Abstract
Vitamin A deficiency is a prevalent health problem in many areas of the world, where cassava genotypes with high pro-vitamin A content have been identified as a strategy to address this issue. In this study, we found a positive correlation between the color of the root pulp and the total carotenoid contents and, importantly, showed how CIELAB color measurements can be used as a non-destructive and fast technique to quantify the amount of carotenoids in cassava root samples, as opposed to traditional methods. We trained several machine learning models using UV-visible spectrophotometry data, CIELAB data and a low-level data fusion of the two. Best performance models were obtained for the total carotenoids contents calculated using the UV-visible dataset as input, with R2 values above 90 %. Using CIELAB and fusion data, values around 60 % and above 90 % were found. Importantly, these results demonstrated how data fusion can lead to a better model performance for prediction when comparing to the use of a single data source. Considering all these findings, the use of colorimetric data associated with UV-visible and HPLC data through statistical and machine learning methods is a reliable way of predicting the content of total carotenoids in cassava root samples.
Keywords: Carotenoids, Cassava genotypes, Chemometrics, CIELAB, Machine learning
1. Introduction
Carotenoids refer to the most important natural pigments, having already been described more than 700 phytochemicals of this class of secondary metabolites, with a broad range of structures and polarities. About 50 carotenoids are vitamin A precursors, however, only three of these represent major sources in the human diet (β-carotene, α-carotene and β-cryptoxanthin) [1].
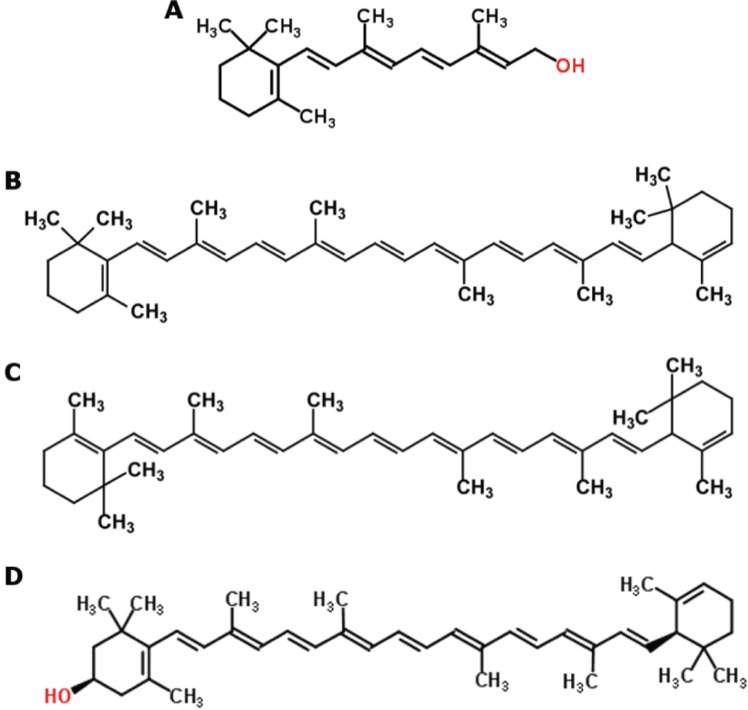
In terms of chemical structure, vitamin A is essentially a half of the β-carotene molecule with a water molecule at the end of the lateral polyene chain (Figure 1), making β-carotene a potent vitamin A precursor, with 100 % of activity assigned to it [2]. Having a broad range of colors, varying from yellow to dark red, carotenoids are responsible for the color of many plant leaves, fruits and flowers, as well as birds, insects, fish, and crustaceans. However, they are only produced by plants, bacteria, fungi and algae, while other organisms can only incorporate them through their diets.
Figure 1:
Structural formula of vitamin A (A) and of the carotenoids β-carotene (B), α-carotene (C) and β-cryptoxanthin (D), the major carotenoid sources in the human diet [3].
The recognized benefits of carotenoid consumption include their correlation with a diminished risk of several degenerative disorders, including various types of cancer, cardiovascular or ophthalmological diseases, as well as their preventive effect associated with their antioxidant activity, protecting cells and tissues from oxidative damage [4].
Though sources of provitamin A carotenoids around the world are abundant, vitamin A deficiency is a leading cause of morbidity and mortality, especially in young children and pregnant and lactating women in less favored countries, affecting around 250 million people worldwide. Therefore, food-based interventions focused on alleviating vitamin A deficiency in susceptible populations have advantages over supplementation and fortification programs, especially in rural areas, because they can provide a sustainable source of a variety of nutrients and other phytochemicals without the recurring transport and administration costs of these other methods [1].
Cassava is the commonly used term to designate the Manihot esculenta species. This tuberous-root plant species offers a wide variety of agronomic advantages, being resilient to droughts, inexpensive, resistant to major diseases and pests, easy to grow and having flexible harvest times, allowing farmers to harvest the roots as needed. It is, therefore, a valuable source of energy for people living in the poorest regions. However, this crop is also a poor source of provitamin A carotenoids, whose deficiency is a major problem in those regions. Thus, the identification of cassava genotypes with high contents of pro-vitamin A carotenoids as a strategy to reduce the prevalence of deficiency of this vitamin is of most importance for the global food security and nutrition [5], [6].
A typical trait of cassava is its starchy root, whose color can vary from white to red. The color is strongly correlated to the presence and contents of several carotenoid pigments and their associations [7]. However, the possibility of adopting the color of roots as an indirect criterion for selection of higher carotene content is questionable, since color is a characteristic of difficult visual evaluation.
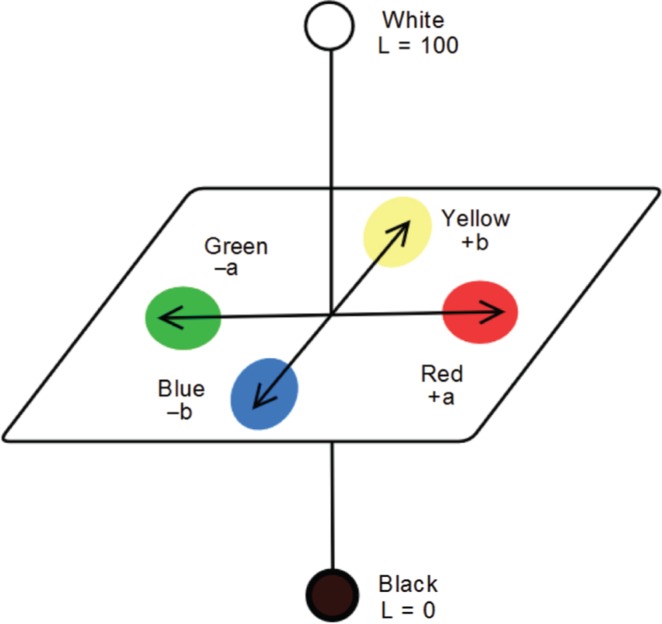
Therefore, the use of a standardized color measurement technique is of most importance. One such technique was adopted by the Commission Internationale de L’Eclairage (CIE) and is based on the Lab color space, which describes mathematically all perceivable colors in three dimensions: L* for lightness and a* and b* for the color opponents green-red and blue-yellow, hence the CIE L* a* b* or CIELAB color scale designation (Figure 2). The values of these three variables are usually absolute, with the L* value representing the darkest black at L* = 0, and the brightest white at L* = 100. On the other hand, the a* value represents red and green opponents at positive and negative values, respectively, while the b* value represents yellow and blue opponents at positive and negative values, respectively. Both color channels, a* and b*, will represent true neutral gray values at a* = 0 and b* = 0 [8], [9].
Figure 2:
Representation of the CIE L* a* b* color space.
CIELAB is currently the most used system for quantitative color description of an object, due to its uniformity, ease of acquisition, very low cost and device independence, having been used for instance in the unique identification of skin color for clinical and scientific purposes [10] and as an optimal color design approach for transforming patients’ perception into color elements [11].
Chemical extraction, followed by the identification and quantification of carotenoid pigments, especially by UV-visible spectrophotometry and high performance liquid chromatography (HPLC) are very accurate, but extremely expensive, also requiring a long time for the analysis. Since the CIELAB color measurement is a non-destructive and very fast technique, that facilitates the acquisition of measurements in the field, while also avoiding the degradation of the compounds, it becomes an appealing approach in comparison to traditionally used methods.
Throughout the last years, there has been a massive increase in available scientific data. While contributing to a more informed community, the large amounts of data can pose a problem when it comes to the analysis, which needs to be automated so that information can be more efficiently retrieved. Machine learning provides systems with the ability to automatically learn and improve from experience without being explicitly programmed, focusing on the development of computer programs that can access and learn for data. The learning process begins with data that is provided and searched for patterns, so that better decisions can be made in the future. This is often a very attractive alternative to manually look for such patterns, and in the last decade the use of machine learning has spread rapidly throughout computer science and beyond [12], [13].
Data fusion is a process of combining data from different sources to improve the performance of prediction models. It deals with association, detection, correlation, and estimation of data to achieve a better information of the system’s state. Low-level fusion is made on a data level, by direct association and combination of raw data, representing measures of the same physical phenomena. After data combination, a feature vector is extracted and used in a machine learning process. It aims to provide more accurate results, assuming proper data association [14].
The aim of this work is to validate a quantification method for carotenoid content estimation in roots of M. esculenta from colorimetric data using the CIELAB color system, assuming that the statistical and machine learning techniques can correlate colorimetric data easily obtained in the field with the contents obtained through traditional techniques such as UV-visible spectrophotometry and HPLC.
The present study employs machine learning, as well as other statistical techniques and bioinformatics tools to detect genotypes of M. esculenta with high contents of carotenoids, while also providing tools that can support the plant-breeding program at Epagri (Agricultural Research Company and Rural Extension of the State of Santa Catarina, http://www.epagri.sc.gov.br/) that aims to obtain genotypes with high levels of pro-vitamin A carotenoids and superior nutritional traits.
2. Materials and Methods
2.1. Selection of Cassava Genotypes
Roots of fifty M. esculenta genotypes harvested in 2015/2016 season from the Epagri’s germplasm bank (Urussanga Experimental Station, 28∘31′18′′S, 49∘19′03′′W, Santa Catarina, southern Brazil) were used in this study due to their economic and social importance.
All genotypes were cultivated under the same soil, climatic conditions and agricultural treatments. Importantly, the investigated genotypes were pre-selected according to their relevance for biofortification projects, due to the presence of carotenoids with provitamin A activity and lycopene (visual selection), low levels of cyanogenic glycosides and suitable agronomic traits (e.g. high yield, resistance to drought and to pests and diseases), being widely cultivated in southern Brazil. In fact, the Epagri plant breeder team indicated fifty genotypes from the germplasm bank that were also preferred by local small farmers for commercial production due to their physiochemical variability.
2.2. Carotenoid Extraction and Quantification
Carotenoids were extracted from fresh cassava roots as described by Rodriguez-Amaya and Kimura [15] using an Ultra-Turrax (Janke & Kunkel IKA - T25 basic) and mixture of acetone: petroleum ether (v/v) as extraction solution.
The absorbances of the organosolvent extracts were then recorded on an UV-visible spectrophotometer (Gold Spectrum lab 53 UV-Vis spectrophotometer, BEL photonics, Brazil) using a spectral window from 200 to 700 ηm. Aliquots (10 μL) of the extracts were also injected into a liquid chromatograph (LC-10A Shimadzu) system equipped with a C18 reversed-phase column (Vydac 201TP54, 250 mm × 4.6 mm, 5 μm ϕ, 35 °C) coupled to a pre-column (C18 Vydac 201TP54, 30 mm × 4.6 mm, 5 μm ϕ) and a spectrophotometric detector (450 nm). A mixture of methanol: acetonitrile (90:10, v/v) was used for elution at a flow rate of 1 mL/min. The identification and quantification of compounds of interest was carried out via co-chromatography and comparison of retention times of samples with those of standard compounds (Sigma-Aldrich, USA) under the same experimental conditions.
The color measurements of the root samples were made immediately after harvest using a colorimeter (CR-400, Minolta®, Japan) and the results expressed according to the CIELAB color space scale [5]. For all fifty samples, three readings were performed at different sites.
2.3. Statistical Analysis
Data relating to the quantification of carotenoids were expressed as the mean (μg carotenoids /g root - dry weight) ± standard deviation and submitted to an analysis of variance (ANOVA) followed by post-hoc Tukey’s honest significant difference test (p < 0.05) for mean comparison.
Spectrophotometric data and the amounts of the target carotenoids determined by HPLC were treated using multivariate statistical analysis and chemometrics techniques supported by scripts written in R language (v. 3.3.1) [16].
All data analysis were supported and structured using the R specmine package [17]. This package was developed within our research group and allows the integrated analysis of metabolomics and spectral data, providing a set of methods for metabolomic data analysis, including the loading of data in different formats, preprocessing, metabolite identification, univariate and multivariate data analysis, machine learning, and feature selection. It is a package that allows conducting a data analysis pipeline, exploring and applying different methods of analysis, while also having functions that provide abundant options for graphical visualization of the results.
2.4. Machine Learning
In order to obtain machine learning models capable of accurately predicting the carotenoid contents in cassava roots, we used regression-derived statistical and machine learning models, such as least absolute shrinkage and selection operator (Lasso), ridge and linear regression, regression trees, random forests, elastic network, partial least squares (PLS), support vector machines (SVM), and K-nearest neighbors models [12], [13].
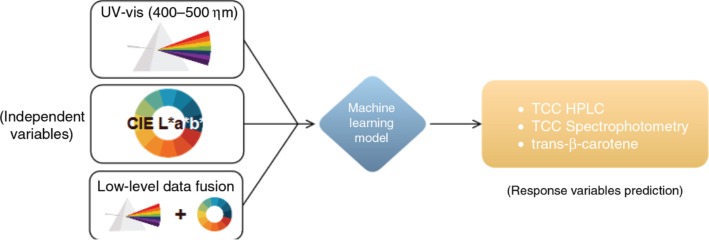
Data from UV-visible spectrophotometry, CIELAB, as well as a fusion of the two were used as inputs to each of the referred machine learning models. The total carotenoid content (TCC) determined by spectrophotometry using the Lambert-Beer law and the TCC determined by HPLC were used as response variables for the prediction. The content of trans-β-carotene determined by HPLC, the most abundant carotene in cassava roots, was also used as an alternative response variable for the models (Figure 3).
Figure 3:
Machine learning approach used. Three different datasets were used as input to the models, namely the UV-visible, CIELAB and fusion datasets. The response variables used for prediction were the total carotenoid content (TCC) determined by spectrophotometry (Lambert-Beer law) and the TCC and trans-β-carotene content determined by HPLC.
This being a regression problem, the chosen evaluation metrics to compare model performance were the root mean square error (RMSE) and the coefficient of determination (R2), since they explicitly show how much the model predictions deviate, on average, from the actual values in the dataset.
2.4.1. UV-Visible Dataset
Considering that most carotenoids exhibit absorption in the visible region of the spectrum, between 400 and 500 ηm, a subset of the original UV-visible dataset was used, with samples belonging to this wavelength interval (101 features). Additionally, missing values contained within this dataset were replaced with the mean of the variables’ values.
Using the different response variables for prediction, we selected the models that showed best performance and the variable importance was calculated. A set of pre-processing methods was applied to the datasets to see whether model performance could be improved, using the models that showed best performance with raw data. These pre-processing methods included smoothing interpolation, scaling, multiplicative scatter correction (MSC), first derivative calculation and background, offset and baseline corrections. The data was also subject to filter-based feature selection (40, 60 and 80 % data filtering) to determine if it could improve model performance.
2.4.2. CIELAB Dataset
The analysis pipeline was similar for the CIELAB dataset, however, linear regression models with feature selection and the data pre-processing and filtering processes were excluded from the analysis pipeline, as it did not make sense to perform these, considering there are only three features in the dataset (L* a* and b* parameters), while pre-processing was meant for spectral data.
2.4.3. Fusion Dataset
For the fusion dataset, which contained 104 variables (absorbance values + L* a* b* parameters), the analysis pipeline was similar to that of the CIELAB dataset, while data filtering was also performed similarly to the UV-visible dataset.
2.4.4. Tools and Reporting
All R scripts, raw data and additional analysis pipelines reports are available as supplementary material at http://darwin.di.uminho.pt/pacbb2017/cassava-carotenoids/, allowing full reproducibility of the experiments. These were made using the already mentioned specmine R package, which provides a number of functions to train, use and evaluate machine learning methods, mostly based in the R package caret [18].
3. Results and Discussion
3.1. Determination of Carotenoid Contents
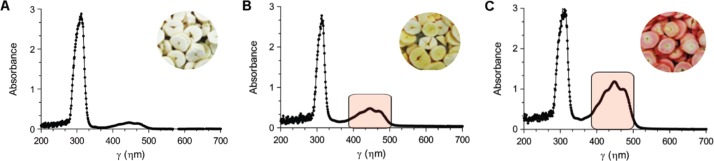
The UV-visible spectrophotometric profiles measured between 200 and 700 ηm clearly allow us to discriminate samples according to their carotenoid content. This is evident when comparing the typical UV-visible spectrophotometric profiles of cassava samples 5, 23 and 74 (Figure 4).
Figure 4:
Typical UV-visible spectrophotometric profiles [λ = 200–700 ηm, acetone: petroleum ether (v/v)] of root parenchymal tissues of three cassava samples: (A) - sample 5, (B) - sample 23 and (C) - sample 74. The 400–500 ηm region of the spectrum is highlighted in cases (B) and (C).
These three samples vary greatly in color, with sample 5 having a cream color, sample 23 a yellow one and the sample 74 a reddish color. In fact, the spectrophotometric profiles differ from each other only at 400–500 ηm region of the spectrum, which is the region where carotenoids typically show absorbance peaks.
The cream colored sample profile (Figure 4A) shows an absence of absorbance peaks between the 400 and 500 ηm region. On the other hand, the yellow colored sample profile (Figure 4B) shows more noticeable peaks in this region, while the reddish colored sample (Figure 4C) presents three peaks of great absorption in this region of the spectrum. It is, therefore, expected that the more colored the root the higher carotenoid content it possesses.
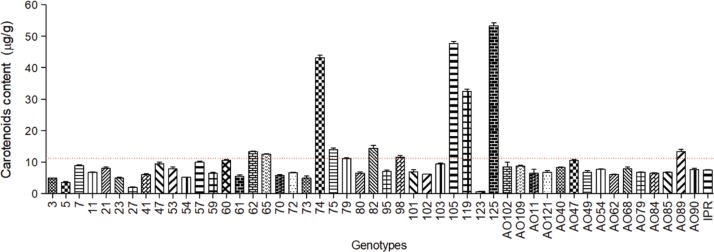
To confirm this possibility, the total carotenoid content was determined by UV-visible spectrophotometry, using the Lambert-Beer formula, and is shown in Figure 5 for each of the fifty fresh root samples.
Figure 5:
Concentration of total carotenoids (μg.g–1 dry weight ± standard deviation, n = 3) in samples of roots of fifty M. esculenta genotypes, determined by UV-visible spectrophotometry (450 ηm, ε =2592 M–1cm–1).
The wide disparity in the observed carotenoid contents reveals the chemical variability among the analyzed genotypes. In the present study, the cream-colored roots showed the lowest concentrations of total carotenoids, with values around 0.57 μg.g–1, while higher concentration values were measured in yellow and reddish pigmented roots i.e, 54.93 μg.g–1. The most abundant carotenoids, trans-β-carotene and cis-β-carotene, had concentration values that ranged from 1.82 to 42.82 μg.g–1 and from 1.19 to 28.86 μg.g–1, respectively. The results from the HPLC carotenoid quantification are available as supplementary material in the metadata file.
These findings altogether are consistent with data reported in the literature that observe a positive correlation between the color of the root pulp and the total carotenoid content [19], [20], [21].
3.2. CIELAB Color Space Interpretation
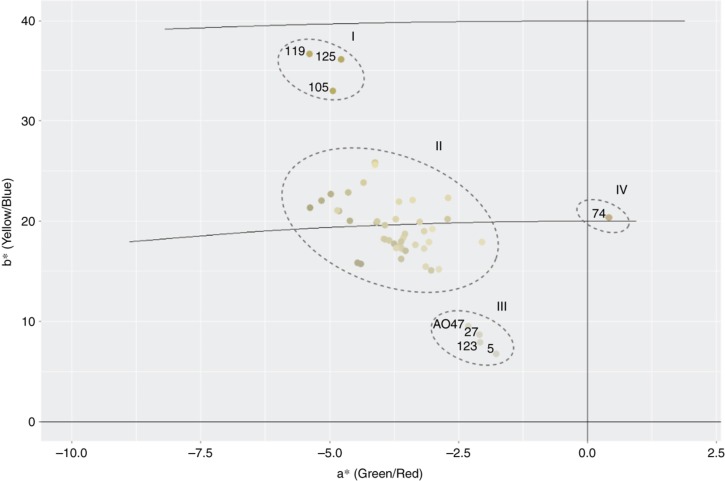
To better understand the correlation between samples and the different types of carotenoids with the CIELAB color space we projected the observed values of L*, a* and b* for each root sample into the CIELAB plane [22]. Figure 6 shows the samples location according to the color of roots in the CIELAB color space, whose visual interpretation is sufficient to verify which samples possess higher carotenoid amounts.
Figure 6:
Location of the cassava samples in the CIELAB color space according to their root pulp colors. The a* value characterizes the coloration in the regions of red (+a*) to green (-a*). The value b* indicates coloring in the range of yellow (+b*) to blue (-b*). Sample identifiers in ellipse II were omitted for easier interpretation of the plot.
Samples 105, 119 and 125 (Figure 6, ellipse I) show high b* values, which stands for the coloration in the yellow range, and these are in fact the samples with the highest carotenoid contents, as it can be observed in Figure 5. Interestingly, sample 74 (Figure 6, ellipse IV) is deviated into the positive axis of a*, which corresponds to the red coloration. In fact, this sample is a reddish root, mostly due to its lycopene content, which confers reddish coloration to the biomass [23]. It is one of the samples with the highest carotenoid concentration (Figure 5).
Samples 123, 27, 05, and AO47 (Figure 6, ellipse III) were grouped in values of b* closer to zero, these being the samples with the lowest carotenoid content (Figure 5). The remaining samples had medium and more similar carotenoid content, being grouped together in a* negative and b* positive values (Figure 6, ellipse II).
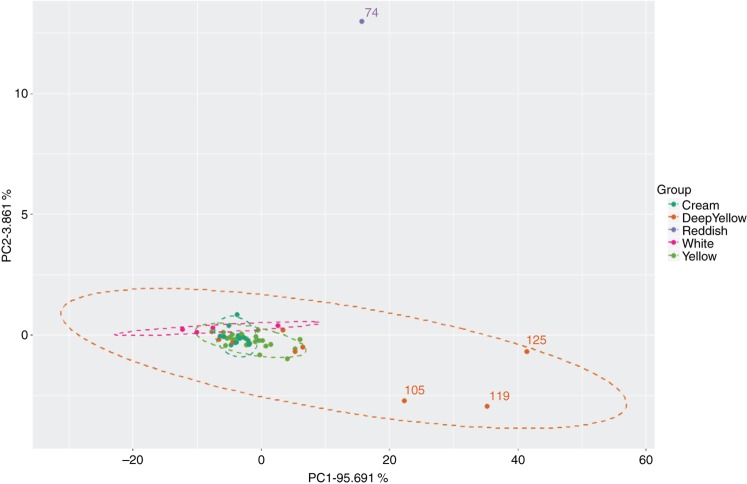
The same similarity patterns of carotenoid composition were found among the evaluated genotypes through a PCA analysis (Figure 7). In their set, PC1 and PC2 explain about 99.5 % of the total variance of the sample population data under this study.
Figure 7:
Scores plot with the distribution of the fifty samples on the first and second PCA components resulting from the UV-visible spectrophotometric data (400–500 ηm) (n =3 replicates). To facilitate the interpretation of the plot, only the sample identifiers for the most relevant samples are shown.
This analysis revealed genotype grouping according to their root pulp coloration, as well as carotenoid quantification, similarly to the CIELAB data, with samples 74, 105, 119, and 125 being the most discrepant within this sample universe.
These results are in accordance with the findings in subsection 3.1 that positively correlate the carotenoid content with the color of the cassava roots.
3.3. Carotenoid Content Prediction Using UV-Visible Data
In Table 1, the performance values obtained by the ML models using UV-visible data (400–500 ηm) as inputs and the total carotenoid content (TCC) determined by spectrophotometry (Lambert-Beer formula), the TCC determined by HPLC and the total content of trans-β-carotene (the most abundant carotene in cassava roots) as response prediction variables are shown.
Table 1:
Performance values (RMSE and R2) obtained for the different machine learning models trained with UV-visible spectrophotometry data (400–500 ηm).
| UV-visible (400–500 ηm) | ||||||
|---|---|---|---|---|---|---|
| TCC Spectrophotometry |
TCC HPLC |
trans-β-carotene |
||||
| RMSE | R 2 | RMSE | R 2 | RMSE | R 2 | |
| Partial least squares (simpls) | 3.492 | 0.9208 | 5.789 | 0.5721 | 4.309 | 0.36296 |
| Support vector machines (el071) | 3.709 | 0.9316 | 5.844 | 0.5975 | 4.218 | 0.39924 |
| Partial least squares (widekernelpls) | 3.732 | 0.9238 | 5.779 | 0.5701 | 4.324 | 0.45308 |
| Random forest | 3.768 | 0.9483 | 7.275 | 0.3596 | 5.753 | 0.23993 |
| Elastic net | 3.793 | 0.9185 | 5.934 | 0.6340 | 4.191 | 0.41274 |
| Partial least squares (pis) | 3.800 | 0.9529 | 5.643 | 0.5971 | 4.265 | 0.47090 |
| Ridge regression (w/FS) | 3.855 | 0.9478 | 5.880 | 0.6038 | 4.159 | 0.35640 |
| Ridge regression | 3.877 | 0.9283 | 7.282 | 0.6163 | 4.407 | 0.31655 |
| Support vector machines (kernlab) | 3.928 | 0.9409 | 5.907 | 0.5892 | 4.230 | 0.46608 |
| Partial least squares (kernelpls) | 4.096 | 0.8962 | 5.878 | 0.5661 | 4.211 | 0.42217 |
| Linear regression (w/Stepwise selection) | 4.158 | 0.9192 | 8.341 | 0.5265 | 6.135 | 0.20603 |
| Linear regression (w/Forward selection) | 4.178 | 0.8883 | 8.783 | 0.4716 | 5.142 | 0.31153 |
| Linear regression (w/Backwards selection) | 4.392 | 0.8711 | 6.373 | 0.5226 | 5.355 | 0.27887 |
| K-Nearest neighbors | 4.732 | 0.9224 | 6.277 | 0.4451 | 4.597 | 0.22467 |
| Lasso | 5.207 | 0.8174 | 17.508 | 0.2494 | 16.145 | 0.18959 |
| Conditional inference random forest | 6.713 | 0.7917 | 6.806 | 0.5588 | 4.703 | 0.36963 |
| Conditional inference tree | 7.363 | 0.7114 | 6.916 | 0.4805 | 4.894 | 0.28851 |
| Decision trees | 7.582 | 0.6833 | 6.795 | 0.4736 | 5.189 | 0.05344 |
The total carotenoid content (TCC) determined by spectrophotometry (Lambert-Beer formula), the TCC determined by HPLC and the total content of trans-β-carotene (the most abundant carotene in cassava roots) were used as response prediction variables. The parenthesis indicate the package specific method chosen for the simulation, with exception to the linear regression models. For each prediction variable used the best performance values are represented in bold.
It is clear that the highest R2 performance values (above 90 %) and lowest RMSE values were obtained when using the TCC determined using spectrophotometric data as response prediction variable. This was expected considering that both input and response data used employ the same physical phenomenon of detection of compounds (absorbance). The models that best performed in this case were partial least squares (PLS) using both simpls and widekernelpls methods, support vector machines (SVMs) and random forests with RMSE performance values of 3.492, 3.732, 3.709 and 3.768, respectively.
Using the TCC determined by HPLC as the response variable, a small decrease in performance values is observed. Here, PLS (widekernelpls and pls methods) and elastic network showed best performance with RMSE values of 5.779, 5.643 and 5.934, respectively, and R2 values around 60 %.
The worst results were obtained by using trans-β-carotene as response variable, with best performance models being PLS (widekernelpls and pls methods) and SVMs, with RMSE values of 4.324, 4.265 and 4.230, respectively, and R2 values around 46 %.
When observed, the values of VIPs (Variable Importance in the Projection) for this analysis (supplementary material), which identify the most relevant variables for the validation of the method, it can be detected that the wavelengths 449, 448 and 450 nm (precisely the wavelength that is used for the quantification of β-carotene through the Lambert-Beer formula) were used in 100, 99.93 and 99.76 % of cross-validation training performance. This result is important because it attests to the robustness of the models in predicting the contents of these compounds in cassava samples.
Applying pre-processing methods to the data, as well as feature selection, showed an overall increase in model performance for most models used (supplementary material). In Table 2 one such case is shown, where using pre-processed UV-visible data as input to Random Forest model, that showed best performance when using raw data, increased even further model performance. By applying smoothing interpolation, background and offset corrections, or background correction alone, RMSE values decreased from 6.194 to 5.773, 5.936 and 6.175, respectively. R2 values also increased from 55 to around 60 % in each case.
Table 2:
Performance values (RMSE and R2) obtained for a random forest model trained with UV-visible spectrophotometry data (400–500 ηm), applying several pre-processing methods to the data.
| UV-visible (400–500 ηm) + Preprocessing, random forest | ||
|---|---|---|
| TCC Spectrophotometry |
||
| RMSE | R 2 | |
| Smoothing interpolation | 5.773 | 0.6053 |
| Background and Offset corrections | 5.936 | 0.5927 |
| Background correction | 6.175 | 0.5956 |
| No preprocessing | 6.194 | 0.5581 |
| Scaling | 6.447 | 0.5740 |
| Background, Baseline and Offset corrections | 9.397 | 0.4780 |
| First derivative | 10.774 | 0.4482 |
| Multiplicative Scatter Correction | 11.621 | 0.3245 |
The total carotenoid content (TCC) determined by HPLC was used as response prediction variable. The best performance values are represented in bold.
3.4. Carotenoid Content Prediction Using CIELAB Data
Table 3 shows the performance values obtained by using CIELAB data as input to the various machine learning models and using the TCC determined by spectrophotometry (Lambert-Beer formula), the TCC determined by HPLC and the total content of trans-β-carotene (the most abundant carotene in cassava roots) as response prediction variables.
Table 3:
Performance values (RMSE and R2) obtained for the different machine learning models trained with CIELAB data.
| CIELAB data | ||||||
|---|---|---|---|---|---|---|
| TCC Spectrophotometry |
TCC HPLC |
trans-β-carotene |
||||
| RMSE | R 2 | RMSE | R 2 | RMSE | R 2 | |
| Partial least squares (simpls) | 6.990 | 0.6022 | 6.789 | 0.4142 | 4.731 | 0.2371 |
| Support vector machines (el071) | 7.015 | 0.5350 | 6.645 | 0.3840 | 4.829 | 0.1506 |
| Partial least squares (widekernelpls) | 7.125 | 0.6221 | 6.696 | 0.3960 | 4.551 | 0.2794 |
| Random forest | 6.647 | 0.5124 | 7.571 | 0.2938 | 5.148 | 0.1532 |
| Elastic net | 6.456 | 0.5785 | 6.534 | 0.4129 | 4.787 | 0.1840 |
| Partial least squares (pis) | 6.939 | 0.5916 | 6.622 | 0.3946 | 4.667 | 0.2446 |
| Ridge regression (w/FS) | 6.638 | 0.5628 | 6.653 | 0.3895 | 4.S02 | 0.2020 |
| Ridge regression | 6.417 | 0.5681 | 6.584 | 0.4213 | 4.886 | 0.1774 |
| Support vector machines (kernlab) | 7.294 | 0.5040 | 6.534 | 0.3662 | 4.878 | 0.2043 |
| Partial least squares (kernelpls) | 7.121 | 0.5827 | 6.756 | 0.4319 | 4.785 | 0.2278 |
| Linear Regression | 6.295 | 0.5933 | 6.749 | 0.4004 | 4.937 | 0.2424 |
| K-Nearest neighbors | 6.636 | 0.5336 | 7.278 | 0.2569 | 4.997 | 0.2036 |
| Lasso | 6.412 | 0.5503 | 6.669 | 0.4110 | 4.826 | 0.1539 |
| Conditional inference random forest | 8.162 | 0.4385 | 6.930 | 0.4085 | 4.667 | 0.2066 |
| Conditional inference tree | 9.388 | 0.3063 | 7.307 | 0.3842 | 4.934 | 0.1105 |
| Decision trees | 9.990 | 0.2679 | 7.641 | 0.3534 | 5.015 | 0.2880 |
The total carotenoid content (TCC) determined by spectrophotometry (Lambert-Beer formula), the TCC determined by HPLC and the total content of trans-β-carotene (the most abundant carotene in cassava roots) were used as response prediction variables. The parenthesis indicate the package specific method chosen for the simulation. For each prediction variable used the best performance values are represented in bold.
Similarly to the results obtained in subsection 3.3, the highest R2 performance values and lowest RMSE values were obtained when using the TCC determined using spectrophotometric data as a response prediction variable. However, there is a noticeable overall decrease in model performance when using all three prediction variables. This is easily explained by the number of features present in the data, considering that in this case only three features are present (L*, a* and b*), while in the previous case there were far more features, about 101 (data measured from 400 to 500 ηm).
Using the TCC determined by spectrophotometry as response variable, the models that showed best performance were linear and ridge regressions and elastic network with RMSE values of 6.295, 6.417 and 6.456, respectively, with R2 values around 60 %.
For the second variable, TCC determined by HPLC, the best models were elastic network, ridge regression and SVMs with RMSE values of 6.534, 6.584 and 6.534, respectively, and R2 values around 40 %.
Lower RMSE values were observed when using trans-β-carotene as response variable, with best performance models being PLS (widekernelpls and pls methods) and conditional inference random forests, with RMSE values of 4.551, 4.667 and 4.667, respectively. However, models showed a decrease in the fitting of the data with an R2 around 25 %.
Looking at the VIPs (supplementary material) it becomes clear which variables played the most important role in the prediction of carotenoid content in the cassava samples. The b* parameter was relevant in 100 % of the cases, which was somewhat expected, considering that the samples are widely distributed across the y axis in Figure 6, which corresponds to the b* parameter. Looking at the same plot we can see that the a* interval in which samples are distributed is not as wide, however, this parameter was relevant in about 56 % of the predictions. The L* parameter was the least relevant of the three, with a VIP of 0 %.
The only pre-processing method applied to CIELAB data was scaling, as the other methods would not make much sense considering they are aimed at spectral data. Scaling the data showed an increase in model performance, however quite limited (results are shown in supplementary material).
3.5. Carotenoid Content Prediction Using Fusion Data
In Table 4 the performance values obtained by using a low-level fusion between UV-visible (400–500 ηm) and CIELAB data as input to the various machine learning models are shown. Similarly to the previous cases, the response prediction variables used were the total carotenoid content (TCC) determined by spectrophotometry (Lambert-Beer formula), the TCC determined by HPLC and the total content of trans-β-carotene.
Table 4:
Performance values (RMSE and R2) obtained for the different machine learning models trained with a fusion between UV-visible spectrophotometry and CIELAB data.
| UV-visible (400–500 ηm) + CIELAB data | ||||||
|---|---|---|---|---|---|---|
| TCC Spectrophotometry |
TCC HPLC |
trans-β-carotene |
||||
| RMSE | R 2 | RMSE | R 2 | RMSE | R 2 | |
| Partial least squares (simpis) | 3.706 | 0.8746 | 6.082 | 0.5032 | 4.685 | 0.2545 |
| Support vector machines (el071) | 3.875 | 0.8887 | 6.379 | 0.5477 | 4.353 | 0.3551 |
| Partial least squares (widekernelpls) | 4.017 | 0.9247 | 6.031 | 0.4448 | 4.592 | 0.2804 |
| Random forest | 3.758 | 0.9444 | 7.114 | 0.3527 | 6.101 | 0.1621 |
| Elastic net | 3.775 | 0.9179 | 6.160 | 0.6031 | 4.450 | 0.3256 |
| Partial least squares (pls) | 3.682 | 0.8931 | 6.187 | 0.4834 | 4.652 | 0.2530 |
| Ridge regression (w/FS) | 3.570 | 0.9298 | 5.981 | 0.5781 | 4.730 | 0.3430 |
| Ridge regression | 4.839 | 0.8510 | 8.469 | 0.4723 | 5.627 | 0.2783 |
| Support vector machines (kernlab) | 4.612 | 0.8800 | 6.299 | 0.5249 | 4.436 | 0.4241 |
| Partial least squares (kernelpls) | 3.804 | 0.8312 | 6.010 | 0.5263 | 4.681 | 0.2745 |
| Linear regression (w/Stepwise selection) | 4.718 | 0.7973 | 8.052 | 0.5295 | 4.909 | 0.2406 |
| Linear regression (w/Forward selection) | 4.829 | 0.8743 | 8.279 | 0.4734 | 4.860 | 0.2492 |
| Linear regression (w/Backwards selection) | 4.479 | 0.8020 | 6.385 | 0.5419 | 5.179 | 0.2966 |
| K-Nearest neighbors | 6.412 | 0.6320 | 7.355 | 0.2622 | 4.996 | 0.1359 |
| Lasso | 4.983 | 0.8076 | 18.784 | 0.2545 | 13.821 | 0.1487 |
| Conditional inference random forest | 6.663 | 0.7671 | 6.531 | 0.5158 | 4.645 | 0.3540 |
| Conditional inference tree | 7.566 | 0.6697 | 6.923 | 0.4351 | 4.870 | 0.2706 |
| Decision trees | 8.021 | 0.6997 | 7.789 | 0.3427 | 5.221 | 0.2181 |
The total carotenoid content (TCC) determined by spectrophotometry (Lambert-Beer formula), the TCC determined by HPLC and the total content of trans-β-carotene (the most abundant carotene in cassava roots) were used as response prediction variables. The parenthesis indicate the package specific method chosen for the simulation, with exception to the linear regression models. For each prediction variable used the best performance values are represented in bold.
The results obtained using fusion data are similar to those explained in subsection 3.3 and subsection 3.4 in the sense that highest R2 performance values and lowest RMSE values were obtained when using the TCC determined using spectrophotometric data as response prediction variable. Overall there is an increase in model performance when comparing to the results obtained for UV-visible data alone.
The best model performance when using the TCC determined by spectrophotometry as response variable was achieved by ridge regression (with feature selection) and PLS (pls and simpls methods) models with RMSE values of 3.570, 3.682 and 3.706, respectively, and R2 values around 90 %.
Using the second variable, TCC determined by HPLC, the models that best performed were ridge regression (with feature selection) and PLS (kernelpls and widekernelpls) models, having RMSE values of 5.981, 6.010 and 6.031, respectively, with R2 values around 50 %.
Lower RMSE values were observed when using trans-β-carotene as response variable, similarly to previous cases, with best performance models being SVMs (e1071 and kernlab methods) and elastic network with RMSE values of 4.353, 4.436 and 4.450, respectively, and R2 values around 30 %.
The VIPs computed for this case (supplementary material) showed that the variables which presented the most important role in the prediction of carotenoid content in the cassava samples were those of wavelength around 170 ηm (VIPs > 99 %). Here, the CIELAB b* parameter was relevant in about 65 % of predictions, while the a* and L* parameters had a VIP close to zero.
The only preprocessing method applied to the fusion data was scaling, as the methods employed in subsection 3.3 are aimed at spectral data. Data filtering was also applied to the data. Both methods contributed to an overall increase in model performance when compared to the performance obtained using raw UV-visible data (supplementary material).
4. Conclusions
The present study has shown how CIELAB color measurement can be used as a fast and non-destructive method to calibrate for the total carotenoid content of cassava genotypes roots with acceptable prediction error. By performing a low-level fusion between UV-visible spectrophotometry and CIELAB data we demonstrated how data fusion can lead to a better model performance for prediction when comparing to the use of a single data source, having similar results already been published [24].
Moreover, the UV-visible spectrophotometric profiles measured between 400–500 ηm and the consequent carotenoid content determination allowed the observation of a positive correlation between the color of the root pulp and the total carotenoid content, which is in accordance with data reported in the literature [19], [20], [21]. This finding was more explicit when observing the projection of the fifty cassava root samples in the CIELAB color space plane, having several clusters been formed, where the highest values of b* (which stands for the yellow coloration) and a* (which stands for the red coloration) were associated to the samples with highest carotenoid content.
In addition, the information obtained by coupling the analysis of pro-vitamin A biochemical markers to bioinformatics tools helps supporting the rational design of biochemically-assisted breeding programs of M. esculenta, that aim to obtain cultivars with high levels of pro-vitamin A carotenoids and superior nutritional traits.
Acknowledgement
To CNPq (National Counsel of Technological and Scientific Development) for financial support (Process n 407323/2013-9), to CAPES (Coordination for the Improvement of Higher Education Personnel (CAPES), and EPAGRI (Agricultural Research and Rural Extension Company of Santa Catarina). The research fellowship from CNPq on behalf of M. Maraschin is acknowledged. The work is partially funded by Project PropMine, funded by the agreement between Portuguese FCT (Foundation for Science and Technology) and Brazilian CNPq.
Conflict of interest statement:
Authors state no conflict of interest. All authors have read the journal’s publication ethics and publication malpractice statement available at the journal’s website and hereby confirm that they comply with all its parts applicable to the present scientific work
References
- [1].Tanumihardjo SA, Palacios N, Pixley KV. Provitamin A carotenoid bioavailability: what really matters? Int J Vitam Nutr Res. 2010;80:336. doi: 10.1024/0300-9831/a000042. [DOI] [PubMed] [Google Scholar]
- [2].Rodriguez-Amaya DB. A guide to carotenoid analysis in foods. Washington, DC: ILSI Press; 2001. [Google Scholar]
- [3].Pence HE, Williams A. ChemSpider: an online chemical information resource. J Chem Educ. 2010;87:1123–4. [Google Scholar]
- [4].Stahl W, Sies H. Antioxidant activity of carotenoids. Mol Aspects Med. 2003;24:345–51. doi: 10.1016/s0098-2997(03)00030-x. [DOI] [PubMed] [Google Scholar]
- [5].La Frano MR, Woodhouse LR, Burnett DJ, Burri BJ. Biofortified cassava increases β-carotene and vitamin A concentrations in the TAG-rich plasma layer of American women. Br J Nutr. 2013;110:310–20. doi: 10.1017/S0007114512005004. [DOI] [PubMed] [Google Scholar]
- [6].Sánchez T, Ceballos H, Dufour D, Ortiz D, Morante N, Calle F, et al. Prediction of carotenoids, cyanide and dry matter contents in fresh cassava root using NIRS and Hunter color techniques. Food Chem. 2014;151:444–51. doi: 10.1016/j.foodchem.2013.11.081. [DOI] [PubMed] [Google Scholar]
- [7].Sánchez T, Chávez AL, Ceballos H, Rodriguez-Amaya DB, Nestel P, Ishitani M. Reduction or delay of post-harvest physiological deterioration in cassava roots with higher carotenoid content. J Sci Food Agric. 2006;86:634–9. [Google Scholar]
- [8].Brockes A. The evaluation of whiteness. Computi Ind Eng. 1982;2:38–39. [Google Scholar]
- [9].Schanda J. Colorimetry: understanding the CIE system. New York, NY: John Wiley & Sons; 2007. [Google Scholar]
- [10].Weatherall IL, Coombs BD. Skin color measurements in terms of CIELAB color space values. J Investig Dermatol. 1992;99:468–73. doi: 10.1111/1523-1747.ep12616156. [DOI] [PubMed] [Google Scholar]
- [11].Liu W, Ji J, Chen H, Ye C. Optimal color design of psychological counseling room by design of experiments and response surface methodology. PLoS One. 2014;9:e90646. doi: 10.1371/journal.pone.0090646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Singh Y, Bhatia PK, Sangwan O. A review of studies on machine learning techniques. Int J Comput Sci Secur. 2007;1:70–84. [Google Scholar]
- [13].Domingos P. A few useful things to know about machine learning. Commun ACM. 2012;55:78–87. [Google Scholar]
- [14].Fourati H. Multisensor data fusion: from algorithms and architectural design to applications. vol. 1. Boca Raton, FL: CRC Press, Taylor & Francis Group LLC; 2016. [Google Scholar]
- [15].Rodriguez-Amaya DB, Kimura M. HarvestPlus handbook for carotenoid analysis. vol. 2. Washington, DC: International Food Policy Research Institute (IFPRI); 2004. [Google Scholar]
- [16].R Core Team. R: a language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2017. Available from: https://www.R-project.org/ [Google Scholar]
- [17].Costa C, Maraschin M, Rocha M. An R package for the integrated analysis of metabolomics and spectral data. Comput Methods Programs Biomed. 2016;129:117–24. doi: 10.1016/j.cmpb.2016.01.008. [DOI] [PubMed] [Google Scholar]
- [18].Kuhn M. Caret package. J Stat Softw. 2008;28:1–26. [Google Scholar]
- [19].Champagne A, Bernillon S, Moing A, Rolin D, Legendre L, Lebot V. Carotenoid profiling of tropical root crop chemotypes from Vanuatu, South Pacific. J Food Compos Anal. 2010;23:763–71. [Google Scholar]
- [20].Chávez AL, Sánchez T, Jaramillo G, Bedoya J, Echeverry J, Bolaños E, et al. Variation of quality traits in cassava roots evaluated in landraces and improved clones. Euphytica. 2005;143:125–33. [Google Scholar]
- [21].Iglesias C, Mayer J, Chavez L, Calle F. Genetic potential and stability of carotene content in cassava roots. Euphytica. 1997;94:367–73. [Google Scholar]
- [22].Kljak K, Grbeša D, Karolyi D. Reflectance colorimetry as a simple method for estimating carotenoid content in maize grain. J Cereal Sci. 2014;59:109–11. [Google Scholar]
- [23].Meléndez-Martínez AJ, Britton G, Vicario IM, Heredia FJ. Relationship between the colour and the chemical structure of carotenoid pigments. Food Chem. 2007;101:1145–50. [Google Scholar]
- [24].Botwey RH, Daskalaki E, Diem P, Mougiakakou SG. Multi-model data fusion to improve an early warning system for hypo-/hyperglycemic events; Engineering in Medicine and Biology Society (EMBC), 2014 36th annual international conference of the IEEE; IEEE; 2014. pp. 4843–4846. [DOI] [PubMed] [Google Scholar]