apoptosis (2), this approach has broad potential for the identification of apoptotic cells in situ. Indeed, it has successfully been applied to a wide variety of organisms and tissues, ranging from alligator teeth (3) to zebrafish tails (4). As with all assays, however, thorough optimization of experimental protocols is necessary to ensure its reliability. This is particularly important for TUNEL, as fixation and labeling variables can strongly impact its efficacy, and false-positive and false-negative results are frequently encountered (5, 6).
We previously developed and validated a whole-mount TUNEL assay for planarians that allowed us to characterize the spatiotemporal dynamics of cell death during tissue homeostasis, regeneration, and degrowth (7). Together with other approaches such as Annexin V labeling, this has enabled significant advances in our understanding of both the molecular mechanisms of planarian apoptosis and its regulation (8–10). However, many questions about planarian cell death have yet to be answered. As TUNEL will likely remain integral to research in this field, we provide detailed instructions for our whole-mount assay here. Some investigators have reported technical difficulties with the protocol (B. Peterson, personal communication), but careful adherence to instructions ensures reliable detection of dying cells by even novice planarian researchers. We also present methods for conducting TUNEL on tissue sections from paraffin-embedded animals and for combining whole-mount TUNEL with immunostaining of endogenous antigens.
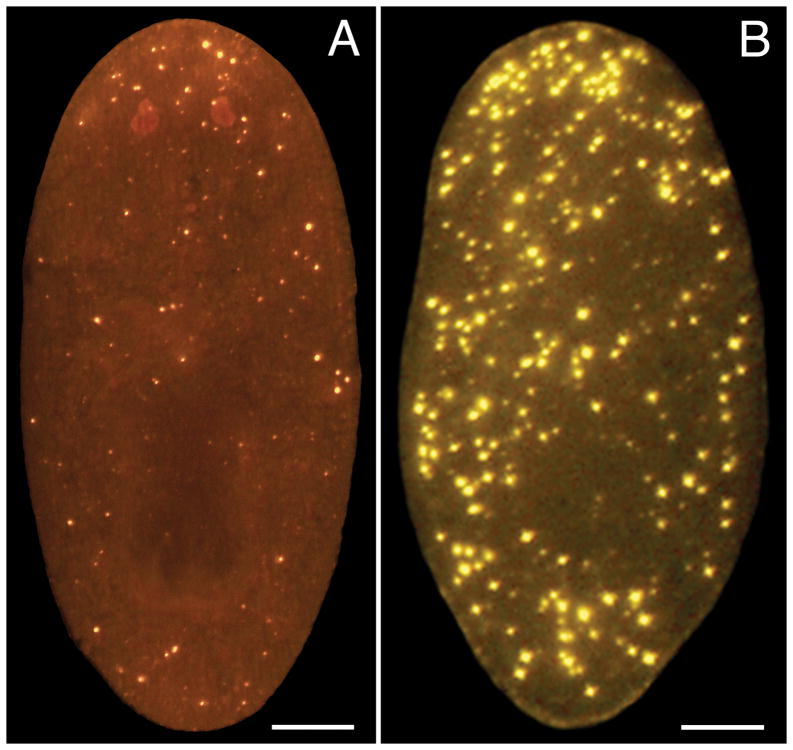
The TUNEL assay is completed in four main steps: 1) a fixation/bleaching step in which animals are killed in N-acetylcysteine, fixed in formaldehyde, and bleached in hydrogen peroxide; 2) a TdT reaction that incorporates digoxigenin-modified dUTP; 3) detection of the incorporated nucleotide with an anti-digoxigenin antibody; and 4) imaging and analysis of labeled animals. We focus here on two protocols that have generated the best results in our experience – an assay utilizing the ApopTag® Red In Situ Apoptosis Detection Kit from EMD Millipore (see Note 1) and a slightly longer, but less expensive assay utilizing individually purchased reaction components and tyramide signal amplification (TSA). We obtain an excellent signal-to-noise ratio with the ApopTag® Red Kit (Fig. 1A), but the use of TSA enables strong signal amplification, potentially affording greater sensitivity (Fig. 1B).
Fig. 1.
Whole-mount TUNEL assays. Images show representative results for intact, 7-day-starved animals labeled with (A) the ApopTag® Red Kit using DIG-dUTP and a rhodamine-conjugated anti-DIG antibody, and (B) 10 units TdT, DIG-dUTP, anti-DIG-POD, and Cy3 TSA. Scale bars = 100 um.
2. Materials
2.1 Fixation/Bleaching
PBS: 1X Phosphate-Buffered Saline
PBST: 0.3% Triton X-100 in PBS
5% NAC: 5% (w/v) N-Acetylcysteine in PBS (prepared fresh)
4% FA: 4% Formaldehyde Solution in PBST (prepared fresh)
6% H2O2: 6% Hydrogen Peroxide in PBST (prepared fresh by dilution of 30% stock)
2.2 TdT Reaction
Individual Reaction Components (or commercially available TUNEL kit – see Note 1): Terminal Transferase (with manufacturer-supplied reaction buffer)
2.5 mM CoCl2 (if not included in reaction buffer)
1 mM Digoxigenin-11-dUTP (DIG-dUTP)
1 mM dATP
2.3 DIG-dUTP Detection
2.4 Imaging and Analysis
Microscope Slides and Coverslips
Mounting Media for Fluorescence Microscopy (e.g., VECTASHIELD®, Vector Labs)
ImageJ (or alternative image-analysis software, if desired – see Note 4)
3. Methods
There are three critical variables to bear in mind when designing a TUNEL experiment – animal size (see Note 5), feeding schedule (see Note 6), and regenerative status (see Note 7). Each of these variables has major quantitative and qualitative impacts on TUNEL results and should be carefully considered prior to beginning an experiment utilizing the procedures described below.
We employ a common set of fixation and bleaching steps for animals to be labeled with either the ApopTag® Red Kit or individually prepared reagents. Both assays begin with a TdT reaction incorporating DIG-dUTP. However, the ApopTag® Red Kit subsequently employs a direct detection approach involving a rhodamine-conjugated anti-DIG antibody, whereas the second assay uses a peroxidase-conjugated anti-DIG antibody (anti-DIG-POD) and TSA. Other detection strategies are also possible; the opportunity to combine various chemically modified deoxynucleotides, fluorochrome- or enzyme-conjugated antibodies, and detection reagents affords substantial flexibility (see Note 3).
3.1 Fixation/Bleaching
Select animals for fixation and place in a petri dish or 15 mL conical tube (see Note 8).
Replace planarian water with 5% NAC and incubate with periodic, gentle agitation (if animals are in a petri dish) or on a Nutator (if animals are in a 15 mL conical tube) for 10 minutes at room temperature. NAC is a mucolytic agent that kills animals on contact and removes their mucus coat (11).
Remove 5% NAC and replace with 4% FA. Incubate animals for 20 minutes at room temperature, with periodic, gentle agitation or on a Nutator, as in Step 2 above. We have also obtained good results with Carnoy’s fixative, though its use in TUNEL is discouraged by some investigators (see Note 9). Following fixation, remove fixative solution and rinse animals in PBST.
Remove PBST and incubate animals overnight in 6% H2O2 in direct light at room temperature. Then, remove 6% H2O2 and rinse animals in PBST. Omitting the bleach step substantially diminishes visible labeling. Animals may be stored in PBST at 4°C prior to beginning the TdT reaction (see Note 10).
Transfer animals to microfuge tubes for labeling. We typically process 2–5 animals per microfuge tube (minor decreases in labeling efficacy can occur with higher numbers, even when reagent volumes are scaled accordingly). Rinse specimens once in PBS prior to starting the TdT reaction.
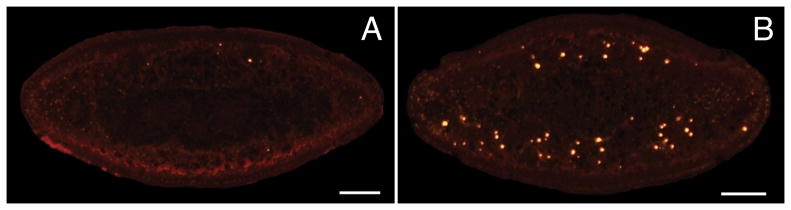
We have successfully applied TUNEL to tissue sections from paraffin-embedded planarians (Fig. 2). For instructions on embedding and sectioning bleached animals, see Note 11.
Fig. 2.
TUNEL in tissue sections. 5 um transverse sections from paraffin-embedded animals were labeled with the ApopTag® Red Kit. (A) Control. (B) 24 hours after a 6,000 rad dose of gamma irradiation, previously shown to induce a pronounced increase in apoptosis (7). Scale bars = 100 um.
3.2 TdT Reaction
Prepare TdT reaction mix. For the ApopTag® Red Kit, mix 6 ul TdT enzyme and 14 ul reaction buffer for each microfuge tube of animals (or for each slide) to be labeled. For individually purchased components, first prepare a working solution of DIG-dUTP by diluting the 1 mM stock 1:50 in 1 mM dATP. Then, mix the following reagents in the indicated volumes for each microfuge tube of animals (or for each slide) to be labeled: 0.8 ul DIG-dUTP working solution, 2 ul 10X reaction buffer (from manufacturer), 2 ul 2.5 mM CoCl2 (if not included in reaction buffer), 10 units TdT, and water to a final volume of 20 ul.
Aspirate PBS and add 20 ul TdT reaction mix to each microfuge tube. Incubate for 4 hours at 37°C. For tissue sections, distribute 20 ul TdT reaction mix evenly over the surface of the specimen using a pipet tip, cover with a Parafilm coverslip (small square of Parafilm cut to extend slightly beyond edges of the specimen), and complete 4-hour 37°C incubation in a humidified chamber (see Note 12). Rinse twice in PBST to stop the TdT reaction (slides can be rinsed in Coplin jars). Immunostaining of endogenous antigens (e.g., phospho-histone H3) can be initiated at this step (see Note 13).
3.3 DIG-dUTP Detection
For the ApopTag® Red Kit, mix 47 ul rhodamine-conjugated antibody with 53 ul blocking solution for every 5 microfuge tubes to be processed. If using individually purchased components, prepare a 1:500 dilution of anti-DIG-POD in PBSTH (or alternative antibody solution – see Note 2). Add 20 ul of antibody solution to each microfuge tube (for tissue sections, distribute 20 ul over the surface of each specimen using a pipet tip, cover with a Parafilm coverslip, and transfer to a humidified chamber – see Note 12). Incubate for 4 hours at room temperature if using the ApopTag® Red Kit, or overnight at room temperature for the anti-DIG-POD antibody.
Following the antibody incubation step, animals should be washed for 2 hours to overnight in several changes of PBST to reduce nonspecific labeling. Tissue sections can be washed in PBST in Coplin jars.
Animals or tissue sections stained with the ApopTag® Red Kit can be directly mounted in fluorescence microscopy mounting media. TUNEL-positive cells in specimens stained with anti-DIG-POD antibody can be visualized by TSA. Specifically, specimens should be incubated for 10 minutes at room temperature in 0.003% H2O2 in PBST, plus 1:500 Cy3-tyramide or 1:3000 FITC-tyramide, and then washed for 2–4 hours at room temperature in PBST before mounting in fluorescence microscopy mounting media.
3.4 Imaging and Analysis
Whole-mounted specimens can be imaged with a fluorescence stereomicroscope to visualize apoptotic cells throughout the animal or with a confocal microscope to visualize apoptotic cells in specific focal planes. Tissue sections can be imaged with either a fluorescence stereomicroscope or a compound fluorescence microscope. Quantitative analysis of TUNEL results can be conducted with a variety of image analysis programs (see Note 4).
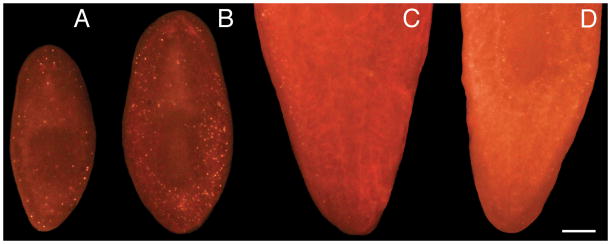
Fig. 3.
Whole-mount TUNEL efficacy declines sharply with increasing animal size. Small (A, B) and large (C, D) animals were fixed and stained using the ApopTag® Red Kit. All animals were imaged under identical conditions at the same magnification (Scale bar = 200 um). Only the tail portions of the large animals are shown here, but the results are also representative for anterior labeling. (A, C) Controls. (B, D) 24 hours after a 6,000 rad dose of gamma irradiation.
Acknowledgments
This work was supported by the New Hampshire IDeA Network of Biological Research Excellence (NH-INBRE) with grants from the National Center for Research Resources (5P20RR030360-03) and the National Institute of General Medical Sciences (8P20GM103506-03), National Institutes of Health. We thank Amber Poirier for conducting irradiations.
Footnotes
Many life sciences manufacturers offer TUNEL assay kits containing all necessary reagents for completing the TdT reaction and antibody incubation/detection step(s). We have obtained particularly good results with the ApopTag® Red In Situ Apoptosis Detection Kit from EMD Millipore (Fig. 1A), which uses DIG-dUTP and a rhodamine-conjugated anti-digoxigenin antibody.
We have used rhodamine-conjugated (Fig. 1A), peroxidase-conjugated (Fig. 1B), and alkaline phosphatase-conjugated anti-digoxigenin antibodies in the whole-mount TUNEL assay with good results. Selection of a suitable antibody will depend on a number of factors, including whether TUNEL will be combined with immunostaining of an endogenous antigen using another fluorophore (see Note 13).
TSA with Cy3-tyramide (Fig. 1B) or FITC-tyramide generates strong fluorescent labeling. We have also used nitro-blue tetrazolium chloride/5-bromo-4-chloro-3′-indolyphosphate p-toluidine salt (NBT-BCIP), commonly employed in colorimetric detection of whole-mount in situ hybridizations (11), to visualize TUNEL-positive cells. Many other detection approaches are possible.
It is important to note that TUNEL provides only limited insight into absolute rates of programmed cell death. Not only do we lack information about the false-positive and false-negative rates for this particular protocol, but DNA fragmentation is a late event in apoptosis (12), so cells initiating apoptosis go undetected. Nevertheless, quantitative analysis of TUNEL results can provide valuable information about relative differences in the rate of apoptosis between experimental conditions. Use of an automated image analysis program can be particularly helpful in this regard. We have used the freely available ImageJ program, as well as commercially available software (7), for quantifying whole-mount TUNEL results. While detailed instructions for these approaches are beyond the scope of this article, we emphasize the importance of utilizing consistent parameters across experimental conditions for both image acquisition and analysis, as TUNEL-positive cells can vary considerably with respect to apparent size and fluorescence intensity (Fig. 1).
Labeling efficacy for whole-mount TUNEL declines precipitously for animals above approximately 2 mm (post-fixation) in length (Fig. 3). We attribute this to permeability issues, but common permeabilization techniques such as Proteinase K treatment (11), SDS treatment (11), reduction (11), or microwaving animals in sodium citrate buffer (13) have not improved labeling of larger animals in our hands (other investigators have reported a benefit to Proteinase K treatment and reduction; 10). Even amputation of larger animals immediately after fixation leads to only a very narrow region of enhanced labeling efficacy within the immediate vicinity of the amputation site. In any event, we obtain excellent results with planarians of approximately 1–1.5 mm (postfixation) in length and routinely maintain stocks of animals within this size range for our TUNEL experiments.
Levels of apoptosis increase during prolonged starvation (7), so it is imperative that all control and experimental animals be fed on the same schedule. We typically fix animals for TUNEL at 1 week post-feeding.
Levels of apoptosis change dramatically during regeneration (7). Animals that have fissioned or that have been amputated within 2 weeks prior to fixation should be avoided, unless regenerative cell death responses are to be analyzed.
Small numbers of animals can be fixed in a petri dish, but a 15 mL conical tube is recommended for greater than 10 animals. To avoid clumping, disperse animals in the petri dish or keep animals in constant motion in the conical tube using a Nutator.
Cross-linking fixatives are generally recommended for TUNEL in the scientific literature because they are thought to prevent extraction of cleaved DNA in apoptotic cells by crosslinking low molecular weight DNA fragments to other cellular components (14). Carnoy’s has been reported to increase background labeling in some instances (e.g., 15), though we have not encountered obvious issues with false-positive artifacts in Carnoy’s-fixed planarians.
We have maintained animals for up to 2 weeks in PBST at 4°C with no apparent decrease in TUNEL efficacy.
For labeling tissue sections, we paraffin embed and section animals according to standard histology protocols. Briefly, animals are fixed and bleached as for whole-mount labeling and then dehydrated through an ethanol series (1 minute each in 40%, 70%, 90%, 95%, and 100% ethanol at room temperature). A final 18-minute incubation in 100% ethanol is conducted at 45°C. Dehydrated animals are incubated in 3 changes of xylene (2 X 1 minute at room temperature, followed by 14 minutes at 45°C) and 3 changes of melted paraffin (2 X 1 minute and then 14 minutes, all at 65°C). Forceps are then used to transfer animals to a mold filled with melted paraffin. After the paraffin hardens, the block containing the embedded animal(s) can be directly inserted into a microtome, or cut and reoriented in a second mold if necessary to correctly position animals for sectioning. We have obtained good results with 5 um – 10 um sections, which are floated onto the surface of clean glass slides (we find silanized slides promote good adhesion without introducing background fluorescence). Finally, slides are dried for 24 hours, deparaffinized in 3 changes of xylene (3 minutes each at room temperature), and rehydrated through an ethanol series (2 X 1 minute in 100% ethanol and then 1 minute each in 95%, 70%, and 40% ethanol, all at room temperature). Following the final ethanol incubation, specimens should be rinsed (and stored for up to 2 hours if desired) in PBST at room temperature prior to beginning the TdT reaction.
A plastic freezer box for microfuge tubes filled approximately halfway with water makes an excellent humidified chamber. Alternatively, line a covered dish with damp paper towels and overlay with slide supports (e.g., serological pipets).
We have combined whole-mount TUNEL with immunostaining of phospho-histone H3 (H3P) using sequential antibody incubation and TSA steps. After the TdT reaction, animals are incubated overnight at room temperature in 1:300 rabbit anti-H3P antibody in PBSTH, washed in several changes of PBST over 2-4 hours on a Nutator, incubated overnight at room temperature in 1:500 anti-DIG-POD antibody in PBSTH, and washed again in PBST. An initial TSA step (see Subheading 3.3.3) with Cy3-tyramide is used to detect TUNEL-positive cells. Then, animals are washed again in PBST and incubated for 1 hour at room temperature in 3% H2O2 in PBST to quench remaining peroxidase activity. Finally, animals are washed in PBST, incubated overnight at room temperature in 1:300 goat anti-rabbit horseradish peroxidase secondary antibody in PBSTH, washed in PBST, subjected to a second TSA step (see Subheading 3.3.3) with FITC-tyramide to detect H3P-positive cells, and washed a final time in PBST prior to microscopic analysis.
Contributor Information
Brad Stubenhaus, Department of Biology, Keene State College, 229 Main Street, Keene, NH 03435.
Jason Pellettieri, Department of Biology, Keene State College, 229 Main Street, Keene, NH 03435.
References
- 1.Gavrieli Y, Sherman Y, Ben-Sasson SA. Identification of programmed cell death in situ via specific labeling of nuclear DNA fragmentation. J Cell Biol. 1992;119:493–501. doi: 10.1083/jcb.119.3.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nagata S. Apoptotic DNA fragmentation. Exp Cell Res. 2000;256:12–18. doi: 10.1006/excr.2000.4834. [DOI] [PubMed] [Google Scholar]
- 3.Wu P, Wu X, Jian TX, Elsey RM, Temple BL, Divers SJ, Glenn TC, Yuan K, Chen MH, Widelitz RB, Chuong CM. Specialized stem cell niche enables repetitive renewal of alligator teeth. Proc Natl Acad Sci USA. 2013;110:E2009–E20018. doi: 10.1073/pnas.1213202110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Robu ME, Larson JD, Nasevicius A, Beiraghi S, Brenner C, Farber SA, Ekker SC. p53 activation by knockdown technologies. PLoS Genet. 2007;3:e78. doi: 10.1371/journal.pgen.0030078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saraste A. Morphologic criteria and detection of apoptosis. Herz. 1999;24:189–195. doi: 10.1007/BF03044961. [DOI] [PubMed] [Google Scholar]
- 6.Walker JA, Quirke P. Viewing apoptosis through a ‘TUNEL’. J Pathol. 2001;195:275–276. doi: 10.1002/path.979. [DOI] [PubMed] [Google Scholar]
- 7.Pellettieri J, Fitzgerald P, Watanabe S, Mancuso J, Green DR, Sánchez Alvarado A. Cell death and tissue remodeling in planarian regeneration. Dev Biol. 2010;338:76–85. doi: 10.1016/j.ydbio.2009.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bender CE, Fitzgerald P, Tait SW, Llambi F, McStay GP, Tupper DO, Pellettieri J, Sánchez Alvarado A, Salvesen GS, Green DR. Mitochondrial pathway of apoptosis is ancestral in metazoans. Proc Natl Acad Sci USA. 2012;109:4904–4909. doi: 10.1073/pnas.1120680109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gaviño MA, Wenemoser D, Wang IE, Reddien PW. Tissue absence initiates regeneration through Follistatin-mediated inhibition of Activin signaling. Elife. 2013;2:e00247. doi: 10.7554/eLife.00247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Almuedo-Castillo M, Crespo X, Seebeck F, Bartscherer K, Salò E, Adell T. JNK controls the onset of mitosis in planarian stem cells and triggers apoptotic cell death required for regeneration and remodeling. PLoS Genet. 2014;10:e1004400. doi: 10.1371/journal.pgen.1004400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pearson BJ, Eisenhoffer GT, Gurley KA, Rink JC, Miller DE, Sánchez Alvarado A. A formaldehyde-based whole-mount in situ hybridization method for planarians. Dev Dyn. 2009;238:443–450. doi: 10.1002/dvdy.21849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kyrylkova K, Kyryachenko S, Leid M, Kioussi C. Detection of apoptosis by TUNEL assay. Methods Mol Biol. 2012;887:41–47. doi: 10.1007/978-1-61779-860-3_5. [DOI] [PubMed] [Google Scholar]
- 13.Labat-Moleur F, Guillermet C, Lorimier P, Robert C, Lantuejoul S, Brambilla E, Negoescu A. TUNEL apoptotic cell detection in tissue sections: critical evaluation and improvement. J Histochem Cytochem. 1998;46:327–334. doi: 10.1177/002215549804600306. [DOI] [PubMed] [Google Scholar]
- 14.Darzynkiewicz Z, Galkowski D, Zhao H. Analysis of apoptosis by cytometry using TUNEL assay. Methods. 2008;44:250–254. doi: 10.1016/j.ymeth.2007.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sasano H, Yamaki H, Nagura H. Detection of apoptotic cells in cytology specimens: an application of TdT-mediated dUTP-biotin nick end labeling to cell smears. Diagn Cytopathol. 1998;18:398–402. doi: 10.1002/(sici)1097-0339(199806)18:6<398::aid-dc3>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]