Abstract
MicroRNAs are a class of small non-coding RNAs that are involved in many important biological processes and the dysfunction of microRNA has been associated with many diseases. The seed region of a microRNA is of crucial importance to its target recognition. Mutations in microRNA seed regions may disrupt the binding of microRNAs to their original target genes and make them bind to new target genes. Here we use a knowledge-based computational method to systematically predict the functional effects of all the possible single nucleotide mutations in human microRNA seed regions. The result provides a comprehensive reference for the functional assessment of the impacts of possible natural and artificial single nucleotide mutations in microRNA seed regions.
Keywords: miRNA seed, Mutation, Gene ontology, miR2GO, miRNA rank
1. Introduction
MicroRNAs (miRNAs) are small non-coding RNAs that play important roles in post-transcriptional regulation. Mature miRNAs are approximately 22 nucleotides long. Nucleotides 2–8 in the miRNA sequences are the seed region that provides guiding information for miRNA target recognition. It has been estimated that more than 60 percent of human genes have at least one conserved miRNA binding site [1]. Strong associations have been found between miRNAs and many diseases such as cancer [2], [3], [4], [5], [6], [7], Type II diabetes [8], [9], cardiovascular diseases [10], autoimmune disorders [11], Alzheimer’s disease [12] and viral diseases [13].
Genetic and somatic mutations in miRNA seed sequences and miRNA target sites can potentially create and disrupt the interactions between miRNAs and their targets. PolymiRTS is a database of naturally occurring polymorphisms in that create or disrupt miRNA-mRNA interactions [14], [15], [16]. About 25,000 SNPs and 1000 INDELs in the miRNA target sites are annotated in PolymiRTS. Among them nearly 1500 SNPs and 700 INDELs are interfering 38 different disease related pathways. A miRNA seed mutation may have big functional impacts [17] because it may disrupt or create several hundreds of miRNA-mRNA interactions. PolymiRTS contains a catalogue of 271 SNPs and 23 INDELs in the miRNA seeds. Some microRNA seed mutations have already been linked to diseases [18], [19].
To investigate the functional impacts of miRNA seed mutations, we developed miR2GO [20], a web-based computational platform. miR2GO is equipped with miRmut2GO [20] module for assessing the impacts of the seed mutations. miRmut2GO compares the functional similarities between the target gene sets of reference and mutated seed sequences. Functional similarities are measured by the semantic similarity of the gene ontology terms that are associated with the target gene sets [21]. For example, a similarity score less than 0.5 indicates less than 50 % functional similarities between the reference and the mutated target sets. The web-based interface of miR2GO allows users to input any set of miRNA seed mutations and evaluate the functional impacts of each mutation. In a subsequent functional analysis of 517 SNPs in miRNA seed regions, miR2GO have assigned very low functional similarity scores for miRNA SNPs +57C>T in hsa-miR-184 and +13G>A in hsa-miR-96 which are associated with the risk of EDICT Syndrome [19] and progressive hearing loss [18], respectively.
We use miR2GO scores for systematic assessment of the functional impacts of seed mutations in human microRNAs. Functional study on all possible seed mutations is important for selecting the seed patterns for designing an artificial miRNA. Artificial miRNA therapy has shown great potential in the treatments of diseases including cancers [22], [23], [24], [25]. A comprehensive list of miR2GO scores from all possible seed mutations would be a valuable resource for determining the best seed pattern for a specific drug therapy. In this work we have systematically evaluated all possible human miRNA seed mutations, assigned miR2GO scores and ranked them based on their probabilities. A detailed gene ontology graph based analysis on a top ranked seed mutation is also featured in the result section.
2. Workflow and Implementation
2.1. Data Collection
Mature miRNA sequences along with their genomic coordinates were downloaded from the miRBase release 21 [26]. Seed sequences and their corresponding genomic coordinates were determined from 2–8 nucleotide locations of the mature miRNAs. SNPs data were downloaded from dbSNPs build 147 of human genome build GRCH38 [27]. In Order to enhance the quality of our analysis we only considered the dbSNP entries from the 1000 Genomes Project. Figure 1 shows the data integration and the analysis workflow.
Figure 1:
Workflow for assessing and ranking all possible seed mutations from miRmut2GO score and mutation probability.
2.2. Assessing all Possible Seed Mutations with the miRmut2GO
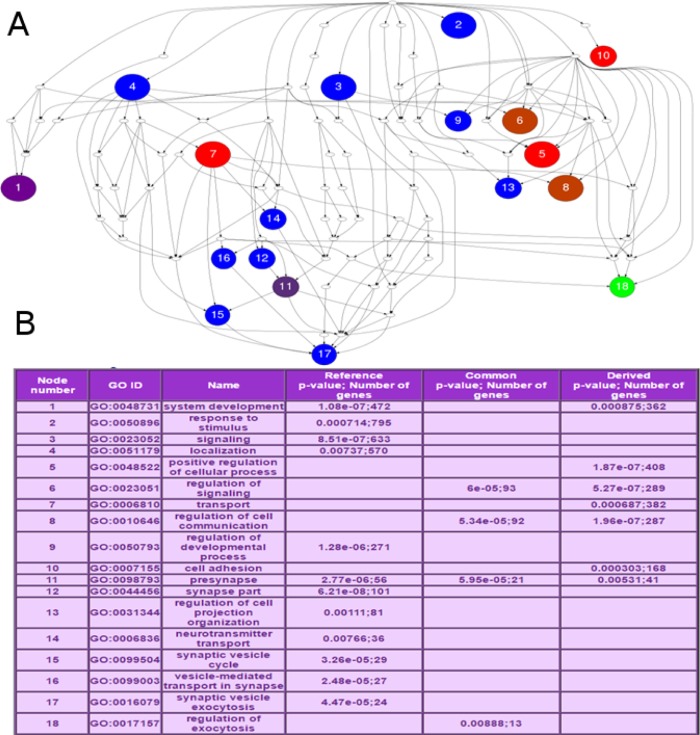
Each unique seed (seed per miRNA family) from miRBase [26] were imputed with all the possible mutations. The mutations were submitted for processing through miRmut2GO interface. miRmut2GO separately predicted the target gene sets for the reference and mutated miRNAs. From miRmut2GO we run both TargetScan [28] and miRanda [29] target prediction algorithms. Common targets of the reference and mutated miRNAs were processed for computing the Gene Ontology based functional similarity scores. miRmut2GO utilized gProfileR [30] package for computing the enriched Gene Ontology terms for each target gene set. Similar Gene Ontology terms were combined based on their hierarchy in the Gene Ontology graph. The default p-value threshold (0.01) was used for the function enrichment test. In the final step, miRmut2GO reported the semantic similarity scores (i.e. miR2GO scores) for the functional similarities between the target gene sets of the reference and the mutated miRNA [31]. miR2GO score is a number between “0” and “1”. miR2GO score for complete functional similarity is “1” and complete functional dissimilarity is “0”. miR2GO score is “NA” when there is no functional enrichment for either the reference or the mutated target gene sets. We discarded all the “NA” scores from our analysis. The detailed Gene Ontology figure (Figure 4) was also generated using miRmut2GO.
Figure 4:
A Gene Ontology graph for C->T mutation at seed position 7 of hsa-miR-615-5p. (A) The Gene Ontology figure in the example shows the distribution of enriched GO terms (with p-value <0.01) in the GO hierarchy by varying the node colors for the enriched terms from reference targets, derived targets and common targets. Number of target genes for reference targets), derived targets and common targets determine the “blue”, “green” and “red” components of nodes color respectively. The node colors are varied for changes in the number of targets to form the color triangle (for details see the HELP page of miR2GO, http://compbio.uthsc.edu/miR2GO/help_GO.php). (B) The Gene Ontology ID and name for each node in A.
2.3. Ranking Seed Mutations Based on Combined Mutation Probabilities
The probability of observing a mutation depends on both the probability of observing the nucleotide change and the probability of observing a mutation at that seed position. For each mutation, first we multiplied the “Mutation Probability in Seed Positions” and “Mutation Type Probability” to report the “Combined Mutation Probability”. Then we sorted the mutations based on the decreasing magnitude of their “Combined Mutation Probability”. The mutations with the same “Combined Mutation Probability” were again sorted based on the increasing order of miR2GO scores. The sorted mutations were then assigned the rank values. Figure 1 describes the complete workflow in details. Mutation Probability in Seed Positions: The genomic coordinates of the miRNA seeds were compared against the dbSNP entries for finding the seed mutations. The seed mutations were then grouped into seven groups, one for each seed position. The probabilities of finding a seed mutation in each seed position were computed from the ratio of the number of seed mutations in each group and total number of seed mutations. Mutation Type Probability: We computed the probabilities of all 21 possible mutations in each miRNA seed.
3. Results and Discussion
We found 2047 unique seed sequences in the 2588 mature human miRNAs from the miRBase (Release 21). Among them 670 seed sequences were found to obtain at least one valid miR2GO score for the mutations. We found a total of 12,401 miR2GO scores from all the possible seed mutations. Figure 2 shows the distribution of miR2GO scores. Approximately 50 % of scores are at the either sides of the middle of “Y” axis (where miR2GO score = 0.5). We also computed the average miR2GO score values for each seed position (Figure 3). The average score for seed position 8 is higher than the others, which indicates that the mutations at base “8” may have lower functional effects than those at the other seed positions.
Figure 2:
Distribution of miR2GO scores for all possible seed mutations.
Figure 3:
Average miR2GO scores by miRNA seed positions (nucleotide bases) from all possible seed mutations.
Interestingly, the ten top ranked seed mutations are all “C->T” (C/U) mutations (Table 1). Supplementary Table 1 listed the complete table of all mutations with miR2GO scores and ranking. Figure 4 shows the miR2GO graph for “C->T” mutation in hsa-miR-615-5p which is the first entry (Rank 1) in Table 1. The nodes in the graph represent the Gene Ontology categories (biological processes) that are enriched among the miRNA target genes. Blue nodes represent the enriched Gene Ontology categories for reference seed sequence “GGGGUCC”. Red nodes represent the enriched Gene Ontology categories for mutated seed sequence “GGGGUUC”. In Figure 4, the enriched categories for reference and mutation are clearly separated in different branches of the Gene Ontology graph. At “node 5” of Figure 4, the Gene Ontology term “GO:0048522; positive regulation of cellular process” is significantly enriched with a p-value of 1.87e−07 among 408 target genes of the mutated hsa-miR-615-5p. On the other hand, at “node 17” a very different Gene Ontology term “GO:0016079; synaptic vesicle exocytosis” is significantly enriched with a p-value of 4.47e−05 among the reference targets of hsa-miR-615-5p.
Table 1:
Top ranked microRNA seed mutations.
| Seed | miRNA | Score | Rank |
|---|---|---|---|
| GGGGU[C/U]C | hsa-miR-615-5p | 0.038 | 1 |
| CUGGG[C/U]A | hsa-miR-5189-5p | 0.084 | 2 |
| GGGGU[C/U]G | hsa-miR-7704 | 0.138 | 3 |
| UCUAG[C/U]C | hsa-miR-1287-3p | 0.145 | 4 |
| CGCCU[C/U]C | hsa-miR-1281 | 0.173 | 5 |
| AAAUU[C/U]G | hsa-miR-10a-3p | 0.187 | 6 |
| AUCAU[C/U]G | hsa-miR-136-3p | 0.189 | 7 |
| AAGAC[C/U]C | hsa-miR-3667-5p | 0.205 | 8 |
| GGGCG[C/U]G | hsa-miR-3178 | 0.209 | 9 |
An intronic region of Hoxc5 is known for transcribing the precursor miRNA, or pre-miRNA of hsa-miR-615-5p [32]. Possible link between developmental processes and hsa-miR-615-5p has been identified [32]. We found a functional category “regulation of developmental process” at “node 9” was significantly enriched among the reference targets of hsa-miR-615-5p with a p-value of 1.28e−06 which was not found enriched among the mutated targets. Our observation indicates the importance of “C” to “T” mutation at the 7th nucleotide of hsa-miR-615-5p.
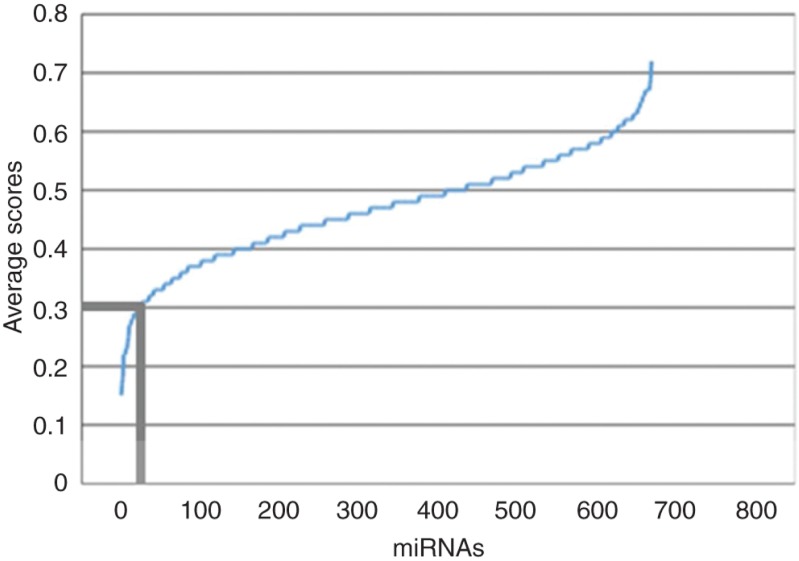
We calculated the average miR2GO score for all the miRNA families. The distribution of the average miR2GO scores of miRNA families is shown in Figure 5. We found that only a small number of miRNA families have low average scores. For example, Only 26 miRNA families have an average miR2GO score 0.3 or less.
Figure 5:
Distribution of average miR2GO scores for miRNA families. There are 26 miRNA families with an average miR2GO score of 0.3 or less.
4. Conclusion
In this article we report our findings from the analysis of all the possible single nucleotide mutations in miRNA seeds. We used our previously designed miR2GO [20] software for systematically predicting the functional effects of mutations. We ranked the miRNA mutations based on their probabilities and functional effects. Most importantly, the results presented here provide a reference for functionally assessing the impacts of all possible natural and artificial single nucleotide mutations in the microRNA seed regions. Scoring and ranking of all miRNA seed mutations may provide a guide for artificial miRNA design and facilitate the selection of a candidate seed [22], [23], [24], [25].
Supporting Information
Supplemental Material
The online version of this article offers supplementary material (DOI: jib-2017-0001).
Contributor Information
Anindya Bhattacharya, Email: anindyamail123@gmail.com.
Yan Cui, Email: ycui2@uthsc.edu.
Conflict of interest statement:
Authors state no conflict of interest. All authors have read the journal’s Publication ethics and publication malpractice statement available at the journal’s website and hereby confirm that they comply with all its parts applicable to the present scientific work.
References
- [1].Siomi H, Siomi MC. Posttranscriptional regulation of microRNA biogenesis in animals. Mol Cell. 2010;38:323–32. doi: 10.1016/j.molcel.2010.03.013. [DOI] [PubMed] [Google Scholar]
- [2].Bhattacharya A, Cui Y. SomamiR 2.0: a database of cancer somatic mutations altering microRNA-ceRNA interactions. Nucl Acids Res. 2016;44:D1005–1010. doi: 10.1093/nar/gkv1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Fu SW, Chen L, Man YG. miRNA biomarkers in breast cancer detection and management. J Cancer. 2011;2:116–22. doi: 10.7150/jca.2.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Luo ZJ, Zhao Y, Azencott R. Impact of miRNA sequence on miRNA expression and correlation between miRNA expression and cell cycle regulation in breast cancer cells. PLoS One. 2014;9:e95205. doi: 10.1371/journal.pone.0095205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Bhattacharya A, Ziebarth JD, Cui Y. SomamiR: a database for somatic mutations impacting microRNA function in cancer. Nucl Acids Res. 2013;41:D977–982. doi: 10.1093/nar/gks1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Ziebarth JD, Bhattacharya A, Cui Y. Integrative analysis of somatic mutations altering MicroRNA targeting in cancer genomes. PLoS One. 2012;7:e47137. doi: 10.1371/journal.pone.0047137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Fan P, Chen Z, Tian P, Liu W, Jiao Y, Xue Y. et al. miRNA biogenesis enzyme drosha is required for vascular smooth muscle cell survival. PLoS One. 2013;8:e60888. doi: 10.1371/journal.pone.0060888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Foley NH, O’Neill LA. miR-107: a Toll-like receptor-regulated miRNA dysregulated in obesity and type II diabetes. J Leukocyte Biol. 2012;92:521–7. doi: 10.1189/jlb.0312160. [DOI] [PubMed] [Google Scholar]
- [9].Chen YY, Wang XJ, Shao XY. A combination of human embryonic stem cell-derived pancreatic endoderm transplant with LDHA-repressing miRNA can attenuate high-fat diet induced type II diabetes in mice. J Diabetes Res. 2015;2015:796912. doi: 10.1155/2015/796912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Nouraee N, Mowla SJ. miRNA therapeutics in cardiovascular diseases: promises and problems. Front Genet. 2015;6:232. doi: 10.3389/fgene.2015.00232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Prabahar A, Natarajan J. MicroRNA mediated network motifs in autoimmune diseases and its crosstalk between genes, functions and pathways. J Immunol Methods. 2017;440:19–26. doi: 10.1016/j.jim.2016.10.002. [DOI] [PubMed] [Google Scholar]
- [12].Shioya M, Obayashi S, Tabunoki H, Arima K, Saito Y, Ishida T. et al. Aberrant microRNA expression in the brains of neurodegenerative diseases: miR-29a decreased in Alzheimer disease brains targets neurone navigator 3. Neuropathol Appl Neurobiol. 2010;36:320–30. doi: 10.1111/j.1365-2990.2010.01076.x. [DOI] [PubMed] [Google Scholar]
- [13].Auvinen E. Diagnostic and prognostic value of MicroRNA in viral diseases. Mol Diagn Ther. 2017;21:45–57. doi: 10.1007/s40291-016-0236-x. [DOI] [PubMed] [Google Scholar]
- [14].Bhattacharya A, Ziebarth JD, Cui Y. PolymiRTS database 3.0: linking polymorphisms in microRNAs and their target sites with human diseases and biological pathways. Nucl Acids Res. 2014;24(Database issue):D86–91. doi: 10.1093/nar/gkt1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Ziebarth JD, Bhattacharya A, Chen A, Cui1 Y. PolymiRTS Database 2.0: linking polymorphisms in microRNA target sites with human diseases and complex traits. Nucl Acids Res. 2012;40:D216–21. doi: 10.1093/nar/gkr1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Bao L, Zhou M, Wu L, Lu L, Goldowitz D, Williams RW. et al. PolymiRTS Database: linking polymorphisms in microRNA target sites with complex traits. Nucl Acids Res. 2007;35:D51–4. doi: 10.1093/nar/gkl797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Siristatidis CS, Gibreel A, Basios G, Maheshwari A, Bhattacharya S. Gonadotrophin-releasing hormone agonist protocols for pituitary suppression in assisted reproduction. Cochrane Database Syst Rev. 2015;11:CD006919. doi: 10.1002/14651858.CD006919.pub4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Mencia A, Modamio-Hoybjor S, Redshaw N, Morín M, Mayo-Merino F, Olavarrieta L. et al. Mutations in the seed region of human miR-96 are responsible for nonsyndromic progressive hearing loss. Nat Genet. 2009;41:609–13. doi: 10.1038/ng.355. [DOI] [PubMed] [Google Scholar]
- [19].Iliff BW, Riazuddin SA, Gottsch JD. A single-base substitution in the seed region of miR-184 causes EDICT syndrome. Invest Ophthalmol Vis Sci. 2012;53:348–53. doi: 10.1167/iovs.11-8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Bhattacharya A, Cui Y. miR2GO: comparative functional analysis for microRNAs. Bioinformatics. 2015;31:2403–5. doi: 10.1093/bioinformatics/btv140. [DOI] [PubMed] [Google Scholar]
- [21].Wang JZ, Du Z, Payattakool R, Yu PS, Chen CF. A new method to measure the semantic similarity of GO terms. Bioinformatics. 2007;23:1274–81. doi: 10.1093/bioinformatics/btm087. [DOI] [PubMed] [Google Scholar]
- [22].Liu C, Wang S, Zhu S, Wang H, Gu J, Gui Z. et al. MAP3K1-targeting therapeutic artificial miRNA suppresses the growth and invasion of breast cancer in vivo and in vitro. Springerplus. 2016;5:11. doi: 10.1186/s40064-015-1597-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Zhan Y, Liu Y, Lin J, Fu X, Zhuang C, Liu L. et al. Synthetic Tet-inducible artificial microRNAs targeting beta-catenin or HIF-1alpha inhibit malignant phenotypes of bladder cancer cells T24 and 5637. Sci Rep. 2015;5:16177. doi: 10.1038/srep16177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Van Vu T, Do VN. Customization of artificial MicroRNA design. Methods Mol Biol. 2017;1509:235–43. doi: 10.1007/978-1-4939-6524-3_21. [DOI] [PubMed] [Google Scholar]
- [25].Tay FC, Lim JK, Zhu HB, Hin LC, Wang S. Using artificial microRNA sponges to achieve microRNA loss-of-function in cancer cells. Adv Drug Deliver Rev. 2015;81:117–27. doi: 10.1016/j.addr.2014.05.010. [DOI] [PubMed] [Google Scholar]
- [26].Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucl Acids Res. 2014;42:D68–73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM. et al. dbSNP: the NCBI database of genetic variation. Nucl Acids Res. 2001;29:308–11. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. eLife. 2015;4:e05005. doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Enright AJ, John B, Gaul U, Tuschl T, Sander C, Marks DS. MicroRNA targets in drosophila. Genome Biol. 2003;5:R1. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Reimand J, Arak T, Adler P, Kolberg L, Reisberg S, Peterson H. et al. g:Profiler-a web server for functional interpretation of gene lists (2016 update) Nucl Acids Res. 2016;44:W83–9. doi: 10.1093/nar/gkw199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Yu GC, Li F, Qin YD, Bo XC, Wu YB, Wang SQ. GOSemSim: an R package for measuring semantic similarity among GO terms and gene products. Bioinformatics. 2010;26:976–8. doi: 10.1093/bioinformatics/btq064. [DOI] [PubMed] [Google Scholar]
- [32].Quah S, Holland PW. The Hox cluster microRNA miR-615: a case study of intronic microRNA evolution. Evodevo. 2015;6:31. doi: 10.1186/s13227-015-0027-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.