Abstract
Objectives
The objective of this systematic review is to identify and summarise studies which examine epigenetic biomarkers in patients with Barrett’s oesophagus (BO) and their association with progression to oesophageal adenocarcinoma (OADC). BO is a precursor lesion for OADC. There is no clinical test to predict patients who are likely to progress to OADC. An epigenetic biomarker could predict patients who are at high risk of progression from BO to OADC which could facilitate earlier diagnosis and spare those unlikely to develop cancer from regular invasive surveillance endoscopy.
Setting
A systematic search was conducted of the following databases: MEDLINE, MEDLINE in Process, EMBASE, Cochrane Central, ISI Conference Proceedings Citation Index and the British Library’s ZETOC. Studies were conducted in secondary and tertiary care settings.
Participants
All studies measuring epigenetic change in patients over 18 years old who progressed from non-dysplastic BO to OADC were included. Genetic, in vitro and studies which did not measure progression in the same patient cohort were excluded. Study inclusion and risk of bias of individual eligible studies were assessed in duplicate by two reviewers using a modified Quality in Prognostic Studies tool.
Results
14 studies met the inclusion criteria. 42 epigenetic markers were identified, and 5 studies developed models aiming to predict progression to OADC.
Conclusions
The evidence from this systematic review is suggestive of a role for p16 as an epigenetic biomarker for the progression of BO to OADC.
Prospero number
CRD42016038654.
Keywords: oesophageal disease, adult surgery, gastrointestinal tumours
Strengths and limitations of this study.
Systematic review conducted following strict Preferred Reporting Items for Systematic Reviews and Meta-Analyses guidelines.
Systematic and reproducible methodology using two independent reviewers.
All mechanisms of epigenetic change included.
Limited meta-analysis resulting from lack of standardisation between studies.
Small patient numbers in the included studies.
Introduction
Barrett’s oesophagus (BO) is defined as an oesophagus in which any portion of the normal distal squamous epithelial lining is replaced by metaplastic columnar epithelium which is clearly visible endoscopically (≥1 cm) above the gastro-oesophageal junction and confirmed histopathologically from oesophageal biopsies.1 BO arises due to long-standing gastro-oesophageal reflux disease (GORD) and chronic inflammation and is a precursor lesion for oesophageal adenocarcinoma (OADC) with progression through the metaplasia-dysplasia-carcinoma sequence.2 The likelihood of developing OADC is increased 1.7 times in patients with GORD, increasing to 10.6 times with BO.3 The incidence of OADC has risen in parallel with increasing obesity and GORD in Western populations.4 Patients with OADC who are diagnosed at an early disease stage benefit from much improved 5-year survival rates of up to 39%5 in comparison to less than 13% with invasive late stage lesions,6 highlighting the importance of early diagnosis and treatment.
Currently, there is no robust way of predicting which patients with BO will progress to OADC. Clinical and histological information is currently the only tools at the clinician’s disposal to aid early detection of OADC. The British Society of Gastroenterology recommends endoscopic surveillance of patients with BO, and the American College of Gastroenterology endorses screening of high-risk patients for BO.1 7 Endoscopic surveillance is invasive and expensive, and despite rigorous biopsy protocols, dysplasia and early cancers can be missed. A recent meta-analysis published in 2012 demonstrated lower risk for progression of non-dysplastic BO (NDBO) to OADC than previously reported with a pooled 0.33% (95% CI 0.28% to 0.38%) annual incidence of OADC in patients with NDBO.8 The annual incidence rate of OADC for patients with BO with high-grade dysplasia (HGD) is 7%–19%.9–11
Epigenetics is an emerging field which describes mechanisms of alteration of gene regulation and expression without changing the genetic code.12 The most widely recognised mechanisms of epigenetic change are covalent modifications and altered gene expression by non-coding RNAs. Covalent modifications alter the structure of DNA and include DNA methylation and histone modification.12 Mechanisms of epigenetic change are discussed in more detail in the protocol for this systematic review.13
Epigenetic changes in ulcerative colitis (UC) are well described14–17 and have been shown to occur before neoplasia occurs at an early stage of UC-associated carcinogenesis.18 UC-associated carcinomas progress in a similar fashion to OADC as a result of chronic inflammation through the metaplasia-dysplasia-adenocarcinoma sequence.19 The Enhanced Neoplasia Detection and Cancer Prevention in Chronic Colitis trial is investigating whether a panel of methylated biomarkers detected in endoscopic biopsy samples can be used as a tool in conjunction with screening colonoscopy to help risk stratify patients who are at higher risk of progressing to carcinoma.20 In light of this, there is a need to consolidate the literature on epigenetic changes in Barrett’s carcinogenesis to determine if such changes provide a method of risk stratifying patients who are at risk of progression to OADC.
A scoping search was performed using MEDLINE, the Cochrane Library and internet sources to identify any systematic reviews or meta-analyses on epigenetic biomarkers in BO and oesophageal cancer (OC). Nine systematic reviews and meta-analyses were identified21–29 which included mixed patient populations with OADC and oesophageal squamous cell carcinoma with only three reviews incorporating patients with BO.24 25 29 Seven reviews concentrated on a single type of epigenetic alteration with four investigating DNA methylation21–23 and three looking at micro RNA (miRNA) expression.25–27 The remaining two reviews investigated genetic alterations in progression of BO to OADC.28 29 No systematic reviews drawing together all aspects of epigenetic change within the field of Barrett’s carcinogenesis were identified.
Aim
To identify and summarise studies which examine epigenetic biomarkers in patients with BO and their association with progression to OADC.
Methods
Details of the methodology were registered on PROSPERO (CRD42016038654) and have also been published.13 A summary is reported here.
Patient and public involvement
This research question was developed to address the issue of BO surveillance. The priority for patients is early diagnosis of OADC and accurate surveillance. In order to achieve this, it is imperative that the correct patient group, that is, those who are at highest risk of progression to OADC, is placed under the most intensive of surveillance, and those at a lower risk can be spared such frequent invasive investigation. Epigenetic biomarkers may provide a robust way of risk stratifying patients for BO surveillance.
Patients and the public were not involved in the development of this systematic review.
Eligibility criteria
Any prospective and retrospective primary studies were eligible for inclusion provided they measured epigenetic markers in patients over the age of 18 years with BO. To be included, the study must have reported on progression from NDBO to BO with HGD or OADC in the same patient cohort. Relevant epigenetic markers are DNA methylation, histone modification, chromatin remodelling and micro and non-coding RNAs. Studies were excluded if they were case reports, narrative reviews, in vitro studies (eg, using cell lines), studies of genetic (rather than epigenetic) mutations, studies using biomarkers to predict a response to treatment (eg, chemotherapy) or animal studies.
Search
A systematic search of the literature to the end of February 2018 was undertaken. Text and index terms relating to the population (BO), the prognostic marker (epigenetic change) and the outcome (BO with HGD or OADC) were combined (see online supplementary appendix 1 for sample search strategy in MEDLINE). No study design, date or language restrictions were applied. MEDLINE, MEDLINE in Process, EMBASE, Cochrane Central, ISI Conference Proceedings Citation Index and the British Library’s ZETOC were searched from inception. Reference lists of identified studies and systematic reviews were screened for any additional relevant primary studies. Registers of clinical trials (ClinicalTrials.gov and ICTRP) were searched for ongoing studies.
bmjopen-2017-020427supp001.pdf (98.4KB, pdf)
Study selection
Two reviewers (TN and CLT) independently screened all titles and abstracts to identify potentially relevant studies using prespecified screening criteria. Full texts of potentially relevant articles were assessed against prespecified eligibility criteria. Eligibility was determined by two reviewers (TN and CLT) independently with any discrepancies resolved by discussion or referral to a third reviewer (OT).
Data extraction
Data extraction of the included studies was carried out by one reviewer (TN) using a standardised data extraction form and checked independently by a second reviewer (OT). Discrepancies were resolved by discussion or referral to a third reviewer (CLT and JD). Data were extracted on study design characteristics, patient characteristics, prognostic marker and outcomes.
Quality assessment
The risk of bias of individual eligible studies was assessed in duplicate by two reviewers (TN and OT) using a modified Quality in Prognostic Studies tool.30 Risk of bias criteria was related to study participation (eg, method of sampling), study attrition, prognostic factor measurement and selection (eg, reliability of epigenetic technique and publication bias), outcome assessment (eg, undertaken in duplicate) and study confounding factors (measured and adjusted for). The main confounders are considered to be age, obesity, smoking and alcohol intake.1 Selected elements from prediction study risk of bias assessment tool (PROBAST) which is currently under development (Wolff R) were used to assess the methodological quality of prognostic models in a similar fashion to Ensor et al’s 2016 systematic review of prognostic models of venous thromboembolism31 including aspects on patient selection, statistical models used and model validation.
Synthesis
Synthesis was narrative, with main findings tabulated. Studies were grouped by individual epigenetic marker or panel of markers. A lack of consistency in reported outcome metrics and heterogeneity relating to study design, length of follow-up, frequency of endoscopy and biopsy and experimental technique precluded any quantitative synthesis. Most studies presented results as percentage methylation or a ratio of differential methylation. Formal assessment of publication bias was not possible.
Reporting
Reporting of this systematic review was according to the Preferred Reporting Items for Systematic Reviews and Meta-Analyses32 guidelines (online supplementary appendix 2)
bmjopen-2017-020427supp002.pdf (202.4KB, pdf)
Systematic review results
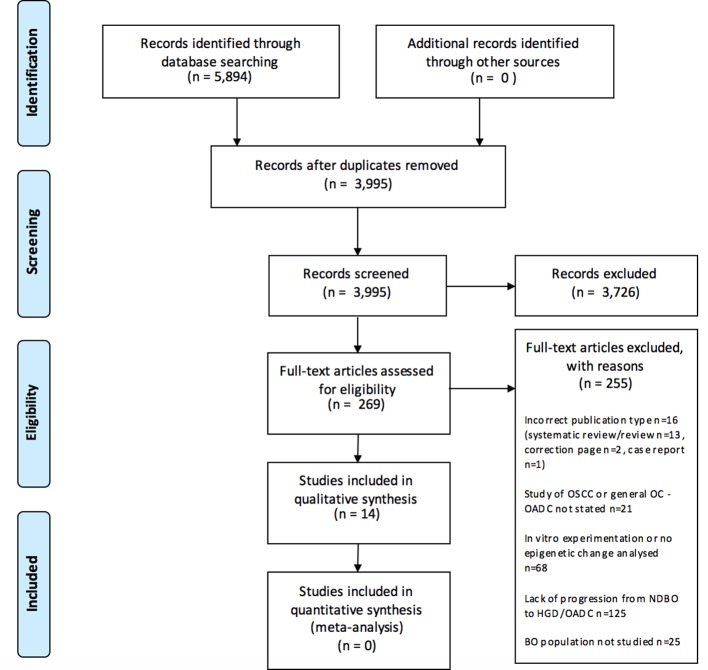
Overall, 3995 records were screened, and 14 studies met the inclusion criteria (see figure 1 for study selection process and reasons for exclusion). The most common reasons for exclusion were lack of progression in the same population from NDBO to HGD or OADC, in vitro experimentation or no epigenetic change analysed in patient samples.
Figure 1.
PRISMA 2009 flow diagram with reasons for exclusion. BO, Barrett’s oesophagus; HGD, high-grade dysplasia; NDBO, non-dysplastic Barrett’s oesophagus; OADC, oesophageal adenocarcinoma; OC, oesophageal cancer; OSCC, oesophageal squamous cell carcinoma; PRISMA, Preferred Reporting Items for Systematic Reviews and Meta-Analyses.
Study characteristics
Table 1 shows the main study characteristics. All included studies were of a similar retrospective cohort design. In total, 12 of 14 studies reported patient numbers. A total of 404 patients were included. Two studies reported total lesions only (n=223). CpG promoter methylation was investigated in 13 studies and miRNA expression in one study.33 Forty-two unique epigenetic markers were reported. No studies investigated histone modification. Thirteen included studies analysed 38 differentially methylated CpG promoter sites, nine used methylation-specific PCR,34–42 two used methylight methylation-specific PCR,43 44 one used bisulfite pyrosequencing45 and one used methylation microarray techniques.46
Table 1.
Summary table of included studies
| Author/year | Study type | Study population | Patient no (P:NP) | Patient demographics (mean age, M:F ratio) | Epigenetic analysis | Markers analysed | Result | |
| Agarwal et al 201246 | Retrospective cohort | Pathology archives at Baltimore Veteran’s Affairs Medical Centre and Johns Hopkins University School of Medicine | 9 (5:4) | Not stated | CpG methylation (244K human CpG microarray) | Extragenic x 3 | 0.45—relative CpG methylation | |
| Intra_BCL11B | 0.17 | |||||||
| Intra_CCDC57 | 0.23 | |||||||
| Intra_F10 | 1.86 | |||||||
| Pro_CKB | 0.83 | |||||||
| Pro_ELAVL3-ZNF653 | 0.6 | |||||||
| Pro_GPR177 | 0.7 | |||||||
| Pro_HOXB7 | 0.67 | |||||||
| Pro_IGF1R | 0.58 | |||||||
| Pro_ITGB8 | 0.55 | |||||||
| Pro_JARID 1B | 0.35 | |||||||
| Pro_JUND | 0.54 | |||||||
| Pro_LAMA5 | 0.45 | |||||||
| Pro_LOC55565 | 0.76 | |||||||
| Pro_MGC35308 | 0.52 | |||||||
| Pro_MMD2 | 1.34 | |||||||
| Pro_TAF10 | 0.43 | |||||||
| Pro_TLX3 | 0.31 | |||||||
| Pro_UBP1 | 0.71 | |||||||
| Pro_WNK4 | 0.76 | |||||||
| Pro_WWC1 | 0.68 | |||||||
| Pro_ZBTB7B | 0.56 | |||||||
| Pro_ZNF358 | 1.25 | |||||||
| Barrett et al 199937 | Retrospective cohort | Fred Hutchinson Cancer Research Centre+University of Washington | 31 (25:6) | Not stated | CpG methylation (methylation-specific PCR) | CDKN2A | Hypermethylation in premalignant samples | |
| Boerwinkel et al 201443 | Retrospective cohort | AMC BO Surveillance | 44 (48:10) | Mean age=6632M:12F | CpG methylation (methylight methylation-specific PCR) | HPP1 | NS | |
| p16 | P hypermethylated | |||||||
| RUNX3 | NS | |||||||
| Clément et al 200635 | Retrospective cohort | Lausanne, Switzerland | 28* (12:16) | Not stated | CpG methylation (methylation-specific PCR) | APC | P hypermethylated | |
| CDKN2A | NS | |||||||
| SFRP1 | Hypermethylated in all samples P and NP | |||||||
| TIMP3 | P hypermethylated | |||||||
| TERT | P hypermethylated | |||||||
| Clément et al 200834 | Retrospective cohort | Lausanne, Switzerland | 31 (16:15) | Not stated | CpG methylation (methylation-specific PCR) | WIF-1 | P hypermethylated | |
| Eads et al 200144 | Retrospective cohort | Norris Comprehensive Cancer Centre | 20 (12:8) | Not stated | CpG methylation (methylight methylation-specific PCR) | CALCA | P hypermethylated | |
| CDKN2A | P hypermethylated | |||||||
| ESR1 | P hypermethylated | |||||||
| MGMT | P hypermethylated | |||||||
| MYOD1 | P hypermethylated | |||||||
| TIMP3 | P hypermethylated | |||||||
| Jin et al 200941 | Case control | Five participating US clinics | 195* (50:145) | P significantly older than NP | CpG methylation (methylation-specific PCR) | HPP1 | P hypermethylated | |
| p16 | P hypermethylated | |||||||
| RUNX3 | P hypermethylated | |||||||
| CDH13 | NS | |||||||
| TAC1 | NS | |||||||
| NELL1 | NS | |||||||
| AKAP12 | NS | |||||||
| SST | NS | |||||||
| Klump et al 199836 | Retrospective cohort | Under routine Barrett’s surveillance | 14 (10:4) | P mean age 57.48M:2F | NP mean age 553M:1F | CpG methylation (methylation-specific PCR) | p16 | P hypermethylated |
| Moinova et al 201242 | Retrospective cohort | Retrospective collection from pathology database | 5 (2:3) | Not stated | CpG methylation (methylation-specific PCR) | VIM | VIM methylation present in NDBO, not relevant in progression | |
| Puertas Canteria 201245 | Retrospective cohort | No details | 55 (6:14) | Not stated | CpG methylation (bisulfite pyrosequencing) | p16 | Methylation P 12.04% versus NP 6.53% | |
| Revilla-Nuin et al 201333 | Retrospective cohort | 1982 trial assessing medical versus surgical therapy for prevention of progression of BO. Randomly selected | 5 (7:17) | Not stated | miRNA expression analysis | miR-192 | Overexpressed AUROC 0.61 | |
| miR-194 | Overexpressed AUROC 0.70 | |||||||
| miR-196a | Overexpressed AUROC 0.80 | |||||||
| miR-196b | Overexpressed AUROC 0.74 | |||||||
| Sato et al 200840 | Retrospective cohort | MAYO and UMD mixed cohort | 62 (28:34) | Not stated | CpG methylation (methylation-specific PCR) | HPP1 | P hypermethylated | |
| p16 | P hypermethylated | |||||||
| RUNX3 | P hypermethylated | |||||||
| Schulmann et al 200539 | Retrospective cohort | Baltimore Veteran’s Affairs hospitals and University of Maryland Hospitals | 53 (8:45) | P mean age 63.34M:0F, segment length 10.0 cm | NP mean age 62.235M:3F, segment length 5.8 cm | CpG methylation (methylation-specific PCR) | RUNX3 | P hypermethylated |
| HPP1 | P hypermethylated | |||||||
| p16 | P hypermethylated | |||||||
| CRBP1 | Hypermethylation not independently associated with P | |||||||
| TIMP3 | Hypermethylation not independently associated with P | |||||||
| APC | Hypermethylation not independently associated with P | |||||||
| Wang et al 200938 | Retrospective cohort | Johns Hopkins under surveillance for GORD/BO | 57 (7:50) | No significant differences in N versus NP. Data not provided | CpG methylation (methylation-specific PCR) | APC | P hypermethylated | |
| p16 | P hypermethylated | |||||||
*Number of lesions, no patient numbers described in the study; number of progressor lesions (P), number of non-progressor lesions (NP).
AUROC, area under receiver operating characteristic; BO, Barrett’s Oesophagus; GORD, gastro-oesophageal reflux disease; M:F, male-to-female ratio; miRNA, micro RNA; NDBO, non-dysplastic BO; NP, non-progressing patients; NS, not statistically significant; P, progressing patients; PLSD, protected least significant difference; ROC, receiver operating characteristic.
Prognostic models
Five studies developed models aiming to predict progression to OADC (table 2).35 38–41 Three models included both epigenetic markers and clinical parameters.39–41
Table 2.
Models aiming to predict progression to OADC
| Author/year | Study type | Patient no (P:NP) | Model used | Result |
| Clément et al 200635 | Retrospective cohort | 28* (12:16) |
APC+TIMP3+TERT | Hypermethylation in P versus NP (P 81% vs NP 26% P<0.0001) |
| Jin et al 200941 | Retrospective cohort | 195* (50:145) |
Biomarker panel (p16, HPP1, RUNX3, CDH13, TAC1, NELL1, AKAP12, SST) | AUROC 0.72 |
| Biomarker panel+age (p16, HPP1, RUNX3, CDH13, TAC1, NELL1, AKAP12, SST) |
AUROC 0.85 | |||
| Sato et al 200840 | Retrospective cohort | 62 (28:34) |
Methylation index (p16, HPP1, RUNX3) | Hypermethylation in P versus NP AUROC 0.75 (no CI stated) |
| Methylation index (p16, HPP1, RUNX3), segment length, pathology | AUROC 0.79 (95% CI 0.6968 to 0.8853) Sensitivity 91.4 Specificity 51.8 |
|||
| Schulmann et al 200539 | Retrospective cohort | 53 (8:45) |
Age, segment length, HPP1, TIMP3, APC, p16, CRBP1, RUNX3 | HPP1, p16, RUNX3 independent risk factors in multivariate analyses Model combined HR index >5 leads to an increased likelihood of progression within 2 years |
| Wang et al 200938 | Retrospective cohort | 57 (7:50) |
P16+APC | Hypermethylation of both APC and p16 OR 14.97 (95% CI 1.73 to ∞, P=0.012) for neoplastic progression |
*Number of lesions, no patient numbers described in the study; number of progressor lesions (P), number of non-progressor lesions (NP).
AUROC, area under receiver operating characteristic; NP, non-progressing patients; OADC, oesophageal adenocarcinoma; P, progressing patients.
Schulmann et al 39 performed a retrospective longitudinal analysis of 53 patients contributing 106 specimens enrolled in a BO surveillance programme using Cox proportional hazards regression. Initially, 10 candidate prognostic marker genes were assessed using cross-sectional data. Six of the genes (HPP1, TIMP3, APC, p16, CRBP1 and RUNX3) which demonstrated hypermethylation in patients with OADC and relative hypomethylation in normal oesophageal tissues were investigated in the longitudinal study. All except APC were associated with progression (HGD and OADC combined) in univariate analysis. A multivariate model including all six genes, age and segment length found evidence that HPP1, p16 and RUNX3 were independently prognostic. The performance of the full model (all eight covariates) was assessed by calculating the exponentiated multiplier of baseline hazard for covariates for each specimen (HR index). For specimens taken within 2 years of progression, the HR index was >5 in progressors compared with <5 in non-progressors. The authors report that the HR index was not predictive for specimens taken over 2 years before progression. The study reports OR but doesn’t explain how as the Cox model produces HRs. The authors note that the sample size is small, the study is retrospective, other potential clinical predictors were unavailable and the model uses a combined end point of HGD and OADC. In univariate analysis, TIMP3 had an OR for progression of 1.68 (95% CI 1.14 to 2.38) but this flipped to 0.5 (95% CI 0.22 to 1.04) in the multivariate analysis. The authors conclude that further validation of the markers (HPP1, p16 and RUNX3), ideally in prospective multicentre trials, is required.
Sato et al 40 performed a retrospective cohort study on 62 patients providing a total of 118 specimens. They developed a model using linear discriminant analysis incorporating both clinical and epigenetic markers. Sex, BO segment length and histological diagnosis were combined with p16, HPP1 and RUNX3 CpG methylation status as well as a methylation index with a score of 0–3 was considered. The best receiver operating characteristic (ROC) curve for a 4-year follow-up in this cohort was generated using segment length, pathology and methylation index with an area under ROC (AUROC) of 0.7910 (95% CI 0.6968 to 0.8853) with a specificity and sensitivity of 91.4% and 51.8%, respectively. Change in AUROC between this model and a model including predictive markers alone was not assessed. Patient selection criteria were not reported, and the small sample size (62 patients), the large number of possible parameter combinations (n=127), and the cut-points for variables were chosen to optimise performance in this dataset results in a high level of uncertainly about the optimal parameter set. No external validation was performed.
Jin et al 41 performed a double-blind multicentre case–control study of 195 tissue specimens using an eight methylation biomarker panel of p16, HPP1, RUNX3, CDH13, TAC1, NELL1, AKAP12 and SST combined with patient age to predict which patients will progress to OADC. When assessed individually, p16, HPP1 and RUNX3 were associated with progression to OADC (P<0.05). When the panel of all eight markers was used compared with age alone as a predictor of progression, the increment in AUROC was 0.114 in a 4-year follow-up (0.630 age alone and 0.753 age+markers), demonstrating a clinically important improvement in the predictive power of the model within the dataset. This study compared clinical factors between progressor and non-progressor groups and found no significant difference in gender, body mass index (BMI), BO segment length, smoking status or alcohol consumption. External validation was not performed.
Clément et al 35 performed a retrospective cohort study of 28 tissue specimens. Eighty-one per cent of patients (n=12) with combined hypermethylation of APC, TIMP3 and TERT progressed to OADC compared with 26% of non-progressors (n=16) (P<0.0001). It was suggested that in combination, these three markers could be used to predict which patients are at higher risk of progression. No sensitivities or specificities were reported, and there was no ROC analysis. There is no indication of independent prognostic value, and no internal or external validation was performed using this model.
Wang et al 38 performed a retrospective cohort study on 7 progressor and 50 non-progressor patients. They reported that hypermethylation in both p16 and APC was a strong predictor of progression to dysplastic BO or OADC. Patients who were negative for both p16 and APC hypermethylation did not progress. Hypermethylation of both APC and p16 yielded an OR of 14.97 (95% CI 1.73 to ∞, P=0.012) for subsequent progression to HGD or OADC. A limitation of this study is the short follow-up time of 4.1 years for the non-progressor group which may not be sufficient time for dysplasia or neoplasia to develop. The reported CI is extremely wide, making it difficult to accurately interpret the OR.
Individual markers analysed
Forty-two individual epigenetic markers were analysed in the 14 included studies. Ten studies investigated one or more of the following five individual markers: p16 (CDKN2A), RUNX3, TIMP3, HPP1 and APC. Details of markers, assessment methods and findings can be found in tables 3–7. All five markers demonstrated CpG hypermethylation in patients who progressed from NDBO to OADC in at least one of the included studies. p16 (a tumour suppressor protein encoded by the CDKN2A gene) offered the most experimental data with 10 studies reporting on 220 progressor samples compared with 332 non-progressor samples. Eight of the 10 studies demonstrated a statistically significant difference in hypermethylation at p16.36 38–41 43–45 Barrett et al 37 demonstrated CpG hypermethylation of CDKN2A in seven progressor samples of a specific genetic clonality; however, no control group was analysed in parallel for this specific marker in this study.
Table 3.
P16/CDKN2A
| Author/year | Patient no (P:NP) | Experimental technique used | Methylation threshold definition | Statistical analysis | Result |
| Barrett et al 199937 | 49 (39:6) | Methylation-specific PCR | Positive or negative PCR on agarose gel | Not stated | Seven progressor patients with specific genetic abnormality (LOH at 17 p and 9 p) displayed hypermethylation at CDKN2A. No numerical value offered |
| Boerwinkel et al 201443 | 44 (48:10) | Methylight methylation-specific PCR | Previously published cut-off values applied to raw MSP data to calculate frequency of hypermethylation: p16 cut-off=0.02 | Mann-Whitney test | Raw MSP values showed Hypermethylated in P P<0.05 |
| Clément et al 200635 | 28* (12:16) | Methylation-specific PCR | Intensity of methylation-specific dot-blot assay compared | Not stated | 0% P versus 12% NP methylated No significant difference between P and NP |
| Eads et al 200144 | 20 (12:8) | Methylight methylation-specific PCR | Intensity of methylated genes compared with controls. ‘PMR’ value of 4 used as cut-off to indicate methylated gene | Fisher’s PLSD | Intensity of PCR band used for quantitative analysis Hypermethylated in P P=0.0048 |
| Jin et al 200941 | 195* (50:145) | Methylation-specific PCR | NMV=amount of methylated DNA compared with control beta-actin DNA generated by PCR | Student’s t test and chi-squared test AUROC |
NMVs P:NP 0.138:0.069 AUROC=0.628 (0.534, 0.722) 90% sensitivity 90% specificity Hypermethylated in P P=0.0066 |
| Klump et al 199836 | 14 (10:4) | Methylation-specific PCR | Yes/no detection of PCR product | Chi-squared test | Hypermethylated in P 8% P versus 0% NP P=0.0001 |
| Puertas Canteria 201245 | 35 (6:14) | Bisulfite pyrosequencing | Quantitative CpG methylation technique | No statistical analysis | Hypermethylated in P Methylation grade 12.04% P versus 6.53% NP |
| Sato et al 200840 | 62 (28:34) | Methylation-specific PCR | NMV=amount of methylated DNA compared with control beta-actin DNA generated by PCR | LDA, LOOCV, AUROC | Hypermethylated in P AUROC increment when added to segment length, histology and global methylation index=0.0335 90% sensitivity 90% specificity P=0.00576 |
| Schulmann et al 200539 | 53 (8:45) | Methylation-specific PCR | NMV=amount of methylated DNA compared with control beta-actin DNA generated by PCR | Cox proportional HRs | Hypermethylated in P OR 1.74 P=0.0005 |
| Wang et al 200938 | 57 (7:50) | Methylation-specific PCR | No threshold described | OR using univariate logistic regression | Hypermethylated in P OR 10.02 P=0.034 |
*Number of lesions, no patient numbers described in the study; number of progressor lesions (P), number of non-progressor lesions (NP).
AUROC, area under receiver operating characteristic; LDA, linear discriminant analysis; LOH, Loss of Heterozygosity; LOOCV, leave-one-out cross-validation; MSP, methylation specific PCR; NMV, normalised methylation value; NP, non-progressing patients; P, progressing patients; PLSD, protected least significant difference; PMR, percent methylated reference.
Table 4.
RUNX3
| Author/year | Patient no (P:NP) | Methods | Methylation threshold definition | Statistical analysis | Result |
| Boerwinkel et al 201443 | 44 (48:10) |
Methylight methylation-specific PCR | Previously published cut-off values applied to raw MSP data to calculate frequency of hypermethylation. RUNX3 cut-off=0.02 | Mann-Whitney test | Raw MSP values compared but no numerical value offered P versus NP not significantly differentially methylated |
| Jin et al 200941 | 195* (50:145) |
Methylation-specific PCR | NMV=amount of methylated DNA compared with control beta-actin DNA generated by PCR | Student’s t test and chi-squared test AUROC |
Hypermethylated in P NMV 0.104:0.063 AUROC=0.671 (0.586, 0.756) 90% sensitivity 90% specificity P=0.0002 |
| Sato et al 200840 | 62 (28:34) |
Methylation-specific PCR | NMV=amount of methylated DNA compared with control beta-actin DNA generated by PCR | LDA, LOOCV, AUROC | Hypermethylated in P AUROC increment when added to segment length, histology and global methylation index=0.0216 90% sensitivity 90% specificity P=0.016 |
| Schulmann et al 200539 | 53 (8:45) |
Methylation-specific PCR | NMV=amount of methylated DNA compared with control beta-actin DNA generated by PCR | Cox proportional HRs | Hypermethylated in P OR=1.80 P=0.0267 |
*Number of lesions, no patient numbers described in the study; number of progressor lesions (P), number of non-progressor lesions (NP).
AUROC, area under receiver operating characteristic; LDA, linear discriminant analysis; LOOCV, leave-one-out cross-validation; MSP, methylation specific PCR; NMV, normalised methylation value; NP, non-progressing patients; P, progressing patients.
Table 5.
HPP1
| Author/year | Patient no (P:NP) | Methods | Methylation threshold definition | Statistical analysis | Result |
| Boerwinkel et al 201443 | 44 (48:10) |
Methylight methylation-specific PCR | Previously published cut-off values applied to raw MSP data to calculate frequency of hypermethylation. HPP1 cut-off=0.05 | Mann-Whitney test | P versus NP not significantly differentially methylated |
| Jin et al 200941 | 195* (50:145) |
Methylation-specific PCR | NMV=amount of methylated DNA compared with control beta-actin DNA generated by PCR | Student’s t test and chi-squared test AUROC |
Hypermethylated in P AUROC=0.647 (0.556, 0.739) P=0.0025 |
| Sato et al 200840 | 62 (28:34) |
Methylation-specific PCR | NMV=amount of methylated DNA compared with control beta-actin DNA generated by PCR | LDA, LOOCV, AUROC | Hypermethylated in P AUROC increment when added to segment length, histology and global methylation index 0.028 90% sensitivity 90% specificity P=0.018 |
| Schulmann et al 200539 | 53 (8:45) |
Methylation-specific PCR | NMV=amount of methylated DNA compared with control beta-actin DNA generated by PCR | Cox proportional HRs | Hypermethylated in P OR=1.77 P=0.0311 |
*Number of lesions, no patient numbers described in the study; number of progressor lesions (P), number of non-progressor lesions (NP).
AUROC, area under receiver operating characteristic; LDA, linear discriminant analysis; LOOCV, leave-one-out cross-validation; MSP, Methylation Specific PCR; NP, non-progressing patients; P, progressing patients.
Table 6.
TIMP3
| Author/year | Patient no (P:NP) | Methods | Methylation threshold definition | Statistical analysis | Result |
| Clément et al 200635 | 28* (12:16) |
Methylation-specific PCR | Intensity of methylation-specific dot-blot assay compared | Not stated | Hypermethylated in P % samples methylated P=91%: NP=23% P<0.0001 |
| Eads et al 200144 | 20 (12:8) |
Methylight methylation-specific PCR | Intensity of methylated genes compared with controls. ‘PMR’ value of 4 used as cut-off to indicate methylated gene | Fisher’s PLSD | Hypermethylated in non-dysplastic tissue in patients with associated dysplasia; however, not statistically significant when comparing P versus NP P=0.13 |
| Schulmann et al 200539 | 53 (8:45) |
Methylation-specific PCR | NMV=amount of methylated DNA compared with control beta-actin DNA generated by PCR | Cox proportional HRs | Hypermethylated in P, but not independent risk factor in multivariate analysis OR=1.68 univariate OR=0.50 multivariate P=0.0109 |
*Number of lesions, no patient numbers described in the study; number of progressor lesions (P), number of non-progressor lesions (NP).
NP, non-progressing patients; P, progressing patients; PLSD, protected least significant difference; PMR, percent methylated reference.
Table 7.
APC
| Author/year | Patient no (P:NP) | Methods | Methylation threshold definition | Statistical analysis | Result |
| Clément et al 200635 | 28* (12:16) |
Methylation-specific PCR | Intensity of methylation-specific dot-blot assay compared | No statistical analysis offered | Hypermethylated in P methylation: P=100% versus NP=36% |
| Schulmann et al 200539 | 53 (8:45) |
Methylation-specific PCR | NMV=amount of methylated DNA compared with control beta-actin DNA generated by PCR | Cox proportional HRs | Hypermethylation not an independent risk factor for P OR=1.00 (95% CI 0.59 to 1.33) P=0.99 |
| Wang et al 200938 | 57 (7:50) |
Methylation-specific PCR | No threshold described | OR using univariate logistic regression | Hypermethylated in P methylation: P=86% versus NP=40% P=0.02 OR=9 (1.01–80.52) P=0.049 |
*Number of lesions, no patient numbers described in the study; number of progressor lesions (P), number of non-progressor lesions (NP).
NP, non-progressing patients; P, progressing patients.
RUNX3 and HPP1 were analysed in four studies39–41 43 as individual markers. Overall, 134 progressors and 234 non-progressors were studied. Three studies39–41 demonstrated statistically significant hypermethylation in both RUNX3 and HPP1 CpG sites in progressor samples; however, one study43 found no difference for either HPP1 or RUNX3.
Three studies investigated TIMP3 CpG methylation35 39 44 (32 progressors and 69 non-progressors). One study showed a statistically significant difference in CpG hypermethylation between progressors and non-progressors.35 One study showed significant hypermethylation and calculated an OR of 1.68 (95% CI 1.14 to 2.38) for progression on univariate analysis39; however, TIMP3 could not be regarded as an independent risk factor as the OR dropped to 0.5 (95% CI 0.22 to 1.04) on multivariate analysis. The final study44 showed significantly increased TIMP3 methylation in non-dysplastic tissues of patients with associated dysplasia elsewhere in the oesophagus; however, there was no difference between progressors and non-progressors.
Three studies analysed APC 35 38 39 (26 progressors and 116 non-progressors). Wang et al 38 demonstrated hypermethylation in progressors. Clément et al 35 reported hypermethylation but did not provide any statistical analyses. Wang et al calculated an OR of 9 (95% CI 1.01 to 88.52) for hypermethylation of APC in progressors. Conversely, Schulmann et al 39 found no evidence of a difference (OR=1.00 (95% CI 0.59 to 1.33)) in progression with or without hypermethylation of APC. All three studies have similar sample size, study design and experimental techniques. These contradictory findings with limited statistical validation and wide CIs call into question the validity of APC as an epigenetic marker of BO progression.
The main methodological limitations across the studies were poor reporting of patient characteristics and study populations, including patient selection. There was little information about loss to follow-up, and it was often unclear exactly which samples were used and at which time-points in longitudinal analyses. There was limited blinding of researchers to the histology, and progressor status of the samples was often not described. Confounding factors were described in six of the studies,33 38–41 46 but only Sato et al 40 adjusted for BO segment length and patient sex in their predictive model. Full details of quality assessment can be found in table 8.
Table 8.
Quality assessment for prognostic factor studies including prognostic models
| Study | Representative patient sample? | Loss to follow-up equal in both arms? | Study conducted prospectively (PRO) or retrospectively (RET)? | Samples collected prospectively (PRO) or retrospectively (RET)? | Sampling method: quadrantic biopsy used? | Sample type? | Validated epigenetic technique used? | Epigenetic analysis undertaken blindly? (without knowledge of histology) | Epigenetic analysis undertaken in duplicate? | Was the threshold defined? | Histology undertaken in duplicate? | Histology undertaken blindly? (without knowledge of epigenetic finding) | Were confounding factors described? (eg, obesity, age, smoking, alcohol) | Were confounding factors adjusted for in design or analysis? | Which confounding factors were adjusted for? |
| Agarwal et al 201246 | Unclear | Unclear | PRO | RET | No | Unclear | Yes | Unclear | Unclear | Yes | Unclear | No | Yes | No | N/A |
| Barrett et al 199937 | Yes | Unclear | PRO | RET | No | Snap frozen | Yes | No | Unclear | Unclear | Unclear | Unclear | No | No | N/A |
| Boerwinkel et al 201443 | Yes | Unclear | PRO | RET | Yes | FFPE | Yes | Unclear | Unclear | Yes | Yes | Yes | No | No | N/A |
| Clément et al 200635 | Unclear | Unclear | PRO | RET | No | FFPE | Yes | No | Unclear | Unclear | Unclear | Unclear | No | No | N/A |
| Clément et al 200834 | Unclear | Unclear | PRO | RET | No | FFPE | Yes | No | Unclear | Unclear | Unclear | Unclear | No | No | N/A |
| Eads et al 200144 | Unclear | Unclear | PRO | RET | No | Snap frozen | Yes | Unclear | Unclear | Yes | Unclear | Unclear | No | No | N/A |
| Jin et al 200941 | Unclear | Unclear | RET | PRO | Yes | Snap frozen | Yes | No | Unclear | Yes | No | Yes | Yes | No | N/A |
| Klump et al 199836 | Unclear | Yes | RET | RET | No | FFPE | Yes | No | Yes | Yes | Unclear | No | No | No | N/A |
| Moinova et al 201242 | Unclear | Unclear | Unclear | Unclear | No | FFPE | Yes | No | Unclear | Yes | Unclear | Unclear | No | No | N/A |
| Revilla-Nuin et al 201333 | Yes | Unclear | PRO | Unclear | Yes | FFPE | Yes | No | Unclear | Yes | Yes | Yes | Yes | Unclear | N/A |
| Puertas Canteria 201245 | Unclear | Unclear | Unclear | Unclear | No details | FFPE | Yes | No | Unclear | Unclear | Unclear | No | No | No | N/A |
| Sato et al 200840 | Unclear | Unclear | Unclear | Unclear | Yes | Snap frozen | Yes | No | Unclear | Yes | Yes | Yes | Yes | Yes | BO segment length |
| Schulmann et al 200539 | Unclear | Unclear | RET | Unclear | Yes | Snap frozen | Yes | No | Unclear | Yes | Yes | Yes | Yes | No | N/A |
| Wang et al 200938 | Yes | Unclear | PRO | RET | Yes | FFPE | Yes | No | Unclear | Yes | No | Yes | Yes | No | N/A |
BO, Barrett’s oesophagus; FFPE, formalin-fixed paraffin-embedded.
Given these methodological uncertainties, the findings need to be viewed with caution.
Discussion
This systematic review of the literature of epigenetic markers and their role in predicting progression of BO to HGD and OADC has revealed a heterogeneous and disparate dataset. Fourteen studies were identified, with five incorporating prognostic models. This is the first systematic review to examine all evidence on epigenetic change and its role in Barrett’s carcinogenesis. It suggests a role for p16 hypermethylation as an individual epigenetic biomarker in predicting progression from BO to OADC.35–41 43–45 However, a paucity of evidence for other epigenetic markers (and combinations), poor reporting of patient characteristics and methods employed and lack of external model validation limit the conclusions that can be drawn.
Only 14 studies were identified with small patient numbers (median 31 (5–195)). The extent of loss to follow-up was usually poorly described. All studies suffered from a paucity of clinical information and a lack of reporting of study patient demographics. Those studies which did report comorbidities and potential confounding factors such as BMI, smoking status, age, histological diagnosis and BO segment length rarely adjusted for these in their analysis. Without adjusting for these factors, it is difficult to assess the incremental predictive clinical value of any epigenetic changes. Future models should explore the predictive ability of epigenetic changes in the context of clinical variables. In their study, Riley et al 47 concluded that provision of individual patient data could have overcome the majority of the reporting issues including poorly reported summary statistics, adjustment factors and outcome measures used.
It is challenging to compare data from different studies due to variable measurement of epigenetic change. Five different experimental techniques were used: miRNA analysis (n=1)33 and CpG methylation including, methylation-specific PCR (n=9),48 methylight methylation-specific PCR (n=2),49 bisulfite pyrosequencing (n=1)45 50 and methylation microarray technology (n=1).46 Early methods of measuring differential DNA methylation involve semiquantitative reporting of results including analysis of intensity of PCR product bands on agarose gel. More advanced techniques use computerised photometry, although differing PCR conditions and equipment may result in variability between laboratories. More advanced techniques such as bisulfite pyrosequencing and methylation microarrays can provide an accurate methylation percentage at individual CpG sites which are suitable for quantitative analysis. The 12 studies using methylation-specific PCR revealed a global pattern of hypermethylation of progressor patients in the CpG regions of interest34–44; however, each used differing methylation thresholds and outcome measures for reporting positive results. One study45 used bisulfite pyrosequencing to analyse p16, but no further markers were analysed on more than one platform. The different methods used, as well as the different outcome metrics reported and lack of clearly reported clinical information, meant that there was insufficient clinical and methodological homogeneity. Meta-analysis was, thus, not appropriate, despite individual epigenetic markers being reported in up to 10 independent studies.
Five models looked at various combinations of epigenetic markers and clinical risk factors. Findings from four of the models suggest a potential role for p16 in combination with HPP1, RUNX3 and/or APC and clinical factors of segment length or age. All models used slightly different combinations of model parameters and reported analyses differently, thus making it difficult to compare findings between models. All suffered from a lack of external validation, and there were uncertainties around the representativeness of the included populations. More research is needed to validate and further explore these initial findings.
P16, a tumour suppressor protein, is the most consistently hypermethylated CpG site in progressors. Hypermethylation leads to reduced p16 production, allowing neoplastic cells to progress from G1 to S phase of cell cycle and replicate uncontrollably. p16 deletions are observed in melanoma, oesophageal, lung, pancreatic, mesothelioma, bladder, head and neck squamous cell carcinoma, breast, lymphocyte, brain, ovarian, osteosarcoma and renal cancer cell lines.51 52 In BO, p16 genetic mutations have been shown to occur early in tumorigenesis and may appear before dysplasia has occurred.53 Timmer et al 54 demonstrated the presence of p16, MYC and aneusomy in a model with age and segment length determined whether patients were at low or high risk of progression from NDBO to OADC. Without further in-depth temporal analysis, it remains unclear whether the index abnormality of Barrett’s carcinogenesis is epigenetic or genetic in origin.
The pattern of hypermethylation in RUNX3 and HPP1 in progressors was less clear with not all studies demonstrating a statistically significant difference between progressor and non-progressor patients. TIMP3 and APC provide variable results in the literature, and much smaller patient groups are analysed. Agarwal et al reported a predominance of hypomethylation in progressors (16 of 19 CpG sites) with only three CpG sites (Pro_MMD2, Pro_ZNF358 and Intra_F10) hypermethylated. The methylation microarray used in this study can analyse 28 700 unique CpG sites, whereas the latest technology offers up to 850 000 sites on a single chip.55 With only a single study using an outdated technique, it is not possible to say whether CpG hypomethylation is a driver in progressive BO. Prior to this technology being available, researchers had to carefully select CpG sites of interest for analysis; selection was based on previous research and literature review giving rise to a positive selection and publication bias. This is a well-recognised phenomenon in prognostic factor research. Sekula et al 56 reviewed published and unpublished work into p53’s role in bladder cancer and discovered that 31% of observational studies were unpublished in a 15-year period. While Sekula’s review is concerned with a different pathology, the type of included studies is of a similar design to those included in this review. A combination of positive publication bias and marker selection bias raises questions as to the validity of the selected markers in studies demonstrating CpG hypermethylation. Selective reporting may also erroneously inflate the importance of individual prognostic markers. Kyzas et al 57 analysed published and unpublished data sought directly from researchers investigating the importance of TP53’s role in head and neck squamous cell carcinoma on patient mortality. They found that if all published and unpublished data were included in their meta-analysis, the risk ratio decreased from 1.38 to 1.16 (95% CI 0.99 to 1.35 P=0.06). They also had difficulty conducting their analysis due to non-standardised definitions and reporting. Heterogeneity in definitions and reporting was also identified within our dataset, and it must also be considered whether selective reporting of results is over stating the importance of individual markers such as p16 in the progression of BO. Similarly, if ever increasing numbers of CpG sites are analysed with microarray technology, there will be an increasing number of negative results which are likely to be under-reported in the literature. As illustrated by Kyzas et al and Sekula et al,56 57 despite a large volume of literature, without standardisation of reporting and outcome measures, the research is often not suitable for translation into clinical use.
The variable quality of prognostic studies has been previously reported in systematic reviews58 59 and improvements suggested for collaboration and standardisation especially with regard to statistical analysis and outcome reporting.60–62 Poor quality of reporting was also a feature in this systematic review. For example, only 7 of 14 included studies documented both the number of patients and samples analysed in each group. Standardised reporting of epigenetic studies in BO should provide basic clinical details for each patient including age, sex, smoking status, BMI and previous OC diagnosis and treatment. Each patient used in the study should have temporal information regarding OGD and biopsies taken, the number of biopsies, BO length, BO islands, hiatus hernia, visible lesions and classification of visible lesions documented according to the British Society of Gastroenterology (BSG) minimum endoscopic reporting dataset.1 In resection specimens, full histological reports should be available or at minimum staging tumour stage, nodal involvement, metastasis (TNM). It should be reported which patient samples have been used to perform epigenetic analysis and which methods of laboratory analysis have been used. Clear reporting of statistical analysis between cohorts must be provided to allow future meta-analysis of studies. Improved standardisation would make meta-analyses more feasible and lead to more informative systematic review results. This in turn would aid the translation of laboratory work into clinical trials.
Conclusion
The evidence from this systematic review is suggestive of a role for p16 as an individual epigenetic biomarker in predicting progression from BO to OADC. Prognostic models incorporating this and other markers also suggest a role for p16 in combination with HPP1, RUNX3 and/or clinical markers. Further large primary studies using current epigenetic techniques and standardised reporting are required to inform future models to further explore the role of epigenetics in progression to HGD and OADC.
Supplementary Material
Footnotes
Contributors: TN, CLT, JD and OT conceived the systematic review protocol. TN, CLT, SB, MJP and JD undertook and reviewed scoping searches and contributed to the methodological development of the protocol with input from OT. TN drafted the initial manuscript. All authors were involved in the critical revision of the manuscript and have given approval to the final version to be published.
Funding: Funding for this systematic review has been kindly provided by the Queen Elizabeth Hospital Charity, Birmingham (grant no 16-3-190).
Competing interests: None declared.
Patient consent: Not required.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data sharing statement: No additional data available.
References
- 1. Fitzgerald RC, di Pietro M, Ragunath K, et al. . British Society of Gastroenterology guidelines on the diagnosis and management of Barrett’s oesophagus. Gut 2014;63:7–42. 10.1136/gutjnl-2013-305372 [DOI] [PubMed] [Google Scholar]
- 2. Haggitt RC, Tryzelaar J, Ellis FH, et al. . Adenocarcinoma complicating columnar epithelium-lined (Barrett’s) esophagus. Am J Clin Pathol 1978;70:1–5. 10.1093/ajcp/70.1.1 [DOI] [PubMed] [Google Scholar]
- 3. Solaymani-Dodaran M, Logan RF, West J, et al. . Risk of oesophageal cancer in Barrett’s oesophagus and gastro-oesophageal reflux. Gut 2004;53:1070–4. 10.1136/gut.2003.028076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Melhado RE, Alderson D, Tucker O. The changing face of esophageal cancer. Cancers 2010;2:1379–404. 10.3390/cancers2031379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. NCI. Surveillance, epidemiology and end results programme database: NC Institute, 2014. [Google Scholar]
- 6. CRUK. Cancer Research UK Oesophageal Cancer Survival Statistics. 2011. http://www.cancerresearchuk.org/health-professional/cancer-statistics/statistics-by-cancer-type/oesophageal-cancer/survival (cited 1st Jun 2016).
- 7. Shaheen NJ, Falk GW, Iyer PG, et al. . ACG Clinical Guideline: Diagnosis and Management of Barrett’s Esophagus. Am J Gastroenterol 2016;111:30–50. quiz 51 10.1038/ajg.2015.322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Desai TK, Krishnan K, Samala N, et al. . The incidence of oesophageal adenocarcinoma in non-dysplastic Barrett’s oesophagus: a meta-analysis. Gut 2012;61:970–6. 10.1136/gutjnl-2011-300730 [DOI] [PubMed] [Google Scholar]
- 9. Rastogi A, Puli S, El-Serag HB, et al. . Incidence of esophageal adenocarcinoma in patients with Barrett’s esophagus and high-grade dysplasia: a meta-analysis. Gastrointest Endosc 2008;67:394–8. 10.1016/j.gie.2007.07.019 [DOI] [PubMed] [Google Scholar]
- 10. Shaheen NJ, Sharma P, Overholt BF, et al. . Radiofrequency ablation in Barrett’s esophagus with dysplasia. N Engl J Med 2009;360:2277–88. 10.1056/NEJMoa0808145 [DOI] [PubMed] [Google Scholar]
- 11. Overholt BF, Lightdale CJ, Wang KK, et al. . Photodynamic therapy with porfimer sodium for ablation of high-grade dysplasia in Barrett’s esophagus: international, partially blinded, randomized phase III trial. Gastrointest Endosc 2005;62:488–98. 10.1016/j.gie.2005.06.047 [DOI] [PubMed] [Google Scholar]
- 12. Egger G, Liang G, Aparicio A, et al. . Epigenetics in human disease and prospects for epigenetic therapy. Nature 2004;429:457–63. 10.1038/nature02625 [DOI] [PubMed] [Google Scholar]
- 13. Nieto T, Tomlinson CL, Dretzke J, et al. . Epigenetic biomarkers in progression from non-dysplastic Barrett’s oesophagus to oesophageal adenocarcinoma: a systematic review protocol. BMJ Open 2016;6:e013361 10.1136/bmjopen-2016-013361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Garrity-Park MM, Loftus EV, Sandborn WJ, et al. . Methylation status of genes in non-neoplastic mucosa from patients with ulcerative colitis-associated colorectal cancer. Am J Gastroenterol 2010;105:1610–9. 10.1038/ajg.2010.22 [DOI] [PubMed] [Google Scholar]
- 15. Dhir M, Montgomery EA, Glöckner SC, et al. . Epigenetic regulation of WNT signaling pathway genes in inflammatory bowel disease (IBD) associated neoplasia. J Gastrointest Surg 2008;12:1745–53. 10.1007/s11605-008-0633-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Moriyama T, Matsumoto T, Nakamura S, et al. . Hypermethylation of p14 (ARF) may be predictive of colitic cancer in patients with ulcerative colitis. Dis Colon Rectum 2007;50:1384–92. 10.1007/10350-007-0302-x [DOI] [PubMed] [Google Scholar]
- 17. Osborn NK, Zou H, Molina JR, et al. . Aberrant methylation of the eyes absent 4 gene in ulcerative colitis-associated dysplasia . Clin Gastroenterol Hepatol 2006;4:212–8. 10.1016/j.cgh.2005.11.009 [DOI] [PubMed] [Google Scholar]
- 18. Sato F, Shibata D, Harpaz N, et al. . Aberrant methylation of the HPP1 gene in ulcerative colitis-associated colorectal carcinoma. Cancer Res 2002;62:6820–2. [PubMed] [Google Scholar]
- 19. Kukitsu T, Takayama T, Miyanishi K, et al. . Aberrant crypt foci as precursors of the dysplasia-carcinoma sequence in patients with ulcerative colitis. Clin Cancer Res 2008;14:48–54. 10.1158/1078-0432.CCR-07-1835 [DOI] [PubMed] [Google Scholar]
- 20. Matthews G. Enhanced Neoplasia Detection and Cancer Prevention in Chronic Colitis (ENDCaP-C): NIHR, 2013. [Google Scholar]
- 21. Xu R, Wang F, Wu L, et al. . A systematic review of hypermethylation of p16 gene in esophageal cancer. Cancer Biomark 2013;13:215–26. 10.3233/CBM-130355 [DOI] [PubMed] [Google Scholar]
- 22. Zhao JJ, Li HY, Wang D, et al. . Abnormal MGMT promoter methylation may contribute to the risk of esophageal cancer: a meta-analysis of cohort studies. Tumour Biol 2014;35:10085–93. 10.1007/s13277-014-2276-3 [DOI] [PubMed] [Google Scholar]
- 23. Yang JZ, Ji AF, Wang JS, et al. . Association between Ras association domain family 1A promoter methylation and esophageal squamous cell carcinoma: a meta-analysis. Asian Pac J Cancer Prev 2014;15:3921–5. 10.7314/APJCP.2014.15.9.3921 [DOI] [PubMed] [Google Scholar]
- 24. Wang Y, Qin X, Wu J, et al. . Association of promoter methylation of RUNX3 gene with the development of esophageal cancer: a meta analysis. PLoS One 2014;9:e107598 10.1371/journal.pone.0107598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fu C, Dong W, Wang Z, et al. . The expression of miR-21 and miR-375 predict prognosis of esophageal cancer. Biochem Biophys Res Commun 2014;446:1197–203. 10.1016/j.bbrc.2014.03.087 [DOI] [PubMed] [Google Scholar]
- 26. Fu W, Pang L, Chen Y, et al. . The microRNAs as prognostic biomarkers for survival in esophageal cancer: a meta-analysis. ScientificWorldJournal 2014;2014:1–8. 10.1155/2014/523979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang Y, Wang Q, Zhang N, et al. . Identification of microRNAs as novel biomarkers for detecting esophageal squamous cell carcinoma in Asians: a meta-analysis. Tumour Biol 2014;35:11595–604. 10.1007/s13277-014-2350-x [DOI] [PubMed] [Google Scholar]
- 28. Findlay JM, Middleton MR, Tomlinson I. A systematic review and meta-analysis of somatic and germline DNA sequence biomarkers of esophageal cancer survival, therapy response and stage. Ann Oncol 2015;26:624–44. 10.1093/annonc/mdu449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Findlay JM, Middleton MR, Tomlinson I. Genetic biomarkers of Barrett’s esophagus susceptibility and progression to dysplasia and cancer: a systematic review and meta-analysis. Dig Dis Sci 2016;61:25–38. 10.1007/s10620-015-3884-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hayden JA, van der Windt DA, Cartwright JL, et al. . Assessing bias in studies of prognostic factors. Ann Intern Med 2013;158:280–6. 10.7326/0003-4819-158-4-201302190-00009 [DOI] [PubMed] [Google Scholar]
- 31. Ensor J, Riley RD, Moore D, et al. . Systematic review of prognostic models for recurrent venous thromboembolism (VTE) post-treatment of first unprovoked VTE. BMJ Open 2016;6:e011190 10.1136/bmjopen-2016-011190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Moher D, Liberati A, Tetzlaff J, et al. . Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. BMJ 2009;339:b2535 10.1136/bmj.b2535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Revilla-Nuin B, Parrilla P, Lozano JJ, et al. . Predictive value of MicroRNAs in the progression of barrett esophagus to adenocarcinoma in a long-term follow-up study. Ann Surg 2013;257:886–93. 10.1097/SLA.0b013e31826ddba6 [DOI] [PubMed] [Google Scholar]
- 34. Clément G, Guilleret I, He B, et al. . Epigenetic alteration of the Wnt inhibitory factor-1 promoter occurs early in the carcinogenesis of Barrett’s esophagus. Cancer Sci 2008;99:46–53. 10.1111/j.1349-7006.2007.00663.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Clément G, Braunschweig R, Pasquier N, et al. . Methylation of APC, TIMP3, and TERT: a new predictive marker to distinguish Barrett’s oesophagus patients at risk for malignant transformation. J Pathol 2006;208:100–7. 10.1002/path.1884 [DOI] [PubMed] [Google Scholar]
- 36. Klump B, Hsieh CJ, Holzmann K, et al. . Hypermethylation of the CDKN2/p16 promoter during neoplastic progression in Barrett’s esophagus. Gastroenterology 1998;115:1381–6. 10.1016/S0016-5085(98)70016-2 [DOI] [PubMed] [Google Scholar]
- 37. Barrett MT, Sanchez CA, Prevo LJ, et al. . Evolution of neoplastic cell lineages in Barrett oesophagus. Nat Genet 1999;22:106–9. 10.1038/8816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wang JS, Guo M, Montgomery EA, et al. . DNA promoter hypermethylation of p16 and APC predicts neoplastic progression in Barrett’s esophagus. Am J Gastroenterol 2009;104:2153–60. 10.1038/ajg.2009.300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Schulmann K, Sterian A, Berki A, et al. . Inactivation of p16, RUNX3, and HPP1 occurs early in Barrett’s-associated neoplastic progression and predicts progression risk. Oncogene 2005;24:4138–48. 10.1038/sj.onc.1208598 [DOI] [PubMed] [Google Scholar]
- 40. Sato F, Jin Z, Schulmann K, et al. . Three-tiered risk stratification model to predict progression in Barrett’s esophagus using epigenetic and clinical features. PLoS One 2008;3:e1890 10.1371/journal.pone.0001890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Jin Z, Cheng Y, Gu W, et al. . A multicenter, double-blinded validation study of methylation biomarkers for progression prediction in Barrett’s esophagus. Cancer Res 2009;69:4112–5. 10.1158/0008-5472.CAN-09-0028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Moinova H, Leidner RS, Ravi L, et al. . Aberrant vimentin methylation is characteristic of upper gastrointestinal pathologies. Cancer Epidemiol Biomarkers Prev 2012;21:594–600. 10.1158/1055-9965.EPI-11-1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Boerwinkel DF, Di Pietro M, Liu X, et al. . Endoscopic TriModal imaging and biomarkers for neoplasia conjoined: a feasibility study in Barrett’s esophagus. Dis Esophagus 2014;27:435–43. 10.1111/j.1442-2050.2012.01428.x [DOI] [PubMed] [Google Scholar]
- 44. Eads CA, Lord RV, Wickramasinghe K, et al. . Epigenetic patterns in the progression of esophageal adenocarcinoma. Cancer Res 2001;61:3410–8. [PubMed] [Google Scholar]
- 45. Puertas Canteria A. Analysis of alterations in epithelial DNA methylation as potential epigenetic biomarkers of neoplastic progression in Barrett’s Esophagus. Virchows Archiv 2012;1:S169–S170. [Google Scholar]
- 46. Agarwal R, Jin Z, Yang J, et al. . Epigenomic program of Barrett’s-associated neoplastic progression reveals possible involvement of insulin signaling pathways. Endocr Relat Cancer 2012;19:L5–L9. 10.1530/ERC-11-0364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Riley RD, Abrams KR, Sutton AJ, et al. . Reporting of prognostic markers: current problems and development of guidelines for evidence-based practice in the future. Br J Cancer 2003;88:1191–8. 10.1038/sj.bjc.6600886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Herman JG, Graff JR, Myöhänen S, et al. . Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci U S A 1996;93:9821–6. 10.1073/pnas.93.18.9821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Eads CA, Danenberg KD, Kawakami K, et al. . MethyLight: a high-throughput assay to measure DNA methylation. Nucleic Acids Res 2000;28:32e 10.1093/nar/28.8.e32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bassil CF, Huang Z, Murphy SK. Bisulfite pyrosequencing. Methods Mol Biol 2013;1049:95–107. 10.1007/978-1-62703-547-7_9 [DOI] [PubMed] [Google Scholar]
- 51. Nobori T, Miura K, Wu DJ, et al. . Deletions of the cyclin-dependent kinase-4 inhibitor gene in multiple human cancers. Nature 1994;368:753–6. 10.1038/368753a0 [DOI] [PubMed] [Google Scholar]
- 52. Kamb A, Gruis NA, Weaver-Feldhaus J, et al. . A cell cycle regulator potentially involved in genesis of many tumor types. Science 1994;264:436–40. 10.1126/science.8153634 [DOI] [PubMed] [Google Scholar]
- 53. Mokrowiecka A, Wierzchniewska-Ławska A, Smolarz B, et al. . p16 gene mutations in Barrett’s esophagus in gastric metaplasia - intestinal metaplasia - dysplasia - adenocarcinoma sequence. Adv Med Sci 2012;57:71–6. 10.2478/v10039-012-0003-0 [DOI] [PubMed] [Google Scholar]
- 54. Timmer MR, Martinez P, Lau CT, et al. . Derivation of genetic biomarkers for cancer risk stratification in Barrett’s oesophagus: a prospective cohort study. Gut 2016;65:1602–10. 10.1136/gutjnl-2015-309642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Pidsley R, Zotenko E, Peters TJ, et al. . Critical evaluation of the Illumina MethylationEPIC BeadChip microarray for whole-genome DNA methylation profiling. Genome Biol 2016;17:208 10.1186/s13059-016-1066-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sekula P, Pressler JB, Sauerbrei W, et al. . Assessment of the extent of unpublished studies in prognostic factor research: a systematic review of p53 immunohistochemistry in bladder cancer as an example. BMJ Open 2016;6:e009972 10.1136/bmjopen-2015-009972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kyzas PA, Loizou KT, Ioannidis JP. Selective reporting biases in cancer prognostic factor studies. J Natl Cancer Inst 2005;97:1043–55. 10.1093/jnci/dji184 [DOI] [PubMed] [Google Scholar]
- 58. Ntzani EE, Ioannidis JP. Predictive ability of DNA microarrays for cancer outcomes and correlates: an empirical assessment. Lancet 2003;362:1439–44. 10.1016/S0140-6736(03)14686-7 [DOI] [PubMed] [Google Scholar]
- 59. Nicholson A, Kuper H, Hemingway H. Depression as an aetiologic and prognostic factor in coronary heart disease: a meta-analysis of 6362 events among 146 538 participants in 54 observational studies. Eur Heart J 2006;27:2763–74. 10.1093/eurheartj/ehl338 [DOI] [PubMed] [Google Scholar]
- 60. Hemingway H, Riley RD, Altman DG. Ten steps towards improving prognosis research. BMJ 2009;339:b4184 10.1136/bmj.b4184 [DOI] [PubMed] [Google Scholar]
- 61. Sauerbrei W. Prognostic factors. Confusion caused by bad quality design, analysis and reporting of many studies. Adv Otorhinolaryngol 2005;62:184–200. 10.1159/000082508 [DOI] [PubMed] [Google Scholar]
- 62. Riley RD, Hayden JA, Steyerberg EW, et al. . Prognosis research strategy (PROGRESS) 2: prognostic factor research. PLoS Med 2013;10:e1001380 10.1371/journal.pmed.1001380 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
bmjopen-2017-020427supp001.pdf (98.4KB, pdf)
bmjopen-2017-020427supp002.pdf (202.4KB, pdf)