Abstract
Microscale separation (e.g., liquid chromatography or capillary electrophoresis) coupled with mass spectrometry (MS) has become the primary tool for advanced proteomics, an indispensable technology for gaining understanding of complex biological processes. In recent decades significant advances have been achieved in MS-based proteomics. However, the current proteomics platforms still face an analytical challenge in overall sensitivity towards nanoproteomics applications for starting materials of less than 1 µg total proteins (e.g., cellular heterogeneity in tissue pathologies). Herein, we review recent advances in microscale separation techniques and integrated sample processing strategies that improve the overall sensitivity and proteome coverage of the proteomics workflow, and their contributions towards nanoproteomics applications.
Keywords: Microscale separations, Nanoproteomics, NanoLC, Capillary electrophoresis, Mass spectrometry
1. Introduction
Proteins are the workhorses of the cell, impacting nearly all aspects of cellular processes. The proteome by its nature is dynamic, and the acute state of the proteome (i.e., the proteotype) depends on both the genotype and external perturbations [1,2]. Therefore, quantitative analysis of the dynamics of the proteome including post-translational modifications (PTMs) and its connection to phenotypes (e.g., diseases) has become indispensable in biological and clinical research [3–5]. Recent advances in mass spectrometry (MS)-based proteomics for both global deep-profiling of the proteome and selected types of PTMs (e.g., phosphorylation) [6–8] and targeted quantification of proteins from specific signaling path-ways [9,10] have greatly expanded our capabilities in performing proteogenomics and systems biology studies for gaining detailed mechanistic insights into physiological and pathological processes.
During the past decade, major advances have been achieved in nearly all areas of the proteomics workflow such as high resolution microscale chromatographic separations, mass spectrometry instrumentation, and bioinformatics data analysis tools to enable large-scale proteome interrogation [11]. Current state-of-the-art MS-based proteomics platforms can afford deep coverage for both the global proteome and selected PTMs in cell or tissue samples. For example, recent studies have reported the identification or quantification of ~10,000 proteins [6,7], >20,000 phosphorylation [12,13] and >15,000 ubiquitination sites [12,14].
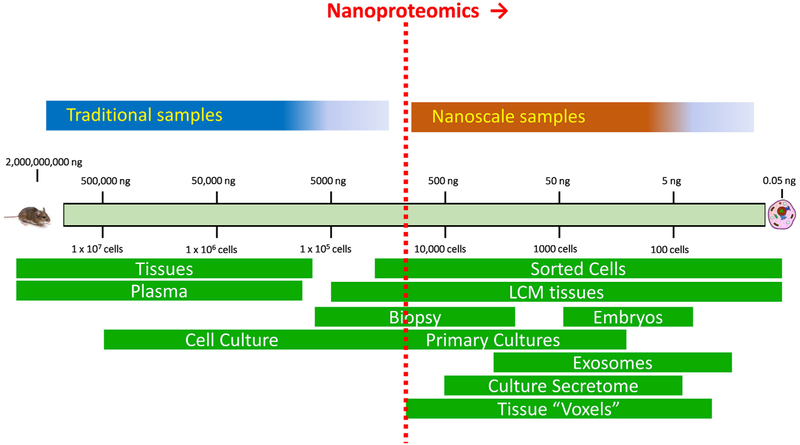
Despite recent advances in improving overall proteome coverage, the current proteomics workflows typically require relatively large amounts of starting materials on the order of millions of cells or 100 s μg of proteins, which excludes many important biological and biomedical applications. The ability to effectively analyze extremely small amounts of protein samples (e.g., nanograms of proteins) from cells or tissues by MS is one of the most significant challenges for current MS-proteomics. Herein, we define sample amounts with less than 1 μg of total protein as “nanoscale” and proteomics analyses of these nanoscale samples as “nanoproteomics” (Fig. 1). The biomedical need for nanoproteomics technologies are compelling, including the analyses of tissue substructures, cellular microenvironments of disease pathologies, rare or small subpopulations of cells, extracellular vesicles, as well as single cell resolution profiling (Fig. 1). Some of these sample types are readily produced by existing technologies such as fluorescence activated cell sorting(FACS) [15], laser capture dissection (LCM) [16,17], and exosome isolation techniques [18]. Moreover, single cell resolution genomics technologies, such as single-cell genomic sequencing [19] and single-cell transcriptomic profiling (RNA-Seq) [20,21], have been making tremendous impact in biological research. However, the current state-of-the-art in MS-based proteomics still falls far short of the sensitivity required for single cell analyses.
Fig. 1.
An illustration of traditional and nanoproteomics domains. The nanoproteomics is defined for dealing with samples containing <1 μg total protein in starting material.
Considerable efforts have been devoted to enhance the overall sensitivity of MS-based proteomics workflow towards enabling analysis of small samples, including the front-end sample processing, microscale separations, and MS instrumentation. Herein, we review recent advances in microscale separation, as well as nanoscale sample processing systems for proteomics analysis. Our focus will be on bottom-up proteomics, and the other important advances in top-down proteomics (measurement of intact proteins) [22] are not covered here.
2. Factors governing overall MS-based proteomics sensitivity
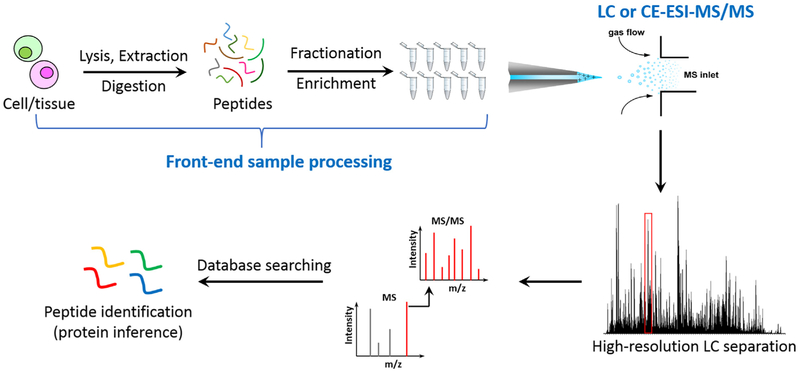
Fig. 2 illustrates a typical MS-based proteomics workflow for protein identification in biological samples. Conceptually, the overall analytical sensitivity of MS-based proteomics depends on the following aspects: a) the efficiency and recovery of front-end sample processing (e.g., protein extraction and protein digestion) and the degree of reducing sample complexity by extensive fractionation and/or enrichment; b) the resolving power of chromatographic or electrophoretic separations when coupled with electrospray ionization (ESI)-MS online in order to achieve deep proteome coverage; and c) the overall sensitivity of MS platforms including ESI and ion transmission efficiency, the resolving power of mass analyzers, and the sensitivity of MS detector.
Fig. 2.
A general workflow of bottom-up MS-based proteomics. It typically starts from cell lysis, protein extraction, and proteolytic digestion to peptide mixtures, followed up by fractionation or enrichment procedures and LC or CE-MS/MS analyses. The experimental spectrum is matched with database to identify peptides or contaminants.
The advances in MS instrumentation during the past decade are perhaps the most significant factor for achieving considerably greater overall sensitivity. Indeed, we have witnessed tremendous technological advancements in MS instrumentation including advanced ion optics (e.g., electrodynamic ion funnel [23]), and the ever increasing scanning speed, resolution, mass accuracy, sensitivity, dynamic range, and the multi-mode fragmentation technologies to facilitate much faster, more accurate, and more sensitive peptide identification and quantification [4,24–26]. Similarly, liquid-phase separations including liquid chromatography (LC) and capillary electrophoresis (CE) systems have become much more robust, reproducible, and sensitive through the use of smaller inner diameter (i.d.) LC columns and operating ESI at very low flow rates [27–30]. In general, the analytical sensitivity of nanoLC-MS and CE-MS is sufficient to detect peptides from a sample with mass equivalent to a single mammalian cell [29,30].
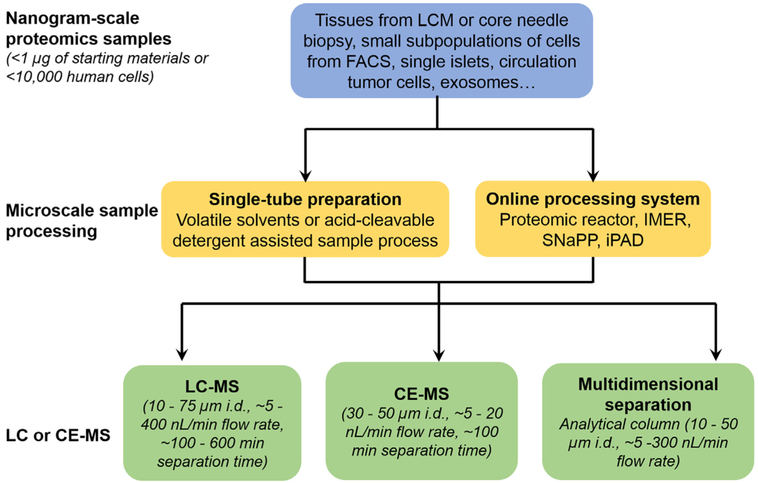
It has become clear that the major bottleneck for applying current MS-based proteomics platforms to nanoscale samples is the front-end sample processing step, where significant sample loss occurs. Further advances in microscale separation-based developments enabling online digestion/separation or miniaturized sample handling hold the promise to make significant contributions in addressing this bottleneck. In the following sections, we review recent advances in individual components of the nanoproteomics workflow (Fig. 3), including LC or CE-MS platforms and front-end microscale sample processing strategies that aim at enhancing the overall sensitivity and proteome coverage towards nanoproteomics applications. We also highlight some recent biological applications enabled by nanoproteomics technologies.
Fig. 3.
An overview of techniques adopted in different components of MS-based nanoproteomics. Generally, nanogram-scale samples are obtained from different sources, and are processed using microscale sample processing techniques such as single-tube, or online processing systems to reduce sample transfer steps for minimum sample loss. Then the digested peptides are subjected to highly sensitive nanoLC-MS or CE-MS analysis. In some cases, multidimensional separation is utilized to increase overall proteome coverage.
3. LC or CE for nanoproteomics
Reversed phase (RP) LC, either in packed columns or monolithic columns, is the most widely used LC separation for bottom-up proteomics due to its relatively high resolving power and ESI friendly mobile phases. CE has also emerged as a powerful separation technique when directly coupled with ESI-MS for proteomic research. One of the foundational discoveries in ESI-MS was that the ESI efficiency can be dramatically enhanced by nearly ~100-fold when ESI was operated at low flow rate (20–40 nL/min) (i.e., nanoESI) compared to conventional flow rate (>1 μl/min) [27,28,31]. This discovery set the general direction of recent LC and CE developments by pushing the limits of operation to nanoESI regimes and at the same time to enhance the overall resolving power of the separations in order to achieve highly sensitive measurements with good proteome coverage. Given that ESI-MS is generally considered as a concentration dependent detector, lower flow rates through a narrower i.d. column separation not only generate higher concentration, but also higher ionization efficiency, and thus higher signal intensity for individual analytes. Herein, we discuss the advances of LC and CE for the analysis of nanoscale samples. The performance metrics for selected LC and CE-MS platforms, including separaration paramters, type of sample, and identification numbers, are summarized in Table 1. Other detailed parameters not listed can be found in the References.
Table 1.
Microscale separation platforms and their performance in nanoproteomics*.
| Column type | L(cm) | i.d. (µm) | F (nL/min) | Sample size | Cp | # proteins | Detection limit | Ref |
|---|---|---|---|---|---|---|---|---|
| Packed | 87 | 15–75 | 20–400 | 0.1 µg yeast digest | ∼1000 | 1345–7 peptides | [32] | |
| LC | 10 | 50 | 500 | 1 µg yeast digest | ∼100 | 288–492 | [106] | |
| 85 | 15 | 20 | 0.5 pg-10 ng bacterium digest | ∼1000 | 3–872 | 75 zmol | [29] | |
| 15 | 75 | 200 | 10,000 LCM breast cells | 76 | [33] | |||
| 25 | 75 | 400 | ∼15,000 LCM pancreatic cells | 900–1300 | [107] | |||
| 10 | 100 | 700 | 0.2–0.8 µg single blastomeres | 644–1466 | [35] | |||
| 10 | 75 | 300 | 500–5000 MCF7 cells | 167–619 | [36] | |||
| 10 | 50 | 50 | single islets (2000–4000 cells) | 2013 | [34] | |||
| 20 | 75 | 300 | 2000 kidney cells (Spintip-based) | 1270 (duplicates) | [108] | |||
| 12 | 75 | 200 | 100,000 cancer cells (Direct lysis-digest) | 2987 phosphosites | [109] | |||
| 75 | 50 | 300 | 4000 LCM lung cells (SNaPP) | 3446 | [17] | |||
| 300 | 100–500 cells (iPAD) | 635–1060 | 200 zmol | [100] | ||||
| 100 | 25 | 5 | 0.2 µg murine cell digest | ∼750 | 3243 | [66] | ||
| Monolithic | 70 | 20 | 40 | 0.5–100 ng bacterium digest | ∼420 | 18–217 | 15 amol | [39] |
| LC | 25 | 10 | 10 | 0.1 µg bacterium digest | 1332 | 5 amol | [42] | |
| 60 | 75 | 100–300 | 0.5 µg yeast digest | ∼313 | 1323 | [40] | ||
| 200 | 100 | 500 | 1 µg HeLa cell digest | ∼360 | 2605 | [110] | ||
| 420 | 10 | 20 | 0.05 µg microbe digest fraction | ∼400 | 566 | <amol | [44] | |
| 320 | 10 | 20 | 45 ng protein (equivalent to 350 cells) | 343 | [43] | |||
| 320(2D) | 10 | 20 | 500 ng cancer cell digest | 850 | [71] | |||
| 800 | 10 | 40 | 1000–10,000 tumor cell digest | 456–1187 | [45] | |||
| 500 | 10 | 40 | 0.5 µg cancer cell digest (OTER) | 1462 | [88] | |||
| CE | 60 | 50 | 6ng rat testis H1 histones digest | 135 peptides | <30 amol | [111] | ||
| 90 | 50 | 400 ng HeLa cell digest | ∼300 | 2100 | [58] | |||
| 50 | 50 | tumor cell digest (1 µL) | 112 | 7 fmol | [54] | |||
| 60 | 50 | 1–100ng bacterium digest | 142–312 | [57] | ||||
| 60 | 50 | <1 µg bacterium digest | 871 | [56] | ||||
| 100 | 20 | 80 ng cancer cell digest | 283 | <2 amol | [59] | |||
| 60 | 50 | 20–40 ng bacterium digest | 179–199 | [60] | ||||
| 90 | 30 | 5–100 ng microbe digest | 370–548 (duplicates) | [112] | ||||
| 40 | 10 | 20 | 400 fg–84 pg bacterium digest | 4–162 (duplicates) | [30] | |||
| 95 (2D) | 50 | 50 ng frog egg digest | 330 | [113] | ||||
| 95 (2D) | 50 | 5.5 ng bacterium digest | 145 | [113] |
L: column length; i.d.: column inner diameter; F: flow rate; Cp: peak capacity.
3.1. LC
The overall performance of LC-ESI in terms of sensitivity and proteome coverage will depend on the i.d. of the capillary column, which determines the optimal flow rate, and the resolving power or peak capacity of LC separations. An early study by Shen et al. has demonstrated that peak capacities close to ~103 (Table 1) can be achieved independent of the column i.d. (15–75 μm) by using 87 cm long columns packed with 3 μm porous particles [32]. LC-ESI-MS sensitivity was significantly increased when the column i.d. decreased from 75 to 15 μm to enable ESI at lower flow rates. When analyzing ~100 ng of a yeast protein digest, the number of detected species was increased from 7 to 1345. In another study with a 15 μm × 80 cm column, a detection limit of 10 zmol for BSA peptide and identification of high abundance proteins from as low as 0.5 pg of bacterium digests were reported [29].
To date, most proteomics applications have been limited to 50–75 μm i.d. capillary columns when commercially available LC systems are used, due to their advantage in operation robustness. These platforms have been applied to nanoscale samples. For instance, Karger et al., reported an early illustration of LC-MS in nanoproteomics in analyzing ~10,000 breast cancer cells isolated by laser capture microdissection (LCM) using a 75 μm × 15 cm column packed with 3 μm particles on a first generation LCQ ion trap platform, leading to identification of 76 proteins [33]. More recently, Waanders et al. reported the identification of ~2000 proteins from single pancreatic islets (2000–4000 cells) using a 50 μm i.d. column packed with 3 μm particles and a long gradient (4-h) gradient separations with ~50 nL/min flowrate on an Orbitrap platform [34].
The sensitivity of 50–75 μm i.d. packed columns seems to be sufficient for analyzing small number of cells, but the achieved proteome coverage typically drops significantly when the starting sample amounts decrease [35,36]. Thus, more sensitive microscale separations are critical for achieving comprehensive proteome coverage of nanogram samples (Table 1). However, there are tremendous challenges for applying packed columns with i.d. smaller than 50 μm including the difficulty in packing, the requirement of ultra-high pressure pump, and limited robustness. Therefore, the development of alternative LC column technologies (e.g., monolithic columns) is an attractive solution.
Monolithic columns can be prepared with either in situ polymerization [37] or a sol-gel process involving the hydrolysis and polycondensation of alkoxysilanes [38]. Due to the small-sized skeletons and relatively large pores in monolithic columns, they can be manufactured in very small i.d. (e.g., 10 μm) with extended length (up to 8 m) and operated at the optimal nanospray ESI flow rates of 20 nL/min for achieving the optimal ESI-MS sensitivity. Luo et al. reported an early demonstration of attomole detection limit of peptide on a 20 μm × 70 cm silica based monolithic column at a flow rate of 40 nL/min when combined with an online SPE column and ESI-MS detection [39]. Follow-up studies by the same group and others have demonstrated the feasibility to identify thousands of proteins from sub-µg of tryptic digests on silica-based monolithic columns [40–42].
Another interesting contribution is the development of polymer monolithic porous layer open tubular (PLOT) columns for ultrasensitive proteomics analyses [43,44]. The PLOT columns displayed good reproducibility in retention times (3% RSD), detection limits of attomole to sub-attomole, and a peak capacity of 400. Rogeberg et al. compared the performance of silica-based and polymer-based monolithic columns and discovered a peak capacity of 950 from a 10 μm × 8 m PS-DVB PLOT column, which meets or surpasses peak capacities of previously reported using packed nanoLC or long silica monolithic columns [45]. With the PLOT column, about 500 and 1200 proteins were identified from an extract corresponding to 1000 and 10,000 cells, respectively [45].
3.2. CE
The potential of CE as a highly sensitive and efficient separation technique is demonstrated by its reputation in deciphering the human genome, but it has not gained as much attention as LC in the field of proteomics. However, some recent developments have made CE-MS based proteomics more promising by allowing for effectively interfacing of CE with MS for peptide detection and high resolution peptide separations.
Although the first sheath-liquid CE-MS interface was developed several decades ago [46], the sheath liquid causes significant dilution of analytes eluted from capillary [47]. Because ESI-MS is a concentration dependent detector, minimal or no sheath-liquid is preferred for high sensitivity CE-ESI and such hyphenation interfaces have been developed in the past decade [48–51]. One successful type of interface that decreases the sheath liquid dilution effect in recent proteomics applications is the nanospray sheath-flow liquid interface, developed by several groups with slightly different ways to pump sheath liquid [48,50,52]. This type of interface has been used for proteomics analyses in both global discovery and targeted quantitation modes [52–60]. Detection limits as low as zmol of protein digest was achieved using a targeted method [53,55]. A high peak capacity of 300 with this interface was reported with a 90 cm length linear polyacrylamide coated capillary with a separation window of 90 min [58]. A single shot CE-MS analysis of 400 ng of HeLa cell lysate digests has resulted in identification of ~10,000 peptides and 2100 proteins, which is approximately 2.5-fold lower than the number from nanoLC-MS using a 300 ng sample. There is 70% overlap of the identified peptides between the CE and LC methods, but CE tends to identify larger peptides than LC [57,58]. More recently, a number of groups developed sheathless CE-MS interfaces where the electroosmotic flow from capillary is the only source of nanospray [49,51].
3.3. Multidimensional separations
Besides the absolute sensitivity, the overall proteome coverage is a critical metric of performance of any proteomics workflow. The resolving power of separations is essential for achieving deep proteome coverage in proteomics analyses. Towards this direction, multidimensional separation platforms have been developed to provide higher peak capacity and applied to achieve deep proteome coverage in complex biological samples. Such multidimensional separation strategies can be operated in either offline or online modes [61–63]. In the offline mode, the fractions from the first dimension separations are typically collected through fraction collectors for later subsequent analyses. For online modes, the eluent fraction from the first dimension will be directly delivered to subsequent dimensions of separations for analysis in a single online system. When compared to online systems, the main limitation of offline multidimensional separations is the potential of sample loss during offline fractionation. On the other hand, a number of studies reported the development and application of online 2D or 3D LC-MS platforms for the analysis of relatively small amounts of digested samples [64–68]. To achieve high efficiency, the small i.d. analytical columns (e.g., 25 μm i.d. × 100 cm) with integrated emitters operating at flow rates of ~10nL/min were typically applied for last dimensional LC-MS, whereas the other dimensions were operated at either 1 or 2 μL/min [64,67]. The peak capacity was enhanced from 750 to ~13,000 for 1D to 3D separation systems. Using their latest 3D separation platform, the interference was largely removed and the quantitation ratios were only 8% compressed compared with 40% using the standard 1D LC-MS/MS [66,68].
Integrated biphasic or multi-phasic columns are also interesting alternatives to achieve the effect of multi-dimensional separations due to their simple configurations with minimum sample exposure surface. The original biphasic multidimensional separation strategy was reported in 2001 by Washburn et al. [63]. Since then, many improvements have been reported [69,70]. For example, Luo et al. demonstrated an integrated triphasic (RP/SCXµ-SPE) trapping column connected to a 10 μm × 3.2 m PLOT analytical column [71]. ~850 proteins were identified from an injection amount equivalent to 1200 cells (~500 ng) from a cervical cancer cell line.
As summarized in Table 1, recent advances in LC or CE-MS clearly make it possible to identify and quantify thousands of protein from sub-µg sample digests. Given this highly sensitive LC or CE-MS platforms being available, the efficiency in microscale front-end sample processing become an extremely critical component for achieving the success of the overall proteomics workflow.
4. Front-end microscale sample processing
As a critical component in the overall workflow, there has been a significant interest in developing microscale sample processing techniques to minimize sample loss and increase processing efficiency. In principle, this would involve processing with minimized liquid volumes and transfer steps. These efforts generally employ two main approaches to reduce sample losses and enhance processing efficiency: single-tube preparation techniques or integrated online processing systems.
The single tube techniques appear to be most straightforward by utilizing volatile solvents or acid-cleavable detergents for protein denaturation and performing all sample manipulations within a single tube to minimize loss [72–76]. This approach has been applied for proteomics analyses of LCM tissue samples [74,75,77]. However, it still falls short of effective processing for nanograms of proteins primarily due to low digestion efficiency when protein and enzyme concentrations get extremely low.
Alternatively, online processing systems integrating with microscale separations become more attractive. Proteomic reactors have been reported by employing a solid phase support to immobilize proteins and/or peptide to allow relatively rapid digestion and buffer exchange without sample transfers compared to traditional in-solution digest [78–80]. However, most proteomic reactor approaches still require many manual sample handling steps and long preparation times [81,82]. Online digestion systems were developed by adapting concepts from microscale separations to carry out proteomics digestion within enclosed systems, i.e. fused silica capillaries, controlled by chromatography pumps and fluidic systems [83–85]. Most commonly, immobilized enzyme reactors (IMER) in a micro-column format are used to achieve rapid protein digestions and eliminate the need for frequent additions of fresh protease [86–92]. Chip-based IMER [93] or tip-based nanoreactors [94] with comparable digestion efficiency were also developed and could be potentially automated and integrated for multiple functions. In general, IMER-based device offers much faster digestion time ranging from several seconds to less than 1 h compared to that of in solution digestion (typically >3 h) [87,92].
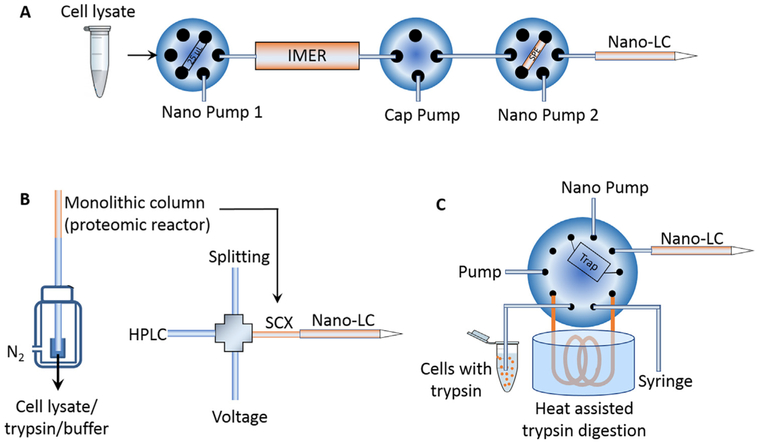
Recently, our group developed a simplified nanoproteomics platform (SNaPP) that integrated an online IMER column for trypsin digestion and solid phase extraction directly coupled to LC-MS (Fig. 4A) [17,91]. The SNaPP system created a simple, sensitive, robust, and reproducible nanoproteomics platform. As a pilot study using this system, 348 proteins were identified from ~20 mouse blastocysts, equal to 100–200 ng samples [91]. More recently, >3400 proteins were identified by SNaPP from only 4000 lung cells isolated by LCM [17]. In another example, Tian et al. integrated a monolithic SCX column-based proteomic reactor that serves for both protein capture/reduction/alkylation/digestion reactor and the first dimensional SCX separation followed by LC-MS (Fig. 4B) [95]. This platform was applied to analyze 500–50,000 human embroyonic stem cells and the identified protein numbers were increased from 69 to 2281 linearly. In an independent study, Zhang et al. reported an integrated proteome analysis device (iPAD) that incorporated a direct injection capillary column based sample loop (100 μm i.d., 40 cm long) and a C8 trap column (75 μm i.d., 10 cm long) and an online nanoLC analytical column (75 μm i.d. C18) (Fig. 4C) [96]. This system allowed direct flow injection of cells in trypsin solution and was able to identify an average 635 proteins from ~100 cells [96]. These examples of integrated online processing systems provided excellent illustrations on the promising aspects of integrating different components into a system for more effective sample processing with a minimal sample loss as well as multidimensional separations towards nanoproteomics applications.
Fig. 4.
Schematics of online integrated sample processing techniques. (A) SNaPP system for global proteomics of nanogram samples. Adpated from Ref [91]. (B) A monolithic column was used as a proteomic reactor and then assembled as the first dimensional SCX separation column. Adapted from Ref [95]. (C) iPAD system for living cell proteome profiling. Adapted with permission from Ref [100]. Copyright (2015) American Chemical Society.
5. Highlights of nanoproteomics applications
Nanoproteomics is an evolving technological capability for enabling analysis of cellular heterogeneity, tissue substructures, and other nanoscale biological or clinical samples at the proteome level when coupled with cell isolation techniques (e.g., LCM [97] and FACS [98]). While we recognized that most of the reports on nanoproteomics were focused on method development or proof-of-concept demonstrations, in this section we highlight some studies to illustrate the potential of biological applications enabled by nanoproteomics.
One of the early interesting nanoproteomics applications was the ability to perform single pancreatic islet proteome profiling by 1D-LC-MS/MS from Waanders et al. [34]. The study utilized a custom RePlay setup which employs a split flow design to create technical replicates and utilizes narrow diameter (50 μm) LC columns to achieve high sensitivity and they were able to quantitatively compare the proteomes of single islets (a type of micro-organ with 2000–4000 cells) with and without glucose treatments. A total of 1482 proteins were quantified with ~140 proteins displaying statistically significant alterations in protein abundances upon glucose stimulation. A general up-regulation of glycolysis, the TCA cycle and ATP translocation was observed [34]. This study illustrates the potential of applying similar nanoproteomics approaches to investigate islet heterogeneity in type 1 or type 2 diabetes [99].
Our group has recently coupled LCM with the SNaPP system to perform spatially resolved quantitative proteome profiling of microdissected lung alveolar tissue that was equivalent to only 4000 cells [17]. A depth of >3400 proteins was achieved on this platform with 2-fold lower quantification variance and 5-fold faster sample processing time when compared to offline sample processing. This study revealed seven defined modules of coordinated transcription factor-signaling molecule expression patterns and suggested the importance of epigenetic regulation in preferentially fine-tuning early processes in lung development. It also demonstrated the unique value of nanoproteomics in providing mechanistic or molecular basis of tissue heterogeneity and dynamic phenotypes.
Another interesting study is the application of a three-dimensional capillary or nanoLC platform to achieve deep proteome coverage of murine embryonic stem cells using only low μg of total cell lysate (12.6 μg) [68]. This study unambiguously quantified 11,352 protein groups that covered ~70% of Swiss-Prot and revealed protein regulation across the full detectable range of high-throughput gene expression and protein translation. We should note that the starting amount of materials was still one order of magnitude higher than the defined upper limit of the nanoproteomics domain. Nevertheless, this does illustrate the potential to achieve deep proteome coverage of nanoproteomics with further advances in sensitivity.
6. Conclusions and perspectives
Tremendous advances in LC- and CE-MS platforms have been achieved in terms of overall sensitivity, proteome coverage, reproducibility, and quantification for global proteome analyses. The absolute sensitivity for LC-MS and CE-MS operating in the nanoflow regime is sufficient for analyzing low ng protein samples or small numbers of cells, and potentially even for single mammalian cells [29,30]. The main bottleneck for the overall sensitivity lies in the sample losses and efficiency in the front-end sample processing. Recent developments in integrated online separation systems for microscale sample processing have demonstrated to be highly promising towards nanoproteomics analyses of small populations of cells [17,91,95,100]. While the current sensitivity of proteomics platforms still falls short of single cell analyses, continuous developments in the front-end integrated sample preparation techniques perhaps will be the most likely path in achieving such level of sensitivity. For instance, automated nanoliter sample handling technology is one potential capability for performing proteomic sample handling in nanoliter volumes by utilizing special patterned surfaces to perform sample manipulations in nanodroplets [101].
While our discussion has been mainly focused on global profiling of protein abundances of nanoscale samples, a uniquely important aspect of proteomics is PTMs, due to their fundamental roles in signal transduction and protein functional regulation. However, the sensitivity challenge for most types of PTM analyses is much more significant compared to global proteome profiling due to sub-stoichiometric and low-abundance nature of PTMs. Using phosphoproteomics as an example, typically hundreds of micrograms of peptides are required even when the most advanced nanoLC-MS platforms were used [102,103]. Recently, online integration of TiO2 enrichment with nanoLC-MS [104,105] or orthogonal 3D separations [64] have significantly improved the overall sensitivity of the phosphoproteomics workflow. However, the current state-of-the-art of phosphoproteomics still falls far short of analyzing nanogram-scale samples [103].
Based on the current performance status of MS-based proteomics, significant advances are still required for effective analyses of both the proteome and multiple types of PTMs in nanoscale samples or even at the single cell resolution in a high throughput manner. Most likely, further breakthroughs in all aspects of the proteomics workflow including MS instrumentation, microscale separations, and front-end sample processing are needed to achieve this long-term goal. With current pace of technological advances in the proteomics field, it is highly likely that nanoproteomics will soon become an indispensable technology for molecular characterization of clinical tissues in biomedical applications. While nanoproteomics is mainly recognized as a discovery technology, it is likely to make an impact on the clinics in facilitating more accurate molecular diagnosis/prognosis provided that further advances in robustness, automation, and throughput are achieved.
Acknowledgements
Portions of this work were supported by the NIH by NIH Grants P41 GM103493, DP3 DK110844, and UC4 DK104167. The experimental work described herein was performed in the Environmental Molecular Sciences Laboratory, a national scientific user facility sponsored by the DOE and located at Pacific Northwest National Laboratory, which is operated by Battelle Memorial Institute for the DOE under Contract DE-AC05–76RL0 1830.
Footnotes
Conflict of interest
None.
References
- [1].Mann M, Kulak NA, Nagaraj N, Cox J, The coming age of complete accurate, and ubiquitous proteomes, Mol. Cell 49 (2013) 583–590. [DOI] [PubMed] [Google Scholar]
- [2].Aebersold R, Mann M, Mass-spectrometric exploration of proteome structure and function, Nature 537 (2016) 347–355. [DOI] [PubMed] [Google Scholar]
- [3].Qian WJ, Jacobs JM, Liu T, Camp DG 2nd, Smith RD, Advances and challenges in liquid chromatography-mass spectrometry-based proteomics profiling for clinical applications, Mol. Cell. Proteomics 5 (2006) 1727–1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Altelaar AF, Munoz J, Heck AJ, Next-generation proteomics: towards an integrative view of proteome dynamics, Nat. Rev. Genet 14 (2013) 35–48. [DOI] [PubMed] [Google Scholar]
- [5].Larance M, Lamond AI, Multidimensional proteomics for cell biology, Nat. Rev. Mol. Cell Biol 16 (2015) 269–280. [DOI] [PubMed] [Google Scholar]
- [6].Zhang H, Liu T, Zhang Z, Payne SH, Zhang B, McDermott JE, Zhou JY, Petyuk VA, Chen L, Ray D, Sun S, Yang F, Chen L, Wang J, Shah P, Cha SW, Aiyetan P, Woo S, Tian Y, Gritsenko MA, Clauss TR, Choi C, Monroe ME, Thomas S, Nie S, Wu C, Moore RJ, Yu KH, Tabb DL, Fenyo D, Bafna V, Wang Y, Rodriguez H, Boja ES, Hiltke T, Rivers RC, Sokoll L, Zhu H, Shih Ie M, Cope L, Pandey A, Zhang B, Snyder MP, Levine DA, Smith RD, Chan DW, Rodland KD, Investigators C, Integrated proteogenomic characterization of human high-grade serous ovarian cancer, Cell 166 (2016) 755–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Mertins P, Mani DR, Ruggles KV, Gillette MA, Clauser KR, Wang P, Wang X, Qiao JW, Cao S, Petralia F, Kawaler E, Mundt F, Krug K, Tu Z, Lei JT, Gatza ML, Wilkerson M, Perou CM, Yellapantula V, Huang KL, Lin C, McLellan MD, Yan P, Davies SR, Townsend RR, Skates SJ, Wang J, Zhang B, Kinsinger CR, Mesri M, Rodriguez H, Ding L, Paulovich AG, Fenyo D, Ellis MJ, Carr SA, Nci C, Proteogenomics connects somatic mutations to signalling in breast cancer, Nature 534 (2016) 55–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Zhang B, Wang J, Wang X, Zhu J, Liu Q, Shi Z, Chambers MC, Zimmerman LJ, Shaddox KF, Kim S, Davies SR, Wang S, Wang P, Kinsinger CR, Rivers RC, Rodriguez H, Townsend RR, Ellis MJ, Carr SA, Tabb DL, Coffey RJ, Slebos RJ, Liebler DC, Nci C, Proteogenomic characterization of human colon and rectal cancer, Nature 513 (2014) 382–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Shi T, Niepel M, McDermott JE, Gao Y, Nicora CD, Chrisler WB, Markillie LM, Petyuk VA, Smith RD, Rodland KD, Sorger PK, Qian WJ, Wiley HS, Conservation of protein abundance patterns reveals the regulatory architecture of the EGFR-MAPK pathway, Sci. Signal 9 (2016) rs6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Bisson N, James DA, Ivosev G, Tate SA, Bonner R, Taylor L, Pawson T, Selected reaction monitoring mass spectrometry reveals the dynamics of signaling through the GRB2 adaptor, Nat. Biotechnol 29 (2011) 653–658. [DOI] [PubMed] [Google Scholar]
- [11].Beck M, Claassen M, Aebersold R, Comprehensive proteomics, Curr. Opin. Biotechnol 22 (2011) 3–8. [DOI] [PubMed] [Google Scholar]
- [12].Mertins P, Qiao JW, Patel J, Udeshi ND, Clauser KR, Mani DR, Burgess MW, Gillette MA, Jaffe JD, Carr SA, Integrated proteomic analysis of post-translational modifications by serial enrichment, Nat. Methods 10 (2013) 634–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Sharma K, D’Souza RC, Tyanova S, Schaab C, Wisniewski JR, Cox J, Mann M, Ultradeep human phosphoproteome reveals a distinct regulatory nature of Tyr and Ser/Thr-based signaling, Cell Rep. 8 (2014) 1583–1594. [DOI] [PubMed] [Google Scholar]
- [14].Kim W, Bennett EJ, Huttlin EL, Guo A, Li J, Possemato A, Sowa ME, Rad R, Rush J, Comb MJ, Harper JW, Gygi SP, Systematic and quantitative assessment of the ubiquitin-modified proteome, Mol. Cell 44 (2011) 325–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Gawad C, Koh W, Quake SR, Single-cell genome sequencing: current state of the science, Nat. Rev. Genet 17 (2016) 175–188. [DOI] [PubMed] [Google Scholar]
- [16].Drummond ES, Nayak S, Ueberheide B, Wisniewski T, Proteomic analysis of neurons microdissected from formalin-fixed, paraffin-embedded Alzheimer’s disease brain tissue, Sci. Rep 5 (2015) 15456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Clair G, Piehowski PD, Nicola T, Kitzmiller JA, Huang EL, Zink EM, Sontag RL, Orton DJ, Moore RJ, Carson JP, Smith RD, Whitsett JA, Corley RA, Ambalavanan N, Ansong C, Spatially-resolved proteomics: rapid quantitative analysis of laser capture microdissected alveolar tissue samples, Sci. Rep 6 (2016) 39223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Livshits MA, Khomyakova E, Evtushenko EG, Lazarev VN, Kulemin NA, Semina SE, Generozov EV, Govorun VM, Isolation of exosomes by differential centrifugation: theoretical analysis of a commonly used protocol, Sci. Rep 6 (2016) 17319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Shapiro E, Biezuner T, Linnarsson S, Single-cell sequencing-based technologies will revolutionize whole-organism science, Nat. Rev. Genet 14 (2013) 618–630. [DOI] [PubMed] [Google Scholar]
- [20].Kalisky T, Blainey P, Quake SR, Genomic analysis at the single-cell level, Annu. Rev. Genet 45 (2011) 431–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Lohr JG, Adalsteinsson VA, Cibulskis K, Choudhury AD, Rosenberg M, Cruz-Gordillo P, Francis JM, Zhang CZ, Shalek AK, Satija R, Trombetta JJ, Lu D, Tallapragada N, Tahirova N, Kim S, Blumenstiel B, Sougnez C, Lowe A, Wong B, Auclair D, Van Allen EM, Nakabayashi M, Lis RT, Lee GSM, Li T, Chabot MS, Taplin ME, Taplin ME, Clancy TE, Loda M, Regev A, Meyerson M, Hahn WC, Kantoff PW, Golub TR, Getz G, Boehm JS, Love JC, Whole-exome sequencing of circulating tumor cells provides a window into metastatic prostate cancer, Nat. Biotechnol 32 (2014) 479–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Toby TK, Fornelli L, Kelleher NL, Progress in top-down proteomics and the analysis of proteoforms, Annu. Rev. Anal. Chem 9 (2016) 499–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Shaffer SA, Tolmachev A, Prior DC, Anderson GA, Udseth HR, Smith RD, Characterization of a new electrodynamic ion funnel interface for electrospray ionization mass spectrometry, Anal. Chem 71 (1999) 2957–2964. [DOI] [PubMed] [Google Scholar]
- [24].Michalski A, Damoc E, Hauschild JP, Lange O, Wieghaus A, Makarov A, Nagaraj N, Cox J, Mann M, Horning S, Mass spectrometry-based proteomics using Q Exactive, a high-performance benchtop quadrupole Orbitrap mass spectrometer, Mol. Cell. Proteomics 10 (2011), 10.1074/mcp.M111.011015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Zhang Y, Fonslow BR, Shan B, Baek MC, Yates JR, Protein analysis by shotgun/bottom-up proteomics, Chem. Rev 113 (2013) 2343–2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Han X, Aslanian A, Yates JR, Mass spectrometry for proteomics, Curr. Opin. Chem. Biol 12(2008) 483–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Wilm M, Mann M, Analytical properties of the nanoelectrospray ion source, Anal. Chem 68(1996) 1–8. [DOI] [PubMed] [Google Scholar]
- [28].Schmidt A, Karas M, Dulcks T, Effect of different solution flow rates on analyte ion signals in nano-ESI MS: or: when does ESI turn into nano-ESI? J. Am. Soc. Mass. Spectr 14 (2003) 492–500. [DOI] [PubMed] [Google Scholar]
- [29].Shen Y, Tolic N, Masselon C, Pasa-Tolic L, Camp DG, Hixson KK, Zhao R, Anderson GA, Smith RD, Ultrasensitive proteomics using high-efficiency on-line micro-SPE-NanoLC-NanoESI MS and MS/MS, Anal. Chem 76 (2004) 144–154. [DOI] [PubMed] [Google Scholar]
- [30].Sun L, Zhu G, Zhao Y, Yan X, Mou S, Dovichi NJ, Ultrasensitive and fast bottom-up analysis of femtogram amounts of complex proteome digests, Angew. Chem. Int. Ed. Engl 52 (2013) 13661–13664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Wilm MS, Mann M, Electrospray and Taylor-Cone theory: dole’s beam of macromolecules at last? Int. J. Mass Spectrom. Ion Process 136 (1994) 167–180. [Google Scholar]
- [32].Shen Y, Zhao R.Berger SJ, Anderson GA, Rodriguez N, Smith RD, High-efficiency nanoscale liquid chromatography coupled on-line with mass spectrometry using nanoelectrospray ionization for proteomics, Anal. Chem 74 (2002) 4235–4249. [DOI] [PubMed] [Google Scholar]
- [33].Zang L, Palmer Toy D, Hancock WS, Sgroi DC, Karger BL, Proteomic analysis of ductal carcinoma of the breast using laser capture microdissection LC-MS, and 160/180 isotopic labeling, J. Proteome Res 3 (2004) 604–612. [DOI] [PubMed] [Google Scholar]
- [34].Waanders LF, Chwalek K, Monetti M, Kumar C, Lammert E, Mann M, Quantitative proteomic analysis of single pancreatic islets, Proc. Natl. Acad. Sci. U. S. A 106 (2009) 18902–18907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Sun L, Dubiak KM, Peuchen EH, Zhang Z, Zhu G, Huber PW, Dovichi NJ, Single cell proteomics using frog (Xenopus laevis) blastomeres isolated from early stage embryos, which form a geometric progression in protein content, Anal. Chem 88 (2016) 6653–6657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Wang N, Xu M, Wang P, Li L, Development of mass spectrometry-based shotgun method for proteome analysis of 500–5000 cancer cells, Anal. Chem 82 (2010) 2262–2271. [DOI] [PubMed] [Google Scholar]
- [37].Ivanov AR, Zang L, Karger BL, Low-attomole electrospray ionization MS and MS/MS analysis of proteintryptic digests using 20-µm-i.d. polystyrene-divinylbenzene monolithic capillary columns, Anal. Chem 75 (2003) 5306–5316. [DOI] [PubMed] [Google Scholar]
- [38].Ishizuka N, Kobayashi H, Minakuchi H, Nakanishi K, Hirao K, Hosoya K, Ikegami T, Tanaka N, Monolithic silica columns for high-efficiency separations by high-performance liquid chromatography, J. Chromatogr. A 960 (2002) 85–96. [DOI] [PubMed] [Google Scholar]
- [39].Luo Q, Shen Y, Hixson KK, Zhao R, Yang F, Moore RJ, Mottaz HM, Smith RD, Preparation of 20 µm-i.d. silica-based monolithic columns and their performance for proteomics analyses, Anal. Chem 77 (2005) 5028–5035. [DOI] [PubMed] [Google Scholar]
- [40].Xie C, Ye M, Jiang X, Jin W, Zou H, Octadecylated silica monolith capillary column with integrated nanoelectrospray ionization emitter for highly efficient proteome analysis, Mol. Cell. Proteomics 5 (2006) 454–461. [DOI] [PubMed] [Google Scholar]
- [41].Luo Q, Page JS, Tang K, Smith RD, MicroSPE-nanoLC-ESI-MS/MS using 10-mu m-i.d. Silica-based monolithic columns for proteomics, Anal. Chem 79 (2007) 540–545. [DOI] [PubMed] [Google Scholar]
- [42].Luo Q, Tang K, Yang F, Elias A, Shen Y, Moore RJ, Zhao R, Hixson KK, Rossie SS, Smith RD, More sensitive and quantitative proteomic measurements using very low flow rate porous silica monolithic LC columns with electrospray ionization-mass spectrometry, J. Proteome Res 5 (2006) 1091–1097. [DOI] [PubMed] [Google Scholar]
- [43].Luo Q, Yue G, Valaskovic GA, Gu Y, Wu SL, Karger BL, On-line 1D and 2D porous layer open tubular/LC-ESI-MS using 10-µm-i.d. poly(styrene-divinylbenzene) columns for ultrasensitive proteomic analysis, Anal. Chem 79 (2007) 6174–6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Yue G, Luo Q, Zhang J, Wu SL, Karger BL, Ultratrace LC/MS proteomic analysis using 10-µm-i.d. Porous layer open tubular poly(styrene-divinylbenzene) capillary columns, Anal. Chem 79 (2007) 938–946. [DOI] [PubMed] [Google Scholar]
- [45].Rogeberg M, Vehus T, Grutle L, Greibrokk T, Wilson SR, Lundanes E, Separation optimization of long porous-layer open-tubular columns for nano-LC-MS of limited proteomic samples, J. Sep. Sci 36 (2013) 2838–2847. [DOI] [PubMed] [Google Scholar]
- [46].Smith RD, Olivares JA, Nguyen NT, Udseth HR, Capillary zone electrophoresis mass-spectrometry using an electrospray ionization interface, Anal. Chem 60 (1988) 436–441. [Google Scholar]
- [47].Mokaddem M, Gareil P, Belgaied JE, Varenne A, New insight into suction and dilution effects in CE coupled to MS via an ESI interface. II–dilution effect, Electrophoresis 30 (2009) 1692–1697. [DOI] [PubMed] [Google Scholar]
- [48].Wojcik R, Dada OO, Sadilek M, Dovichi NJ, Simplified capillary electrophoresis nanospray sheath-flow interface for high efficiency and sensitive peptide analysis, Rapid Commun. Mass Spectrom 24 (2010) 2554–2560. [DOI] [PubMed] [Google Scholar]
- [49].Moini M, Simplifying CE-MS operation. 2. Interfacing low-flow separation techniques to mass spectrometry using a porous tip, Anal. Chem 79 (2007) 4241–4246. [DOI] [PubMed] [Google Scholar]
- [50].Zhong X, Maxwell EJ, Chen DD, Mass transport in a micro flow-through vial of a junction-at-the-tip capillary electrophoresis-mass spectrometry interface, Anal. Chem 83 (2011) 4916–4923. [DOI] [PubMed] [Google Scholar]
- [51].Wang CW, Her GR, Sheathless capillary electrophoresis electrospray ionization-mass spectrometry interface based on poly(dimethylsiloxane) membrane emitter and thin conducting liquid film, Electrophoresis 34 (2013) 2538–2545. [DOI] [PubMed] [Google Scholar]
- [52].Wang C, Lee CS, Smith RD, Tang K, Ultrasensitive sample quantitation via selected reaction monitoring using CITP/CZE-ESI-triple quadrupole MS, Anal. Chem 84 (2012) 10395–10403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Li Y, Wojcik R, Dovichi NJ, Champion MM, Quantitative multiple reaction monitoring of peptide abundance introduced via a capillary zone electrophoresis-electrospray interface, Anal. Chem 84 (2012) 6116–6121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Zhu G, Sun L, Yang P, Dovichi NJ, On-line amino acid-based capillary isoelectric focusing-ESI-MS/MS for protein digests analysis, Anal. Chim. Acta 750 (2012) 207–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Sun L, Li Y, Champion MM, Zhu G, Wojcik R, Dovichi NJ, Capillary zone electrophoresis-multiple reaction monitoring from 100pg of RAW 264.7 cell lysate digest, Analyst 138 (2013) 3181–3188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Yan X, Essaka DC, Sun L, Zhu G, Dovichi NJ, Bottom-up proteome analysis of E. coli using capillary zone electrophoresis-tandem mass spectrometry with an electrokinetic sheath-flow electrospray interface, Proteomics 13 (2013) 2546–2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Zhu G, Sun L, Yan X, Dovichi NJ, Single-shot proteomics using capillary zone electrophoresis-electrospray ionization-tandem mass spectrometry with production of more than 1 250 Escherichia coli peptide identifications in a 50 min separation, Anal. Chem 85 (2013) 2569–2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Sun L, Hebert AS, Yan X, Zhao Y, Westphall MS, Rush MJP, Zhu G, Champion MM, Coon JJ, Dovichi NJ, Over 10,000 peptide identifications from the HeLa proteome by using single-shot capillary zone electrophoresis combined with tandem mass spectrometry, Angew. Chem. Int. Ed. Engl 53 (2014) 13931–13933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Sun L, Zhu G, Mou S, Zhao Y, Champion MM, Dovichi NJ, Capillary zone electrophoresis-electrospray ionization-tandem mass spectrometry for quantitative parallel reaction monitoring of peptide abundance and single-shot proteomic analysis of a human cell line, J. Chromatogr. A 1359 (2014) 303–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Zhu G, Sun L, Yan X, Dovichi NJ, Bottom-up proteomics of Escherichia coli using dynamic pH junction preconcentration and capillary zone electrophoresis-electrospray ionization-tandem mass spectrometry, Anal. Chem 86 (2014) 6331–6336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Shi T, Fillmore TL, Sun X, Zhao R, Schepmoes AA, Hossain M, Xie F, Wu S, Kim JS, Jones N, Moore RJ, Pasa-Tolic L, Kagan J, Rodland KD, Liu T, Tang K, Camp DG, Smith RD, Qian WJ, Antibody-free, targeted mass-spectrometric approach for quantification of proteins at low picogram per milliliter levels in human plasma/serum, Proc. Natl. Acad. Sci. U. S. A 109 (2012) 15395–15400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Kulak NA, Geyer PE, Mann M, Loss-less nano-fractionator for high sensitivity, high coverage proteomics, Mol. Cell. Proteomics 16 (2017) 694–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Washburn MP, Wolters D, Yates JR, Large-scale analysis of the yeast proteome by multidimensional protein identification technology, Nat. Biotechnol 19 (2001) 242–247. [DOI] [PubMed] [Google Scholar]
- [64].Ficarro SB, Zhang Y, Carrasco-Alfonso MJ, Garg B, Adelmant G, Webber JT, Luckey CJ, Marto JA, Online nanoflow multidimensional fractionation for high efficiency phosphopeptide analysis, Mol. Cell. Proteomics 10 (2011) 011064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Zhou F, Cardoza JD, Ficarro SB, Adelmant GO, Lazaro JB, Marto JA, Online nanoflow RP-RP-MS reveals dynamics of multicomponent Ku complex in response to DNA damage, J. Proteome Res 9 (2010) 6242–6255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Zhou F, Lu Y, Ficarro SB, Webber JT, Marto JA, Nanoflow low pressure high peak capacity single dimension LC-MS/MS platform for high-throughput, in-depth analysis of mammalian proteomes, Anal. Chem 84 (2012) 5133–5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Zhou F, Sikorski TW, Ficarro SB, Webber JT, Marto JA, Online nanoflow reversed phase-strong anion exchange-reversed phase liquid chromatography-tandem mass spectrometry platform for efficient and in-depth proteome sequence analysis of complex organisms, Anal. Chem 83 (2011) 6996–7005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Zhou F, Lu Y, Ficarro SB, Adelmant G, Jiang WY, Luckey CJ, Marto JA, Genome-scale proteome quantification by DEEP SEQ mass spectrometry, Nat. Commun 4 (2013), 10.1038/ncomms3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Dai J, Shieh CH, Sheng QH, Zhou H, Zeng R, Proteomic analysis with integrated multiple dimensional liquid chromatography/mass spectrometry based on elution of ion exchange column using pH steps, Anal. Chem 77 (2005) 5793–5799. [DOI] [PubMed] [Google Scholar]
- [70].Motoyama A, Yates JR, Multidimensional LC separations in shotgun proteomics, Anal. Chem 80 (2008) 7187–7193. [DOI] [PubMed] [Google Scholar]
- [71].Luo Q, Gu Y, Wu S, Rejtar T, Karger BL, Two-dimensional strong cation exchange/porous layer open tubular/mass spectrometry for ultratrace proteomic analysis using a 10 µm id poly(styrene-divinylbenzene) porous layer open tubular column with an on-line triphasic trapping column, Electrophoresis 29 (2008) 1604–1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Wang N, Xu M, Wang P, Li L, Development of mass spectrometry-based shotgun method for proteome analysis of 500–5000 cancer cells, Anal. Chem 82 (2010) 2262–2271. [DOI] [PubMed] [Google Scholar]
- [73].Wang H, Qian WJ, Mottaz HM, Clauss TRW, Anderson DJ, Moore RJ, Camp DG, Khan AH, Sforza DM, Pallavicini M, Smith DJ, Smith RD, Development and evaluation of a micro- and nanoscale proteomic sample preparation method, J. Proteome Res 4 (2005) 2397–2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Braakman RB, Tilanus-Linthorst MM, Liu NQ, Stingl C, Dekker LJ, Luider TM, Martens JW, Foekens JA, Umar A Optimized nLC-MS workflow for laser capture microdissected breast cancer tissue, J. Proteomics 75 (2012) 2844–2854. [DOI] [PubMed] [Google Scholar]
- [75].Liu NQ, Braakman RB, Stingl C, Luider TM, Martens JW, Foekens JA, Umar A, Proteomics pipeline for biomarker discovery of laser capture microdissected breast cancer tissue, J. Mammary Gland Biol. Neoplasia 17 (2012) 155–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Kulak NA, Pichler G, Paron I, Nagaraj N, Mann M, Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells, Nat. Methods 11 (2014) 319–324. [DOI] [PubMed] [Google Scholar]
- [77].De Marchi T, Braakman RB, Stingl C, van Duijn MM, Smid M, Foekens JA, Luider TM, Martens JW, Umar A, The advantage of laser-capture microdissection over whole tissue analysis in proteomic profiling studies, Proteomics 16 (2016) 1474–1485. [DOI] [PubMed] [Google Scholar]
- [78].Chen W, Wang S, Adhikari S, Deng Z, Wang L, Chen L, Ke M, Yang P, Tian R, Simple and integrated spintip-based technology applied for deep proteome profiling, Anal. Chem 88 (2016) 4864–4871. [DOI] [PubMed] [Google Scholar]
- [79].Wisniewski JR, Ostasiewicz P, Mann M, High recovery FASP applied to the proteomic analysis of microdissected formalin fixed paraffin embedded cancer tissues retrieves known colon cancer markers, J. Proteome Res 10 (2011) 3040–3049. [DOI] [PubMed] [Google Scholar]
- [80].Ethier M, Hou WM, Duewel HS, Figeys D, The proteomic reactor: a microfluidic device for processing minute amounts of protein prior to mass spectrometry analysis, J. Proteome Res 5 (2006) 2754–2759. [DOI] [PubMed] [Google Scholar]
- [81].Liebler DC, Ham AJL, Spin filter-based sample preparation for shotgun proteomics, Nat. Methods 6 (2009) 785–786. [DOI] [PubMed] [Google Scholar]
- [82].Nel AJ, Garnett S, Blackburn JM, Soares NC, Comparative reevaluation of FASP and enhanced FASP methods by LC-MS/MS, J. Proteome Res 14 (2015) 1637–1642. [DOI] [PubMed] [Google Scholar]
- [83].Tian R, Alvarez-Saavedra M, Cheng HY, Figeys D, Uncovering the proteome response of the master circadian clock to light using an AutoProteome system, Mol. Cell. Proteomics 10 (2011), 10.1074/mcp.M110.007252, M110.007252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Lopez-Ferrer D, Petritis K, Robinson EW, Hixson KK, Tian Z, Lee JH, Lee SW, Tolic N, Weitz KK, Belov ME, Smith RD, Pasa-Tolic L, Pressurized pepsin digestion in proteomics, Mol. Cell. Proteomics 10 (2011), 10.1074/mcp.M110.001479, M110.001479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Lopez-Ferrer D, Petritis K, Lourette NM, Clowers B, Hixson KK, Heibeck T, Prior DC, Pasa-Tolic L, Camp DG, Belov ME, Smith RD, On-line digestion system for protein characterization and proteome analysis, Anal. Chem 80 (2008) 8930–8936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Hustoft HK, Brandtzaeg OK, Rogeberg M, Misaghian D, Torsetnes SB, Greibrokk T, Reubsaet L, Wilson SR, Lundanes E, Integrated enzyme reactor and high resolving chromatography in sub-chip dimensions for sensitive protein mass spectrometry, Sci. Rep 3 (2013) 3511, 10.1038/srep03511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Yamaguchi H, Miyazaki M, Enzyme-immobilized reactors for rapid and efficient sample preparation in MS-based proteomic studies, Proteomics 13 (2013) 457–466. [DOI] [PubMed] [Google Scholar]
- [88].Hustoft HK, Vehus T, Brandtzaeg OK, Krauss S, Greibrokk T, Wilson SR, Lundanes E, Open tubular lab-on-column/mass spectrometry for targeted proteomics of nanogram sample amounts, PLoS One 9 (2014) e106881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Jiang S, Zhang Z, Li L, A one-step preparation method of monolithic enzyme reactor for highly efficient sample preparation coupled to mass spectrometry-based proteomics studies, J. Chromatogr. A 1412 (2015) 75–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Colquhoun DR, Feild BJ, Automated, Online sample preparation for LC-MS analyses affinity capture, digestion, and clean-up, in: Schiel JE, Davis DL, Borisov OV (Eds.), State-of-the-Art and Emerging Technologies for Therapeutic Monoclonal Antibody Characterization Volume 3 Defining the Next Generation of Analytical and Biophysical Techniques, American Chemical Society, 2015, pp. 335–356. [Google Scholar]
- [91].Huang E, Piehowski PD, Orton DJ, Moore RJ, Qian WJ, Casey CP, Sun X, Dey SK, Burnum-Johnson KE, Smith RD, SNaPP: simplified nanoproteomics platform for reproducible global proteomic analysis of nanogram protein quantities, Endocrinology 157 (2016) 1307–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Safdar M, Sproß J, Jänis J, Microscale immobilized enzyme reactors in proteomics: latest developments,J. Chromatogr. A 1324 (2014) 1–10. [DOI] [PubMed] [Google Scholar]
- [93].Jonsson A, Svejdal RR, Bogelund N, Nguyen TTTN, Flindt H, Kutter JP, Rand KD, Lafleur JP, Thiolene monolithic pepsin microreactor with a 3D-printed interface for efficient UPLC-MS peptide mapping analyses, Anal. Chem 89 (2017) 4573–4580. [DOI] [PubMed] [Google Scholar]
- [94].Yan L, Qiao L, Ji J, Li Y, Yin X, Lin L, Liu X, Yao J, Wang Y, Liu B, Qian K, Liu B, Yang P, In-tip nanoreactors for cancer cells proteome profiling, Anal. Chim. Acta 949 (2017) 43–52. [DOI] [PubMed] [Google Scholar]
- [95].Tian R, Wang S, Elisma F, Li L, Zhou H, Wang L, Figeys D, Rare cell proteomic reactor applied to stable isotope labeling by amino acids in cell culture (SILAC)-based quantitative proteomics study of human embryonic stem cell differentiation, Mol. Cell. Proteomics 10 (2011), M110.000679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Chen Q, Yan G, Gao M, Zhang X, Ultrasensitive proteome profiling for 100 living cells by direct cell injection, online digestion and nano-LC-MS/MS analysis, Anal. Chem 87 (2015) 6674–6680. [DOI] [PubMed] [Google Scholar]
- [97].Espina V, Wulfkuhle JD, Calvert VS, VanMeter A, Zhou W, Coukos G, Geho DH, Petricoin EF, Liotta LA, Laser-capture microdissection, Nat. Protoc 1 (2006) 586–603. [DOI] [PubMed] [Google Scholar]
- [98].Bonner WA, Hulett HR, Sweet RG, Herzenberg LA, Fluorescence activated cell sorting, Rev. Sci. Instrum 43 (1972) 404–409. [DOI] [PubMed] [Google Scholar]
- [99].Rodriguez-Calvo T, Suwandi JS, Amirian N, Zapardiel-Gonzalo J, Anquetil F, Sabouri S, von Herrath MG, Heterogeneity and lobularity of pancreatic pathology in type 1 diabetes during the prediabetic phase, J. Histochem. Cytochem 63 (2015) 626–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Chen Q, Yan G, Gao M, Zhang X, Ultrasensitive proteome profiling for 100 living cells by direct cell injection, online digestion and nano-LC-MS/MS analysis, Anal. Chem 87 (2015) 6674–6680. [DOI] [PubMed] [Google Scholar]
- [101].Zhu Y, Zhu LN, Guo R, Cui HJ, Ye S, Fang Q, Nanoliter-scale protein crystallization and screening with a microfluidic droplet robot, Sci. Rep 4 (2014) 5046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].de Graaf EL, Giansanti P, Altelaar AF, Heck AJ, Single-step enrichment by Ti4(+)-IMAC and label-free quantitation enables in-depth monitoring of phosphorylation dynamics with high reproducibility and temporal resolution, Mol. Cell. Proteomics 13 (2014) 2426–2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Chan CY, Gritsenko MA, Smith RD, Qian WJ, The current state of the art of quantitative phosphoproteomics and its applications to diabetes research, Expert Rev. Proteomics 13 (2016) 421–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Lemeer S, Pinkse MWH, Mohammed S, van Breukelen B, den Hertog J, Slijper M, Heck AJR, Online automated in vivo zebrafish phosphoproteomics: from large-scale analysis down to a single embryo, J. Proteome Res 7 (2008) 1555–1564. [DOI] [PubMed] [Google Scholar]
- [105].Pinkse MW, Mohammed S, Gouw JW, van Breukelen B, Vos HR, Heck AJ, Highly robust automated, and sensitive online TiO2-based phosphoproteomics applied to study endogenous phosphorylation in Drosophila melanogaster, J. Proteome Res 7 (2008) 687–697. [DOI] [PubMed] [Google Scholar]
- [106].Shen Y, Strittmatter EF, Zhang R, Metz TO, Moore RJ, Li FM, Udseth HR, Smith RD, Unger KK, Kumar D, Lubda D, Making broad proteome protein measurements in 1–5 min using high-speed RPLC separations and high-accuracy mass measurements, Anal. Chem 77 (2005) 7763–7773. [DOI] [PubMed] [Google Scholar]
- [107].Zhu J, Nie S, Wu J, Lubman DM, T arget proteomic profiling of frozen pancreatic CD24+ adenocarcinoma tissues by immuno-laser capture microdissection and nano-LC-MS/MS, J. Proteome Res 12 (2013) 2791–2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Chen W, Wang S, Adhikari S, Deng Z, Wang L, Chen L, Ke M, Yang P, Tian R, Simple and integrated spintip-based technology applied for deep proteome profiling, Anal. Chem 88 (2016) 4864–4871. [DOI] [PubMed] [Google Scholar]
- [109].Liu F, Ye M, Pan Y, Zhang Y, Bian Y, Sun Z, Zhu J, Cheng K, Zou H, Integration of cell lysis protein extraction, and digestion into one step for ultrafast sample preparation for phosphoproteome analysis, Anal. Chem 86 (2014) 6786–6791. [DOI] [PubMed] [Google Scholar]
- [110].Horie K, Kamakura T, Ikegami T, Wakabayashi M, Kato T, Tanaka N, Ishihama Y, Hydrophilic interaction chromatography using a meter-scale monolithic silica capillary column for proteomics LC-MS, Anal. Chem 86 (2014) 3817–3824. [DOI] [PubMed] [Google Scholar]
- [111].Faserl K, Sarg B, Kremser L, Lindner H, Optimization and evaluation of a sheathless capillary electrophoresis-electrospray ionization mass spectrometry platform for peptide analysis: comparison to liquid chromatography-electrospray ionization mass spectrometry, Anal. Chem 83 (2011) 7297–7305. [DOI] [PubMed] [Google Scholar]
- [112].Wang Y, Fonslow BR, Wong CC, Nakorchevsky A, Yates JR 3rd, Improving the comprehensiveness and sensitivity of sheathless capillary electrophoresis-tandem mass spectrometry for proteomic analysis, Anal. Chem 84 (2012) 8505–8513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Zhang Z, Zhu G, Peuchen EH, Dovichi NJ, Preparation of linear polyacrylamide coating and strong cationic exchange hybrid monolith in a single capillary, and its application as an automated platform for bottom-up proteomics by capillary electrophoresis-mass spectrometry, Microchim. Acta 184 (2017) 921–925. [Google Scholar]