Abstract
Tumor initiating cells (TIC) represent a subset of tumor cells with increased self-renewal capability. TICs display resistance to frontline cancer treatment and retain the ability to repopulate a tumor after therapy, leading to cancer relapse. NOTCH signaling has been identified as an important driver of the TIC population, yet mechanisms governing regulation of this pathway in cancer remain to be fully elucidated. Here, we identify a novel mechanism of NOTCH regulation and TIC induction in breast cancer, via the miR-106b-25 miRNA cluster. We show that the miR-106b-25 cluster upregulates NOTCH1 in multiple breast cancer cell lines, representing both estrogen receptor (ER+) and triple negative breast cancer (TNBC), through direct repression of the E3 ubiquitin ligase, NEDD4L. We further show that upregulation of NOTCH1 is necessary for TIC induction downstream of miR-106b-25 in both ER+ and TNBC breast cancer cells, and that re-expression of NEDD4L is sufficient to reverse miR106b-25-mediated NOTCH1 upregulation and TIC induction. Importantly, we demonstrate a significant positive correlation between miR-106b-25 and NOTCH1 protein, yet a significant inverse correlation between miR-106b-25 and NEDD4L mRNA in human breast cancer, suggesting a critical role for the miR106b-25/NEDD4L/NOTCH1 axis in the disease. Further, we show for the first time that NEDD4L expression alone is significantly associated with a better relapse free prognosis for breast cancer patients. These data expand our knowledge of the mechanisms underlying NOTCH activation and TIC induction in breast cancer, and may provide new avenues for the development of therapies targeting this resistant subset of tumor cells.
Keywords: miR-106b-25, miRNA, breast cancer, NOTCH1, tumor-initiating cell
Introduction
Tumor initiating cells (TIC) are a population of cells strongly implicated in breast cancer progression that are defined by their increased capability to seed new tumors when compared to the general population of tumor cells(1) (2). In breast cancer, TICs are rare and can be enriched by cell surface markers CD44+/CD24−/low (1), or by increased aldehyde dehydrogenase activity (ALDH) (3). Both in vitro and in vivo studies demonstrate that TICs not only possess the ability to self-renew, but can also generate cells of multiple lineages to give rise to a heterogeneous tumor. Importantly, TICs have been shown to drive tumor initiation, mediate metastasis, and harbor resistance to standard chemotherapies and targeted therapeutics(4).
A number of signaling pathways have been implicated in maintaining the “stemness” of TIC populations, including WNT, HEDGEHOG (Hh), and TGF-β pathways, all of which are also important in stem cells during development(5). Additionally, the evolutionary conserved NOTCH signaling pathway, which is critical for cell fate determination, stem cell maintenance, differentiation, proliferation and survival during development has been heavily associated with TIC populations in breast cancer(6). In mammals, the NOTCH signaling pathway consists of five transmembrane ligands (DELTA-like1, 3, and 4 and JAGGED1 and 2), and four transmembrane receptors, NOTCH 1–4. The receptor is triggered via cell-to-cell contact when its extracellular domain binds to a ligand on a neighboring cell. This binding event elicits a sequential two-step cleavage of the NOTCH1 receptor to produce the NOTCH1 intracellular domain (NICD). The first cleavage event is mediated by the disintegrin and metalloproteinase protease family members, ADAM10 or ADAM17, followed by γ-secretase complex-mediated cleavage, ultimately leading to cytoplasmic release of the NICD. The NICD then translocates to the nucleus and, together with the DNA binding protein CBF-1/suppressor of hairless/Lag1 (CSL) and a family of Mastermind-like genes (MAML), acts as a canonical transcription factor to upregulate a number of target genes, including members of the hairy enhancer of split gene families, HES and HEY (7). CSL binding sites have also been confirmed in many other NOTCH target genes including CDKN1A (p21), c-MYC, and SLUG(8).
The NOTCH1 receptor was initially identified as an oncogene when chromosomal fusions were discovered between it and T-cell receptor β (TCRβ) in T-cell Acute Lymphoblastic Leukemia (T-ALL), resulting in a constitutively active form of NOTCH1(9). The pathway has now been implicated in a variety of solid and hematopoietic tumors(10), with most studies corroborating a tumor promotional role for the NOTCH pathway. In breast cancer, constitutively active forms of NOTCH1, NOTCH3, and NOTCH4 result in mammary tumors in mice(10, 11). Given the pro-tumorigenic role of NOTCH signaling in many cancers, targeting the NOTCH pathway has become a desirable cancer therapeutic avenue. While the importance of NOTCH signaling in breast cancer is undisputed, the precise mechanisms by which it is regulated remain unclear (12).
Here we demonstrate a novel mode of regulation of NOTCH1 in breast cancer, via miR-106b-25 mediated repression of the E3 ubiquitin ligase NEDD4L. The miR-106b-25 cluster, which resides in the 13th intron of the MCM7 gene on chromosome 7, is highly conserved across vertebrates and is overexpressed in many types of cancers including gastric, hepatocellular, prostate, lung, and breast cancer(13–19). miR-106b-25 is pro-tumorigenic/metastatic in numerous contexts, in part via increasing cell proliferation and decreasing apoptosis, effects that are mediated by its ability to downregulate PTEN, p21, BIM, and the TGF-β negative regulator Smad7(15, 16, 20). Work from our and other laboratories previously implicated the miR-106b-25 cluster in the regulation of TICs, although the mechanism by which it does so remained largely unexplored (20–23). Herein, we demonstrate that miR-106b-25 also activates NOTCH signaling, and that its ability to increase NOTCH1 is critical for its TIC function. We show for the first time that all three miRNAs target NEDD4L, and that miR-106b-25-mediated repression of NEDD4L leads to enhanced NOTCH signaling, and is required for miR-106b-25/NOTCH-induced TIC phenotypes. We further show that expression of miR-106b-25 positively correlates with NOTCH1 mRNA expression and negatively correlates with NEDD4L expression in human breast cancer, suggesting that miR-106b-25 mediated regulation of NOTCH signaling is conserved in the human disease. Furthermore, we demonstrate for the first time that low expression of NEDD4L significantly correlates with decreased time to relapse in breast cancer patients. Together, these data support a role for NEDD4L in TIC induction in breast cancers and highlight a new pharmacological avenue for suppression of TICs in breast cancer.
Results
The miR-106b-25 miRNAs regulate NOTCH1
Our group, and others, have previously uncovered a role for the miR-106b-25 cluster of microRNAs in enhancing the TIC population in breast cancers(20, 24). While the mechanism by which miR-106b-25 induces a TIC phenotype was unknown, we identified a number of genes upregulated by the cluster that have established roles in stem cell maintenance, growth, and differentiation, including NOTCH1(20). Given the previously established pro-tumorigenic role for NOTCH signaling in breast cancer, as well as the link of NOTCH signaling to breast cancer stem cells(25), we examined whether this signaling pathway may be a critical mediator of miR-106b-25 induced TIC phenotypes. To this end, we overexpressed the miR-106b-25 cluster, through stable integration of a viral plasmid containing the cluster genomic region, in 5 different breast cancer cell lines spanning various breast cancer subtypes. Levels of all three microRNAs in all cell lines were confirmed using qRT-PCR (Supplemental Figure 1). We found that overexpression of the miR-106b-25 cluster led to a marked increase in the levels of full-length NOTCH1 (FL-NOTCH1) and/or NOTCH1-ICD, when compared to the non-silencing controls (containing a scrambled non-targeting miRNA sequence -NS), in 4 of the 5 cell lines, with a more modest increase in the 5th line, the T47D cells. (Figure 1A). To investigate the impact of each individual miRNA in the cluster, we transfected individual miRNA mimics into MCF7 cells and found that miR-106b and miR-93 upregulate NOTCH1-ICD, while FL-NOTCH1 was upregulated only by miR-93. Importantly, all three miRNAs expressed together upregulate both forms of NOTCH1 (Supplemental Figure 2). To establish whether increased NOTCH1, in response to miR-106b-25 expression, results in an increase in NOTCH transcriptional activity, we performed a luciferase reporter assay with a plasmid containing tandem repeats of the RBP-Jk transcriptional response element, a NOTCH responsive sequence (26). In line with the increased expression of the NOTCH-ICD observed in response to miR-106b-25, we observed a significant increase in NOTCH luciferase reporter activity in the same 4 cell lines in which NOTCH1-ICD levels were markedly elevated by miR-106b-25, and again a slight increase (although not significant) in the T47D cells (Figure 1B). Because overexpression of miR-106b-25 led to increased expression of NOTCH1-ICD and transcriptional activity in numerous breast cancer cell lines, demonstrating a consistent phenotype across different breast cancer subtypes, we chose to move forward with two cell lines from different breast cancer subtypes for mechanistic studies; luminal MCF7 cells and triple negative Sum-159PT cells. We then examined whether targets of NOTCH1 were increased in these two cell lines in response to overexpression of the miR-106b-25 cluster, focusing on a subset of canonical NOTCH1 target genes including HEY2, HES5, HES1, HEYL. Using qRT-PCR analysis, we demonstrate that NOTCH1 target mRNA levels are indeed upregulated in response to miR-106b-25 in both systems (Figure 1C).
Figure 1. Overexpression of miR-106b-25 upregulates Notch1 signaling.
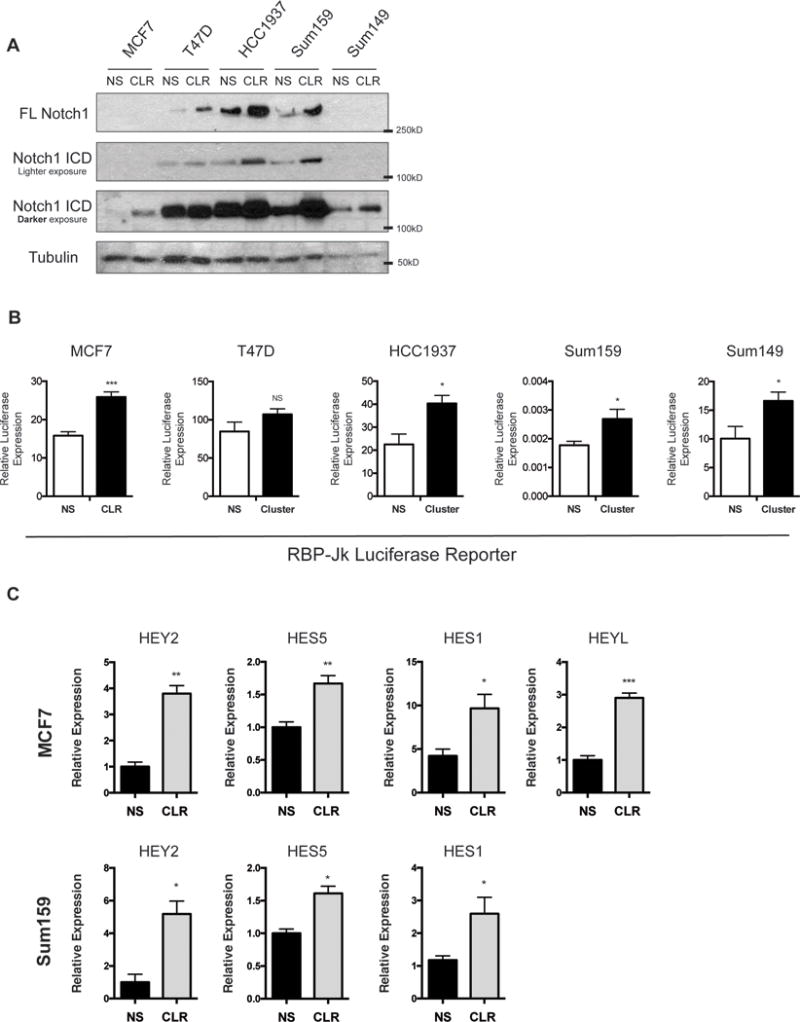
(A) Western blot analysis for NOTCH1-ICD and Full length (FL) NOTCH1 in a panel of breast cancer cell lines with stable overexpression of the miR-106b-25 cluster (CLR) including miR-106b, miR-93, and miR-25 or a non-silencing control (NS). (B) Notch Luciferase reporter assay using the RBP-Jk reporter transfected into MCF7, T47D, HCC1937, Sum159, and Sum149 NS and Cluster expressing lines. Statistical significance assessed by a student T-test on triplicate samples of a representative experiment (n=3). (C) qRT-PCR analysis in MCF7 and Sum159 NS and CLR cells showing an increase in NOTCH1 direct target genes with CLR overexpression. Statistical significance assessed by a student T-test on six replicate samples of a representative experiment (n=2).
NOTCH1 is required for miR-106b-25 induced TIC phenotypes
Given our demonstration that miR-106b-25 regulates NOTCH1 levels and transcriptional activity (based on NOTCH1 target upregulation), we asked whether NOTCH1 can contribute to the previously reported TIC phenotypes induced by the aforementioned cluster of miRs (20, 24). To address this question, we first measured the effects of miR-106b-25 overexpression on CD44 and CD24, as CD44+/CD24low/− cells are known to have increased TIC activity both in vitro and in vivo(1). MCF7 cells transiently overexpressing miR106b-25 mimics have an increased percentage of CD44+/CD24low/− cells compared to cells transfected with a non-silencing mimic (Figure 2A). This effect is abrogated when the cells are treated with an siRNA-targeting NOTCH1 (Figure 2A). To perform a more functional assay for TIC activity using both models, we measured the ability of MCF7 and Sum159 cells with or without stable miR-106b-25 cluster overexpression (CLR) to form mammospheres in the presence or absence of the γ-secretase inhibitor, DAPT (27). We confirmed that this dose of DAPT (5μM) inhibited NOTCH1 activation as measured by a decrease in NOTCH1-ICD in the MCF7 cells (Supplemental Figure 3A). Our data show that DAPT significantly decreases miR-106b-25 induced mammosphere formation in both the MCF7 and Sum-159 cells, suggesting a requirement for NOTCH1 signaling downstream of miR-106b-25 to mediate TIC function (Figure 2B).
Figure 2. miR-106b-25 increases TIC characteristics in-vitro and in-vivo in a Notch dependent manner.
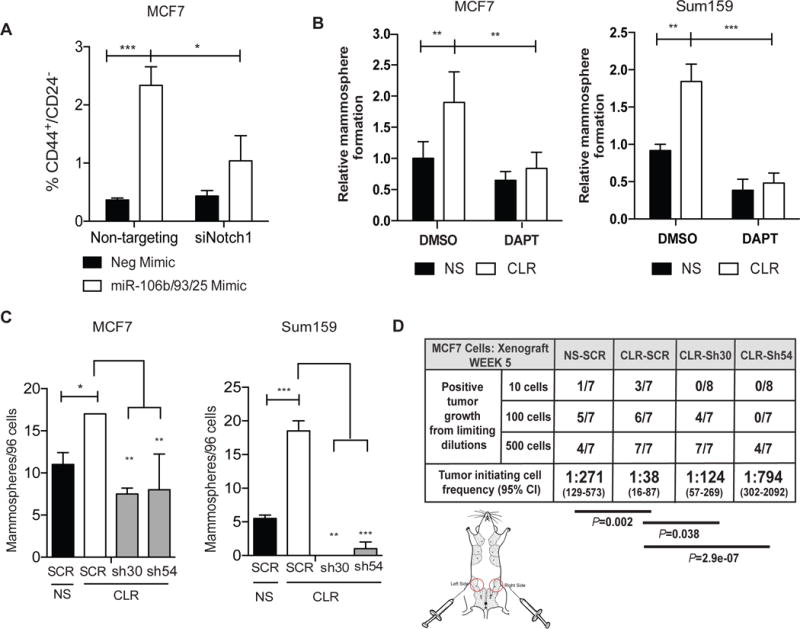
(A) Flow cytometry analysis in MCF7 cells treated with 30nM of either non-targeting siRNAs or pooled siRNAs that target NOTCH1 and transiently transfected with either a negative mimic or a combination of miR-106b, miR-93, and miR-25 mimics (20 nM total) to assess the percentage of CD44+/CD24− cells. (B) Secondary mammosphere assay in MCF7- and Sum159-NS and Cluster (CLR) stable cell lines in the presence of DAPT (5μM) or a DMSO vehicle control. (C) Left: Secondary mammosphere assay in MCF7 NS and CLR stable cell lines with stable shRNA knock down of NOTCH1. Right: Primary mammosphere assay in Sum-159 NS and Cluster stable cell lines with stable shRNA knock down of NOTCH1. Statistical significance on all mammosphere assays was assessed by one-way ANOVA followed by a Tukey post-test on triplicate samples of a representative experiment (n=3). (D) MCF7-NS and CLR cells with stable shRNA KD of Notch1 or a scramble control (SCR) were transplanted into the 4th mammary fat pad of NOG/SCID mice at limiting dilutions, data represents presence of tumors at week 5 post tumor cell implantation.
To more directly investigate the role of NOTCH1 in miR-106b-25-mediated TIC induction, we generated MCF7-CLR and Sum-159-CLR cells in which we knocked down NOTCH1 (resulting in both decreased FL-NOTCH1 and NOTCH-ICD) using two separate NOTCH1 shRNAs (sh30 and sh54) (Supplemental Figure 3B–D). Knockdown of NOTCH1 with both shRNAs is sufficient to significantly reverse the cluster-induced increases in mammosphere formation in both MCF7 and Sum-159 cells (Figure 2C). Of note, unlike MCF7-CLR cells with NOTCH1 KD, which formed sufficient primary mammospheres to enable secondary mammosphere assays, too few mammospheres formed with NOTCH1 KD in Sum-159 cells to perform secondary assays and thus primary assays are shown.
We further tested the ability of our MCF7-CLR NOTCH1 KD cells to form tumors using a limiting dilution assay in vivo, a critical test for TIC functionality. As previously observed, MCF7-CLR cells are significantly more efficient at forming tumors at limiting dilution when compared to MCF7-NS cells(20). This increase in TIC capacity is significantly reversed when NOTCH1 is knocked down in the MCF7-CLR cells (sh30 and sh54) (Figure 2D). Further, the level of NOTCH1 KD correlates highly with tumor initiating capacity, as KD of NOTCH1 in the MCF7-CLR-sh30 cells was lost over time (Supplemental Figure 3D), and these cells retained more TIC capability than the MCF7-CLR-sh54 cells, in which NOTCH1 KD was maintained.
The miR-106b-25 miRNAs enhance NOTCH1 protein stability
We next examined how the miR-106b-25 cluster upregulates NOTCH1. Interestingly, miR-106b-25 expression does not significantly upregulate NOTCH1 mRNA in all cell lines in which increased NOTCH1 protein is observed (Supplemental Figure 4). This result suggested that NOTCH1 is likely primarily regulated post-transcriptionally. To explore whether NOTCH1 protein stability is influenced by the miR-106b-25 cluster, we treated cells with the protein synthesis inhibitor cycloheximide (CHX), and measured NOTCH1 protein levels at various time points post CHX treatment. Both MCF7 and Sum-159 cells exhibit a clear increase in NOTCH1 protein stability in the presence of miR-106b-25 as compared to the NS control (Figure 3A). Indeed, the calculated half-life for the NOTCH1 protein was increased 3–5 fold in the presence of miR-106b-25 when compared to control cells (Figure 3B). Therefore, overexpression of miR-106b-25 leads to enhanced NOTCH1 protein stability, in both MCF7 and Sum-159 cells.
Figure 3. The miR-106b-25 cluster produces an increase in Notch1-ICD stability.
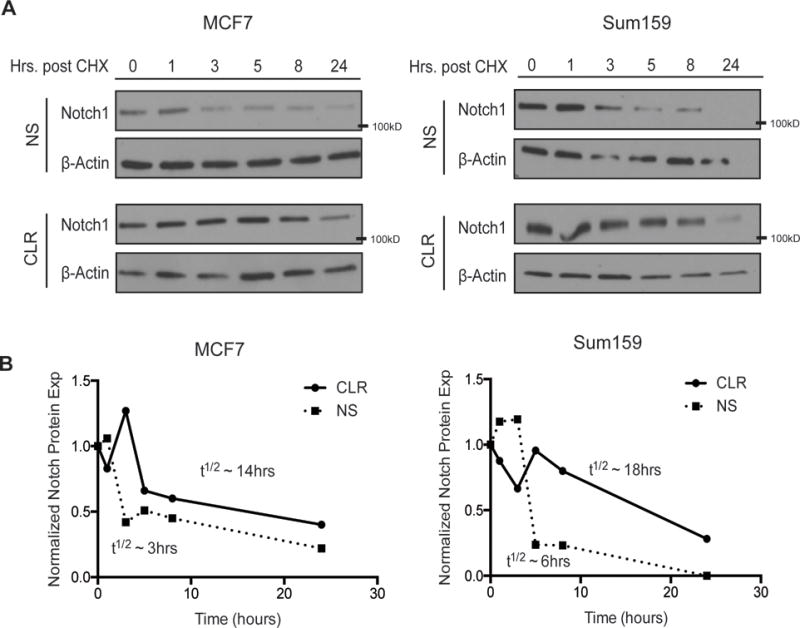
(A–B) Top: Western blot analysis in (A) MCF7 and (B) Sum159 NS and Cluster (CLR) cell lines after treatment with cyclohexamide (CHX - 20μM) for indicated periods of time. Bottom: Quantification of the above representative western blots (n=3), and approximate half-lives (representative data of 3 experiments). The half-lives shown were approximated from the graphs shown from one experiment and are representative of all 3 experiments.
The miR-106b-25 cluster targets the E3 ubiquitin ligase NEDD4L
To account for miR-106b-25 mediated increases in NOTCH1 protein stability, we asked whether the miR-106b-25 cluster might target a protein involved in NOTCH1 turnover. Using miRNA target prediction software (TargetScan.com), we found that all three miRNAs in the miR-106b-25 cluster are predicted to target the HECT domain E3 ubiquitin ligase NEDD4L, which contains two miR-93/miR-106b sites, and one miR-25 site. All three sites are present in the 3′UTR of NEDD4L, and are all remarkably well conserved across species (Figure 4A, Supplemental Figure 5). NEDD4L belongs to the NEDD4 family of ubiquitin ligases, and is the closest homologue to NEDD4 itself (28). Two NEDD4 family members, including NEDD4 and ITCH, have previously been identified to ubiquitinate the NOTCH1 protein, leading to its degradation(29, 30), however NEDD4L has not previously been linked to NOTCH1. Given the similarity of NEDD4L to its family members, we reasoned that it may similarly regulate NOTCH1 protein stability, and may thus be the means through which the miR-106b-25 cluster regulates NOTCH1 protein levels. Thus, we examined whether the miR-106b-25 cluster indeed targets NEDD4L. Our data demonstrate that miR-106b-25 expression leads to a reduction of both NEDD4L mRNA and protein in MCF7 and Sum-159 cells (Figure 4B,C). To test whether the miR-106b-25 cluster directly regulates NEDD4L, we constructed a luciferase vector harboring the 3′UTR of NEDD4L and measured the luciferase activity of this construct in the presence of each individual miRNA, and all three miRNAs together, using transfection of miRNA mimics. Normalized luciferase activity demonstrates that each miRNA of the miR-106b-25 cluster has the ability to target and repress the 3′UTR of NEDD4L (Figure 4D). Further, miR-106b-25 mediated repression of NEDD4L is abrogated when the three miRNA seed sites are mutated (Nd4L_Mut). Similar results were seen with stable miR-106b-25 cluster expression in both MCF7 and Sum-159 cells (Figure 4E). These data demonstrate that all members of the miR-106b-25 cluster can directly repress the expression of NEDD4L by binding to its 3′UTR.
Figure 4. The miR-106b-25 cluster directly targets the NEDD4L.
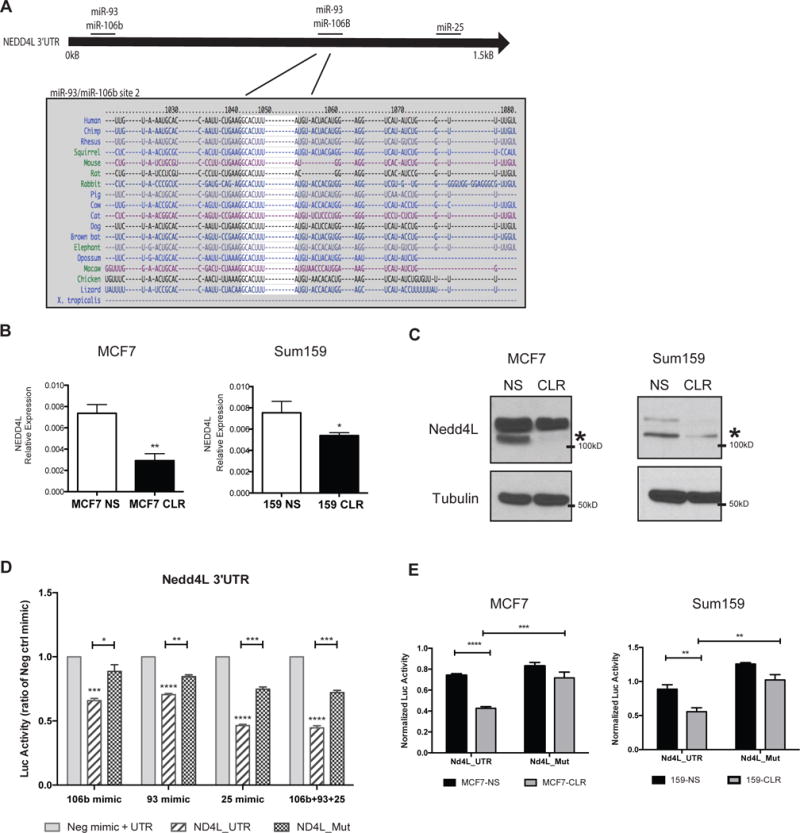
(A) Top: A schematic of the NEDD4L-3′UTR indicating the 3 predicted binding sites for the cluster of microRNAs. Bottom: Conservation analysis across species for a miR-106b/93 binding site. (B) qRT-PCR for NEDD4L expression performed on cDNA from MCF7 and Sum159 NS and CLR cell lines. Statistical significance was assessed by performing a student T-test on triplicate samples of a representative experiment (representative data of 3 experiments). (C) Western blot analysis in MCF7 and Sum159 NS and Cluster cell lines for NEDD4L protein expression. D) Luciferase assay in HEK293 cells after transient co-transfection with miRNA mimics (20nM) and a luciferase construct containing the full NEDD4L-3′UTR (ND4L_UTR) or a construct containing the full NEDD4L-3′UTR with all three microRNA target sites mutated (ND4L_Mut). UTR sequences sit behind a Renilla Luciferase gene, with an internal firefly luciferase for transfection control. Data represents Renilla Luc normalized to firefly Luc. Statistical significance was assessed using One-way ANOVA followed by a Tukey post-test on triplicate samples of a representative experiment (representative data of 2 experiments). (E) MCF7 and Sum159 NS and Cluster cell lines were co-transfected with the full NEDD4L-3′UTR luciferase construct (Nd4L_UTR), or the full NEDD4L-3′UTR with all three of the predicted microRNA binding sites mutated (Nd4L_Mut), similar to above. Statistical significance was assessed using One-way ANOVA followed by a Tukey post-test on triplicate samples of a representative experiment (representative data of 2 experiments).
NEDD4L mediates ubiquitination of NOTCH1 and is necessary for miR-106b-25 induced TIC characteristics
To investigate the role of NEDD4L in controlling NOTCH1 and thus miR-106b-25-mediated TIC phenotypes, we generated MCF7-CLR and Sum159-CLR cell lines with rescued expression of NEDD4L (ND4L) by stable integration of an HA tagged NEDD4L plasmid lacking it’s 3′UTR (Figures 5A). HA-NEDD4L is rapidly turned over in MCF7 cells and its overexpression can only be detected in the presence of the proteasome inhibitor, MG132 (Figure 5A, upper panel). In Sum159 cells, we were able to observe rescued NEDD4L in the absence of the addition of a proteasome inhibitor (Fig. 5A, lower panel). Reintroduction of NEDD4L in miR-106b-25 overexpressing cells leads to reduced NOTCH1-ICD protein levels, demonstrating that NEDD4L is involved in miR-106b-25 cluster-mediated regulation of NOTCH1-ICD (Figure 5B). To determine whether the miR/NEDD4L/NOTCH1 axis is intact in an endogenous setting in which the miR levels have not been manipulated, we knocked down the cluster of microRNAs using a stable lentiviral miRNA knockdown system (miRzip) to inhibit all three miRNAs in the cluster simultaneously in MDA-MB-231 cells (which exhibit high basal miR-106b-25 expression). Inhibition of the miR-106b-25 cluster led to a clear upregulation of NEDD4L and downregulation of both FL-NOTCH1 and NOTCH1-ICD (Supplemental Figure 6).
Figure 5. NEDD4L ubiquitinates NOTCH1 and its re-expression is sufficient to reverse miR-106b-25 induced TIC characteristics.
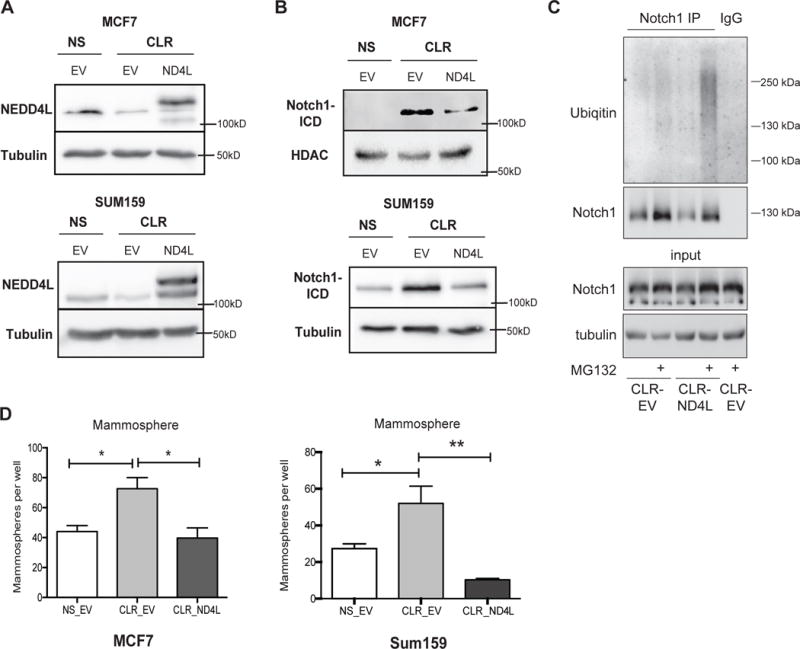
(A) Western blot analysis, using a NEDD4L antibody, in MCF7 and Sum159 NS and CLR cell lines with stable HA-NEDD4L expression (ND4L). Due to the rapid turnover of the exogenous protein in MCF7 cells, MG132 (25μM) was added to MCF7 NS and CLR cells for 4hrs to stabilize HA-NEDD4L and allow its detection (B) Western blot analysis showing that Notch-ICD levels are decreased in MCF7 and Sum159 Cluster cell lines with stable rescue of NEDD4L (C) Western blot analysis to identify polyubiquitinated NOTCH1 species in MCF7-CLR-EV (low NEDD4L) vs MCF7-CLR-ND4L (high NEDD4L) cells after NOTCH1 immunoprecipitation. (D) Quantification of the number of secondary mammospheres after 7 days in culture in MCF7 and Sum159 NS, CLR, and CLR-ND4L rescue lines. Statistics were assessed by ANOVA followed by a Tukey post-test of triplicate samples of a representative experiment (n=2) and by a Fisher’s LSD test of triplicate samples from a representative experiment (n=2), respectively.
To test if NEDD4L mediates the decrease in NOTCH1 levels via altering its ubiquitination, and thus subsequent degradation, we immunoprecipitated NOTCH1-ICD in the presence and absence of the proteasome inhibitor MG-132 in order to assess polyubiquitinated NOTCH1 species. We found that MCF7-CLR cells with NEDD4L add-back (MCF7-CLR-ND4L) resulted in an increased ubiquitination compared to MCF7-CLR-EV cells (in which NEDD4L levels are low), supporting the notion that NEDD4L targets NOTCH1 for ubiquitination, and suggesting that this increased NEDD4L-dependent ubiquitination results in increased NOTCH1 degradation (Figure 5C). To determine whether NEDD4L restoration in breast cancer cells reverses the ability of miR-106b-25 to induce TIC phenotypes, we performed mammosphere assays with the MCF7 and Sum-159 NS, CLR, and CLR-ND4L cells, demonstrating that reintroduction of a non-targetable NEDD4L reverses the cluster-induced increases in mammosphere formation (Figure 5D). These data, coupled with data in Fig. 2, clearly demonstrate that miR-106b-25 mediated downregulation of NEDD4L and resultant induction of NOTCH1 is critical for its ability to induce TIC phenotypes.
The miR-106b-25/NOTCH1/NEDD4L axis is conserved in human breast tumors
To determine whether the miR-106b-25 cluster is linked to NEDD4L repression and NOTCH1 upregulation in human breast cancers, we interrogated the breast invasive carcinoma (BRCA) TCGA dataset. Two members of the cluster, miR-106b and miR-93, are significantly upregulated in breast tumors (n=762) vs. normal breast tissue (n=87), a finding that is in line with previous studies demonstrating overexpression of these miRNAs in other cancers (14, 15, 17, 19) (Figure 6A). Similar results are seen when tumors are compared to their corresponding matched normal tissue (Figure 6B). Importantly, all three miRNAs also have a significant negative correlation with NEDD4L mRNA expression in breast tumors (n=760) (Figure 6C), and a significant positive correlation with NOTCH1 protein in human breast tumors (n=638) (Figure 6C). To determine whether the miR/NOTCH1 axis is prevalent in certain breast cancer subtypes, we examined the correlation between each miRNA and Notch1 protein in different subtypes. We found that miR106b-25 expression significantly correlates with NOTCH1 protein expression in Luminal A cancers, a trend which is maintained in HER2, and Basal-like tumors (likely not statistically significant due to low sample numbers). However, there is not a significant correlation between the miRs and NOTCH1 in Luminal B subtypes (Supplemental Figure 7). These data demonstrate that the miR/NOTCH1 axis is present across multiple breast cancer subtypes, similar to what we observed in our cell line work. Lastly, we also identified a significant negative correlation between NEDD4L mRNA and NOTCH1 protein expression in breast tumors (R=−0.121, P=0.0003) (Figure 6D). Together, these data suggest that the connection between the miR-106b-25 miRNAs, NOTCH1, and NEDD4L in breast cancer cell lines extends to human breast cancer. Previously, our group has demonstrated that high miR-106b-25 expression in breast tumors significantly correlates with decreased time to relapse (20), a clinical outcome that is associated with elevated levels of TICs (4). Given our novel finding that NEDD4L controls TIC phenotypes downstream of miR-106b-25 through controlling NOTCH1 levels, we asked whether NEDD4L itself is associated with poor prognosis in human breast cancers. Using the BRCA TCGA dataset, we found that NEDD4L is significantly downregulated in breast tumors vs. normal tissue, with an even further decrease in metastatic tissue (Figure 6E). Importantly, patients whose breast tumors express low levels of NEDD4L (below the median) have a significantly shorter time to relapse, than those that express above the median levels of NEDD4L (Figure 6F). Together, these data suggest an important role for NEDD4L in breast cancer progression, and support our finding that the miR-106b-25 miRNA/NEDD4L/NOTCH1 axis may be critical for the induction of TIC properties in human breast cancers.
Figure 6. The miR-106b-25/NOTCH1/NEDD4L axis is relevant in human breast tumors.
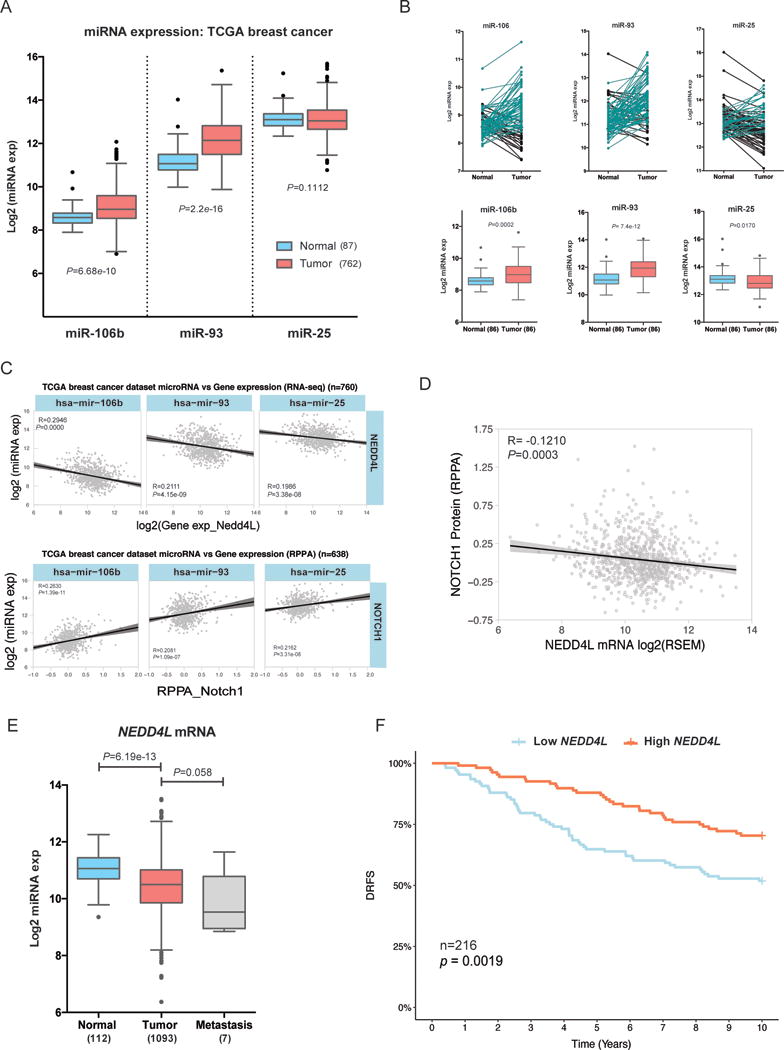
(A–C) In the TCGA breast invasive carcinoma dataset we plotted (A) the normalized log2 miRNA expression for each microRNA in normal adjacent breast tissue (n=87) vs. breast tumor tissue (n=762), statistics derived from KS test. (B) Top: The normalized log2 expression of each miRNA in matched normal and tumor samples are plotted, with black lines corresponding to samples where expression is decreased in matched tumor tissue and aqua lines corresponding to samples where expression is increased in matched tumor tissue. Bottom: Box and whisker plots of the above data with statistics derived from KS test C) Pearson correlations between individual miRNAs within the cluster (log2 normalized) and NEDD4L mRNA expression extracted from TCGA RNAseqV2 (Log2 normalized) (top), and Pearson correlations between miRNA expression (miRNA-seq) and NOTCH1 protein expression (from TCGA RPPA) (bottom). (D) Pearson correlation demonstrates a negative association between NEDD4L mRNA expression and NOTCH1 RPPA protein expression in the TCGA breast cancer dataset (E) Differential expression of NEDD4L mRNA (RSEM value) in normal vs. breast tumor vs. metastatic tissue in same TCGA dataset as A–C (F) Kaplan-Meier of Distant Relapse Free Survival (DRFS) in 216 individuals with early primary breast cancer, compiled from GEO dataset (GSE22219), stratified into NEDD4L high (above the median) vs. low (below the median).
Because miR-106b-25 mRNAs has been associated with stem cell characteristics (both within this manuscript and elsewhere(20–24), we investigated the association of these miRNAs with breast cancer metastasis. Previous data identified a connection between miR-106b and the development of site-specific lung metastasis from breast cancer (31). To determine whether these miRs may also strongly correlate with overall metastasis in breast cancer, we examined an available dataset containing matched primary breast tumor and metastatic tissue with global miRNA expression profiling (32). Of interest, expression of all three microRNAs was higher in breast cancer metastasis when compared to matched normal tissue, supporting a role for this miR cluster in more aggressive disease (Figure 7).
Figure 7. The miR-106b-25 cluster is increased in human breast cancer metastasis.
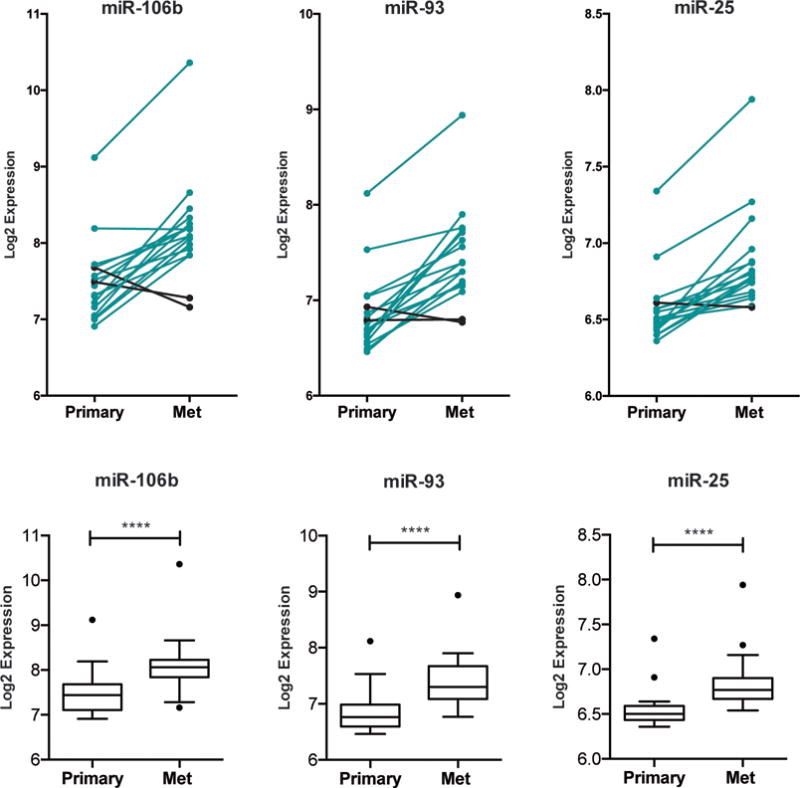
Data was extracted from GSE37407, which contains miRNA expression profiling on primary breast tumors vs. matching metastatic tissue, and was then interrogated for expression of each miRNA in the miR-106b-25 cluster. Top: The normalized log2 expression of each miRNA in matched tumor and metastatic (mets) sample is plotted, with black lines corresponding to samples where expression is decreased in matched mets and aqua lines corresponding to samples where expression is increased in matched mets. Bottom: Box and whisker plots of the above data with statistics represented as KS test.
Discussion
Our data demonstrate that the miR-106b-25 cluster positively regulates NOTCH1 protein expression, resulting in increased NOTCH signaling. We further demonstrate that overexpression of this cluster of miRNAs increases TIC phenotypes in vitro and in vivo, via its ability to induce NOTCH1 through repression of NEDD4L. Our findings suggest that this cluster induces NOTCH1, and TIC phenotypes, across different subtypes of breast cancer, including the Sum159 triple negative and MCF7 luminal breast cancer cell lines. These findings are in contrast to previous evidence that has demonstrated differential TIC phenotypes with overexpression of miR-93 in luminal vs. basal breast cancer lines, where TIC characteristics are increased in luminal breast cancer lines, but decreased in basal breast cancer lines, including the Sum159 cells used in our study (24). Our opposing results, however, may reflect the difference in studying only specific members of the miR-106b-25 cluster in isolation, as compared to studying the effects of their coordinate expression. Indeed, other studies have reported a tumor suppressive role for members of the miR-106b-25 cluster (24, 33–35), but most of these studies have examined the role of only a single miRNA within the cluster. Our findings are in line with many other published studies that demonstrate a tumor promotional role for the miR-106b-25 cluster as a whole, as it has been shown to increase proliferation and invasion in hepatocellular (36, 37), lung(17), endometrial(38), esophageal(39), and gastric cancers(18), and to increase tumor progression in prostate cancer(19). Our results further solidify an oncogenic role for the miR-106b-25 miRNA cluster in breast cancer, particularly as the cluster is co-expressed behind a single promoter for the MCM7 gene and its effects are thus likely mediated by all three members in coordination.
Importantly, we show for the first time that the miR-106b-25 cluster robustly upregulates NOTCH1 protein, both the full length and intracellular forms, and increases signaling downstream of NOTCH1. While our overexpression studies show that miR106b-25 is sufficient to affect the NEDD4L/NOTCH1 axis, we also show that it is necessary to mediate alterations in NEDD4L and NOTCH1 via the use of miR-Zip constructs in MDA-MB-231 cells (Supplemental Figure 6). Induction of this pathway largely explains how miR-106b-25 regulates TIC phenotypes, as inhibition of NOTCH1 reverses miR-106b-25 induced TIC characteristics (Figure 2). As NOTCH signaling has been linked to enhanced stem cell phenotypes in both normal and cancerous contexts(25, 40–42), its regulation by miR-106b-25 may have implications not only in a cancerous context, but also in the context of normal stem cells.
We further demonstrate that the miR-106b-25 cluster can directly target the E3 ubiquitin ligase NEDD4L, leading to its repression both on the mRNA and protein level. The 3′UTR of NEDD4L contains three evolutionary conserved microRNA-binding sites, two targeted by miR-106b/93 and one targeted by miR-25. Interestingly, a recent publication demonstrates that miR-93 targets NEDD4L in lung cancer(43), supporting our data that this miRNA can target NEDD4L in breast cancer. However, it was not previously known that all the miRNAs within the cluster are capable of targeting NEDD4L. We also present the novel finding that overexpression of NEDD4L leads to a decrease in NOTCH1-ICD protein (Figure 5), via its ability to ubiquitinate and target NOTCH1 for proteasome-mediated degradation. In Drosophila, other NEDD4 family members, including Nedd4 and Suppressor of Deltdex [Su(dx)] have been shown to negatively regulate NOTCH endocytosis and signaling(30, 44). In mammals, the E3 ubiquitin ligase and NEDD4 family member, ITCH, has been shown to ubiquitinate NOTCH1 resulting in proteasome-mediated degradation(29). Thus, our results implicate yet another member of this family, NEDD4L, in mediating NOTCH1 degradation. Further, we show for the first time that NEDD4L is dramatically downregulated in human primary breast cancers and metastasis when compared to normal tissue, and those patients whose breast tumors have low expression of NEDD4L have a significantly higher chance of tumor relapse (Figure 6). These data highlight for the first time the importance of NEDD4L as a tumor suppressor in breast cancer, and open up a new therapeutic avenue, with particular emphasis on targeting stem cell populations in cancer.
This cluster of microRNAs, particularly miR-106b has also recently been implicated in metastasis, as hepatocellular carcinoma cells with overexpression of miR-106b showed increased metastatic potential(45). Importantly, we demonstrate that metastatic breast cancer patient samples have increased expression of all three microRNAs within the miR106b-25 cluster when compared to matched normal tissue (Figure 7). As outlined above, we also show that NEDD4L mRNA expression is decreased in patient metastasis, further implicating members of this axis in tumor progression/ metastasis (Figure 6E).
Pathways that regulate TICs are complex, and crosstalk between numerous pathways is important for the TIC phenotype(46). In particular, the miR-106b-25 cluster was previously shown to enhance TGF-β signaling(20), and crosstalk between NOTCH and TGF-β signaling has been shown in numerous contexts (47–49). While our data suggest that miR-106b-25 regulation of NOTCH1 is critical for its ability to regulate TIC phenotypes, it is highly likely that the cluster’s regulation of both NOTCH and TGF-β are together critical for its tumor promotional effects. We and others have previously reported that the miR-106b-25 cluster can mediate the switch in TGF-β signaling from tumor suppressive to tumor promotional by targeting the TGF-β negative regulator SMAD7(20, 22, 50). Intriguingly, recent reports have shown that miR-93 mediated regulation of NEDD4L leads to increased TGF-β signaling and EMT in lung cancer(43), and thus effects of the miRs within the miR106b-25 cluster on NEDD4L may be critical for enhancing stem-like and EMT characteristics via control of both NOTCH and TGF-β signaling. However, NEDD4L has also been shown to target the TGF-β activating Smads, SMAD2/3, for proteasome-mediated degradation, resulting in decreased TGF-β signaling(51). These studies highlight the complexities of signaling networks that regulate TIC maintenance, and remind us that it is important to consider the cell and tissue context when investigating key pathways controlling stem-like characteristics.
Overall, our data demonstrate that the miR-106b-25/NOTCH1/NEDD4L axis plays a tumor promotional role in breast cancer via stimulating TIC phenotypes. Evidence that this axis is important in human breast cancer is supported by our findings that increased expression of the cluster correlates with increased levels of NOTCH1 and decreased expression of NEDD4L in human breast cancer. It is increasingly recognized that cancer therapies designed to target and eliminate the TIC population may have high therapeutic efficacy. Our results highlight potential new therapeutic targets to inhibit NOTCH signaling and TIC phenotypes in breast cancer, via inhibiting the miR-106b-25 cluster or activating NEDD4L.
Materials and Methods
Cell culture, constructs, and inhibitors
Cell growth conditions are per ATCC guidelines. The T47D cells were a generous gift from Dr. Jennifer Richer (University of Colorado AMC). The Sum-149 and Sum-159 cells were generous gifts from Dr.Stephen Ethier (Medical University of South Carolina). The HCC1937 and MCF7 cells have been cultured long term in the Ford Laboratory. All cell lines were authenticated (STR analysis performed 3/2017) and monitored for mycoplasma contamination every 6–8 months. Generation of the MCF7-NS and Cluster lines was described previously(20) and the Sum159, HCC-1937, T47D, and Sum-149 NS and Cluster lines were made similarly. When indicated, microRNA miRIDIAN mimics for miR-106b, miR-93, and miR-25 (Dharmacon, Lafayette, CO, USA) were transiently transfected into parental cells at 20nM concentration. MCF7 and Sum159-NS and Cluster lines with stable NOTCH1 knockdown were generated with PLKO.1 vectors (Sigma Aldrich, St. Louis, MO, USA) containing the NOTCH1 targeting sequences CCGGCCGGGACATCACGGATCATATCTCGAGATATGATCCGTGATGTCCCGGTTTTTG or CCGGAGGGAAGTTGAACGAGCATAGCTCGAGCTATGCTCGTTCAACTTCCCTTTTTTG (obtained from the University of Colorado Functional Genomics shared resource). Rescue of NEDD4L either transiently (MCF7-NS and CLR) or stably (Sum159-NS and CLR) was achieved with pCI HA NEDD4L, a gift from Joan Massague (Addgene plasmid # 27000)(51). DAPT, Cycloheximide, and SB431542 were purchased from Sigma-Aldrich. To functionally knock down the cluster of microRNAs, we used a lentiviral miRZIP construct targeting all three miRNAs (System Biosciences, Palo Alto, CA) that we transduced into MDA-MB-231 cells.
Western blot Analysis
Whole cell extracts were isolated as previously described (52) and normalized with a Lowry Protein Assay (BioRad, Hercules, CA, USA). Proteins were electrophoresed (on 8% or 10% gels) and transferred to polyvinylidene difluoride (PVDF) membranes followed by a 1 hour blocking incubation at room temperature in 5% milk in Tris-buffered saline with 0.1% tween 20 (TBST). Membranes were then incubated in primary antibody overnight at 4°C. Primary antibodies against the following proteins were used: NOTCH1 (D6F11) (1:1000, Cell Signaling Technologies #4380, Danvers, MA, USA), phospho-Smad3 (1:500, Cell Signaling Technologies, #9520S), β-Actin (1:10,0000, Sigma-Aldrich, #A5316), β-Tubulin (1:1000, Sigma-Aldrich #T4026), and NEDD4L (1:1000, Bethyl Laboratories Inc. #A302-513A, Montgomery, TX, USA). Membranes were then washed in TBST and incubated for 1 hour at room temperature in HRP-anti-mouse (1:10,000, Sigma-Aldrich #A9044) or HRP-anti-rabbit (1:10,000, Sigma-Aldrich #A9169) secondary antibodies. All western blots in the manuscript were replicated in the laboratory at minimum three times, and representative blots are shown.
Immunoprecipitation and ubiquitination assay
MCF7 cells were lysed in RIPA buffer (150 mM NaCl, 50 mM Tris-HCl pH 8.0, 5 mM EDTA, 1% (v/v) IGEPAL CA-630 (NP-40 substitute), 0.5% (w/v) sodium deoxycholate, 0.1% (w/v) SDS, 0.2–0.45 μm filtered) supplemented with protease and phosphatase inhibitors. Following ultrasonic disruption (30 s at 3 W) lysates were cleared by centrifugation (20 000 g at 4°C for 25 min). Sample aliquots (1 mg of total protein in 1 ml) were subjected to immunoprecipitation with either 5 μl of anti-NOTCH1 antibody (Cell Signaling Technologies, #4380) or non-specific rabbit IgG (Santa Cruz Biotechnology, sc-3888). Precipitated complexes immobilized on Protein G Dynabeads (Thermo Fisher Scientific, 10003D) were washed twice with RIPA buffer, twice with immunoprecipitation wash buffer (500 mM LiCl, 100 mM Tris-HCl pH 8.5, 1% (v/v) IGEPAL CA-630 (NP-40 substitute), 1% (w/v) sodium deoxycholate. 0.2–0.45 μm filtered), and again twice with RIPA buffer. Pelleted beads were incubated with 4× SDS loading buffer (400 mM Tris-HCl pH 7.5, 4% (w/v) SDS, 40% (w/v) glycerol, 4% (v/v) mercaptoethanol, 0.04% (w/v) bromophenol blue) at 95°C for 5 min to release protein complexes. Proteins resolved with SDS-PAGE were transferred to a PVDF membrane and detected using the following antibodies: Ubiquitin (Cell Signaling Technologies, #3936), NOTCH1 (Abcam, ab65297), and HRP-conjugated secondary antibodies (Rockland, TrueBlot 18-8817-30, Cell Signaling Technologies, #3678). The ubiquitin assay was reproduced three times with two separate NOTCH1 antibodies, Figure 5C shows a representative experiment with the Abcam NOTCH1 antibody.
qRT-PCR
miRNeasy isolation kits (Qiagen, Germantown, MD, USA) were used to extract total RNA, including microRNAs. The miScript II RT (BioRad) CDNA synthesis kit was used for both mRNA and microRNA qRT-PCR. The ssoFast Evagreen supermix (BioRad) was used for all qRT-PCR reactions. miScript primer assays (Qiagen) were used for miRNA specific qRT-PCR.
Luciferase Reporter Assays
The RBP-Jk reporter plasmid was acquired from SABiosciences (a Qiagen company). The reporter was transfected with Xtremegene siRNA (Sigma Aldrich), and 48hr lysates were analyzed using the dual luciferase assay kit (Promega, Madison, WI, USA). RBP-jK luciferase are representative experiments from two reproducible experiements for each cell lines, and show technical replicates of 3. For 3′UTR experiments, the 3′UTR of NEDD4L was cloned into the psi-Check2 luciferase reporter (Promega), and transfection and analysis was performed as above. The miR mimics with UTR experiment was reproduced twice, with representative experiment shown in Figure 4D. Figure 4E was reproduced three times, with representative experient shown. To mutate the three microRNA binding sites in the NEDD4L 3′UTR we performed site directed mutagenesis using the QuickChange Lightning kit (Agilent Technologies, Santa Clara, CA, USA) and the following oligonucleotide sequences: gcaaaagtctgttaggcaaatgcaatatttcaagcagaacttgcttgaagaacaa, tgttgtaaatgcaccaattctgaaggaaatatatgtactacatggaggtcatatctg, ccagattcacaattgatagatacttctccggaaagatgtgtgcagggaa.
TIC Assays
Flow cytometry analysis and mammosphere formation assays were performed as described previously(53). Mammosphere assays show triplicate samples of representative experiment (n=3). In vivo TIC assays were done and analyzed as described previously(20), with the exceptions that we used 6–8 week old female NOG/SCID mice, kindly gifted by Paul Jedlicka, and had 7–8 mice per dilution per cohort. Sample size was determined from a power analysis at 80% power with 5% error, with consideration of effect and standard deviation from a pilot experiment with 5 animals for each group. No animals were excluded from analysis, and samples were randomized by injection into both the left and right flank of animals. Determination of tumor appearance was done by an individual who was blinded to treatment groups in the animal at the time of measurement. All animal studies were performed according to protocols reviewed and approved by the Institutional Animal Care and Use Committee at the University of Colorado Anschutz Medical Campus.
Datamining
Breast invasive carcinoma (BRCA) data was downloaded from the TCGA archive 2016_01_28 (http://gdac.broadinstitute.org). The data included Level 3 Illumina Hiseq expression data for microRNA and mRNA expression and Level 3 gene annotated RPPA data for protein intensity values. Boxplots and scatterplots were generated using the R library “ggplot2” and Pearson correlations and statistics were generated using the hmisc package in R. Gene expression profiling by array and DRFS information was downloaded from GEO [series GSE22219]. Samples were stratified based on median expression of NEDD4L into high and low groups. The R libraries “survival” and “survminer” were used to calculate statistics and draw survival plots. miR106b-25 expression in breast primary vs. metastatic tissue was assessed using the dataset from GSE37407.
Statistical Analysis
All the assays performed fall under a normal distribution and the correct statistical test is used based on the number of comparisons and replicates. In every case the variance between groups that are compared is similar. Each statistical test and replicate number is defined in the figure legend.
Supplementary Material
Acknowledgments
The authors thank Paul Jedlicka for the generous gift of mice for limiting dilution experiments. We also want to thank the University of Colorado Cancer center flow cytometry shared resource for performing flow cytometry experiments needed for this work (P30CA046934), the Functional Genomics Facility for shRNA constructs, and The Cancer Genome Atlas (TCGA) for use of their breast cancer patient dataset. This work was supported by R01CA095277 (H.L.F.), R01CA117907 (J.M.E.), the METAvivor Foundation (H.L.F.), the Department of Defense Breast Cancer Research Program W81XWH-10-1-0296 (A.L.G.), the Ruth L. Kirschstein National Service Research Award (F32) 1F32CA199716-01 (A.L.G.), the Ruth L. Kirschstein National Service Research Award (F31) 5F31CA210622-02 (M.U.J.O.), the NCI fellow transition award F99 CA223023 (MUJO), UC Denver AMC Molecular Biology Program T32 training grant T32 GM008730 (C.G.T.), NIH-RO1 Diversity Supplement to R01-CA157790 (C.G.T.), the UNCF/MERCK Graduate Fellowship (C.G.T), and the T32 Cancer Biology training grant CA190216-1 A1 (C.G.T.).
Financial support: This work was supported by R01CA095277 (H.L.F.), R01CA117907 (J.M.E.), the METAvivor Foundation (H.L.F.), the Department of Defense Breast Cancer Research Program W81XWH-10-1-0296 (A.L.G), the Ruth L. Kirschstein National Service Research Award (F32) 1F32CA199716-01 (A.L.G), the Ruth L. Kirschstein National Service Research Award (F31) 5F31CA210622-02 (M.U.J.O), UC Denver AMC Molecular Biology Program T32 training grant (C.G.T), NIH-RO1 Diversity Supplement to R01-CA157790 (C.G.T), the UNCF/MERCK Graduate Fellowship (C.G.T), and the T32 training grant CA190216-1 A1 (C.G.T)
Footnotes
Conflict of Interest: The authors declare no conflict of interest.
Supplementary information accompanies the paper on the Oncogene website (http://www.nature.com/onc).
References
- 1.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100(7):3983–8. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Visvader JE. Cells of origin in cancer. Nature. 2011;469(7330):314–22. doi: 10.1038/nature09781. [DOI] [PubMed] [Google Scholar]
- 3.Ginestier C, Hur MH, Charafe-Jauffret E, Monville F, Dutcher J, Brown M, et al. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell. 2007;1(5):555–67. doi: 10.1016/j.stem.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brooks MD, Burness ML, Wicha MS. Therapeutic Implications of Cellular Heterogeneity and Plasticity in Breast Cancer. Cell Stem Cell. 2015;17(3):260–71. doi: 10.1016/j.stem.2015.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang F, Xu J, Tang L, Guan X. Breast cancer stem cell: the roles and therapeutic implications. Cell Mol Life Sci. 2016 doi: 10.1007/s00018-016-2334-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pires BR, IS DEA, Souza LD, Rodrigues JA, Mencalha AL. Targeting Cellular Signaling Pathways in Breast Cancer Stem Cells and its Implication for Cancer Treatment. Anticancer Res. 2016;36(11):5681–91. doi: 10.21873/anticanres.11151. [DOI] [PubMed] [Google Scholar]
- 7.Pursglove SE, Mackay JP. CSL: a notch above the rest. Int J Biochem Cell Biol. 2005;37(12):2472–7. doi: 10.1016/j.biocel.2005.06.013. [DOI] [PubMed] [Google Scholar]
- 8.Capaccione KM, Pine SR. The Notch signaling pathway as a mediator of tumor survival. Carcinogenesis. 2013;34(7):1420–30. doi: 10.1093/carcin/bgt127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ellisen LW, Bird J, West DC, Soreng AL, Reynolds TC, Smith SD, et al. TAN-1, the human homolog of the Drosophila notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasms. Cell. 1991;66(4):649–61. doi: 10.1016/0092-8674(91)90111-b. [DOI] [PubMed] [Google Scholar]
- 10.Espinoza I, Miele L. Notch inhibitors for cancer treatment. Pharmacol Ther. 2013;139(2):95–110. doi: 10.1016/j.pharmthera.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hu C, Dievart A, Lupien M, Calvo E, Tremblay G, Jolicoeur P. Overexpression of activated murine Notch1 and Notch3 in transgenic mice blocks mammary gland development and induces mammary tumors. Am J Pathol. 2006;168(3):973–90. doi: 10.2353/ajpath.2006.050416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ntziachristos P, Lim JS, Sage J, Aifantis I. From fly wings to targeted cancer therapies: a centennial for notch signaling. Cancer Cell. 2014;25(3):318–34. doi: 10.1016/j.ccr.2014.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Petrocca F, Visone R, Onelli MR, Shah MH, Nicoloso MS, de Martino I, et al. E2F1-regulated microRNAs impair TGFbeta-dependent cell-cycle arrest and apoptosis in gastric cancer. Cancer Cell. 2008;13(3):272–86. doi: 10.1016/j.ccr.2008.02.013. [DOI] [PubMed] [Google Scholar]
- 14.Li Y, Tan W, Neo TW, Aung MO, Wasser S, Lim SG, et al. Role of the miR-106b-25 microRNA cluster in hepatocellular carcinoma. Cancer Sci. 2009;100(7):1234–42. doi: 10.1111/j.1349-7006.2009.01164.x. [DOI] [PubMed] [Google Scholar]
- 15.Kan T, Sato F, Ito T, Matsumura N, David S, Cheng Y, et al. The miR-106b-25 polycistron, activated by genomic amplification, functions as an oncogene by suppressing p21 and Bim. Gastroenterology. 2009;136(5):1689–700. doi: 10.1053/j.gastro.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Poliseno L, Salmena L, Riccardi L, Fornari A, Song MS, Hobbs RM, et al. Identification of the miR-106b~25 microRNA cluster as a proto-oncogenic PTEN-targeting intron that cooperates with its host gene MCM7 in transformation. Sci Signal. 2010;3(117):ra29. doi: 10.1126/scisignal.2000594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lo Sardo F, Forcato M, Sacconi A, Capaci V, Zanconato F, Di Agostino S, et al. MCM7 and its hosted miR-25, 93 and 106b cluster elicit YAP/TAZ oncogenic activity in lung cancer. Carcinogenesis. 2017;38(1):64–75. doi: 10.1093/carcin/bgw110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang R, Li F, Wang W, Wang X, Li S, Liu J. The effect of antisense inhibitor of miRNA 106b approximately 25 on the proliferation, invasion, migration, and apoptosis of gastric cancer cell. Tumour Biol. 2016;37(8):10507–15. doi: 10.1007/s13277-016-4937-x. [DOI] [PubMed] [Google Scholar]
- 19.Choi N, Park J, Lee JS, Yoe J, Park GY, Kim E, et al. miR-93/miR-106b/miR-375-CIC-CRABP1: a novel regulatory axis in prostate cancer progression. Oncotarget. 2015;6(27):23533–47. doi: 10.18632/oncotarget.4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith AL, Iwanaga R, Drasin DJ, Micalizzi DS, Vartuli RL, Tan AC, et al. The miR-106b-25 cluster targets Smad7, activates TGF-beta signaling, and induces EMT and tumor initiating cell characteristics downstream of Six1 in human breast cancer. Oncogene. 2012;31(50):5162–71. doi: 10.1038/onc.2012.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brett JO, Renault VM, Rafalski VA, Webb AE, Brunet A. The microRNA cluster miR-106b~25 regulates adult neural stem/progenitor cell proliferation and neuronal differentiation. Aging (Albany NY) 2011;3(2):108–24. doi: 10.18632/aging.100285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu D, Shin HS, Lee YS, Lee YC. miR-106b modulates cancer stem cell characteristics through TGF-beta/Smad signaling in CD44-positive gastric cancer cells. Lab Invest. 2014;94(12):1370–81. doi: 10.1038/labinvest.2014.125. [DOI] [PubMed] [Google Scholar]
- 23.Qian S, Ding JY, Xie R, An JH, Ao XJ, Zhao ZG, et al. MicroRNA expression profile of bronchioalveolar stem cells from mouse lung. Biochem Biophys Res Commun. 2008;377(2):668–73. doi: 10.1016/j.bbrc.2008.10.052. [DOI] [PubMed] [Google Scholar]
- 24.Liu S, Patel SH, Ginestier C, Ibarra I, Martin-Trevino R, Bai S, et al. MicroRNA93 regulates proliferation and differentiation of normal and malignant breast stem cells. PLoS Genet. 2012;8(6):e1002751. doi: 10.1371/journal.pgen.1002751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Z, Li Y, Banerjee S, Sarkar FH. Emerging role of Notch in stem cells and cancer. Cancer Lett. 2009;279(1):8–12. doi: 10.1016/j.canlet.2008.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137(2):216–33. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Geling A, Steiner H, Willem M, Bally-Cuif L, Haass C. A gamma-secretase inhibitor blocks Notch signaling in vivo and causes a severe neurogenic phenotype in zebrafish. EMBO Rep. 2002;3(7):688–94. doi: 10.1093/embo-reports/kvf124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goel P, Manning JA, Kumar S. NEDD4-2 (NEDD4L): the ubiquitin ligase for multiple membrane proteins. Gene. 2015;557(1):1–10. doi: 10.1016/j.gene.2014.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chastagner P, Israel A, Brou C. AIP4/Itch regulates Notch receptor degradation in the absence of ligand. PLoS One. 2008;3(7):e2735. doi: 10.1371/journal.pone.0002735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sakata T, Sakaguchi H, Tsuda L, Higashitani A, Aigaki T, Matsuno K, et al. Drosophila Nedd4 regulates endocytosis of notch and suppresses its ligand-independent activation. Curr Biol. 2004;14(24):2228–36. doi: 10.1016/j.cub.2004.12.028. [DOI] [PubMed] [Google Scholar]
- 31.Schrijver WA, van Diest PJ, Dutch Distant Breast Cancer Metastases C. Moelans CB. Unravelling site-specific breast cancer metastasis: a microRNA expression profiling study. Oncotarget. 2017;8(2):3111–23. doi: 10.18632/oncotarget.13623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gravgaard KH, Lyng MB, Laenkholm AV, Sokilde R, Nielsen BS, Litman T, et al. The miRNA-200 family and miRNA-9 exhibit differential expression in primary versus corresponding metastatic tissue in breast cancer. Breast Cancer Res Treat. 2012;134(1):207–17. doi: 10.1007/s10549-012-1969-9. [DOI] [PubMed] [Google Scholar]
- 33.Shyamasundar S, Lim JP, Bay BH. miR-93 inhibits the invasive potential of triple-negative breast cancer cells in vitro via protein kinase WNK1. Int J Oncol. 2016;49(6):2629–36. doi: 10.3892/ijo.2016.3761. [DOI] [PubMed] [Google Scholar]
- 34.Liu Y, Zhang J, Sun X, Li M. EMMPRIN Down-regulating miR-106a/b Modifies Breast Cancer Stem-like Cell Properties via Interaction with Fibroblasts Through STAT3 and HIF-1alpha. Sci Rep. 2016;6:28329. doi: 10.1038/srep28329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ni X, Xia T, Zhao Y, Zhou W, Wu N, Liu X, et al. Downregulation of miR-106b induced breast cancer cell invasion and motility in association with overexpression of matrix metalloproteinase 2. Cancer Sci. 2014;105(1):18–25. doi: 10.1111/cas.12309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu X, Liu Z, Wang J, Ling Q, Xie H, Guo H, et al. miRNA profiles in livers with different mass deficits after partial hepatectomy and miR-106b~25 cluster accelerating hepatocyte proliferation in rats. Sci Rep. 2016;6:31267. doi: 10.1038/srep31267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yen CS, Su ZR, Lee YP, Liu IT, Yen CJ. miR-106b promotes cancer progression in hepatitis B virus-associated hepatocellular carcinoma. World J Gastroenterol. 2016;22(22):5183–92. doi: 10.3748/wjg.v22.i22.5183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen S, Chen X, Sun KX, Xiu YL, Liu BL, Feng MX, et al. MicroRNA-93 Promotes Epithelial-Mesenchymal Transition of Endometrial Carcinoma Cells. PLoS One. 2016;11(11):e0165776. doi: 10.1371/journal.pone.0165776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li C, Ding C, Chen T, Chen J, Xu Z, Lei Z, et al. Micro ribonucleic acid-93 promotes proliferation and migration of esophageal squamous cell carcinoma by targeting disabled 2. Thorac Cancer. 2015;6(4):524–33. doi: 10.1111/1759-7714.12242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Espinoza I, Miele L. Deadly crosstalk: Notch signaling at the intersection of EMT and cancer stem cells. Cancer Lett. 2013;341(1):41–5. doi: 10.1016/j.canlet.2013.08.027. [DOI] [PubMed] [Google Scholar]
- 41.Koch U, Lehal R, Radtke F. Stem cells living with a Notch. Development. 2013;140(4):689–704. doi: 10.1242/dev.080614. [DOI] [PubMed] [Google Scholar]
- 42.Sansone P, Storci G, Giovannini C, Pandolfi S, Pianetti S, Taffurelli M, et al. p66Shc/Notch-3 interplay controls self-renewal and hypoxia survival in human stem/progenitor cells of the mammary gland expanded in vitro as mammospheres. Stem Cells. 2007;25(3):807–15. doi: 10.1634/stemcells.2006-0442. [DOI] [PubMed] [Google Scholar]
- 43.Qu MH, Han C, Srivastava AK, Cui T, Zou N, Gao ZQ, et al. miR-93 promotes TGF-beta-induced epithelial-to-mesenchymal transition through downregulation of NEDD4L in lung cancer cells. Tumour Biol. 2016;37(4):5645–51. doi: 10.1007/s13277-015-4328-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mazaleyrat SL, Fostier M, Wilkin MB, Aslam H, Evans DA, Cornell M, et al. Down-regulation of Notch target gene expression by Suppressor of deltex. Dev Biol. 2003;255(2):363–72. doi: 10.1016/s0012-1606(02)00086-6. [DOI] [PubMed] [Google Scholar]
- 45.Yau WL, Lam CS, Ng L, Chow AK, Chan ST, Chan JY, et al. Over-expression of miR-106b promotes cell migration and metastasis in hepatocellular carcinoma by activating epithelial-mesenchymal transition process. PLoS One. 2013;8(3):e57882. doi: 10.1371/journal.pone.0057882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414(6859):105–11. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 47.Guo S, Liu M, Gonzalez-Perez RR. Role of Notch and its oncogenic signaling crosstalk in breast cancer. Biochim Biophys Acta. 2011;1815(2):197–213. doi: 10.1016/j.bbcan.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guo X, Wang XF. Signaling cross-talk between TGF-beta/BMP and other pathways. Cell Res. 2009;19(1):71–88. doi: 10.1038/cr.2008.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kolch W, Halasz M, Granovskaya M, Kholodenko BN. The dynamic control of signal transduction networks in cancer cells. Nat Rev Cancer. 2015;15(9):515–27. doi: 10.1038/nrc3983. [DOI] [PubMed] [Google Scholar]
- 50.Gong C, Qu S, Liu B, Pan S, Jiao Y, Nie Y, et al. MiR-106b expression determines the proliferation paradox of TGF-beta in breast cancer cells. Oncogene. 2015;34(1):84–93. doi: 10.1038/onc.2013.525. [DOI] [PubMed] [Google Scholar]
- 51.Gao S, Alarcon C, Sapkota G, Rahman S, Chen PY, Goerner N, et al. Ubiquitin ligase Nedd4L targets activated Smad2/3 to limit TGF-beta signaling. Mol Cell. 2009;36(3):457–68. doi: 10.1016/j.molcel.2009.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Micalizzi DS, Christensen KL, Jedlicka P, Coletta RD, Baron AE, Harrell JC, et al. The Six1 homeoprotein induces human mammary carcinoma cells to undergo epithelial-mesenchymal transition and metastasis in mice through increasing TGF-beta signaling. J Clin Invest. 2009;119(9):2678–90. doi: 10.1172/JCI37815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Farabaugh SM, Micalizzi DS, Jedlicka P, Zhao R, Ford HL. Eya2 is required to mediate the pro-metastatic functions of Six1 via the induction of TGF-beta signaling, epithelial-mesenchymal transition, and cancer stem cell properties. Oncogene. 2012;31(5):552–62. doi: 10.1038/onc.2011.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


