Abstract
Here we present the analysis of alternative splicing events on an example of glioblastoma cell culture samples using a set of computer tools in combination with database integration. The gene expression profiles of glioblastoma were obtained from cell culture samples of primary glioblastoma which were isolated and processed for RNA extraction. Transcriptome profiling of normal brain samples and glioblastoma were done by Illumina sequencing. The significant differentially expressed exon-level probes and their corresponding genes were identified using a combination of the splicing index method. Previous studies indicated that tumor-specific alternative splicing is important in the regulation of gene expression and corresponding protein functions during cancer development. Multiple alternative splicing transcripts have been identified as progression markers, including generalized splicing abnormalities and tumor- and stage-specific events. We used a set of computer tools which were recently applied to analysis of gene expression in laboratory animals to study differential splicing events. We found 69 transcripts that are differentially alternatively spliced. Three cancer-associated genes were considered in detail, in particular: APP (amyloid beta precursor protein), CASC4 (cancer susceptibility candidate 4) and TP53. Such alternative splicing opens new perspectives for cancer research.
Keywords: transcriptome, glioblastoma, alternative splicing, differential splicing, cancer stem cells
1. Introduction
Multiple layers of post-transcriptional gene regulation significantly impact protein output [1]. Alternative splicing is a critical mechanism for expanding regulatory and functional diversity from a limited number of genes. It is particularly complex in the mammalian brain [2].
Gliomas are the most common type of brain tumor and are often fast growing with a poor prognosis for patient [3], [4], [5]. Diffuse gliomas are the most common type of intracranial malignant neoplasm, and account for >60 % of all brain tumors [6]. The World Health Organization classified gliomas in 2016 using molecular parameters in addition to their histological and immunohistochemical resemblance to presumed cells of origin and graded them by increasing degrees of undifferentiation, anaplasia, and aggressiveness (i.e. mitotic figures, necrosis, and vascular endothelial hyperplasia) [6]. Based on the classification of nervous system tumors, diffuse gliomas are classified into several principal categories by 4 grades: diffuse astrocytoma (grade II), oligodendroglioma (grade II), oligoastrocytoma (grade II), anaplastic astrocytoma, anaplastic oligodendroglioma and oligoastrocytoma (grade III) and glioblastoma (GBM, grade IV) [6]. GBM is the most common and aggressive type of primary brain tumor, accounting for 80 % of malignant astrocytomas. Because GBM cells vary in size and shape, it was frequently called glioblastoma multiforme, a term no longer in use. GBM may develop rapidly without the diagnosis of a less malignant precursor lesion, and this is termed primary or de novo GBM. It may also develop slowly through progression from a pre-existing low-grade glioma, in which case it is termed a secondary GBM. Despite advances in neurosurgery, chemotherapy and radiotherapy, glioma commonly has a poor prognosis. Therefore, it is critical that the genetic pathways underlying the development of this type of cancer are defined. From a molecular point of view, GBM is a highly heterogeneous tumor.
In the current study, the gene expression profiles of GBM and normal brain (NB) were obtained on primary cell culture samples from patients of the same age and sex. The cell culture samples were isolated following [7] and processed for RNA extraction. The significant differentially expressed exon-level probes and their corresponding genes were identified using a combination of the splicing index (SI) method. A previous study indicated that tumor-specific alternative splicing is important in the regulation of gene expression and corresponding protein functions during cancer development [8]. Multiple alternative splicing transcripts have been identified as progression markers, including generalized splicing abnormalities and tumor- and stage-specific events.
2. Alternative Splicing in Glioma
Alternative splicing is important part of differentiation and functioning of the genes of nervous tissue of higher eukaryotes [9], [10]. The process of specialization of tissue-specific gene expression has several levels, including the processes of replication, transcription and splicing, as well as regulation of factors involved in the regulation of splicing of neural genes by micro-RNA (miRNA).
Overall, dysregulation of alternative splicing (AS) is one of the molecular hallmarks of cancer, with splicing alteration of numerous genes in cancer patients. However, studying splicing mis-regulation in cancer is complicated by the large noise generated from tissue-specific splicing.
The following neurospecific splicing enhancers simultaneously modify the structure of the mRNA of large number of gene targets: NOVA1/2, FOX1/2, nSR100/SRM4 [10], [11] and silencers PTB1/2 [11]. At the same time, it was shown that the dynamic competition of these enhancers and the repressor PTB1 differ in the brain areas and development stages [10].
Unusually long length of neurospecific genes is a distinctive feature [12] that confirms the presence of alternative splicing. The degree of splicing intensity, in turn, is related to the length of the genes, to the length of introns and the number of exons [13].
Epigenetic factors, such as histone modifications and DNA methylation, can be involved in the splicing process [10].
There are 5 main classes of alternative transcription (splicing) events. The most frequent are:
Alternative initiation of transcription – 40 % of all splicing events (alternative initiation, AI);
Alternative termination – 30 % of all splicing events (Alternative termination, AT);
Alternation of inclusion (skip) of internal exon (20 %; Exon Skipping, ES);
Intron retention - occurs less frequently, for vertebrates about 4 % (Intron retention, IR). For plants and invertebrates, the relative frequency of IR is much greater, since IR depends on the average length of introns in the genome;
Alternative 5′ and 3′ splice sites, as well as complex variants that occur in no more than 6 %.
One type of exon skipping is the so-called Mutually Exclusive Exons (MXE), in which one of the set of similar and usually closely spaced exons is selected. An example is a unique neurospecific cell adhesion gene D. melanogaster DSCam1 (Down syndrome cell adhesion molecule 1) in which about 19,000 isoforms can be generated due to two cassettes of 16 and 18 exons (as well as two small cassettes in 3 and 5 exons). Extremely large number of isoforms was also reported in human brain-specific genes [14].
In human several alternative exon usage patterns were reported as significantly associated with glioblastoma patient survival. G-protein coupled receptor 98 (Gpr98) and epidermal growth factor (Egf) were among the genes exhibiting alternative exon usage with 30 and 9 exons associated with GBM survival, respectively [15]. Heterogeneous nuclear ribonucleoprotein (hnRNP) A1 splicing factor promotes glycolytic gene expression and conferring significantly shorter survival in patients [16]. HnRNPA1 promotes splicing of a transcript encoding the Myc-interacting partner Max, generating Delta Max, an enhancer of Myc-dependent transformation. Splicing factor hnRNPH, which is upregulated in gliomas, controls splicing event for Insuloma-Glucagonoma protein 20 (IG20) to generate MADD isoform (MAP-kinase activating death domain protein – MADD), which effectively redirects TNF-α/TRAIL-induced death signaling to cell survival and proliferation instead of triggering apoptosis [17]. Splicing factor hnRNPH also mediates switching to a ligand-independent, constitutively active Recepteur d’Origine Nantais (RON) tyrosine kinase receptor variant that promotes migration and invasion. It was shown by antineoplastic assay that A2BP1 (ataxin 2 binding protein 1, Rbfox1), an RNA-binding and splicing regulator is deleted in 10 % of GBM cases. A2BP1 serves to regulate TPM1’s alternative splicing to promote cytoskeletal organization and terminal differentiation and suppress malignancy [18].
3. Workflow
Primary cell cultures from surgical samples of GBM and normal brain were obtained from male patients aged 60 years. The cell cultures have been isolated and propagated in F12/DMEM medium supplemented by fetal bovine serum under standard conditions. At three passages the culture were stained of neuronal- and astrocyte-specific markers to confirm the tissue specificity. The cell culture samples were processed for RNA extraction. RNA-seq sequencing of cell samples was done using Illumina HiSeq 1500. The RNA quality was tested on Bioanalyzer 2100 (Agilent) following the Illumina protocol. Sequencing depth consisted of at least 10 mol reads for each sample.
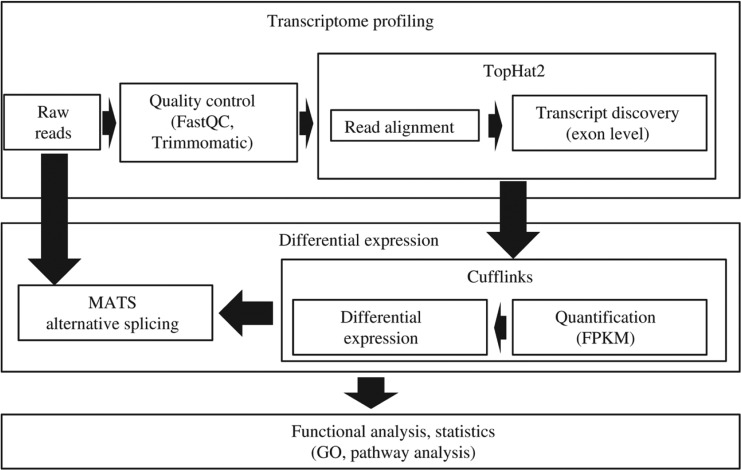
We used a set of computer tools applied recently to analysis of gene expression in laboratory animals [2]. The algorithm of the pipeline consists of several stages: (1) pre-processing of the reads; (2) mapping reads to the reference genome; (3) identification of differential expression and cases of alternative splicing; (4) annotation of obtained results. A similar approach for the analysis of RNA-seq data is used in web-based platform GALAXY [19]. The files obtained in “fastq” format were used for the reads mapping onto the human reference genome GRCh38 using Tophat2 aligner. Input reads were filtered and trimmed by Trimmomatic software [20]. Trimmomatic omits technical sequences and sequences with low quality from input data. Low quality sequences, technical sequences and reads with low length were omitted. We used FastQC program for quality control [21]. Nowadays there are a number of mapping programs such as GSNAP [22], STAR [23], Bowtie [24], and TopHat2 [25] (Figure 1).
Figure 1:
Scheme of transcriptome profiling analysis: from read alignment and mapping to differential gene expression and alternative splicing analysis.
In the work we have used TopHat2 program because of it allow taking into account intron-exon structure and is compatible with Cufflinks. We used the Tophat2 program [26] for mapping reads on the reference genomes. Tophat2 is built on the fast, memory-efficient program Bowtie [27] and can identify splice junctions. Files with genome annotations were downloaded from UCSC genome browser. The bam-files received from Tophat output were used for detection of differentially expressed genes and alternative splicing analysis in the samples. There are two types of programs for gene expression analysis: programs that use known transcripts like RSEM [28] and eXpress [29]; programs for novel transcript discovery. One of the more popular programs for novel transcript discovery is Cufflinks [26]. We have analyzed gene expression by Cufflinks v2.0.2 programs [26]. The Cufflinks tool provides information about differently expressed genes between the samples. Gene expression levels were estimated in FPKM (fragments per kilobase per million reads) values. The detection of splice variants and differential splicing was performed using the rMATS software [30]. The overrepresentation of GO terms was evaluated with DAVID (https://david-d.ncifcrf.gov/) and the GOseq package [31].
To analyze alternative splicing events in the transcriptomics data MATS (multivariate analysis of transcript splicing) and rMATS (replicate MATS) have shown to be effective tools [32]. We implemented a Python-based application to tackle rMATs annotation. The rMATs output lacks the information on the short isoforms spectra of particular exon. We implemented an add-on that based on 3 subsequent exons locations used in alternative exons identification retrieves from the RefGene database short isoforms IDs lacking the exon. While it is not possible to relate an exon skipping event to the particular full length isoform due to the short length of sequencing reads, we can still speculate on tissue specificity of the exon skipping events. The rMATS program [30] was used to assess the difference in alternative splicing profiles between Normal brain (NB) tissue and secondary glioblastoma (GBM) tissue. The pipeline – described above – was implemented using the bash language and integrated into Linux RedHat 10 system of high-performance computing cluster SB RAS (www2.sscc.ru) and Shared Facility Center “Bioinformatics”.
Thus, for the analysis of work of genes of glioma tissues, a system – based on modern approaches to the analysis of RNA-seq data – was created, which makes it possible to identify differentially expressed genes and cases of alternative splicing.
4. Results
We revealed a set of differently expressed genes of the nuclear pore complex (NPC) in normal brain and GBM cell cultures. Most of them are expressed higher in the GBM cells. Alterations in NPC genes were linked to several human neoplastic diseases as reported in [33]. The antisense RNA transcript of Nup50 (Nup50-AS1) is the only one among NPC-related genes that is under expressed in tumor less in comparison to normal brain. At the same time, expression of Nup50 gene is not distinguished in tumor and normal brain cell cultures. We may suggest that there is a higher level of Nup50 protein in tumor cells and it is regulated by antisense RNA transcript. We found set of hormone transporter genes overexpressed in the glioblastoma cell culture. SLCO1C1 mediates the Na+-independent high affinity transport of organic anions such as the thyroid hormones thyroxine (T4) and rT3. Other potential substrates, such as triiodothyronine (T3), 17-beta-glucuronosyl estradiol, estrone-3-sulfate and sulfobromophthalein are transported with much lower efficiency. It may play a significant role in regulating T4 flux into and out of the brain. SLCO2A1 may mediate the release of newly synthesized prostaglandins from cells, the transepithelial transport of prostaglandins, and the clearance of prostaglandins from the circulation. Transports PGD2, as well as PGE1, PGE2 and PGF2A. SLCO3A1 mediates the Na+-independent transport of organic anions such as estrone-3-sulfate [34]. It mediates transport of prostaglandins (PG) E1 and E2, thyroxine (T4), deltorphin II, BQ-123 and vasopressin, but not DPDPE (a derivative of enkephalin lacking an N-terminal tyrosine residue), estrone-3-sulfate, taurocholate, digoxin nor DHEAS [35]. SLCO4A1 mediates the Na+-independent transport of organic anions such as the thyroid hormones T3 (triiodo-L-thyronine), T4 (thyroxine) and rT3, and of estrone-3-sulfate and taurocholate [34].
We used text mining tools such as ANDvisio [36], [37] and ANDSystem (Associative Network Discovery System) [38], [39] for literature mining of genes by terms “glioma”, “glioblastoma”, alternative splicing”. We found 51 genes from the differentially expressed as related to “glioma” term in text mining approach. 73 of the differentially expressed genes were found in OMIM as related to glioma (of 193 loci) including GLI1, GLI3 (GLI-Kruppel family member 3); GLM4 (GLIOMA SUSCEPTIBILITY 4), GLTSCR1 (Glioma tumor suppressor candidate region gene 1) and others.
To study alternative splicing in GBM we selected 69 gene loci with highly significant differential isoforms expression in our experiment by the software tools described in previous section (Supplementary Table 1). This alternatively spliced gene list was constructed by rMATS program overlapping in part with the larger list of all differentially expressed genes.
The total number of significant alternative splicing (exon skipping) events comprised 107 instances with FDR < 0.01 (160 with FDR < 0.05). Gene ontology categories for differentially alternatively spliced genes were related to neuronal tissues and tumor progression terms. The major GO entries were cytoskeleton and intracellular (cytoplasmic) related genes (Table 1).
Table 1:
GO categories (compartments) for differential isoforms.
| Category | Term | p-value | Fold | Bonferroni corrected |
|---|---|---|---|---|
| SP_PIR_KEYWORDS | cytoskeleton | 1.51E | 6.5 | 2.65E-04 |
| GOTERM_CC_FAT | GO:0005856∼ cytoskeleton | 2.77E | 3.5 | 4.92E-04 |
| GOTERM_CC_FAT | GO:0043232 ∼intracellular non-membrane- bounded organelle | 6.67E | 2.5 | 0.001 |
| GOTERM_CC_FAT | GO:0043228∼non-membrane-bounded organelle | 6.67E | 2.5 | 0.001 |
| SP_PIR_KEYWORDS | Cytoplasm | 1.74E | 2.5 | 0.003 |
Here we consider a case of APP isoforms expression alteration. One of the target genes was amyloid beta precursor (APP) implicated in Alzheimer disease and a range of other neural disorders. We elucidated this gene in detail. We maintained the expression profile of 9 alternatively spliced isoforms for this gene based on 4 exon skipping events [exon 2 (E2), exon 7 (E7), exon 8 (E8), exon 15 (E15)]. All of the skipped events were described in previous studies and are known to modify the function of the protein [40], [41].
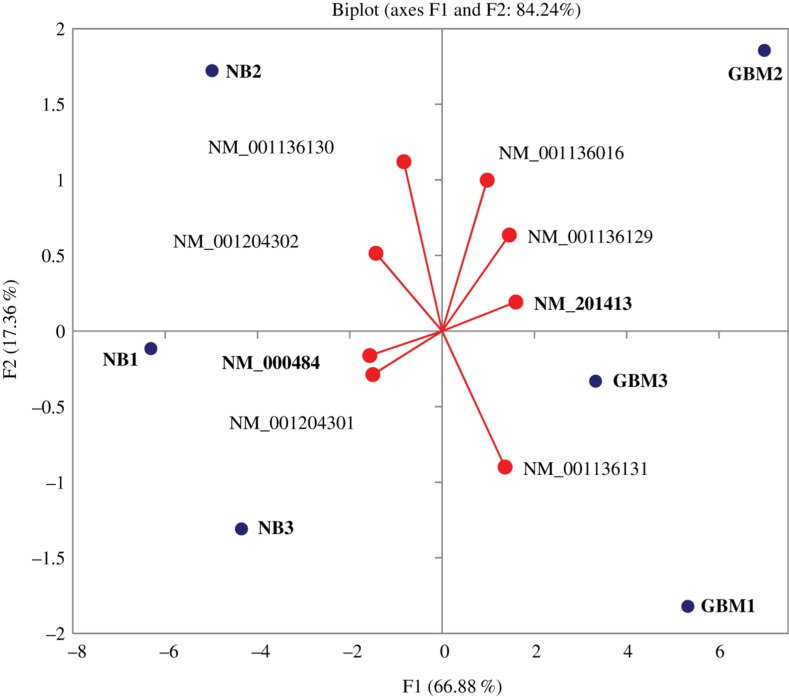
We observed distinctly different overall splicing landscape of APP between Normal Brains and glioblastoma tissues (Figure 2). The short isoforms were preferably expressed in the GBM cells, while normal cells produce mostly long isoforms.
Figure 2:
Distribution of generalized vectors of isoform expressions against samples (NB and GBM) for APP gene. The plot was constructed in the course of Principal Component analysis of isoforms expression profiles (FPKM matrix). The most pronounced differentially expressed isoforms are given in bold. All isoforms on the right are shorter than their closest counterpart on the left (Table 2).
Table 2:
Pairs of closest by structure isoforms of APP gene ordered by expression rate difference (See Figure 3).
| Normal brain | Exons skipped | NB, fpkm | GBM, fpkm | Glioblastoma | Exons skipped | NB, fpkm | GBM, fpkm |
|---|---|---|---|---|---|---|---|
| NM_000484 | – | 43.46 | 0 | NM_201413 | (E7, E8) | 60.29 | 146.9 |
| NM_001204302 | (E7,E8, E15) | 15.28 | 29.07 | NM_001136131 | (E7, E8, alt 5′ CDS) | 4.85 | 20.77 |
| NM_001204301 | (E8, E15) | 35.69 | 0 | NM_001136016 | (E8, short TSS) | 0 | 0.49 |
| NM_001136130 | (E2) | 184.39 | 167.71 | NM_001136129 | (E2, E7, 8) | 0 | 0.02 |
The most discordant isoforms are shown in bold.
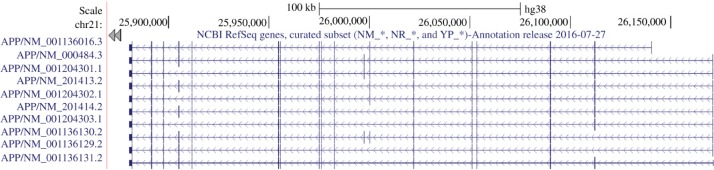
Figure 3:
Nine isoforms of APP gene (retrieved from UCSC browser, chr 21).
It has been previously reported that cancer often causes mis-splicing of significant amount of genes due to spliceosome defects resulted in overall depressed splicing rate, leading to aberrant transcripts [42], [43]. In a significant range of cases the general splicing downregulation was caused by altered expression of key alternative splicing regulators, such as FOX2 [9] leading to a massive exon skipping instances. It is also confirmed by repression of the synapse specialized genes abundant in alternative splicing [6] and overexpression of cell cycle genes. Accordingly, we also observe the expression of amyloid precursor gene with massive exon skipping events in GBM. As APP is an excreted protein it may jeopardize the malicious nature of glioblastoma for other brain tissues.
During the analysis of differential splicing events we found significant differences in splicing of three cancer-associated genes, in particular: APP (amyloid beta precursor protein), CASC4 (cancer susceptibility candidate 4) TP53 and corresponding inducible genes. For TP53 the whole range of p53-related transcripts specific for GBM cells were observed, distinct from NB cells. In particular non-coding RNA NR_015381 [TP53 target 1 (non-protein coding) TP53TG1] was attributable for GBM cells.
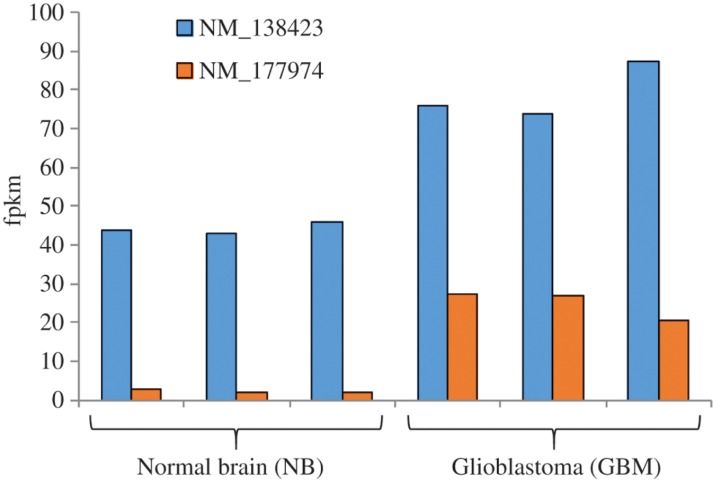
The increased expression level of the CASC4 gene is associated with HER-2/neu proto-oncogene overexpression. It was found overexpressed in breast cancer, sarcoma, and ovarian cancer [44]. While no survival time correlation was observed with the expression of this gene, we found that the alternative short isoform with a missing exon at 3′ CDS is elevated 6-fold in expression for our data, in comparison with 1.7-fold expression rate between normal and tumor tissues for long isoforms (Figure 4). Overall, it confirms the previously observed proportional increase of cancer-related gene isoforms observed previously for CD97 gene [45]. Thus, this observation needs further investigation to estimate the prognostic value of CASC4 gene expression.
Figure 4:
Distributed isoforms expression of CASC4 gene measured by fpkm values calculated by cuffnorm application. The short isoform (NM_177974) is expressed about 6 fold higher in GBM cells, while the longest isoform (NM_138423) maintains increased expression as well.
For CASC4 the short isoform (NM_177974) is expressed 6-fold higher in GBM cells, while the longest isoform (NM_138423) maintains an increased expression level as well. Overexpression of CASC4 is linked to Her2 proto oncogene-increased expression observed in ovarian and breast cancers [43], [44].
For APP the shortest isoform (NM_201413) is highly expressed in GBM cells while the longest isoform (NM_000484) is specifically expressed in NB cells. For CASC4 the short isoform (NM_177974) is expressed 6-fold higher in GBM cells, while the longest isoform (NM_138423) maintains increased expression as well. Observed overexpression of CASC4 is previously reported to be linked to Her2 proto oncogene increased expression observed in ovarian and breast cancers [43], [44]. Finally, TP53 – a well-known tumor suppressor gene – is quite distinct for spectra of related transcripts in GBM compared to NB cells, in particular featuring unusual non-coding RNA NR_015381 (TP53TG1) abundant expression.
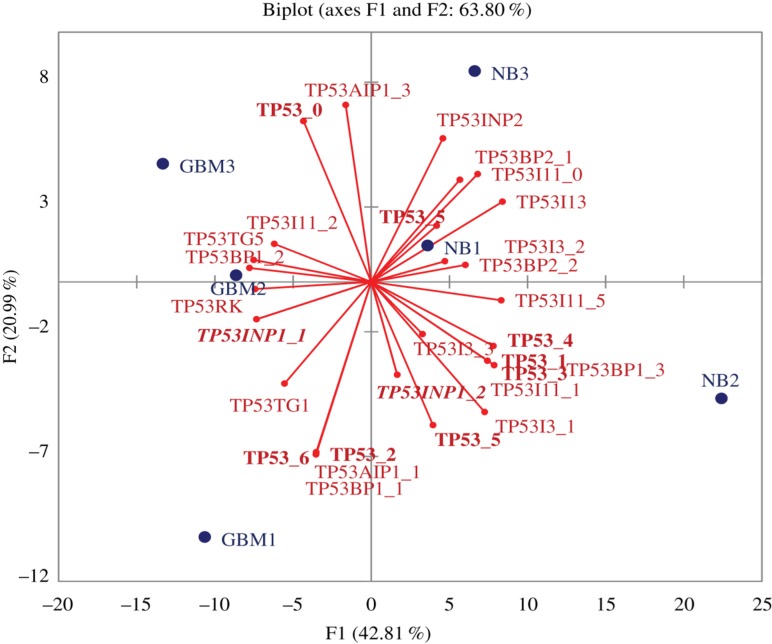
Using Principal component analysis of RNA-Seq data, we found that a range of isoforms are distinct for GBM samples (Figure 5). We compared expression of 43 transcripts for 16 p53-related genes (Table 3): TP53 (8 transcripts), TP53AIP1 (4 transcripts, correspondingly), TP53BP1 (3), TP53BP2 (2), TP53I11 (6), TP53I13, TP53I3 (3), TP53INP1 (2), TP53INP2, TP53RK, TP53TG1, TP53TG3 (3), TP53TG3B (3), TP53TG3C (2), TP53TG3D (2) and TP53TG5 (See Figure 5).
Figure 5:
Distributed expression of TP53 related genes measured by fpkm values calculated by cuffnorm application.
Table 3:
RefSeq accession numbers and gene ID for the TP53 related genes used.
| Status | Length, kb | Accession | gene_id |
|---|---|---|---|
| GBM | 19 | NM_001276761 | TP53_0 |
| NB | 19 | NM_001126113 | TP53_1 |
| GBM | 19 | NM_001126114 | TP53_2 |
| NB | 7 | NM_001126117 | TP53_3 |
| NB | 19 | NM_001126118 | TP53_4 |
| NB | 19 | NM_000546 | TP53_5 |
| NB | 7 | NM_001276697 | TP53_5 |
| GBM | 7 | NM_001276698 | TP53_6 |
| GBM | 8 | NM_001251964 | TP53AIP1_1 |
| GBM | 2 | NM_001195194 | TP53AIP1_3 |
| GBM | 86 | NM_001141979 | TP53BP1_1 |
| GBM | 86 | NM_001141980 | TP53BP1_2 |
| NB | 103 | NM_005657 | TP53BP1_3 |
| NB | 66 | NM_001031685 | TP53BP2_1 |
| NB | 66 | NM_005426 | TP53BP2_2 |
| NB | 19 | NM_006034 | TP53I11_0 |
| NB | 18 | NM_001258320 | TP53I11_1 |
| GBM | 19 | NM_001258321 | TP53I11_2 |
| NB | 19 | NM_001258324 | TP53I11_5 |
| NB | 4 | NM_138349 | TP53I13 |
| NB | 7 | NM_001206802 | TP53I3_1 |
| NB | 8 | NM_004881 | TP53I3_2 |
| NB | 8 | NM_147184 | TP53I3_3 |
| GBM | 23 | NM_001135733 | TP53INP1_1 |
| NB | 23 | NM_033285 | TP53INP1_2 |
| NB | 9 | NM_021202 | TP53INP2 |
| GBM | 5 | NM_033550 | TP53RK |
| GBM | 20 | NR_015381 | TP53TG1 |
| GBM | 4 | NM_014477 | TP53TG5 |
Transcripts NM_014477 (TP53TG5), NM_001141980 (TP53BP1, transcript variant 1), NM_001141979 (TP53BP1, transcript variant 2), NM_033550 (TP53RK), NM_001135733 (TP53INP1), NR_015381 (TP53TG1), NM_001126114 (TP53, transcript variant 9), NM_001276698 (TP53, transcript variant 6), NM_001251964 (TP53AIP1) are specific for GBM tissues (left half of the plot – GBM1, GBM2, GBM3).
The whole range of transcripts-specific form GBM cells were observed, distinct from NB cells. In particular non-coding NR_015381 was attributable for GBM cells.
5. Discussion
The RNA-seq analysis of the cell cultures of normal brain and GBM confirmed the association of some NPCs genes with tumor progression. The nuclear pore complexes are essential not only for regulating nuclear-cytoplasm transport but also for controlling genome organization and expression [33]. Therefore the NPC is linked with many diseases including cancers. Nucleoporins have been directly implicated in cancers via three routes: chromosomal translocations generating fusion proteins, changes in protein expression levels and single point mutations. The protein Nup50 is a member of the FG-repeat containing nucleoporins and functions as a soluble cofactor in importin-alpha:beta-mediated nuclear protein import. It has been recently reported that Nup50 is required for cell differentiation and exhibits nuclear dynamics which is dependent on active transcription by RNA polymerase II. The expression of gene Nup50-As1 increases in the human neocortex during development period spanning infancy to adulthood [46]. Our finding demonstrates the complicated role of nuclear pore complex proteins in tumor progression.
The overexpression of hormone transporter genes in glioblastoma cell culture provide an experimental basis for the observation that hypothyroidism induction by administration of propylthiouracil is associated with improved survival in glioblastoma patients [47]. Hypothyroidism may improve survival in animal models of cancer and clinical studies of tyrosine kinase inhibitor (TKI) treatment of renal cell carcinoma patients have suggested that the side effect of hypothyroidism induced by the TKIs contributes to improved outcomes [48].
The work on alternative splicing opens new perspectives for cancer research in glioma continuing studies [9]. p53 was originally found as an essential tumor suppressor preventing cell cycle progression and inducing apoptosis [48]. External and internal stress signals, including DNA damage, oncogene activation, hypoxia, and nutrient stress, induce p53 expression at transcriptional and post-transcriptional levels and regulate its activation through various kinds of modification, including phosphorylation, ubiquitination, acetylation, methylation, glycosylation, sumoylation, and ADP ribosylation [48], [49]. To conclude, note that therapeutic targets depend on the molecular mechanisms of tumors demanding more detailed classification by molecular parameters. The development of glioma involves many factors and signaling pathways, including, p53, retinoblastoma, and receptor tyrosine kinase, playing crucial roles in glioblastoma development.
Other examples of glioma-associated isoforms include truncated glioma-associated oncogene homolog 1 (TGLI1) that promotes tumor cell migration and invasion [50].
Transcriptome sequencing data analysis from more than 1000 patients in The Cancer Genome Atlas project have shown a set of alternative splicing events significantly altered across multiple cancer types [51]. These cancer-specific alternative splicing events are highly conserved, are more likely to maintain protein reading frame, and mainly function in cell cycle, cell adhesion/migration, and insulin signaling pathways. These events can serve as new molecular biomarkers to distinguish cancer from normal tissues, and to separate cancer subtypes.
Recent works uncovered alternative splicing in Huntington’s disease [52], studied alternative splicing on multiple tissues, including brain, heart, liver, and muscle [53]. In the future, additional problems of transcription analysis in the human genome should be addressed. Examples are the transposon activity, non-coding gene transcription, complex transcription landscape including antisense transcripts [54], [55], [56].
New tools for RNA-seq analysis present comparison of the workflows for the differential expression studies underlying importance of the correct tool selection [56], [57]. The future analysis of alternative splicing landscapes in tumors will play a fundamental role and have to be investigated using different tools demanding data integration including epigenetic factors [58].
Supporting Information
Acknowledgements
The work was supported by RFBR, ICG SB RAS budget project (0324-2016-0003 for NVG, GVV) and (0324-2016-0008 for IVC, IVM, YLO).
Supplemental Material
The online version of this article offers supplementary material (DOI: jib-2017-0022).
Contributor Information
Irina V. Medvedeva, Email: brukaro@gmail.com.
Yuriy L. Orlov, Email: orlov@bionet.nsc.ru.
Conflict of interest statement:
Authors state no conflict of interest. All authors have read the journal’s Publication ethics and publication malpractice statement available at the journal’s website and hereby confirm that they comply with all its parts applicable to the present scientific work.
References
- [1].Andreev DE, O’Connor PB, Loughran G, Dmitriev SE, Baranov PV, Shatsky IN. Insights into the mechanisms of eukaryotic translation gained with ribosome profiling. Nucleic Acids Res. 2017;45:513–26. doi: 10.1093/nar/gkw1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Babenko VN, Bragin AO, Spitsina AM, Chadaeva IV, Galieva ER, Orlova GV. et al. Analysis of differential gene expression by RNA-seq data in brain areas of laboratory animals. J Integr Bioinform. 2016;13:292. doi: 10.2390/biecoll-jib-2016-292. [DOI] [PubMed] [Google Scholar]
- [3].Westphal M, Lamszus K. The neurobiology of gliomas: from cell biology to the development of therapeutic approaches. Nat Rev Neurosci. 2011;12:495–508. doi: 10.1038/nrn3060. [DOI] [PubMed] [Google Scholar]
- [4].Cloughesy TF, Cavenee WK, Mischel PS. Glioblastoma: from molecular pathology to targeted treatment. Annu Rev Pathol. 2014;9:1–25. doi: 10.1146/annurev-pathol-011110-130324. [DOI] [PubMed] [Google Scholar]
- [5].Pereira M, Klamt F, Thomé C, Worm P, de Oliveira D. Metabotropic glutamate receptors as a new therapeutic target for malignant gliomas. Oncotarget. 2017 Mar 27; doi: 10.18632/oncotarget.15299. 10.18632/oncotarget.15299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Yu F, Fu W-M. Identification of differential splicing genes in gliomas using exon expression profiling. Mol Med Rep. 2015;11:843–50. doi: 10.3892/mmr.2014.2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Seidel S, Garvalov BK, Acker T. Isolation and culture of primary glioblastoma cells from human tumor specimens. Methods Mol Biol. 2015;1235:263–75. doi: 10.1007/978-1-4939-1785-3_19. [DOI] [PubMed] [Google Scholar]
- [8].Song SW, Fuller GN, Zheng H, Zhang W. Inactivation of the invasion inhibitory gene IIp45 by alternative splicing in gliomas. Cancer Res. 2005;65:3562–7. doi: 10.1158/0008-5472.CAN-04-3392. [DOI] [PubMed] [Google Scholar]
- [9].Venables JP, Klinck R, Koh C, Gervais-Bird J, Bramard A, Inkel L. et al. Cancer-associated regulation of alternative splicing. Nat Struct Mol Biol. 2009;16:670–6. doi: 10.1038/nsmb.1608. [DOI] [PubMed] [Google Scholar]
- [10].Raj B, Blencowe BJ. Alternative splicing in the mammalian nervous system: recent insights into mechanisms and functional roles. Neuron. 2015;87:14–27. doi: 10.1016/j.neuron.2015.05.004. [DOI] [PubMed] [Google Scholar]
- [11].Makeyev EV, Zhang J, Carrasco MA, Maniatis T. The microRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol Cell. 2007;27:435–48. doi: 10.1016/j.molcel.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Belyakin SN, Babenko VN, Maksimov DA, Shloma VV, Kvon EZ, Belyaeva ES. et al. Gene density profile reveals the marking of late replicated domains in the Drosophila melanogaster genome. Chromosoma. 2010;119:589–600. doi: 10.1007/s00412-010-0280-y. [DOI] [PubMed] [Google Scholar]
- [13].Shepard PJ, Hertel KJ. Embracing the complexity of pre-mRNA splicing. Cell Res. 2010;20:866–88. doi: 10.1038/cr.2010.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Yap K, Xiao Y, Friedman BA, Je HS, Makeyev EV. Polarizing the neuron through sustained co-expression of alternatively spliced isoforms. Cell Rep. 2016;15:1316–28. doi: 10.1016/j.celrep.2016.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Sadeque A, Serão NV, Southey BR, Delfino KR, Rodriguez-Zas SL. Identification and characterization of alternative exon usage linked glioblastoma multiforme survival. BMC Med Genomics. 2012;5:59. doi: 10.1186/1755-8794-5-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Babic I, Anderson ES, Tanaka K, Guo D, Masui K, Li B. et al. EGFR mutation-induced alternative splicing of Max contributes to growth of glycolytic tumors in brain cancer. Cell Metab. 2013;17:1000–8. doi: 10.1016/j.cmet.2013.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Lefave CV, Squatrito M, Vorlova S, Rocco GL, Brennan CW, Holland EC. et al. Splicing factor hnRNPH drives an oncogenic splicing switch in gliomas. EMBO J. 2011;30:4084–97. doi: 10.1038/emboj.2011.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Hu J, Ho AL, Yuan L, Hu B, Hua S, Hwang SS. et al. From the Cover: Neutralization of terminal differentiation in gliomagenesis. Proc Natl Acad Sci USA. 2013;110:14520–7. doi: 10.1073/pnas.1308610110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Goecks J, Nekrutenko A, Taylor J. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 2017;11:R86. doi: 10.1186/gb-2010-11-8-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Bolger M, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–20. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Andrews S. FastQC: a quality control tool for high throughput sequence data. 2010 Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc.
- [22].Wu TD, Nacu S. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics. 2010;26:873–81. doi: 10.1093/bioinformatics/btq057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–9. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR. et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–78. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Roberts A, Pachter L. Streaming fragment assignment for real-time analysis of sequencing experiments. Nat Methods. 2013;10:71–3. doi: 10.1038/nmeth.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Shen S, Park JW, Lu ZX, Lin L, Henry MD, Wu YN. et al. rMATS: robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc Nat Acad Sci USA. 2014;111:E5593–601. doi: 10.1073/pnas.1419161111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Young MD, Wakefield MJ, Smyth GK, Oshlack A. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 2010;11:R14. doi: 10.1186/gb-2010-11-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Shen S, Park JW, Huang J, Dittmar KA, Lu ZX, Zhou Q. et al. MATS: a Bayesian framework for flexible detection of differential alternative splicing from RNA-Seq data. Nucleic Acids Res. 2012;40:e61. doi: 10.1093/nar/gkr1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Nofrini V, Di Giacomo D, Mecucci C. Nucleoporin genes in human diseases. Eur J Hum Genet. 2016;24:1388–95. doi: 10.1038/ejhg.2016.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Tamai I, Nezu J, Uchino H, Sai Y, Oku A, Shimane M. et al. Molecular identification and characterization of novel members of the human organic anion transporter (OATP) family. Biochem Biophys Res Commun. 2000;273:251–60. doi: 10.1006/bbrc.2000.2922. [DOI] [PubMed] [Google Scholar]
- [35].Huber RD, Gao B, Sidler Pfändler MA, Zhang-Fu W, Leuthold S, Hagenbuch B. et al. Characterization of two splice variants of human organic anion transporting polypeptide 3A1 isolated from human brain. Am J Physiol Cell Physiol. 2007;292:13. doi: 10.1152/ajpcell.00597.2005. [DOI] [PubMed] [Google Scholar]
- [36].Sommer B, Tiys ES, Kormeier B, Hippe K, Janowski SJ, Ivanisenko TV. et al. Visualization and analysis of a cardio vascular disease- and MUPP1-related biological network combining text mining and data warehouse approaches. J Integr Bioinform. 2010;7:148. doi: 10.2390/biecoll-jib-2010-148. [DOI] [PubMed] [Google Scholar]
- [37].Demenkov PS, Ivanisenko TV, Kolchanov NA, Ivanisenko VA. ANDVisio: a new tool for graphic visualization and analysis of literature mined associative gene networks in the ANDSystem. In Silico Biol. 2011–2012;11:149–61. doi: 10.3233/ISB-2012-0449. [DOI] [PubMed] [Google Scholar]
- [38].Ivanisenko VA, Saik OV, Ivanisenko NV, Tiys ES, Ivanisenko TV, Demenkov PS. et al. ANDSystem: an Associative Network Discovery System for automated literature mining in the field of biology. BMC Syst Biol. 2015;9(Suppl 2):S2. doi: 10.1186/1752-0509-9-S2-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Saik OV, Ivanisenko TV, Demenkov PS, Ivanisenko VA. Interactome of the hepatitis C virus: literature mining with ANDSystem. Virus Res. 2016;218:40–8. doi: 10.1016/j.virusres.2015.12.003. [DOI] [PubMed] [Google Scholar]
- [40].Alam S, Suzuki H, Tsukahara T. Alternative splicing regulation of APP exon 7 by RBFox proteins. Neurochem Int. 2014;78:7–17. doi: 10.1016/j.neuint.2014.08.001. [DOI] [PubMed] [Google Scholar]
- [41].Nguyen KV. The human β-amyloid precursor protein: biomolecular and epigenetic aspects. Biomol Concepts. 2015;6:11–32. doi: 10.1515/bmc-2014-0041. [DOI] [PubMed] [Google Scholar]
- [42].Wong J. Altered expression of RNA splicing proteins in Alzheimer’s disease patients: evidence from two microarray studies. Dement Geriatr Cogn Dis Extra. 2013;3:74–85. doi: 10.1159/000348406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Inoue K, Fry EA. Aberrant splicing of estrogen receptor, HER2, and CD44 genes in breast cancer. Genet Epigenet. 2015;7:19–32. doi: 10.4137/GEG.S35500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Oh JJ, Grosshans DR, Grosshans SG, Slamon DJ. Identification of differentially expressed genes associated with HER-2/neu overexpression in human breast cancer cells. Nucleic Acids Res. 1999;27:4008–17. doi: 10.1093/nar/27.20.4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Safaee M, Fakurnejad S, Bloch O, Clark AJ, Ivan ME, Sun MZ. et al. Proportional upregulation of CD97 isoforms in glioblastoma and glioblastoma-derived brain tumor initiating cells. PLoS One. 2015;10:e0111532. doi: 10.1371/journal.pone.0111532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Lipovich L, Tarca AL, Cai J, Jia H, Chugani HT, Sterner KN. et al. Developmental changes in the transcriptome of human cerebral cortex tissue: long noncoding RNA transcripts. Cereb Cortex. 2014;24:1451–9. doi: 10.1093/cercor/bhs414. [DOI] [PubMed] [Google Scholar]
- [47].Hercbergs AH, Lin HY, Davis FB, Davis PJ, Leith JT. Radiosensitization and production of DNA double-strand breaks in U87MG brain tumor cells induced by tetraiodothyroacetic acid (tetrac) Cell Cycle. 2011;10:352–7. doi: 10.4161/cc.10.2.14641. [DOI] [PubMed] [Google Scholar]
- [48].Kruiswijk F, Labuschagne CF, Vousden KH. p53 in survival, death and metabolic health: a lifeguard with a licence to kill. Nat Rev Mol Cell Biol. 2015;16:393–405. doi: 10.1038/nrm4007. [DOI] [PubMed] [Google Scholar]
- [49].Kondo T. Molecular mechanisms involved in gliomagenesis. Brain Tumor Pathol. 2017;34:1–7. doi: 10.1007/s10014-017-0278-8. [DOI] [PubMed] [Google Scholar]
- [50].Zhu H, Carpenter RL, Han W, Lo HW. The GLI1 splice variant TGLI1 promotes glioblastoma angiogenesis and growth. Cancer Lett. 2014;343:51–61. doi: 10.1016/j.canlet.2013.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Tsai YS, Dominguez D, Gomez SM, Wang Z. Transcriptome-wide identification and study of cancer-specific splicing events across multiple tumors. Oncotarget. 2015;6:6825–39. doi: 10.18632/oncotarget.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Lin L, Park JW, Ramachandran S, Zhang Y, Tseng YT, Shen S. et al. Transcriptome sequencing reveals aberrant alternative splicing in Huntington’s disease. Hum Mol Genet. 2016;25:3454–66. doi: 10.1093/hmg/ddw187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Badr E, ElHefnawi M, Heath LS. Computational identification of tissue-specific splicing regulatory elements in human genes from RNA-Seq data. PLoS One. 2016;11:e0166978. doi: 10.1371/journal.pone.0166978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Babenko V, Chadaeva I, Orlov Y. Genomic landscape of CpG rich elements in human genome. BMC Evol Biol. 2017;17(Suppl 1):19. doi: 10.1186/s12862-016-0864-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Grinchuk OV, Jenjaroenpun P, Orlov YL, Zhou J, Kuznetsov VA. Integrative analysis of the human cis-antisense gene pairs, miRNAs and their transcription regulation patterns. Nucleic Acids Res. 2010;38:534–47. doi: 10.1093/nar/gkp954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Abnizova I, te Boekhorst R, Orlov Y. Computational errors and biases of short read next generation sequencing. J Proteomics Bioinform. 2017;10:1–17. [Google Scholar]
- [57].Williams CR, Baccarella A, Parrish JZ, Kim CC. Empirical assessment of analysis workflows for differential expression analysis of human samples using RNA-Seq. BMC Bioinformatics. 2017;18:38. doi: 10.1186/s12859-016-1457-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Wang J, Ye Z, Huang TH, Shi H, Jin VX. Computational methods and correlation of exon-skipping events with splicing, transcription, and epigenetic factors. Methods Mol Biol. 2017;1513:163–70. doi: 10.1007/978-1-4939-6539-7_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.