Abstract
ApoE expression status was proved to be a highly specific marker of energy metabolism rate in the brain. Along with its neighbor, Translocase of Outer Mitochondrial Membrane 40 kDa (TOMM40) which is involved in mitochondrial metabolism, the corresponding genomic region constitutes the neuroenergetic hotspot. Using RNA-Seq data from a murine model of chronic stress a significant positive expression coordination of seven neighboring genes in ApoE locus in five brain regions was observed. ApoE maintains one of the highest absolute expression values genome-wide, implying that ApoE can be the driver of the neighboring gene expression alteration observed under stressful loads. Notably, we revealed the highly statistically significant increase of ApoE expression in the hypothalamus of chronically aggressive (FDR < 0.007) and defeated (FDR < 0.001) mice compared to the control. Correlation analysis revealed a close association of ApoE and proopiomelanocortin (Pomc) gene expression profiles implying the putative neuroendocrine stress response background of ApoE expression elevation therein.
Keywords: ApoE, Tomm40, App, Pomc, brain lipid metabolism, gene expression profile, social stress mouse model, RNA-Seq data, differentially expressed genes, brain regions, hypothalamus
1. Introduction
APOE is featured as a major apolipoprotein that mediates the cholesterol and lipid metabolism upon binding the (very) low-density lipoprotein receptors, including low density lipoprotein receptor (LDLR), very low density lipoprotein receptor (VLDR), and some of the low density lipoprotein receptor-related protein (LRP1-12) genes family [1]. It binds lipids and thus forms intermediate density lipoproteins further cleared from the plasma into the liver by receptor-mediated endocytosis. The liver and brain are major APOE secretion organs in the vertebrates [2]. Liver originated plasma APOE cannot pass the blood brain barrier (BBB), making brain APOE synthesis and metabolism independent [2].
Notably, ApoE expression in the rodent brain is two orders of magnitude higher than any other apolipoprotein gene as observed from contemporary RNA-Seq data. While basal ApoE gene expression can be observed in virtually all cell types, the cells maintaining the highest ApoE expression, other than hepatocytes in the liver, are brain astrocytes and microglia [3], [4], [5].
The fact that astrocytes synthesize lipids and at the same time highly express ApoE – which mediates cholesterol as well as amyloid and lipid metabolism – underlines the ultimate significance of ApoE functioning specifically in the brain. Notably, astrocytes shed “mini energy stations,” large membrane vesicles comprised of lipids, mitochondria, and ATPs [6]. It was also shown that ApoE eps4 allele carriers exhibit altered energy metabolism pathways in the brain [7]. The role of brain APOE in cholesterol and amyloid beta homeostasis and its implication in Alzheimer’s disease (AD) was presented as a new paradigm in a recent review by [8]. Similarly, the “Triad” of ultimate factors such as ApoE, mitochondrial haplotype and sexual dimorphism as major risk factors of AD etiology was pointed out in [9]. All of these observations underscore vital APOE neuroenergetic components in neural diseases such as AD. Along with ApoE involvement in phagocytosis of cell corpses [10] and astrocyte mediated synapse elimination in the brain [11], ApoE stands as a vital gene in brain catabolism.
While cholesterol rich lipoproteins are required in neurons to support synaptogenesis and maintenance of synaptic connections, another function of APOE is delivering Amyloid beta (APP/ABeta)-APOE complexes to cellular catabolism via neuronal APOE receptors [1]. With many APOE/ABeta complexes entering the cytoplasm via neuronal LDLR receptors [12], another mediator is the pro-neurotrophin receptor, sortilin (SORT1), which also proved to be one of the major endocytic pathways for clearance of APOE/ABeta complexes in neurons [3], [13]. It was shown that a lack of Sort1 receptor expression in mice results in accumulation of APOE and ABeta in the brain and in aggravated plaque burden [13].
It is also necessary to mention the signalling role of APOE protein within the brain. It was shown that APOE suppresses food intake via a mechanism enhancing proopiomelanocortin (POMC) signalling in the hypothalamus [14]. ApoE takes part in RhoA [15] and NMDA mediated [16] pathways. It also plays a distinct role in microvasculature maintenance [15]. Previous studies revealed that CNS neurons in mice express ApoE in response to excitotoxic injury [17], implying brain defense functions. ApoE gene is also expressed in primary cultured human CNS astrocytes [5] and in many human neuronal cell lines, including SY-5Y, Kelly, and NT2 cells [18].
The human ApoE gene E4 (ApoE4) allele is the major known genetic risk factor for AD. In most clinical studies, ApoE4 carriers account for 65–80 % of all AD cases. ApoE mouse models are widely used in neurobiology and in the pathogenesis of AD [3]. Thus, ApoE locus and closely located neighboring genes are the subject of close attention both in mice and in humans [19]. In addition to ApoE, particular attention has been drawn to its neighbor, Tomm40 [19]. Tomm40 has been reported to be involved in the predisposition to AD and non-pathologic cognitive decline based on “mitochondrial cascade hypothesis,” which proposes a genetic contribution toward mitochondrial durability and function in the brain in particular [19], [20], [21]. Due to high linkage disequilibrium between ApoE and Tomm40 in humans, however [21], it is hard to elucidate an independent impact of Tomm40 on AD development.
The high linkage disequilibrium rate in ApoE extended locus region and its high evolutionary conservation across mammalian species [22] imply that there might be some impact on gene expression coordination within the locus. In the current study, we analyzed the joint gene expression rate in the vicinity of ApoE in mice for 8 genes.
A specific ApoE implication in stress response metabolism was first reported in 1996 [23]. Further studies indicate a significant interaction between ApoE expression status and stressful life events [24], [25]. Stress is a factor of enhanced risk for AD [26]. In a series of animal knockout studies [27], [28] it has been postulated that the effects of stress are mediated by glucocorticoids and depend on the presence of APOE. The effects of ApoE expression rate variation may alter susceptibility to environmental factors such as stress. The threshold at which the stress can result in damage to neurons may differ depending on ApoE isoforms proportions [29]. CNS injury is supposed to induce neuronal expression of ApoE to participate in neuronal repair or to protect neurons from injury [3], [17].
The present research is designed to study the interference of gene expression variation in the ApoE locus at stressful loads in brain regions of male mice. We used the model of chronic social conflicts in daily agonistic interactions, featuring strong stress induction in the mice. The gene expression in the ApoE locus was primarily studied in the hypothalamus, the main brain region of stress regulation, as well as in other brain regions – the hippocampus, ventral tegmental area, striatum, and midbrain raphe nuclei.
We pursued the issue of the consistency of ApoE expression assessment and its coordination with the genomic context by considering the neighboring genes. Additionally, we targeted the small subset of genes which are the partners of ApoE in metabolic processes and signal transduction to assess their possible interaction in brain regions. Finally, as APOE metabolism involves lipoprotein receptors, we considered a list of lipoprotein receptors to elucidate their performance specifics in various brain regions and their relation to ApoE expression rate.
2. Materials and Methods
2.1. Animals
Adult male mice of the C57BL/6J strain from Animal Breeding Facility, Branch of Institute of Bioorganic Chemistry of the RAS (ABF BIBCh, RAS) (Pushchino, Moscow region) were used. The animals were housed under standard conditions (12:12 h light/dark regime, switch-on at 8:00 a.m.; food (pellets) and water available (ad libitum). Mice were weaned at one month of age and housed in groups of 8 to 10 in plastic cages (36 × 23 × 15 cm). Experiments were performed on mice 10 to 12 weeks of age. All procedures were in compliance with the European Communities Council Directive 2010/63/EU on September 22, 2010. The study was approved by Scientific Council N 9 of the Institute of Cytology and Genetics SD RAS of March 24, 2010, N 613.
2.2. Generation of Stressful Loads Under Chronic Agonistic Interactions in Male Mice
Stressful loads were induced by daily agonistic interactions in male mice in situations of chronic social conflicts, which were accompanied by positive and negative social experience (wins and defeats) [30], [31]. Pairs of weight-matched animals were each placed in a steel cage (28 × 14 × 10 cm), bisected by a perforated, transparent partition that allowed the animals to see, hear, and smell each other, but prevented physical contact. The animals were left undisturbed for 2 or 3 days to adapt to new housing conditions and sensory contact before they were exposed to encounters. Every afternoon (14:00–17:00 p.m. local time), the cage lid was replaced with a transparent one. Five minutes later, (the period necessary for individuals’ activation), the partition was removed for 10 min to encourage agonistic interactions. The superiority of one of the mice was firmly established within two or three encounters with the same opponent. The superior mouse behaved by attacking, biting, and chasing another mouse, who in response displayed only defensive behaviors (sideways postures, upright postures, withdrawal, lying on the back, or freezing). If aggressive interaction between males lasted for more than 3 min, or less in some cases, the attack was discontinued by lowering the partition to prevent damage to the loser. Each defeated mouse (loser) was exposed to the same winner for three days, while afterwards, each loser was placed, once a day after the fight, in an unfamiliar cage with an unfamiliar winner behind the partition. Each victorious mouse (winners, aggressive mice) remained in its original cage. This procedure was performed once a day for 20 days and yielded an equal number of winners and losers.
In the behavioral study, the following three groups of animals were used: (a) defeated mice – chronically defeated mice after 20 days of agonistic interactions; (b) aggressive mice – chronically aggressive mice that exhibited daily aggression during twenty days of agonistic interactions; (c) controls – mice without a consecutive experience of agonistic interactions.
Aggressive and defeated mice with the most prominent behavioral phenotypes were selected for the transcriptome analysis. All mice were simultaneously decapitated, 24 h after the last agonistic interaction and the control animals. The brain regions were dissected by one experimenter according to the map presented in the Allen Mouse Brain Atlas (http://mouse.brain-map.org/static/atlas). All biological samples were placed in RNAlater solution (Life Technologies, USA) and were stored at −70 °C until sequencing. In the current study we did not pursue the task of elucidating the differential background between the aggressive and defeated mice, but treated the samples as replicas of affected mice, gaining the insight on their common features.
The brain regions were selected for the analysis based on their functions and localization of neurons of neurotransmitter systems. These areas are as follows: the midbrain raphe nuclei, which comprises a multifunctional brain region and contains the majority of serotonergic neuronal bodies; the ventral tegmental area (VTA), which contains the bodies of dopaminergic neurons, is widely implicated in the natural reward circuitry of the brain, and is important in cognition, motivation, drug addiction, and emotions related to several psychiatric disorders; the striatum, which is responsible for the regulation of motor activity and stereotypical behaviors and is also potentially involved in various processes; the hippocampus, which belongs to the limbic system and is essential for memory consolidation and storage, and plays an important role in neurogenesis, and emotional mechanisms; and the hypothalamus, which regulates the stress reaction and many other physiological functions.
Thus, we maintained 45 samples comprised of 3 groups (control, aggressive mice, defeated mice) and 5 brain regions (hypothalamus, striatum, ventral tegmental area, hippocampus, midbrain raphe nuclei). Each group was represented with 3 replicas.
Cuffdiff software was used [32] to elucidate differentially expressed genes in the region.
2.3. RNA-Seq
The collected samples were sequenced at JSC Genoanalytica (http://genoanalytica.ru/, Moscow, Russia), and the mRNA was extracted using a Dynabeads mRNA Purification Kit (Ambion, Thermo Fisher Scientific, Waltham, MA, USA). cDNA libraries were constructed using the NEBNext mRNA Library PrepReagent Set for Illumina (New England Biolabs, Ipswich, MA, USA) following the manufacturer’s protocol and were subjected to Illumina sequencing. More than 20 million reads were obtained for each sample. The resulting “fastq” format files were used to align all reads to the GRCm38.p3 reference genome using the TopHat aligner [32]. DAVID Bioinformatics Resources 6.7 (http://david.ncifcrf.gov) was used for the description of differentially expressed gene ontology. The Cufflinks program was used to estimate the gene expression levels in FPKM (fragments per kilobase of transcript per million mapped reads) and subsequently identify the differentially expressed genes in the analyzed and control groups. Each brain area was considered separately in nine animals total. Only annotated gene sequences were used in the following analysis. Genes were considered differentially expressed at p ≤ 0.05. q < 0.05 was also taken into consideration.
We have previously conducted studies of gene expression in males in similar experiments using the RT-PCR method with larger samples for each compared experimental group, i.e. aggressive and defeated mice (>10 animals). The direction and extent of changes in expressions of the Tph2, Slc6a4, Bdnf, Creb1, and Gapdh genes in the midbrain raphe nuclei of defeated or aggressive males compared with the control produced by two methods, including RT-PCR [33], [34] and RNA-Seq, are generally consistent. This finding suggests that the transcriptome analyses of the data provided by the Company Genoanalitika have been verified, and this method reflects the actual processes that occur in the brain under our experimental paradigm.
2.4. Previously Published Expression Data in Mouse Brain
In order to cross-validate the results obtained, we employed the unique resource from Barres Lab, Stanford University, USA [4], [5]. The brain cell type specific RNA-Seq data proved to be highly concordant with our RNA-Seq data pool when we averaged them to use as a “cell mix,” represented in our brain region samples. In particular, when we compared average FPKM values of 32 apolipoprotein (Apo*) genes expression, we achieved Pearson correlation coefficient to be r = 0.9943; df = 31; p < 10−5 , which underlines significant consistency in expression variation. It was also consistent for other genes as will be shown in the results section.
2.4.1. Mouse RNA-Seq Data of Barres Lab in Seven Cortex Cell Types
For mouse cortex, 7 cell types were retrieved with original methods [4], [5]: astrocytes, neurons, oligodendrocyte precursor cells, newly formed oligodendrocytes, myelinating oligodendrocytes, microglia, and endothelial cells.
Brain cortex cell types FPKM expression data was downloaded from series description text file: GSE73721_Human_and_mouse_table.zip, available at GEO repository (www.ncbi.nlm.nih.geo/ GSE73721) [5].
2.5. Genes of Interest in the ApoE Locus
Based on the Hi-C data on chromatin domains [35] and linkage disequilibrium in humans, we selected 7 genes in the vicinity of ApoE, that are Bcam, Pvrl2, Tomm40, ApoE, Apoc1, Apoc2, Apoc4, Clptm1 ordered by DNA strand. The short description of the genes is presented in Table 1. All are located on the same DNA strand. As was reported previously [22], the gene order within the 1mb of the locus is highly conservative throughout mammals, which implies evolutionary constrains on the locus [36].
Table 1:
Description of the genes used in the study within the vicinity of ApoE.
| Genes | Description | Location |
|---|---|---|
| Bcam | Basal cell adhesion molecule precursor | Membrane bound |
| Pvrl2 | Poliovirus receptor-related 2 | Membrane protein |
| Tomm40 | Mitochondrial import receptor subunit TOM40 homolog | Cytoplasm |
| ApoE | Apolipoprotein E precursor | Secreted protein |
| Apoc1 | Apolipoprotein C-I precursor | Secreted protein |
| Apoc4 | Apolipoprotein C-IV precursor | Secreted protein |
| Apoc2 | Apolipoprotein C-II precursor | Secreted protein |
| Clptm1 | Cleft lip and palate transmembrane protein 1 | Membrane bound |
We also targeted low-density lipoprotein receptors expression rates to assess the correlation with ApoE (Table 2). Additionally, we included Pomc, which is a hypothalamus-specific gene involved in neuroendocrine stress response and ApoE-mediated signal transduction, as reported previously [14]. The large molecule of POMC is the source of several important, biologically active substances which may be connected with regulation of lipid metabolism, stress, and psychoemotional state. All of them changed expression rate in aggressive and defeated mice relative to the controls.
Table 2:
Description of lipoprotein receptors annotated in mouse genome and assessed for expression coordination with ApoE.
| Genes | Description | # isoforms |
|---|---|---|
| Lrp1 | Low-density lipoprotein receptor-related protein 1, mRNA | 3 |
| Lrp10 | Low-density lipoprotein receptor-related protein 10, mRNA | 1 |
| Lrp11 | Low-density lipoprotein receptor-related protein 11, mRNA | 3 |
| Lrp12 | Low-density lipoprotein-related protein 12, mRNA | 2 |
| Lrp1b | Low-density lipoprotein-related protein 1B (deleted in tumors), mRNA | 6 |
| Lrp2 | Low-density lipoprotein receptor-related protein 2, mRNA | 3 |
| Lrp3 | Low-density lipoprotein receptor-related protein 3, mRNA | 2 |
| Lrp4 | Low-density lipoprotein receptor-related protein 4, mRNA | 4 |
| Lrp5 | Low-density lipoprotein receptor-related protein 5, mRNA | 3 |
| Lrp6 | Low-density lipoprotein receptor-related protein 6, mRNA | 2 |
| Lrp8 | Low-density lipoprotein receptor-related protein 8, apolipoprotein e receptor, non-coding RNA | 6 |
| Ldlr | Low-density lipoprotein receptor, mRNA | 5 |
| Vldlr | Very low-density lipoprotein receptor, mRNA | 2 |
| Sort1 | Sortilin 1, mRNA | 2 |
Also incorporated into analysis were Amyloid precursor protein App gene, which is bound by ApoE in amyloid plagues [3], and Sortilin 1 (Sort1) gene, involved in App catabolism [13].
2.6. Statistical Analysis
Pearson correlation coefficient was used in all instances of correlation analysis. The significance rates for Pearson correlation values were derived based on Student t-test with corresponding degrees of freedom.
Principal component (PC) analysis has been carried out using XLStat software (www.xlstat.com). Pearson correlation coefficient has been used as a similarity metric during pairwise similarity matrix construction in PC analysis.
The multi-dimensional scaling (MDS) plots were built with XLStat software as well on the basis of Pearson pairwise similarity matrix. The target functional for minimization was Kruskal’s stress statistic. Initial configuration was set to random. Number of repetitions was set to 5. Stop conditions were: convergence step less that 1E−5; maximal number of iterations: 500.
ANOVA analysis has been carried out by XLStat software (www.xlstat.com). Fisher test was used for elucidation of variation significance in ANOVA.
3. Results and Discussion
3.1. ApoE Expression Locus Rate Evaluation and Data Assessment
The 7 adjacent genes at ApoE region may be grouped according to their location relative to cell compartments (Table 1). Bcam and Clptm1 genes are cell membrane receptors, Pvrl2 is a cell membrane protein, Tomm40 is a component of mitochondrial import translocase complex providing import of proteins into mitochondria, and ApoE, Apoc1, Apoc2, and Apoc4 are secreted proteins.
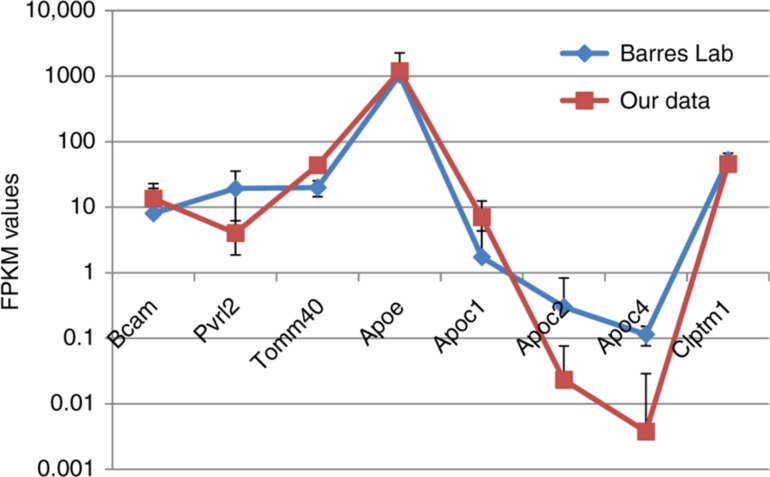
To evaluate the consistency of ApoE gene expression rate in mouse brain regions, we selected 7 consequent genes within ApoE location vicinity and assessed their joint average expression rate from our data (Supplementary Table 1), as well as from Barres Lab ([4], [5]; Supplementary Table 2). The compilation of averaged FPKM values across correspondent samples, as well as their standard deviation in mouse brain, for 2 projects are presented in Table 3/Figure 1. As it can be seen (Table 3; Figure 1), while genes from two projects maintain the same order of FPKM values, the ApoE expression rate is more than an order of magnitude higher than that of the neighboring genes. As the data was drawn from various brain regions/cell types, we present the overall compiled data set in Supplementary Table 1.
Table 3:
FPKM expression data (averaged) from 2 projects on mouse.
| Genes | Zhang et al. [5] |
Our data |
||
|---|---|---|---|---|
| Averaged | SD | Averaged | SD | |
| Bcam | 8.04 | 14.91 | 13.51 | 5.85 |
| Pvrl2 | 19.26 | 16.01 | 4.03 | 2.17 |
| Tomm40 | 19.89 | 5.38 | 43.59 | 8.39 |
| ApoE | 1047.7 | 1218.85 | 1189.92 | 317.58 |
| Apoc1 | 1.73 | 2.62 | 7.02 | 5.43 |
| Apoc2 | 0.3 | 0.53 | 0.02 | 0.05 |
| Apoc4 | 0.11 | 0.04 | 0.004 | 0.025 |
| Clptm1 | 52.61 | 14.11 | 45.60 | 7.45 |
Figure 1:
Assessment of 8 consequent genes expression by FPKM values in two projects. Average FPKM values, across 45 samples and 7 cell lines, from our data and previous data from Barres Lab [4], correspondingly, are presented for eight genes. Bars represent standard deviation.
As we witnessed tight coordination within apolipoprotein cluster (Table 4), and at the same time negligible expression values of Apoc2 and Apoc4 in data considered (Table 3), we omitted from further consideration Apoc2, Apoc4 genes. Thus, we further considered Bcam, Pvrl2, Tomm40, ApoE, Apoc1 and Clptm1 genes.
Table 4:
Correlation coefficients between genes of ApoE region in our data based on 45 samples.
| Variables | Bcam | Pvrl2 | Tomm40 | ApoE | Apoc1 | Clptm1 |
|---|---|---|---|---|---|---|
| Bcam | 1 | 0.880 | 0.550 | 0.717 | 0.806 | 0.482 |
| Pvrl2 | 0.880 | 1 | 0.446 | 0.725 | 0.864 | 0.367 |
| Tomm40 | 0.550 | 0.446 | 1 | 0.665 | 0.202 | 0.566 |
| ApoE | 0.717 | 0.725 | 0.665 | 1 | 0.606 | 0.592 |
| Apoc1 | 0.806 | 0.864 | 0.202 | 0.606 | 1 | 0.326 |
| Clptm1 | 0.482 | 0.367 | 0.566 | 0.592 | 0.326 | 1 |
Values in bold are different from 0 with a significance level alpha = 0.05.
3.2. ApoE Expression Rate in the Control, Aggressive and Defeated Mice Based on RNA-Seq Sequencing
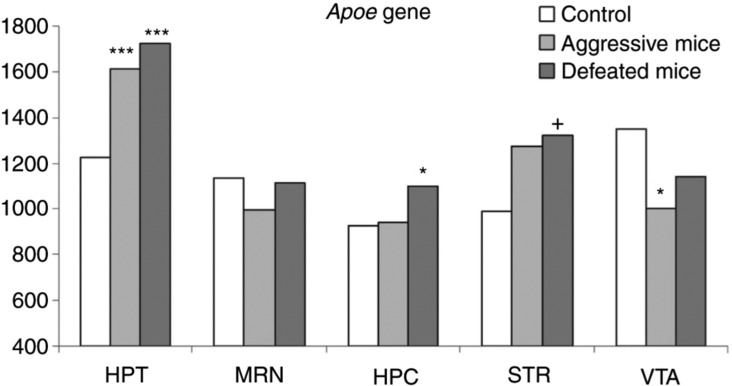
The differential gene expression list obtained with CuffDiff analysis and corresponding FPKM values are presented in Supplementary Table 3. We assessed ApoE expression rate in the control mice, and the mice with the chronic agonistic interactions experience which was accompanied by stress, pooled into 3 different groups (see Materials and Methods; Supplementary Tables 1 and 3). We observed the consistent significant elevation of ApoE expression rate between the control and affected groups (Figure 2; Supplementary Table 3). The pairwise expression correlation values are presented in Table 4.
Figure 2:
Expression level of ApoE in 5 brain regions of the control, aggressive and defeated mice. Hypothalamus ApoE expression rate is elevated up to 30 % in affected mice compared to controls and exceeds the average across the regions. HPC, hippocampus; HPT, hypothalamus; STR, striatum; MRN, midbrain raphe nuclei; VTA, ventral tegmental area. *FDR < 0.05; ***FDR < 0.007; +p < 0.053.
3.3. Proopiomelanocortin (Pomc) and App Genes are Positively Correlated with ApoE Expression in Mouse Brain, While Sort1 is Antagonistic to ApoE
As mentioned above, ApoE is grossly associated with AD etiology. Based on literature mining of other AD-associated genes, we selected 3 genes that were reported to associate with AD, and also maintain highly non-random expression covariation with ApoE in our data (Table 5). Their descriptions are presented below.
Table 5:
Pairwise correlation of 4 genes based on 45 samples in our data.
| Variables | App | ApoE | Pomc | Sort1 |
|---|---|---|---|---|
| App | 1 | 0.659 | 0.094 | −0.420 |
| ApoE | 0.659 | 1 | 0.618 | −0.446 |
| Pomc | 0.094 | 0.618 | 1 | −0.578 |
| Sort1 | −0.420 | −0.446 | −0.578 | 1 |
Values in bold are different from 0 with a significance level alpha = 0.05.
Pomc gene is the source of up to 10 enzymatically derived hormone related peptides involved in pain and energy homeostasis, melanocyte stimulation, and immune modulation and is involved in neuroendocrine stress response [37]. Pomc function in the hypothalamus neurons differs from that of the pituitary gland, another major Pomc source [37]. In particular, Pomc gene is known for the regulation of food intake in rats in the presence of ApoE [14]. These facts underline the involvement of the ApoE-Pomc tandem in specific energy metabolism signalling cascades in the brain.
APP, the main constituent of Amyloid plagues, is targeted by APOE [3]. It was elucidated recently that APOE epsilon 2, 3, 4 isoforms differentially stimulate APP expression in neurons [38]; Supplementary Figure 1), APOE eps4 isoform being the highest stimulator.
Sortilin 1 (SORT1) is also considered the APOE- related receptor responsible to major uptake of APOE-APP complexes [13] and is specifically expressed in astrocytes [4], [5] (Supplementary Figure 1). One of the closest homologues of Sort1 involved in the process is sorting protein-related receptor (SORL1) which is associated with AD pathology in human [39] and is preferentially expressed in microglia (compared to Sort1) [4], [5] (Supplementary Figure 1).
We found that both Pomc and App expression rates are significantly correlated with ApoE expression (r = 0.618; p < 3E−6 and r = 0.659; p < 4E−7, correspondingly; df = 44; Table 5). Notably, though the correlation values of Pomc and App with ApoE are similar, they constitute significant differences in gene pathways. In particular, we observed Pomc expression solely in the hypothalamus of the mice studied, consistent with previous research studies [14], [37]. In contrast, non-basal App, as well as ApoE expression, was observed throughout all 45 samples considered. On the neural cells level, ApoE and App maintain antagonistic expression throughout 7 cell types [4] (Supplementary Figure 1). Pearson correlation rate across the cells is r = −0.91; df = 6; p < 6E−6). The highest ApoE expression is observed in microglia and astrocytes with App maintaining minimal expression within them. So the ApoE-App expression correlation observed across brain regions reflects coordinated inter-cellular interaction between asrocytes and neurons reported in [38]. Thus, it could be interpreted that ApoE-App covariation represents the metabolic interaction background observed in all brain regions, while Pomc pairing with Apoe apparently implies a specific (signalling) pathway, particularly in the hypothalamus.
Consistent with [13], Sort1 is negatively correlated with all other genes considered (Table 5): ApoE (r = −0.45; p < 1E−3), App (r = −0.42; p < 2E−3) and Pomc (r = −0.578; p < 1.5E−5). Considering Sort1/Sorl1 genes interaction, their expression was highly correlated. It was observed in all samples, and both genes’ expression was significantly down in the hypothalamus featuring the highest ApoE/App expression. The significant negative association of Sorl1 with ApoE (r = −0.49; df = 44; p < 3E−4; not shown) in the brain regions may mediate Sorl1 highly confident AD association reported previously [13], [39] (Table 5). We did not manage to provide any additional interpretation on the Sort1/Sorl1 negative correlation with Pomc, ApoE, App observed in this instance besides partly presented in [13].
3.4. Lipoprotein Receptors and ApoE Expression Covariation in Brain Regions
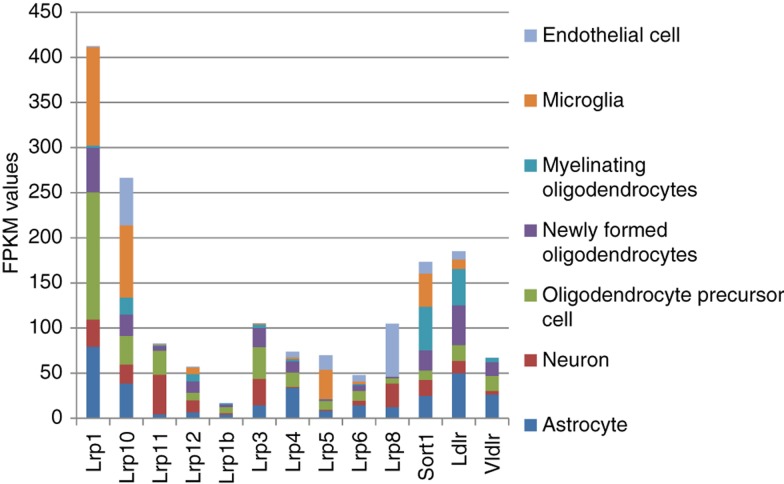
We assessed expression rate of available (with average FPKM > 0.2) 10 Lrp* receptors in ours and 10 Lrp* receptors in [4] projects, and also added to the list LDLR, VLDLR, and Sort1 receptors (Figure 3). As can be seen from Figure 3 based on [4] data, 4 receptors (Lrp1, Lrp10, Ldlr and Sort1) manifest major receptor expression profile in neural cell types by expression rate.
Figure 3:
FKPM values of Lrp receptors in 7 brain cell types (1). The highest expressed Lrp receptor genes in brain cell types are Lrp1, Lrp10, and Sort1 [4]. Lrp11 manifests a neuron cell-specific receptor preference. Lrp8 is a largely endothelial cell-specific receptor.
In our data we observed similar major receptors’ expression pattern (Supplementary Table 1). Also, we found that Sort1 expression rate is significantly elevated in the hippocampus and downregulated in the hypothalamus, being contrary to ApoE. Lrp11 also maintains the hippocampus-specific expression, while Lrp1 can be considered a hypothalamus-specific receptor (Supplementary Table 1).
Overall expression rate of lipoprotein receptors considered was 170 ± 7 FPKM units in four brain regions, and in the hippocampus it is 200.4. Thus, overall lipoprotein receptors’ expression homeostasis proved to be rather conserved across brain regions, except for in the hippocampus due to the Sort1 expression outburst (Supplementary Table 1).
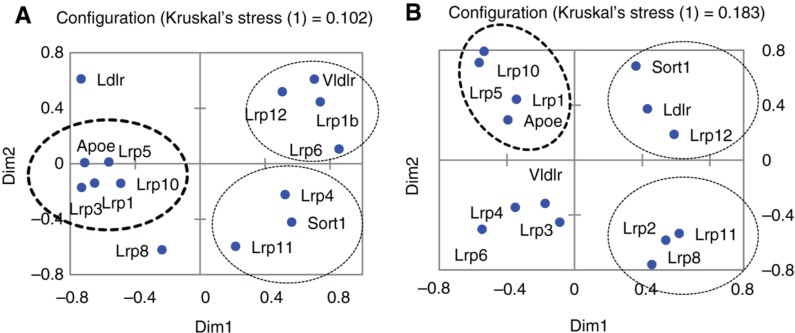
To elaborate on the relations between ApoE and lipoprotein receptors expressed in mouse brain, we built up multi-dimensional scaling (MDS) plots based on pairwise Pearson correlations of 17 Ldr* receptors and the ApoE gene (Figure 4A, B). We observed 3 statistically significant clusters, which consisted of one ApoE-synergistic and 2 ApoE-antagonistic ones.
Figure 4:
MDS plot based on Pearson pairwise correlation values of 14 lipoprotein receptors and ApoE expression profiles (A) across 45 samples; (B) across 7 neural cell types [4]. Lrp receptor genes that manifest positive and negative correlations with ApoE expression were observed in accordance with encircled clusters. Bold circles correspond to ApoE – synergistic genes; otherwise, circles correspond to ApoE antagonistic gene clusters. Ldlr and Lrp8 were uncorrelated with ApoE expression rate in (A).
Lrp1 expression rate is highly correlated with ApoE expression profile across 45 samples (r = 0.858; df = 44; p < 7.6E−7; Figure 4A), next is Lrp10 (r = 0.767; df = 44; p < 1.2E−5; Figure 4A). It was also proved that Lrp1 is differentially expressed in aggressive and defeated mice, compared to the control in the hypothalamus manifesting the same regression pattern (Table 3). Thus, Lrp1 is confirmed as the major partner of ApoE in the brain, as reported previously [40].
Data from [4] confirms the high correlation rate elucidated through our data (Figure 4A) between ApoE and Lrp1 (r = 0.925; df = 11; p < 1.2E−5), and Lrp5 (r = 0.65; df = 11; p < 0.011) across various brain cell types (Figure 4B), though other clusters are distinct from brain regions’ ones.
We also performed an ANOVA analysis of brain region-specific variation of ApoE and lipoprotein receptors to assess non-randomness of lipoprotein expression rate. R2 residuals’ values correspond to non-randomness of the brain-specific expression. As can be seen from the resulting Table 6, most lipoprotein receptors maintain highly non-random brain region-specific expression. Its specificity ultimately bifurcates between the hypothalamus and hippocampus/striatum clusters (Table 6). Based on the results, we can assume that each brain region is characterized with specific lipoprotein receptors’ expression profile, some of which (Lrp1, Lrp10, Lrp5) respond to ApoE expression rate alteration.
Table 6:
ANOVA results (with Fisher test) for Lipoprotein receptors in the 5 brain regions.
| Lrp1 | Lrp10 | Lrp11 | Lrp12 | Lrp1b | Sort1 | Ldlr | Vldlr | ApoE | Lrp3 | Lrp4 | Lrp5 | Lrp6 | Lrp8 | Lrp2 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| R2 | 0.59 | 0.28 | 0.87 | 0.28 | 0.57 | 0.98 | 0.89 | 0.71 | 0.33 | 0.40 | 0.91 | 0.07 | 0.41 | 0.78 | 0.65 |
| F | 14.51 | 3.79 | 68.18 | 3.88 | 13.19 | 445.60 | 80.59 | 24.08 | 4.94 | 6.73 | 105.38 | 0.72 | 6.94 | 35.11 | 18.64 |
| Pr > F | <0.0001 | 0.011 | <0.0001 | 0.009 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | 0.002 | 0.000 | <0.0001 | 0.587 | 0.000 | <0.0001 | <0.0001 |
| Major region* | HPT | HPT | HPC | STR | STR | HPC | ^HPC, ^STR | STR | HPT | HPT | HPC, STR | – | HPC, STR | HPC | HPT |
HPC, hippocampus; HPT, hypothalamus; STR, striatum. ‘Major region’ row denotes the preferred expression brain region. ^ Sign means ‘not’.
4. General Discussion
Bearing in mind the distinct, confirmed involvement of the ApoE locus in neuropathologic etiology, including AD, we aimed at an analysis of the ApoE gene and its genomic and pathway contexts using the expression rate co-variation assessment across the sample of 45 brain region RNA-Seq sets. The contemporary RNA-Seq data [4], [5] have unambiguously confirmed that the astrocytes and microglia are the major sources of ApoE (Supplementary Table 2), achieving as high as 32540.3 FPKM units for certain exons’ peak instances in microglia [4], [41] (Supplementary Table 4). Thus, one overlooked issue concerning the ApoE phenomenon is its high expression rate in the mouse brain. This, nevertheless, obviously corroborates the fact that the brain is the “fattiest” organ in the human body and is composed of roughly 60 % fat, while APOE is needed for lipid metabolism in the cells. According to the human protein atlas resource, APOE protein expression maintains the highest score in brain tissues (http://www.proteinatlas.org/ENSG00000130203-APOE/tissue), and it is even higher than in the liver for protein expression. This fact underscores the intense transcription of ApoE which belongs to the top 20 highest expressed genes in mouse brain regions.
The overall expression rate of ApoE gene in all mouse brain regions considered proved to be extraordinarily high, and, in our case, it averaged 1100 FPKM units (Table 7) across brain regions. To get a more elaborated view and underline ApoE high expression rate, we compiled the list of genes which maintain average expression rate more than 700 in FPKM values in our data. The results are presented in Table 7.
Table 7:
Most highly expressed genes (FPKM > 700) in our RNA-Seq data.
| Genes | FPKM avg | Description |
|---|---|---|
| Aldoa | 855.7 | Aldolase A, fructose-bisphosphate, mRNA |
| ApoE | 1189.9 | Apolipoprotein E, mRNA |
| Atp1a3 | 886.8 | ATPase, Na+/K+ transporting, alpha 3 polypeptide, mRNA |
| Calm3 | 800.2 | Calmodulin 3, mRNA |
| Ckb | 957.3 | Creatine kinase, brain, mRNA |
| Cst3 | 1255.6 | Cystatin C, mRNA |
| Eef1a1 | 751.5 | Eukaryotic translation elongation factor 1 alpha 1, mRNA |
| Gm20594 | 2085.8 | Predicted gene, 20594, mRNA |
| Gnas | 918.5 | GNAS (guanine nucleotide binding protein, alpha stimulating) complex locus, mRNA |
| Hexb | 2170.4 | Hexosaminidase B, mRNA |
| Hspa8 | 813.2 | Heat shock protein 8, mRNA |
| Lars2 | 4047.1 | Leucyl-tRNA synthetase, mitochondrial, mRNA |
| Mbp | 2627.1 | Myelin basic protein, mRNA |
| Plp1 | 807.4 | Proteolipid protein (myelin) 1, mRNA |
| Ppia | 721.1 | Peptidylprolyl isomerase A, mRNA |
| Snap25 | 814.7 | Synaptosomal-associated protein 25, mRNA |
| Tsr1 | 1097.6 | TSR1 20S rRNA accumulation, mRNA |
| Rpl38 | 1002.4 | Ribosomal protein L38, mRNA |
| Rpl41 | 820.8 | Ribosomal protein L41, mRNA |
| Rplp1 | 1063.2 | Ribosomal protein, large, P1, mRNA |
| Rps29 | 918.3 | Ribosomal protein S29, mRNA |
The expression rate of the ApoE gene enters the list of the top 21 highest, expressed both in our sample (Table 7) and one from top 17 genes from mouse brain cell types data [5] (Table 8). Discarding ribosomal and other non-specific genes, we may state that ApoE expression rate in mouse is uniquely high.
Table 8:
Most highly expressed genes (average FPKM > 700) in the [5] study (see Materials and Methods).
| Genes | FPKM avg | Description |
|---|---|---|
| ApoE | 1047.7 | Apolipoprotein E, mRNA |
| Bsg | 1200.5 | Basigin, mRNA |
| Cldn11 | 796.9 | Claudin 11, mRNA |
| Cnp | 1160.4 | 2′,3′-cyclic nucleotide 3′ phosphodiesterase, transcript variant 2, mRNA |
| Cst3 | 8162.7 | Cystatin C, mRNA |
| Ctsd | 1548.9 | Cathepsin D, mRNA |
| Eef1a1 | 867.6 | Eukaryotic translation elongation factor 1 alpha 1, mRNA |
| Fos | 908.6 | FBJ osteosarcoma oncogene, mRNA |
| Fth1 | 1951.6 | Ferritin heavy chain 1, transcript variant 2, non-coding RNA |
| Ftl1 | 777.1 | Ferritin light chain 1, mRNA |
| Gm1821 | 974.1 | Predicted gene 1821, non-coding RNA |
| Mbp | 17509.4 | Myelin basic protein, mRNA |
| Plp1 | 2886.1 | Proteolipid protein (myelin) 1, mRNA |
| Sparc | 988.3 | Secreted acidic cysteine rich glycoprotein, mRNA |
| Tmsb4x | 2162.9 | Thymosin, beta 4, X chromosome, mRNA |
| Tuba1a | 921.8 | Tubulin, alpha 1A, mRNA |
| Ubc | 888.4 | Ubiquitin C, mRNA |
Genes marked in bold are replicated in our data (Table 7).
The preliminary analysis of highly expressed gene ratio revealed that some of the genes represent non-specific, ubiquitous proteins such as ribosomal proteins and others. We replicated the results for previously published data [4], [5] (Table 8), and found that 5 genes are replicated in both lists (Table 7 and Table 8; marked in bold) underscoring highly non-random replication of highly expressed genes in the brain for distinct transcriptome projects (p < 1.5E−11 for random null hypothesis of 5 genes coincidence based on binomial test for 20 top genes pool with p = 0.001).
A class of highly expressed genes is reported to maintain specific means for their regulation and features [42]. In particular, ribosomal proteins encoding genes maintain specific transcription subunit TATA-box-binding protein-related factor 2 (TRF2) as a core subunit for their transcription [43], [44]. A significant part of highly expressed genes maintain miRNA and other noncoding RNA within their vicinity [45]. Also, CTCF binding sites and remote enhancers are common for their expression regulation [46]. We also observed a range of CTCF binding sites along the ApoE region.
Interestingly, while ApoE expression rate differs significantly between astrocytes and cortical neurons (FPKM values of 3006.2 vs. 200.4, correspondingly; Supplementary Table 2; Figure 1) [4], the amount of highly expressed genes proves to be the closest in these two cell types compared to other 5 cell types [41], revealing that they are structurally similar by that metric (highly expressed genes number; HEGN) [41]. While close co-variation between ApoE and its neighbors has been observed throughout all brain regions considered (Table 4), the highest ApoE elevation rate of affected groups related to the controls was observed in the hypothalamus (Figure 5).
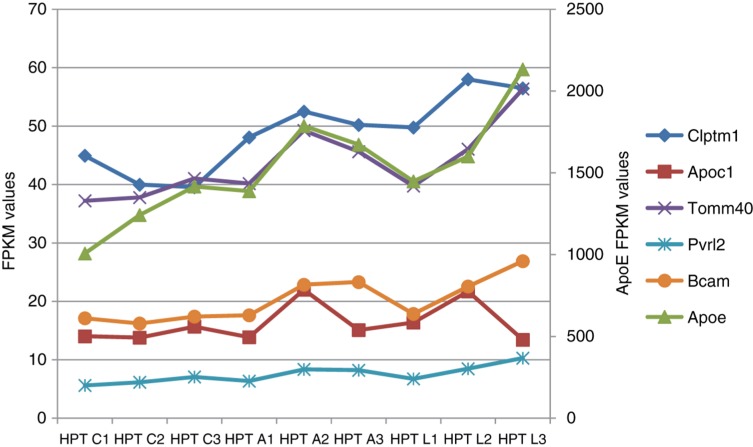
Figure 5:
Expression profiles of 5 genes in the vicinity of the ApoE locus in the hypothalamus (mouse, our data). Ordinate on the right corresponds to ApoE expression level, ordinate on the left corresponds to all other genes. Three animals in each experimental groups (numbered 1–3) were considered for each experiment. HPT, hypothalamus; C1, C2, C3, control group; A1, A2, A3, aggressive mice; L1, L2, L3, defeated mice.
The genes coordination rate has been the highest therein, as well as across brain regions (Supplementary Table 3). Notably, one of the highest expression covariation rates with ApoE manifests Tomm40 (r = 0.964 df = 8; p < 1.3E−13; Figure 5; Supplementary Table 3). Tomm40 expression modulation might affect the mitochondrial metabolism homeostasis reported previously [20].
The issue of gene co-expression and the tendency for similar expressed genes to cluster in eukaryotes have been studied throughout the genome research era [47]. Still, no implicit evidence for cystronic-like organization for the majority of eukaryotic genes exists and such cases are majorly confined to closely located paralogous gene clusters [48].
We should herein distinguish the functional and non-specific co-expression coordination. While the co-regulation issue discussed in the previous paragraph refers to the former instance, in the case of ApoE, we observed non-specific co-regulation based on disproportional expression rates of genes located densely at the chromosome [22] while maintaining focal highly expressed ApoE gene. Non-specific co-expression may thus arise from an abundance of transcription factors and chromatin accessibility within the region due to high transcription intensity at ApoE locus.
Based on the covariation analysis, we also found that ApoE turnover by lipoprotein receptors manifests highly complicated, brain region-specific homeostasis which may affect its proper function in a case of misbalance. While stated earlier [40], this is the first report of highly significant Apoe-LRP1 covariation based on RNA-Seq data, elucidating the major receptor counterpart for ApoE across brain regions.
Considering the mouse model of chronic agonistic interactions, we observed the highly statistically significant elevation (FDR < 0.007 in aggressive mice and FDR < 0.001 in defeated mice) of ApoE gene expression in the hypothalamus of affected mice, but not in other brain regions. The observation was additionally supported by a concordant expression shift of surrounding genes. The high correlation between neighboring genes expression in the vicinity of the ApoE gene was observed in all brain regions (Table 4). The increased expression of the ApoE gene might represent a metabolism rate increase featuring the behaviour affection. In particular, the stabilization of ApoE expression in the course of aging by means of aerobic exercise was proved to prevent astrocyte dysfunction in mice [49]. Notably, the absence of ApoE (ApoE deficient mice) did not reveal that relation [49]. High elevation of expression of the ApoE gene in the hypothalamus, both in the defeated and the aggressive mice, directly indicates the effects of social stress and anxiety on both participants of agonistic interactions, as has been shown previously [50], [51].
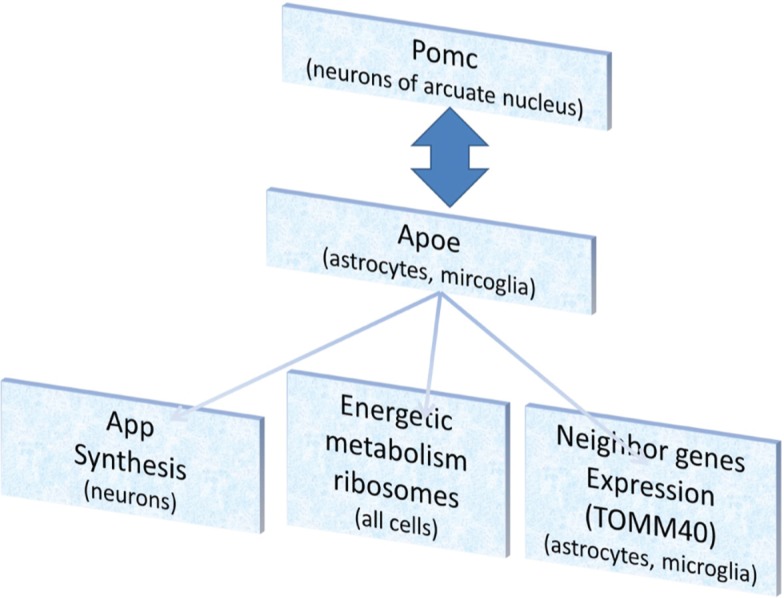
Overall, the causes and consequences of ApoE expression elevation we observed in hypothalamus RNA-Seq data can be interpreted as it is depicted in Figure 6.
Figure 6:
Schematic diagram of APOE mediated pathways elucidated from RNA-Seq data in current study.
The major cause of ApoE elevation in the hypothalamus can be the neuroendocrine stress response, which induces Pomc expression and adrenocorticotropic hormone in particular [52]. We observed high Pomc elevation in stressed mice compared to mice in the control group (Supplementary Table 1; p < 2E−5 for stress groups merged).
The interaction of Pomc and ApoE has been reported previously [14], [53], and we observe their covariation with high confidence (r = 0.618; p < 3E−6). It may be considered both ways, since intracerebroventricular administration of APOE was reported to increase Pomc expression [14].
To summarize the causes and consequences of ApoE expression elevation (Figure 6), we would like to stress the issues we observed and reported in the results:
We report significant covariation of ApoE and App expression (r = 0.659; p < 2E−7). It was elucidated in a recent paper [38] that APOE induction rate of APP is isoform dependent, with APOE eps4 inducing App expression the most, which might explain the APOE allelic effect on AD etiology.
Previously we observed a metabolic increase in the hypothalamus of stress-induced mice based on Rpl/Rps cytoplasmic ribosomal genes expression elevation [54]. The overall index of covariation of ApoE with Rps, Rpl genes was equal to 0.65 (p < 1E−6).
The Apoe expression elevation in mice under social stress (Supplementary Table 1) impacts the neighboring genes apparently in a non-specific manner. The covariation of Bcam, Pvrl2, Tomm40, Apoc1, and Clptm1, listed in Table 4, results in the following corresponding p-values: 1.37E−08, 7.88E−09, 2.96E−07, 4.85E−06, 8.63E−06. In the hypothalamus only, the corresponding values are: 3.14E−08, 4.29E−14, 1.35E−13, 0.226, 0.003.
Due to the sufficient independent replications number in RNA-Seq data, we were able to derive highly confident covariation assessments. Coordination with brain specific cell type data [4], [5] accommodated cross-validation of our data.
Supporting Information
Acknowledgements
This study was supported by the Russian Science Foundation (Nо. 14-15-00063).
Supplemental Material
The online version of this article offers supplementary material (DOI: jib-2017-0024).
Conflict of interest statement
Authors state no conflict of interest. All authors have read the journal’s Publication ethics and publication malpractice statement available at the journal’s website and hereby confirm that they comply with all its parts applicable to the present scientific work
References
- [1].Holtzman DM, Herz J, Bu G. Apolipoprotein E and apolipoprotein E receptors: normal biology and roles in Alzheimer disease. Cold Spring Harb Perspect Med. 2012;2:a006312. doi: 10.1101/cshperspect.a006312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Liu M, Kuhel DG, Shen L, Hui DY, Woods SC. Apolipoprotein E does not cross the blood-cerebrospinal fluid barrier, as revealed by an improved technique for sampling CSF from mice. Am J Physiol Regul Integr Comp Physiol. 2012;303:R903–8. doi: 10.1152/ajpregu.00219.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Huang Y. Roles of apolipoprotein E4 (ApoE4) in the pathogenesis of Alzheimer’s disease: lessons from ApoE mouse models. Biochem Soc Trans. 2011;39:924–32. doi: 10.1042/BST0390924. [DOI] [PubMed] [Google Scholar]
- [4].Zhang Y, Chen K, Sloan SA, Bennett ML, Scholze AR, O’Keeffe S. et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci. 2014;34:11929–47. doi: 10.1523/JNEUROSCI.1860-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Zhang Y, Sloan SA, Clarke LE, Caneda C, Plaza CA, Blumenthal PD. et al. Purification and characterization of progenitor and mature human astrocytes reveals transcriptional and functional differences with mouse. Neuron. 2016;89:37–53. doi: 10.1016/j.neuron.2015.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Falchi AM, Sogos V, Saba F, Piras M, Congiu T, Piludu M. Astrocytes shed large membrane vesicles that contain mitochondria, lipid droplets and ATP. Histochem Cell Biol. 2013;139:221–31. doi: 10.1007/s00418-012-1045-x. [DOI] [PubMed] [Google Scholar]
- [7].Perkins M, Wolf AB, Chavira B, Shonebarger D, Meckel JP, Leung L. et al. Altered energy metabolism pathways in the posterior cingulate in young adult apolipoprotein E epsilon 4 carriers. J Alzheimers Dis. 2016;53:95–106. doi: 10.3233/JAD-151205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Wolf AB, Caselli RJ, Reiman EM, Valla J. APOE and neuroenergetics: an emerging paradigm in Alzheimer’s disease. Neurobiol Aging. 2013;34:1007–17. doi: 10.1016/j.neurobiolaging.2012.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Wang YW, Brinton RD. Triad of risk for late onset Alzheimer’s: mitochondrial haplotype, APOE genotype and chromosomal sex. Front Aging Neurosci. 2016;8:232. doi: 10.3389/fnagi.2016.00232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Grainger DJ, Reckless J, McKilligin E. Apolipoprotein E modulates clearance of apoptotic bodies in vitro and in vivo, resulting in a systemic proinflammatory state in apolipoprotein E-deficient mice. J Immunol. 2004;173:6366–75. doi: 10.4049/jimmunol.173.10.6366. [DOI] [PubMed] [Google Scholar]
- [11].Chung WS, Verghese PB, Chakraborty C, Joung J, Hyman BT, Ulrich JD. et al. Novel allele-dependent role for APOE in controlling the rate of synapse pruning by astrocytes. Proc Natl Acad Sci USA. 2016;113:10186–91. doi: 10.1073/pnas.1609896113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Castellano JM, Deane R, Gottesdiener AJ, Verghese PB, Stewart FR, West T. et al. Low-density lipoprotein receptor overexpression enhances the rate of brain-to-blood Abeta clearance in a mouse model of beta-amyloidosis. Proc Natl Acad Sci USA. 2012;109:15502–7. doi: 10.1073/pnas.1206446109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Carlo AS, Gustafsen C, Mastrobuoni G, Nielsen MS, Burgert T, Hartl D. et al. The pro-neurotrophin receptor sortilin is a major neuronal apolipoprotein E receptor for catabolism of amyloid-beta peptide in the brain. J Neurosci. 2013;33:358–70. doi: 10.1523/JNEUROSCI.2425-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Shen L, Tso P, Woods SC, Clegg DJ, Barber KL, Carey K. et al. Brain apolipoprotein E: an important regulator of food intake in rats. Diabetes. 2008;57:2092–8. doi: 10.2337/db08-0291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Casey CS, Atagi Y, Yamazaki Y, Shinohara M, Tachibana M, Fu Y. et al. Apolipoprotein E inhibits cerebrovascular pericyte mobility through a RhoA protein-mediated pathway. J Biol Chem. 2015;290:14208–17. doi: 10.1074/jbc.M114.625251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].May P, Rohlmann A, Bock HH, Zurhove K, Marth JD, Schomburg ED. et al. Neuronal LRP1 functionally associates with postsynaptic proteins and is required for normal motor function in mice. Mol Cell Biol. 2004;24:8872–83. doi: 10.1128/MCB.24.20.8872-8883.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Xu Q, Bernardo A, Walker D, Kanegawa T, Mahley RW, Huang Y. Profile and regulation of apolipoprotein E (ApoE) expression in the CNS in mice with targeting of green fluorescent protein gene to the ApoE locus. J Neurosci. 2006;26:4985–94. doi: 10.1523/JNEUROSCI.5476-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Dupont-Wallois L, Soulie C, Sergeant N, Wavrant-de Wrieze N, Chartier-Harlin MC, Delacourte A. et al. ApoE synthesis in human neuroblastoma cells. Neurobiol Dis. 1997;4:356–64. doi: 10.1006/nbdi.1997.0155. [DOI] [PubMed] [Google Scholar]
- [19].Payton A, Sindrewicz P, Pessoa V, Platt H, Horan M, Ollier W. et al. A TOMM40 poly-T variant modulates gene expression and is associated with vocabulary ability and decline in nonpathologic aging. Neurobiol Aging. 2016;39:217e1–7. doi: 10.1016/j.neurobiolaging.2015.11.017. [DOI] [PubMed] [Google Scholar]
- [20].Roses A, Sundseth S, Saunders A, Gottschalk W, Burns D, Lutz M. Understanding the genetics of APOE and TOMM40 and role of mitochondrial structure and function in clinical pharmacology of Alzheimer’s disease. Alzheimers Dement. 2016;12:687–94. doi: 10.1016/j.jalz.2016.03.015. [DOI] [PubMed] [Google Scholar]
- [21].Swerdlow RH, Khan SM. The Alzheimer’s disease mitochondrial cascade hypothesis: an update. Exp Neurol. 2009;218:308–15. doi: 10.1016/j.expneurol.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Grimwood J, Gordon LA, Olsen A, Terry A, Schmutz J, Lamerdin J. et al. The DNA sequence and biology of human chromosome 19. Nature. 2004;428:529–35. doi: 10.1038/nature02399. [DOI] [PubMed] [Google Scholar]
- [23].Gordon I, Ben-Eliyahu S, Rosenne E, Sehayek E, Michaelson DM. Derangement in stress response of apolipoprotein E-deficient mice. Neurosci Lett. 1996;206:212–4. doi: 10.1016/s0304-3940(96)12470-8. [DOI] [PubMed] [Google Scholar]
- [24].Dose J, Huebbe P, Nebel A, Rimbach G. APOE genotype and stress response – a mini review. Lipids Health Dis. 2016;15:121. doi: 10.1186/s12944-016-0288-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Sheffler J, Moxley J, Sachs-Ericsson N. Stress, race, and APOE: understanding the interplay of risk factors for changes in cognitive functioning. Aging Ment Health. 2014;18:784–91. doi: 10.1080/13607863.2014.880403. [DOI] [PubMed] [Google Scholar]
- [26].Peskind ER, Wilkinson CW, Petrie EC, Schellenberg GD, Raskind MA. Increased CSF cortisol in AD is a function of APOE genotype. Neurology. 2001;56:1094–8. doi: 10.1212/wnl.56.8.1094. [DOI] [PubMed] [Google Scholar]
- [27].de Kloet ER, Grootendorst J, Karssen AM, Oitzl MS. Gene x environment interaction and cognitive performance: animal studies on the role of corticosterone. Neurobiol Learn Mem. 2002;78:570–7. doi: 10.1006/nlme.2002.4079. [DOI] [PubMed] [Google Scholar]
- [28].Grootendorst J, Kempes MM, Lucassen PJ, Dalm S, de Kloet ER, Oitzl MS. Differential effect of corticosterone on spatial learning abilities in apolipoprotein E knockout and C57BL/6J mice. Brain Res. 2002;953:281–5. doi: 10.1016/s0006-8993(02)03399-1. [DOI] [PubMed] [Google Scholar]
- [29].Peavy GM, Lange KL, Salmon DP, Patterson TL, Goldman S, Gamst AC. et al. The effects of prolonged stress and APOE genotype on memory and cortisol in older adults. Biol Psychiatry. 2007;62:472–8. doi: 10.1016/j.biopsych.2007.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Kudryavtseva NN. A sensory contact model for the study of aggressive and submissive behavior in male-mice. Aggress Behav. 1991;17:285–91. [Google Scholar]
- [31].Kudryavtseva NN, Smagin DA, Kovalenko IL, Vishnivetskaya GB. Repeated positive fighting experience in male inbred mice. Nat Protoc. 2014;9:2705–17. doi: 10.1038/nprot.2014.156. [DOI] [PubMed] [Google Scholar]
- [32].Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–11. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Boyarskikh UA, Bondar NP, Filipenko ML, Kudryavtseva NN. Downregulation of serotonergic gene expression in the Raphe nuclei of the midbrain under chronic social defeat stress in male mice. Mol Neurobiol. 2013;48:13–21. doi: 10.1007/s12035-013-8413-y. [DOI] [PubMed] [Google Scholar]
- [34].Smagin DA, Boyarskikh UA, Bondar NP, Filipenko ML, Kudriavtseva NN. Reduction of serotonergic gene expression in the raphe nuclei of the midbrain under positive fighting experience in male mice. Adv Biosci Biotechnol. 2013;4:36–44. [Google Scholar]
- [35].Rao SS, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT. et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159:1665–80. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Semon M, Duret L. Evolutionary origin and maintenance of coexpressed gene clusters in mammals. Mol Biol Evol. 2006;23:1715–23. doi: 10.1093/molbev/msl034. [DOI] [PubMed] [Google Scholar]
- [37].Jenks BG. Regulation of proopiomelanocortin gene expression an overview of the signaling cascades, transcription factors, and responsive elements involved. Ann NY Acad Sci. 2009;1163:17–30. doi: 10.1111/j.1749-6632.2008.03620.x. [DOI] [PubMed] [Google Scholar]
- [38].Huang YW, Zhou B, Wernig M, Sudhof TC. ApoE2, ApoE3, and ApoE4 differentially stimulate APP transcription and A beta secretion. Cell. 2017;168:427.e21–441. doi: 10.1016/j.cell.2016.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Yin RH, Yu JT, Tan L. The role of SORL1 in Alzheimer’s disease. Mol Neurobiol. 2015;51:909–18. doi: 10.1007/s12035-014-8742-5. [DOI] [PubMed] [Google Scholar]
- [40].Bell RD, Winkler EA, Singh I, Sagare AP, Deane R, Wu Z. et al. Apolipoprotein E controls cerebrovascular integrity via cyclophilin A. Nature. 2012;485:512–6. doi: 10.1038/nature11087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Trakhtenberg EF, Pho N, Holton KM, Chittenden TW, Goldberg JL, Dong L. Cell types differ in global coordination of splicing and proportion of highly expressed genes. Sci Rep. 2016;6:32249. doi: 10.1038/srep32249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Hurst LD, Ghanbarian AT, Forrest AR, Huminiecki L, Consortium F. The constrained maximal expression level owing to haploidy shapes gene content on the mammalian X chromosome. PLoS Biol. 2015;13:e1002315. doi: 10.1371/journal.pbio.1002315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Wang YL, Duttke SH, Chen K, Johnston J, Kassavetis GA, Zeitlinger J. et al. TRF2, but not TBP, mediates the transcription of ribosomal protein genes. Gen Dev. 2014;28:1550–5. doi: 10.1101/gad.245662.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Zehavi Y, Kedmi A, Ideses D, Juven-Gershon T. TRF2: TRansForming the view of general transcription factors. Transcription. 2015;6:1–6. doi: 10.1080/21541264.2015.1004980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Engreitz JM, Haines JE, Perez EM, Munson G, Chen J, Kane M, McDonel PE, Guttman M, Lander ES. Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature. 2016;539:452–5. doi: 10.1038/nature20149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Eldholm V, Haugen A, Zienolddiny S. CTCF mediates the TERT enhancer- promoter interactions in lung cancer cells: identification of a novel enhancer region involved in the regulation of TERT gene. Int J Cancer. 2014;134:2305–13. doi: 10.1002/ijc.28570. [DOI] [PubMed] [Google Scholar]
- [47].Ghanbarian AT, Hurst LD. Neighboring genes show correlated evolution in gene expression. Mol Biol Evol. 2015;32:1748–66. doi: 10.1093/molbev/msv053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Lan X, Pritchard JK. Coregulation of tandem duplicate genes slows evolution of subfunctionalization in mammals. Science. 2016;352:1009–13. doi: 10.1126/science.aad8411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Soto I, Graham LC, Richter HJ, Simeone SN, Radell JE, Grabowska W. et al. APOE stabilization by exercise prevents aging neurovascular dysfunction and complement induction. PLoS Biol. 2015;13:e1002279. doi: 10.1371/journal.pbio.1002279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Kudryavtseva NN, Avgustinovich DF. Behavioral and physiological markers of experimental depression induced by social conflicts (DISC) Aggress Behav. 1998;24:271–86. [Google Scholar]
- [51].Kudryavtseva NN, Bondar NP, Avgustinovich DF. Association between experience of aggression and anxiety in male mice. Behav Brain Res. 2002;133:83–93. doi: 10.1016/s0166-4328(01)00443-0. [DOI] [PubMed] [Google Scholar]
- [52].Hoeve ES, Kelly G, Luz S, Ghanshani S, Bhatnagar S. Short-term and long-term effects of repeated social defeat during adolescence or adulthood in female rats. Neuroscience. 2013;249:63–73. doi: 10.1016/j.neuroscience.2013.01.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Raber J, Akana SF, Bhatnagar S, Dallman MF, Wong D, Mucke L. Hypothalamic-pituitary-adrenal dysfunction in Apoe(-/-) mice: possible role in behavioral and metabolic alterations. J Neurosci. 2000;20:2064–71. doi: 10.1523/JNEUROSCI.20-05-02064.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Smagin DA, Kovalenko IL, Galyamina AG, Orlov YL, Babenko VN, Kudryavtseva NN. Heterogeneity of brain ribosomal genes expression following repeated experience of aggression in male mice as revealed by RNA-Seq. Mol Neurobiol. 2017 doi: 10.1007/s12035-016-0327-z. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.