Abstract
Puberty involves a series of morphological, physiological and behavioral changes during the last part of the juvenile period that culminates in the attainment of fertility. The activation of the pituitary-gonadal axis by increased hypothalamic secretion of Gonadotropin Releasing Hormone (GnRH) is an essential step in the process. The current hypothesis postulates that a loss of transsynaptic inhibition together with a rise in excitatory inputs are responsible for the activation of GnRH release. Similarly, a shift in the balance in the expression of puberty activating (PA) and puberty inhibitory (PI) genes exists during the pubertal transition. In addition, recent evidence suggests that the epigenetic machinery controls this genetic balance, giving rise to the tantalizing possibility that epigenetics serves as a relay of environmental signals known for many years to modulate pubertal development. Here we review the contribution of epigenetics as a regulatory mechanism in the hypothalamic control of female puberty.
Keywords: GnRH neurons, KNDy neurons, kisspeptin neurons, arcuate nucleus, female puberty, chromatin modifications, transcriptional repression, transcriptional activation, epigenetic regulators, Polycomb group, Trithorax complex
Introduction
Puberty is the process through which a child maturates into an adult capable of sexual reproduction. This process is accompanied by body growth as well as development of secondary sexual characteristics [1]. Form a purely neuroendocrine perspective, the onset of puberty requires the activation of hypothalamic secretory neurons that produce and release GnRH [2] in pulsatile fashion. Pulsatile GnRH release activity depends on excitatory and/or inhibitory transsynaptic and glial inputs originated from either neuronal subsets or glial cells interconnected to the GnRH neuronal network [2–5].
For a long time, studies have acknowledged that puberty has a strong genetic component and in recent years, epigenetics has been recognized as a key regulatory mechanism by which GnRH release is first kept in place before puberty, and later it becomes in charge of modulating the increased GnRH secretion in order to initiate the pubertal process. In this review, we will discuss the concept supporting epigenetics as a fundamental mechanism providing gene-specific gatekeeper functions [6] and the plasticity needed to temporary modify gene expression [7;8] during pubertal development. It is well-known that these mechanisms are essential to an adequate development and differentiation of distinct cell lineages in the organism. These processes can also be modified by exogenous influences, and as such, can contribute to or be result of the environmental alterations of the phenotype. Epigenetic programing has a decisive role in modulating pluripotency genes that become inactive during differentiation [9–11]. This machinery is also indispensable for a number of neural processes, including sexual differentiation of the brain [12;13], learning and memory formation [7;14], neuronal plasticity [15], dendritic development [16], and glial-neuronal interactions [17]. In addition, we will focus our discussion on the emerging evidence that supports the idea of a dynamic epigenetic machinery that regulates the neuroendocrine core process controlling the initiation of female puberty.
Neuroendocrine regulation of puberty: Neuronal circuits, GnRH pulsatility and LH release
In humans and non-human primates, during early postnatal life, Luteinizing Hormone (LH) pulsatility is high in a period called “mini puberty” [18;19]. In rodents, mini puberty is manifested by a transient increase in testosterone during early postnatal hours [20;21] in males. Whereas, in female rats and mice there is a transient GnRH dependent increase in Follicle-Stimulating Hormone (FSH) release around postnatal day 12, responsible for enhancing the development of preantral follicles as well as rescuing early antral follicles from apoptotic death [22–26]. This phase is followed by infancy, characterized by a marked quiescence of the gonadotropic axis. During the late juvenile period in rodents, or early pubertal period in humans and primates, there is an awakening of the GnRH pulse generator and therefore increased LH pulsatility [18;19].
It is well established that the first endocrine manifestation of puberty in humans is an increase in nocturnal pulsatile frequency of plasma LH, detected during the early stages of pubertal development [27]. Subsequent studies on other mammalian species have also evidenced an increase of pulsatile patterns of LH although during daytime, this is the case for nonhuman primates, rodents and sheep [2].
GnRH neurons are controlled by counteracting excitatory and inhibitory inputs that, within defined areas of the hypothalamus, regulate GnRH secretion. Strong excitatory inputs to GnRH release are provided by glutamatergic neurons [2;3] as well as a family of peptides known as kisspeptins [28–31]. These peptides are originated by a same kisspeptin precursor which are the product of the KiSS-1 Metastasis-Suppressor (KISS1/Kiss1) gene [32;33]. The presence and function of kisspeptins is vital for puberty to occur [34]. On the other hand, inhibitory transsynaptic regulation of GnRH neurons is mostly provided by gamma-aminobutyric acid (GABAergic) neurons [35;36], opiatergic neurons [35], and arginine and amidated phenylalanine (RFamide)-related peptide-containing neurons [37;38]. Finally, glial cells also collaborate to the hypothalamic control of puberty [4;5], where astrocytes and tanycytes expedite GnRH secretion by i) releasing small molecules like adenosine triphosphate (ATP), estradiol (E2), prostaglandin E2 (PGE2), and growth factors, and ii) via cell-cell interactions which require direct glia-GnRH neuron contact [5;39;40].
The transsynaptic mechanisms underlying GnRH pulsatility proceeds from the coordinated activities of a set of neurons located in the Arcuate Nucleus (ARC) of the hypothalamus. These neurons are known as KNDy because they secrete kispeptin, neurokinin B (NKB) and dynorphin [41;42]. While NKB stimulates kisspeptin release from other KNDy neurons in a positive feedback loop, the phase shifted inhibitory action of dynorphin on KNDy neurons produces an oscillatory behavior [42;43].
Besides the ARC, other populations of kisspeptin neurons have been found in the rostral periventricular area of humans and rodents, and in the anteroventral periventricular nucleus (AVPV) of rodents [44–46]. Kisspeptin neurons in the ovine brain show a similar pattern of distribution [47]. These kisspeptin neurons do not produce NKB or dynorphin and they do not contribute to the modulation of GnRH pulsatility, but they are necessary for the preovulatory surge of gonadotropins after the pubertal process has started [48].
The initiation of puberty takes place after the removal of a central restraint that keeps GnRH release in check [49–52]. Up to this point, during the prepubertal or infantile period, the secretory activity of GnRH neurons is under a predominant transsynaptic inhibitory control. Once puberty is underway, the inhibition lock is lifted, there is a concomitant increase in excitatory inputs to the GnRH network provided in part by activated kisspeptin neurons [48;53], ultimately resulting in increased GnRH release. Overall, it is now accepted that a joint decrease in inhibitory inputs together with an increase in excitatory neurotransmission are essential antagonistic mechanisms through which the pubertal process gets initiated [54–65]. Recent evidence shows that this inter-cellular mechanism of excitatory vs. inhibitory balance is mirrored at the genomic level. Three gene groups have emerged: as either PI genes (e.g. GAD67 [35;36], PDYN [35], GnIH [37;38]), PA genes (e.g. GnRH [66;67], GLS [2;3], KISS1 [32;33], TTF1 [68;69]) or genes with dual effects depending on hormonal milieu and cell identity (e.g.TAC3 [42;70–72], EAP1 [73;74]).
Decoding pubertal gene networks
The well-organized output of biological signals that influence the time of puberty are heavily affected by a variety of genes that work in coordinated networks operating in the hypothalamus. The Tumor Related Genes (TRG) is a good example of a network of genes operating within the hypothalamus, that regardless of bearing a wide-range of cellular functions, are all related to tumor suppression or tumor formation [75]. In silico studies in rats and non-human primates have suggested that TRGs show a well-defined network structure with central node members, named hubs, which are the core components, and a number of subordinated genes placed peripherally that are transcriptionally regulated by the central nodes. The five putative central nodes of the TRG are strongly connected between them and to other upper-echelon TRG genes (EAP1, TTF1 and OCT2) suggested also to be implicated in the transcriptional regulation of the pubertal process [68;73;76]. Experimental validation has been demonstrated for the upper-echelon gene EAP1 [77], which is part of a repressive complex that modulates apoptosis in breast cancer [78], and for the subordinate gene KISS1 [79] previously known as a metastasis suppressor gene [80]. TSLC1 (also known as SynCAM1) has an important role in glia-GnRH neuron adhesive communication during development, and also in astrocytic facilitation of GnRH secretion towards a correct reproductive function [81;82].
Other transcriptional gene networks related to puberty have been described in cattle [83]. Network connectivity analysis of core components have shown similar TRG-related target genes like NELL2 and NRG1, the latter encoding for an astrocytic erbB4 receptor ligand. The Trithorax group (TrxG) member Mll3 was identified as part of the top 10% most connected transcription factors during bovine puberty [83]. Mll3, expressed in the bovine hypothalamus, serves as a central hub by regulating the expression of 39 target genes [83].
As new gene interactions are unveiled, new network models become available and modified according to the current information. For example, a Genome Wide Association Study (GWAS) performed on a cohort of Japanese population discovered a Single-Nucleotide Polymorphism (SNP) associated to precocious menarche. This SNP was near the TTF1 locus, known as Nkx2.1 [84]. Moreover, network connectivity analysis showed that TTF1 and EAP1 serve as central hubs to a number of genes associated to menarche [85–87] including: GAB2, NR4A2, ZNF462, ZNF483, TMEM18, PAX8, KDM3B, KISS1 and KISS1R (for an extended gene list see [69]). These menarche-related genes are connected to two or more TRG central nodes as determined by GeneMANIA network software [88] and available databases. Genes connected to more than one TRG become potential regulators of gene expression. This is the case of lysine demethylase KDM3B, which is implicated in the removal of repressive histone marks, monomethylation and dimethylation of histone 3 at lysine 9 (H3K9me1/me2) [89;90]. Overall, these studies show the existence of strong interrelationships in expression between TRG central nodes, puberty genes, and menarche-related genes discovered using different platforms. Notwithstanding the possible role of these gene regulatory networks on pubertal development, evidence is accumulating suggesting an epigenetic layer of regulation of the neurons involved in stimulating GnRH release.
Mechanisms of epigenetic control
Epigenetics refers to hereditable modifications that are not due to changes in the DNA sequence. These alterations ultimately modify the DNA accessibility and chromatin structure, thus controlling different patterns of gene expression. There are three well-known mechanisms of epigenetic regulation: First, by chemical alterations of the DNA via DNA methylation and hydroxymethylation. Second, by modifications of the chromatin structure caused by posttranslational modifications (PTMs) of histones, which carry the protein component of the nucleosome, the core unit of chromatin. The third mechanism of epigenetic control is through non-coding RNAs (ncRNAs), providing epigenetic information as either microRNAs (miRNAs) or as long-intergenic noncoding RNAs (lincRNAs).
DNA methylation and hydroxymethylation
The predominant epigenetic tag in the DNA of differentiated mammalian cells is the covalent attachment of a methyl group to the C5 position of cytosine residues. This modification targets the dinucleotide sequence CpG [91;92]. DNA is methylated by DNA methyltransferases (DNMTs), resulting in the formation of 5-methylcytosine (5-mC). On the other hand, oxidation of 5-mC by the TET family of dio-oxygenase enzymes yields 5-hydroxymethylcytosine (5-hmC) [93;94]. Overall, increased abundance of 5-mc DNA is associated with transcriptional repression, whereas hypomethylation (i.e. less 5-mc and more 5-hmc) is linked with the activation of gene transcription [95;96]. The equilibrium of both tags at a defined genomic region will depend on DNMTs and TET enzymes activity. While DNMT1 is responsible of maintaining the basal levels of DNA methylation, DNMT3A and DNMT3B are in charge for de novo methylation of DNA that is either unmethylated or hemimethylated [91]. It’s important to emphasize that both tags, 5-mc and 5-hmc, occur simultaneously throughout the genome. However, while 5-mc is more abundant in silenced genes and compacted chromosomal regions that display heterochromatin (i.e. closed-state or condensed chromatin), 5-hmc is associated with more accessible regions or euchromatin (i.e. open-state or exposed chromatin) at enriched promoter and/or enhancer regions of active genes [95].
Histone Post-Translational Modifications (PTMs)
Histones can be post-translationally modified to restructure the chromatin in multiple ways. PTMs occur on N-terminal tails of histones (H2A, H2B, H3, and H4) that make up the core of the nucleosome surrounded by two superhelix turns of DNA [97;98]. These tails are the most accessible region of the histone proteins and can be subjected to acetylation, methylation, phosphorylation, ubiquitination, and sumoylation among others [97]. PTMs can create new recruitment sites for specific factors, or alter the existing ones in order to eliminate previous interactions [99]. In addition, PTMs are capable to recruit enzymes that can ‘write’, ‘erase’ or ‘read’ modifications. There is a large repertoire of such histone modifiers (around 150 enzymes in humans) [100].
Acetylation and methylation of lysine at histone tails are the two most common PTMs, with distinct distributions along both heterochromatin and euchromatin [101]. Generally, acetylation is related to activation of gene transcription, and deacetylation with transcriptional repression [97]. Lysine acetylation is mediated by histone acetyltransferase enzymes (HATs) and deacetylation by histone deacetylases (HDACs) [97;102;103]. The positive charge of lysine residues binds strongly to the negatively charged DNA condensing nucleosomes and making chromatin inaccessible to transcription factors. Acetylation removes positive charges, reducing the affinity between DNA and histones, and exposing the compacted chromatin to give the transcriptional machinery an easier access to promoter/enhancer regions [102]. In contrast, histone methylation is related to either transcriptional activation or repression depending on where methylation takes place [101]. Different lysine residues can by modified by adding one, two or three methyl groups. The effect on the chromatin state and gene function will depend on the specific lysine modified and its degree of methylation. In general, methylation of lysine 9 and 27 of H3 (H3K9me and H3K27me) is associated with transcriptional quiescence, whereas trimethylation of H3 at lysine 4 (H3K4me3) is mostly present at active promoter and enhancer regions [97;104].
Noncoding RNAs as epigenetic modulators
Contrary to former impressions, recent studies have showed that most of the human genome is transcribed into ncRNAs, instead of protein-encoding messenger (m)RNAs [105;106]. An astonishing variety of biological functions are accomplished by ncRNAs: they regulate gene expression at the levels of transcription, RNA processing and translation [107]. ncRNAs are classified in two main groups: small RNAs (sRNAs) and long noncoding RNAs (lncRNAs). sRNAs are 20–30 nucleotides long and lncRNAs are longer than 200 nucleotides [106;108]. Three types of sRNAs can be found: miRNAs [105;108], endo-small inhibitory RNAs (endo siRNAs) [109], and piwiRNAs (piRNAs) [110;111], all of them are implicated in epigenetic silencing [105]. The most well known in terms of epigenetic regulation are the miRNAs [112], since they target mRNAs encoding proteins required for both DNA methylation and histone PTMs [112]. Some of the miRNA targets include DNMTs, TETs, EZH2 (enzyme responsible for the H3K27me3 mark), and HDAC1, 4 and 6 [112].
The lncRNAs epigenetic contribution is reasonably more complex. They undergo polyadenylation despite they do not code for any protein [106], and most of them, named lincRNAs, are generated from long gene-free (i.e. intergenic) regions within the genome. These lincRNAs bind to chromatin-modifying complexes and guide them to genomic regions implicated in the control of gene expression [106;113].
Thousands of lincRNAs have been epigenetically identified in the genome because of two signature marks, H3K4me3 at regulatory regions of genes transcribed by RNApol II, together with the trimethylation of H3 at lysine 36 (H3K36me3) throughout the transcribed segment. Both marks result in a chromatin configuration known as bivalency [114].
Epigenetic mechanisms behind the hypothalamic control of reproductive development
Recent studies have highlighted the contribution of epigenetics at three levels of hypothalamic regulation; throughout the development of the GnRH neuronal network, within the AVPV kisspeptin neurons responsible of the preovulatory surge of gonadotropins and in ARC KNDy neurons responsible for the control of pulsatile GnRH secretion (Summarized in Table 1).
Table 1.
Epigenetic enzymes characterized in the hypothalamic control of pubertal development.
| Enzyme | Function | Epigenetic mark | Target | Effect on normal puberty | Reference |
|---|---|---|---|---|---|
| DNMTs | Writer | CpG methylation | GnRH | Repression | [117] |
| DNMTs | Writer | CpG methylation | Cbx7 and Eed promoters | Activation | [123] |
| DICER | miRNA’s processing | miR-200 miR-155 |
Zeb1/GnRH Cebpb |
Activation | [122] |
| Polycomb (EED and CBX7) | Writer | H3K27me3 | Kiss1 promoter | Repression | [123] |
| GATAD1 | Reader | H3K4me2 | Kiss1 promoter | Repression | [134] |
| KDM1 | Eraser | H3K4me2 | Kiss1 promoter | Repression | [134] |
| MLL1 | Writer | H3K4me3 | Kiss1 promoter | Activation | [128] |
| HATs | Writer | H3K9ac H3K14ac |
Kiss1 promoter | Activation | [123] |
| MLL3 | Writer | H3K4me3 | Kiss1 enhancer | Activation | [128] |
| P300/CBP | Writer | H3K27ac | Kiss1 enhancer | Activation | [128] |
Sexual differentiation of the hypothalamus
Neuronal sex differences take place at many levels and are determined by steroid hormones. Testosterone from the gonads, converted into estradiol (E2) in the brain, is the main factor causing sexual differentiation, affecting adult reproductive physiology and behavior. The development of the preoptic area (POA), involved in sexual and maternal behavior, is strongly coordinated by E2. A number of epigenetic modifications within the POA, mostly DNA methylation and histone PTM patterns of the estrogen receptor alpha (ERα) gene have shown to influence the estrogen-dependent sexual differentiation of this region in rodents [115]. While a segment of the ERα promoter displays a pattern of DNA methylation throughout development that does not correlate with ERα gene expression [116], another segment exhibits DNA methylation with a good correlation [117], suggesting that overall changes are subtle and confined, and only limited to some CpGs in specific portions of the 5′ flanking regulatory region of ERα. On the other hand, histone PTMs have a more crucial role than DNA methylation regarding the masculinization of the POA, since brain-targeted inhibition of HDAC activity in neonatal rats show a reduced male behavior when adults [118]. In addition, HDAC2 and HDAC4, apparently in association with ERα, are required for proper male behavior.
The expression of Kiss1 in the AVPV area is sexually dimorphic [119]. Despite the fact that Kiss1 expression is considerable higher in females than males [12], its promoter methylation levels are significantly larger in females [12]. This difference, although counterintuitive, may reflect the DNA methylation ability to block the recruitment of repressive transcriptional factors under certain conditions [12]
GnRH neuron maturation: DNA methylation and miRNA processing
Whole-Genome Bisulfite Sequencing (WGBS) studies together with RNA-sequencing (RNA-seq) performed on goat hypothalamus support the concept that gene expression patterns, influenced by changes in DNA methylation, regulate the onset of puberty [120]. Several genomic regions, including promoters, introns and untranslated regions (UTRs) have shown significant differences in expression of methylation profiles throughout development [120]. In addition, studies performed in non-human primate cultured GnRH neurons evidenced a decrease in methylation levels at multiple specific CpGs upstream the GNRH transcription starting site (TSS) throughout development, suggesting that demethylation at regulatory regions of the GNRH gene removes the repressive influence facilitating GNRH transcription [121]. In fact, GnRH mRNA levels increase significantly between 14 and 18 days in vitro, when the peptide release reaches a maximum, coinciding with decreasing DNA methylation.
In an elegant study, Messina and colleagues [122] determined the role of miRNAs as epigenetic regulators of the pubertal onset and reproductive maturation. By crossing Dicer-flox with GnRH-Cre animals, the authors generated a GnRH specific Dicer knockout mouse model, in order to determine the role of miRNAs in GnRH neuron maturation and reproductive competence. These mice showed impaired miRNA synthesis that lead to reduced plasma LH and FSH, hypogonadotropic hypogonadism and infertility due to a loss in GnRH expression. These animals displayed a reduction in transcriptional activation of the Gnrh gene and an increase in Zeb-1 and Cebpp, two potent transcriptional repressors of Gnrh. Altogether, this study suggests that there is a multilayered miRNA-operated switch during the infantile-to-juvenile transition within GnRH neurons, where two key components, miR-200 and miR-155, regulate Zeb1, Cebpb, and Gnrh expression in order to modulate the balance between activating and repressive signals that ultimately regulate the neuroendocrine control of reproduction.
ARC Kiss1 and the onset of puberty
The presence of a transcriptional repressive mode of control regulating PA genes was first proposed by our studies of the TRG network whose central nodes were able to repress the transcriptional activity of Kiss1 [73;77]. Subsequently, conclusive evidence demonstrated that the Polycomb Group (PcG) of transcriptional repressors prevents the premature initiation of puberty by inhibiting Kiss1 transcription in KNDy neurons within the ARC [123]. Here, cDNA arrays and methylation array studies were performed on hypothalamic tissue during the prepubertal developmental phases in female rats. The three stages examined were: postnatal day (PND) 21 (early juvenile), PND 28 (late juvenile, the day when diurnal pulsatile LH is increased [124;125]) and PND 32–36 (late proestrus, the day of the first preovulatory surge of gonadotropins [126]). Using Kiss1 as a prototype, it was shown that two central members of the PcG, Eed and Cbx7, are expressed within ARC kisspeptin neurons, and their protein products are associated to the Kiss1 promoter during prepubertal development [123]. When the juvenile development period is approaching the end, the methylation of Eed and Cbx7 promoter regions increase in the ARC at the same time the expression of both genes get significantly reduced. Interestingly, as Eed and Cbx7 lower their expression, EED and CBX7 become evicted from the Kiss1 promoter in an estrogen independent manner. Importantly, the loss of PcG components from the promoter is accompanied with a reorganization of the chromatin status, showing increased levels of the epigenetic marks H3K9ac, H3K14ac and H3K4me3, which are modifications associated with gene activation. Finally, and as predicted by these epigenetic changes, Kiss1 mRNA expression is increased in the ARC.
The Kiss1 promoter contains a bivalent region where both repressive and activating marks coexist. This allows the promoter to be poised for activation in response to diverse incoming inputs [127]. Despite the fact that the content of repressive H3K27me3 mark does not decrease at the same speed as the activating H3K4me3 mark increases, this repressive mark diminishes on the day of the pre-ovulatory surge of gonadotropins [128].
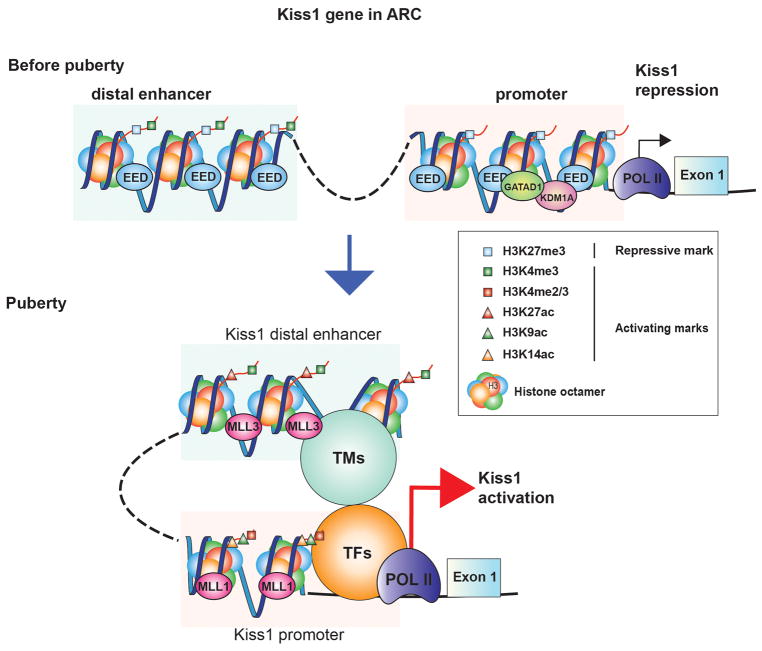
The fact that the Kiss1 promoter is subjected to increasing epigenetic-activating marks like trimethylation and acetylation of H3 during pre-pubertal development denotes the activity of an activating transcriptional complex occurring simultaneously to the loss of PcG inhibition. The candidates for this activating role are members of the TrxG complex due to its well-known PcG-antagonizing activity [129;130] (Figure 1).
Figure 1. The epigenetic control of Kiss1 expression in the ARC.
This model postulates that during prepubertal development the transcriptional activity of the Kiss1 gene is repressed by a series of repressors, exemplified by the PcG as well as the GATAD1/KDM1A complex. By depositing histone PTMs associated with gene silencing (e.g. H3K27me3, blue squares) at Kiss1 promoter and enhancer sites, PcG proteins (epitomized by EED) act as ‘writers’ of a repressive chromatin configuration. Furthermore, the GATAD1 ‘reader’ protein recruits KDM1A, the ‘eraser’ of H3K4me2 further repressing chromatin accessibility. As puberty advances, these ‘writers’ and ‘readers’ are removed from these regulatory regions, and as a consequence of this loss, the amount of H3K27me3 is reduced, while H3K4me2/3 increases. As this transformation takes place, another set of writers, typified by the TrxG-activating multiprotein complex is engaged causing the deposition of histone PTMs linked to transcriptional activation at two sites: a) H3K4me2/3 (red squares) and H3K9,14ac (green and orange triangles) are added at promoter regions and, b) H3K4me3 and H3K27ac (red triangles) at enhancer sites characterized by their high and sustained content in H3K4me1 (green squares). This chromatin looping also brings a series of unknown transcription factors (TFs) like pioneer, determining and collaborative TFs, as well as transcriptional mediators (TMs) like Histone Acetyltransferases, DNA demethylation enzymes, Chromosomal looping factors, RNA processing factors, non-coding RNAs, etc [153]. An outcome of these changes is that the activated enhancer is attracted to the promoter region enhancing polymerase II (POLII) transcriptional activity, thus increasing Kiss1 mRNA transcription.
MLL1 and MLL3, two members of the TrxG complex, acting on promoter and enhancer regions of the Kiss1 gene respectively, provide the trans-activational influence at the time when the inhibitory influence of the PcG complex is fading [128]. siRNA mediated ARC specific down regulation of Mll1 expression, inhibits Kiss1 expression and delays puberty [128]. Furthermore, the use of a CRISPR-CAS9 epigenetic remodeling approach that inhibits the action of Mll3 on the Kiss1 enhancer region, also blocks the peripubertal rise in Kiss1 expression and delays puberty [128]. In addition, the TrxG member UTX may assist the PcG eviction by demethylating the repressive histone H3K27me3 mark [131;132] allowing for an increase in H3K27ac, a hallmark of an active enhancer [128]. By implementing these essential key histone PTMs, the TrxG is capable of changing the epigenetic control of Kiss1 from a repressive to an active state around the time of puberty (Figure 1). A translational view for the role of TrxG in the control of puberty was evidenced by the fact that inactive mutations of CHD7, which in normal conditions antagonizes the PcG activity by binding to activating marks H3K4me2/me3 via its chromodomain, results in hypothalamic hypogonadism in humans [133].
Recently, our lab provided evidence that members of the zinc finger (ZNF) family of transcriptional repressors are also important components of the system that keeps the GnRH pulse generator in check during the pre-pubertal development, by preventing the premature re-awakening of GnRH secretion in non-human primates [134]. While KISS1 and TAC3 mRNA expression increases, GATAD1 expression decreases in the MBH of both ovary-intact females and agonadal male rhesus monkeys during the juvenile-pubertal transition. In addition, when overexpressing GATAD1 in the ARC of immature rats, the time of puberty becomes significantly delayed and the estrous cyclicity gets seriously compromised [134]. GATAD1, a well-known chromatin reader, recruits the histone eraser KDM1A, inducing the loss of activating H3K4me3/2 marks at gene regulatory regions [135;136]. Through in vitro studies, we showed that KDM1A recruitment increases when GATAD1 is overexpressed, and the association of H3K4me2 to both, KISS1 and TAC3 promoters gets significantly reduced. These results are consistent with the idea that GATAD1 attenuates KISS1 and TAC3 gene activity, in part by facilitating the loss of the H3K4me2 mark from PA gene promoters via gain of KDM1A (Figure 1). The decrease in GATAD1 and KDM1A association to the KISS1 and TAC3 promoters and the concomitant increase in H3K4me2 abundance observed in the monkey MBH at the time of puberty, strongly support the in vitro results and the notion that there is a mechanism of epigenetic repression that is lifted during the re-awakening of the GnRH pulsatility determined in the monkey infantile-juvenile transition.
Kiss1 activation and the preovulatory surge of gonadotrophins
It is known that both populations of kisspeptin neurons are differentially regulated by E2. While E2 inhibits Kiss1 expression in KNDy neurons of the ARC, it enhances its expression in kisspeptin neurons of the AVPV. The mechanisms involved in this differential effect are not yet understood. Recent evidence has showed that E2 has an epigenetic role in the control of kisspeptin neurons within the AVPV [137]. Here, E2 induces acetylation of H3 at the promoter region of Kiss1, increasing its expression. In contrast, the acetylation of H3 is significantly reduced in ARC Kiss1 regulatory region and therefore Kiss1 expression is reduced [137]. In addition, E2 induces ERα binding to the Kiss1 promoter exclusively in the AVPV area. Because no changes were observed in the DNA methylation pattern of Kiss1 promoter either in the AVPV or ARC, it is suggested that DNA methylation does not contribute as an epigenetic cue to the modulation of Kiss1 expression at the time when the preovulatory surge of gonadotrophins takes place. Surprisingly, an estrogen-responsive enhancer region located in the intergenic 3′ region of Kiss1 gene was discovered in kisspeptin neurons from the AVPV but not from the ARC [137]. Despite these results, unveiling an epigenetic contribution for E2 positive feedback in the AVPV, it is not clear whether there is a similar epigenetic mechanism for estrogen inhibitory action on ARC Kiss1 expression (Figure 2).
Figure 2. The epigenetic control of Kiss1 expression in the AVPV.
During prepubertal development the transcriptional activity of the Kiss1 gene in the AVPV is repressed. After ARC kisspeptin induced GnRH pulsatilitiy and thus LH release is increased, ovarian maturation takes place increasing estrogen production. When serum estrogen reaches pre-ovulatory levels, AVPV kisspeptin expression is induced trough ERα binding to ERE sites (green rectangles), enhancing Histone 3 acetylation (red triangles), as well as formation of 3D chromatin rearrangements bringing transcription factors (TFs) and transcriptional mediators (TMs) like: Histone Acetyltransferases, DNA demethylation enzymes, Chromosomal looping factors, RNA processing factors, non-coding RNAs, etc [153] in close proximity to the RNA polymerase 2 (Pol II).
Genomic imprinting and mutations at the Makorin 3 (MKRN3) gene
Recent studies [138–148], have found heterozygous loss of function mutations of MKRN3 in patients with central precocious puberty. Moreover, GWAS found that a specific SNP near the paternal allele of the MKRN3 gene is associated with earlier age at menarche [149;150]. MKRN3 is an intronless gene located in the Prader-Willy syndrome region of chromosome 15q11.2. and is maternally imprinted, meaning that patients inherit the mutation from their paternal allele [138]. A gender dimorphism appears evident since MKRN3 mutations seem to affect females more severely than males [138;139]. Mkrn3, which has only been identified in therian mammals, belongs to a highly conserved family of makorin genes [151]. Makorins are zinc finger proteins with C3HC4 motifs called RING domains associated with E3 ubiquitin ligase activity [151]. These enzymes are involved in transferring ubiquiting from an E2-ubiquiting-conjugating enzyme to target proteins. Makorins are also implicated in the regulation of RNA polymerase II activity, suggesting they could be involved in transcriptional and/or epigenetic modes of regulation [152]. The exact mechanism by which MKRN3 mutations affect the timing of puberty is currently unknown, but Mkrn3 is highly expressed in the ARC nucleus of juvenile mice, and its expression decreases as puberty progresses, suggesting that it functions as a strong repressor of the GnRH network [138]. More research is needed to verify the role of this gene or the involvement of genomic imprinting in the pathogenesis of central precocious puberty.
Conclusions
Nowadays it is well established that epigenetics plays a major role in the regulation of reproductive development and the initiation puberty. The interaction between epigenetic activation and repression of gene transcription seems to be the core of a process by which epigenetic mechanisms are able to modulate the development of the pubertal process. Much more remains to be unveiled in order to characterize components and comprehend signal pathways triggered by epigenetic mechanisms that regulate gene expression in a coordinated fashion throughout prepubertal neuroendocrine development. Moreover, studies that identify how epigenetic pathways convey information from a wide range of stimuli (e.g. nutrition, environment, circadian activity) to hypothalamic neurons regulating the onset of puberty are still in need. For that, it is important to emphasize that the epigenetic code, linking DNA methylation, histone PTMs and ncRNAs is extremely dynamic and likely to diverge from different cell populations and at different developmental times. New technologies and methods available, like the possibility of isolating specific cellular types from the neuroendocrine hypothalamus together with micro methods for genome-wide characterization of histone landscapes and/or DNA methylation profiles, will help determine the epigenetic status of defined cell populations at specific times of development, or after the exposure to stimuli.
While this research is still in its infancy, we have made great advances to the field by opening new avenues in our understanding of this complex and multilayered developmental process. We envision the future use of epigenetic biomarkers as a readout of normal developmental patterns that could also serve as early detectors of deranged pubertal development.
Acknowledgments
This work was supported by National Institute of Health (NIH) grants 1R01HD084542 to A.L. and 8P51OD011092 for the operation of the Oregon National Primate Research Center. C.A.T was supported by NIH grants F32-HD-86904, T32-HD-007133 and T32-DK-7680.
Acronyms used
- 5-mC
5-Methylcytosine
- 5-hmC
5-Hydroxymethylcytosine
- ARC
Arcuate Nucleus
- ATP
Adenosine Triphosphate
- AVPV
Anteroventral Periventricular Nucleus
- CBX7
Chromobox Homolog 7
- Cebpb
CCAAT/Enhancer-Binding Protein Beta
- CHD7
Chromodomain Helicase DNA Binding Protein 7
- DNA
Deoxyribonucleic Acid
- DNMT1
DNA Methyltransferase 1
- DNMT3A
DNA Methyltransferase 3A
- DNMT3B
DNA Methyltransferase 3B
- E2
Estradiol
- EAP1
Enhanced at Puberty 1
- EED
Embryonic Ectoderm Development
- ERα
Estrogen Receptor Alpha
- erbB4
Erb-B2 Receptor Tyrosine Kinase 4
- EZH2
Enhancer of Zeste 2
- FSH
Follicle-Stimulating Hormone
- GABA
Gamma-Aminobutyric Acid
- GAB2
Growth Factor Receptor Bound Protein 2-Associated Protein 2
- GAD67
Glutamate Decarboxylase 67
- GATAD1
GATA Zinc Finger Domain Containing 1
- GLS
Glutaminase
- GnIH
Gonadotropin-Inhibitory Hormone
- GnRH
Gonadotropin-Releasing Hormone
- GWAS
Genome Wide Association Study
- H2A
Histone 2A
- H2B
Histone 2B
- H3
Histone 3
- H4
Histone 4
- H3K4me3
Histone 3 Trimethylated at Lysine 4
- H3K9ac
Histone 3 Acetylated at Lysine 9
- H3K14ac
Histone 3 Acetylated at Lysine 14
- H3K9me1
Histone 3 Monomethylated at Lysine 9
- H3K9me2
Histone 3 Dimethylated at Lysine 9
- H3K27ac
Histone 3 Acetylated at Lysine 27
- H3K27me
Histone 3 Monomethylated at Lysine 27
- H3K36me3
Histone 3 Trimethylated at Lysine 36
- HATs
Histone Acetyltransferase Enzymes
- HDAC1
Histone Deacetylase 1
- HDAC2
Histone Deacetylase 2
- HDAC4
Histone Deacetylase 4
- HDAC6
Histone Deacetylase 6
- KDM1A
Lysine Demethylase 1A
- KDM3B
Lysine Demethylase 3B
- KISS1
KiSS-1 Metastasis-Suppressor
- KISS1R
KISS1 Receptor
- KNDy
Kisspeptin/Neuokinin B/Dynorphin
- LH
Luteinizing Hormone
- lncRNA
Long-Noncoding RNA
- lincRNAs
Long-Intergenic Noncoding RNA
- MKRN3
Makorin Ring Finger Protein 3
- mRNA
Messenger RNA
- miRNA
MicroRNA
- Mll1
Mixed-Lineage Leukemia 1
- Mll3
Mixed-Lineage Leukemia 3
- NELL2
Neural Epidermal Growth Factor-Like Like 2
- ncRNA
Non-Coding RNA
- NKB
Neurokinin B
- NR4A2
Nuclear Receptor Subfamily 4 Group A Member 2
- NRG1
Neuregulin-1
- OCT2
Organic Cation Transporter-2
- PA
Puberty Activating
- PAX8
Paired Box 8
- PDYN
Prodynorphin
- PGE2
Prostaglandin E2
- PI
Puberty Inhibitory
- piRNA
PiwiRNA
- PcG
Polycomb Group
- PND
Postnatal Day
- POA
Pre-Optic Area
- PTM
Posttranslational Modifications
- RFamide
Arginine and Amidated Phenylalanine
- RING
Really Interesting New Gene
- RNA
Ribonucleic Acid
- RNA-seq
RNA-sequencing
- RNApol II
RNA polymerase II
- SNP
Single-Nucleotide Polymorphism
- sRNA
Small RNA
- SynCAM
Synaptic Cell Adhesion Molecule
- TAC3
Tachykinin 3
- TET
Ten-Eleven Translocation Methylcytosine Dioxygenase
- TMEM18
Transmembrane Protein 18
- TRG
Tumor Related Genes
- TrxG
Trithorax Group
- TSLC1
Tumor-Suppressor Gene in Human Non-Small-Cell Lung Cancer
- TTF1
Transcription Termination Factor 1
- UTR
Untranslated Regions
- UTX
Ubiquitously Transcribed Tetratricopeptide Repeat-X
- WGBS
Whole-Genome Bisulfite Sequencing
- Zeb1
Zinc Finger E-Box Binding Homeobox 1
- ZNF462
Zinc Finger Protein 462
- ZNF483
Zinc Finger Protein 483
References
- 1.Plant TM. 60 YEARS OF NEUROENDOCRINOLOGY: The hypothalamo-pituitary-gonadal axis. J Endocrinol. 2015;226:T41–T54. doi: 10.1530/JOE-15-0113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ojeda SR, Skinner MK. Puberty in the rat. In: Neill JD, editor. The Physiology of Reproduction. 3. San Diego: Academic Press/Elsevier; 2006. pp. 2061–2126. [Google Scholar]
- 3.Plant TM, Witchel SF. Puberty in nonhuman primates and humans. In: Neill JD, editor. The Physiology of Reproduction. 3. San Diego: Academic Press/Elsevier; 2006. pp. 2177–2230. [Google Scholar]
- 4.Ojeda SR, Ma YJ, Dziedzic B, Prevot V. Astrocyte-neuron signaling and the onset of female puberty. In: Bourguignon J-P, Plant TM, editors. The Onset of Puberty in Perspective. Amsterdam: Elsevier Science B.V; 2000. pp. 41–57. [Google Scholar]
- 5.Prevot V. Glial-neuronal-endothelial interactions are involved in the control of GnRH secretion. J Neuroendocrinol. 2002;14:247–255. doi: 10.1046/j.0007-1331.2001.00772.x. [DOI] [PubMed] [Google Scholar]
- 6.Garcia-Bassets I, Kwon YS, Telese F, Prefontaine GG, Hutt KR, Cheng CS, Ju BG, Ohgi KA, Wang J, Escoubet-Lozach L, Rose DW, Glass CK, Fu XD, Rosenfeld MG. Histone methylation-dependent mechanisms impose ligand dependency for gene activation by nuclear receptors. Cell. 2007;128:505–518. doi: 10.1016/j.cell.2006.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miller CA, Sweatt JD. Covalent modification of DNA regulates memory formation. Neuron. 2007;53:857–869. doi: 10.1016/j.neuron.2007.02.022. [DOI] [PubMed] [Google Scholar]
- 8.Metivier R, Gallais R, Tiffoche C, Le PC, Jurkowska RZ, Carmouche RP, Ibberson D, Barath P, Demay F, Reid G, Benes V, Jeltsch A, Gannon F, Salbert G. Cyclical DNA methylation of a transcriptionally active promoter. Nature. 2008;452:45–50. doi: 10.1038/nature06544. [DOI] [PubMed] [Google Scholar]
- 9.Liu J, Casaccia P. Epigenetic regulation of oligodendrocyte identity. Trends Neurosci. 2010;33:193–201. doi: 10.1016/j.tins.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taranger CK, Noer A, Sorensen AL, Hakelien AM, Boquest AC, Collas P. Induction of dedifferentiation, genomewide transcriptional programming, and epigenetic reprogramming by extracts of carcinoma and embryonic stem cells. Mol Biol Cell. 2005;16:5719–5735. doi: 10.1091/mbc.E05-06-0572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tang WW, Kobayashi T, Irie N, Dietmann S, Surani MA. Specification and epigenetic programming of the human germ line. Nat Rev Genet. 2016;17:585–600. doi: 10.1038/nrg.2016.88. [DOI] [PubMed] [Google Scholar]
- 12.Semaan SJ, Dhamija S, Kim J, Ku EC, Kauffman AS. Assessment of epigenetic contributions to sexually-dimorphic kiss1 expression in the anteroventral periventricular nucleus of mice. Endocrinology. 2012;153:1875–1886. doi: 10.1210/en.2011-1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McCarthy MM, Auger AP, Bale TL, De Vries GJ, Dunn GA, Forger NG, Murray EK, Nugent BM, Schwarz JM, Wilson ME. The epigenetics of sex differences in the brain. J Neurosci. 2009;29:12815–12823. doi: 10.1523/JNEUROSCI.3331-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fischer A, Sananbenesi F, Wang X, Dobbin M, Tsai LH. Recovery of learning and memory is associated with chromatin remodelling. Nature. 2007;447:178–182. doi: 10.1038/nature05772. [DOI] [PubMed] [Google Scholar]
- 15.Kumar A, Choi KH, Renthal W, Tsankova NM, Theobald DE, Truong HT, Russo SJ, Laplant Q, Sasaki TS, Whistler KN, Neve RL, Self DW, Nestler EJ. Chromatin remodeling is a key mechanism underlying cocaine-induced plasticity in striatum. Neuron. 2005;48:303–314. doi: 10.1016/j.neuron.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 16.Wu JI, Lessard J, Olave IA, Qiu Z, Ghosh A, Graef IA, Crabtree GR. Regulation of dendritic development by neuron-specific chromatin remodeling complexes. Neuron. 2007;56:94–108. doi: 10.1016/j.neuron.2007.08.021. [DOI] [PubMed] [Google Scholar]
- 17.Shen S, Sandoval J, Swiss VA, Li J, Dupree J, Franklin RJM, Casaccia-Bonnefil P. Age-dependent epigenetic control of differentiation inhibitors is critical for remyelination efficiency. Nature Neuroscience. 2008;11:1024–1034. doi: 10.1038/nn.2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grumbach MM, Styne DM. Puberty: Ontogeny, neuroendocrinology, physiology, and disorders. In: Williams RH, Foster DW, Kroenenberg H, Larsen PR, Zorab R, editors. Williams Textbook of Endocrinology. 9. Philadelphia: W.B. Saunders; 1998. pp. 1509–1625. [Google Scholar]
- 19.Guatelli-Steinberg D, Boyce J. The postnatal endocrine surge and its effects on subsequent sexual growth. In: Preedy V, editor. Handbook of Growth and Growth Monitoring in Health and Disease. Springer; New York, NY: 2011. pp. 663–680. [Google Scholar]
- 20.Corbier P, Edwards DA, Roffi J. The neonatal testosterone surge: a comparative study. Arch Int Physiol Biochim Biophys. 1992;100:127–131. doi: 10.3109/13813459209035274. [DOI] [PubMed] [Google Scholar]
- 21.Li R, Vannitamby A, Yue SSK, Handelsman D, Hutson J. Mouse minipuberty coincides with gonocyte transformation into spermatogonial stem cells: a model for human minipuberty. Reprod Fertil Dev. 2017;29:2430–2436. doi: 10.1071/RD17100. [DOI] [PubMed] [Google Scholar]
- 22.Kragt CL, Dahlgren J. Development of neural regulation of follicle-stimulating hormone (FSH) secretion. Neuroendocrinology. 1972;9:30–40. doi: 10.1159/000122035. [DOI] [PubMed] [Google Scholar]
- 23.Dahl KD, Jia XC, Hsueh JW. Bioactive follicle-stimulating hormone levels in serum and urine of male and female rats from birth to prepubertal period. Biol Reprod. 1988;39:32–38. doi: 10.1095/biolreprod39.1.32. [DOI] [PubMed] [Google Scholar]
- 24.Dohler KD, Wuttke W. Changes with age in levels of serum gonadotropins, prolactin and gonadal steroids in prepubertal male and female rats. Endocrinology. 1975;97:898–907. doi: 10.1210/endo-97-4-898. [DOI] [PubMed] [Google Scholar]
- 25.Kamberi IA, de VJ, Bacleon ES, Inglish D. Hormonal patterns of the hypothalamo-pituitary-gonadal axis in the rat during postnatal development and sexual maturation. Endokrinologie. 1980;75:129–140. [PubMed] [Google Scholar]
- 26.McGee EA, Hsueh AJ. Initial and cyclic recruitment of ovarian follicles. Endocr Rev. 2000;21:200–214. doi: 10.1210/edrv.21.2.0394. [DOI] [PubMed] [Google Scholar]
- 27.Boyar R, Finkelstein J, Roffwarg H, Kapen S, Weitzman E, Hellman L. Synchronization of augmented luteinizing hormone secretion with sleep during puberty. N Engl J Med. 1972;287:582–586. doi: 10.1056/NEJM197209212871203. [DOI] [PubMed] [Google Scholar]
- 28.d’Anglemont dT X, Colledge WH. The role of kisspeptin signaling in reproduction. Physiology (Bethesda ) 2010;25:207–217. doi: 10.1152/physiol.00009.2010. [DOI] [PubMed] [Google Scholar]
- 29.Oakley AE, Clifton DK, Steiner RA. Kisspeptin signaling in the brain. Endocr Rev. 2009;30:713–743. doi: 10.1210/er.2009-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shahab M, Mastronardi C, Seminara SB, Crowley WF, Ojeda SR, Plant TM. Increased hypothalamic GPR54 signaling: A potential mechanism for initiation of puberty in primates. Proc Natl Acad Sci U S A. 2005;102:2129–2134. doi: 10.1073/pnas.0409822102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kauffman AS, Clifton DK, Steiner RA. Emerging ideas about kisspeptin-GPR54 signaling in the neuroendocrine regulation of reproduction. Trends Neurosci. 2007;30:504–511. doi: 10.1016/j.tins.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 32.Kotani M, Detheux M, Vandenbogaerde A, Communi D, Vanderwinden JM, Le Poul E, Brezillon S, Tyldesley R, Suarez-Huerta N, Vandeput F, Blanpain C, Schiffmann SN, Vassart G, Parmentier M. The metastasis suppressor gene KiSS-1 encodes kisspeptins, the natural ligands of the orphan G protein-coupled receptor GPR54. J Biol Chem. 2001;276:34631–34636. doi: 10.1074/jbc.M104847200. [DOI] [PubMed] [Google Scholar]
- 33.Ohtaki T, Shintani Y, Honda S, Matsumoto H, Hori A, Kanehashi K, Terao Y, Kumano S, Takatsu Y, Masuda Y, Ishibashi Y, Watanabe T, Asada M, Yamada T, Suenaga M, Kitada C, Usuki S, Kurokawa T, Onda H, Nishimura O. !Lost Data: Metastasis suppressor gene KiSS-1 encodes peptide ligand of a G-protein-coupled receptor. Nature. 2001;411:613–617. doi: 10.1038/35079135. [DOI] [PubMed] [Google Scholar]
- 34.Seminara SB, Messager S, Chatzidaki EE, Thresher RR, Acierno JS, Jr, Shagoury JK, Bo-Abbas Y, Kuohung W, Schwinof KM, Hendrick AG, Zahn D, Dixon J, Kaiser UB, Slaugenhaupt SA, Gusella JF, O’Rahilly S, Carlton MB, Crowley WF, Jr, Aparicio SA, Colledge WH. The GPR54 gene as a regulator of puberty. N Engl J Med. 2003;349:1614–1627. doi: 10.1056/NEJMoa035322. [DOI] [PubMed] [Google Scholar]
- 35.Terasawa E, Fernandez DL. Neurobiological mechanisms of the onset of puberty in primates. Endocr Rev. 2001;22:111–151. doi: 10.1210/edrv.22.1.0418. [DOI] [PubMed] [Google Scholar]
- 36.Herbison AE, Moenter SM. Depolarising and hyperpolarising actions of GABA(A) receptor activation on gonadotrophin-releasing hormone neurones: towards an emerging consensus. J Neuroendocrinol. 2011;23:557–569. doi: 10.1111/j.1365-2826.2011.02145.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gibson EM, Humber SA, Jain S, Williams WP, III, Zhao S, Bentley GE, Tsutsui K, Kriegsfeld LJ. Alterations in RFamide-related peptide expression are coordinated with the preovulatory luteinizing hormone surge. Endocrinology. 2008;149:4958–4969. doi: 10.1210/en.2008-0316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ducret E, Anderson GM, Herbison AE. RFamide-related peptide-3, a mammalian gonadotropin-inhibitory hormone ortholog, regulates gonadotropin-releasing hormone neuron firing in the mouse. Endocrinology. 2009;150:2799–2804. doi: 10.1210/en.2008-1623. [DOI] [PubMed] [Google Scholar]
- 39.Clasadonte J, Poulain P, Hanchate NK, Corfas G, Ojeda SR, Prevot V. Prostaglandin E2 release from astrocytes triggers gonadotropin-releasing hormone (GnRH) neuron firing via EP2 receptor activation. Proc Natl Acad Sci U S A. 2011;108:16104–16109. doi: 10.1073/pnas.1107533108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lomniczi A, Ojeda SR. A role for glial cells of the neuroendocrine brain in the central control of female sexual development. In: Parpura V, Haydon P, editors. Astrocytes in (Patho)Physiology of the Nervous System. Springer; NY: 2009. pp. 487–511. [Google Scholar]
- 41.Lehman MN, Coolen LM, Goodman RL. Minireview: kisspeptin/neurokinin B/dynorphin (KNDy) cells of the arcuate nucleus: a central node in the control of gonadotropin-releasing hormone secretion. Endocrinology. 2010;151:3479–3489. doi: 10.1210/en.2010-0022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Navarro VM, Gottsch ML, Wu M, Garcia-Galiano D, Hobbs SJ, Bosch MA, Pinilla L, Clifton DK, Dearth A, Ronnekleiv OK, Braun RE, Palmiter RD, Tena-Sempere M, Alreja M, Steiner RA. Regulation of NKB Pathways and Their Roles in the Control of Kiss1 Neurons in the Arcuate Nucleus of the Male Mouse. Endocrinology. 2011;152:4265–4275. doi: 10.1210/en.2011-1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wakabayashi Y, Nakada T, Murata K, Ohkura S, Mogi K, Navarro VM, Clifton DK, Mori Y, Tsukamura H, Maeda K, Steiner RA, Okamura H. Neurokinin B and dynorphin A in kisspeptin neurons of the arcuate nucleus participate in generation of periodic oscillation of neural activity driving pulsatile gonadotropin-releasing hormone secretion in the goat. J Neurosci. 2010;30:3124–3132. doi: 10.1523/JNEUROSCI.5848-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hrabovszky E, Ciofi P, Vida B, Horvath MC, Keller E, Caraty A, Bloom SR, Ghatei MA, Dhillo WS, Liposits Z, Kallo I. The kisspeptin system of the human hypothalamus: sexual dimorphism and relationship with gonadotropin-releasing hormone and neurokinin B neurons. Eur J Neurosci. 2010;31:1984–1998. doi: 10.1111/j.1460-9568.2010.07239.x. [DOI] [PubMed] [Google Scholar]
- 45.Clarkson J, d’Anglemont dT X, Colledge WH, Caraty A, Herbison AE. Distribution of kisspeptin neurones in the adult female mouse brain. J Neuroendocrinol. 2009;21:673–682. doi: 10.1111/j.1365-2826.2009.01892.x. [DOI] [PubMed] [Google Scholar]
- 46.Gottsch ML, Cunningham MJ, Smith JT, Popa SM, Acohido BV, Crowley WF, Seminara S, Clifton DK, Steiner RA. A role for kisspeptins in the regulation of gonadotropin secretion in the mouse. Endocrinology. 2004;145:4073–4077. doi: 10.1210/en.2004-0431. [DOI] [PubMed] [Google Scholar]
- 47.Pompolo S, Pereira A, Estrada KM, Clarke IJ. Colocalization of kisspeptin and gonadotropin-releasing hormone in the ovine brain. Endocrinology. 2006;147:804–810. doi: 10.1210/en.2005-1123. [DOI] [PubMed] [Google Scholar]
- 48.Pinilla L, Aguilar E, Dieguez C, Millar RP, Tena-Sempere M. Kisspeptins and reproduction: physiological roles and regulatory mechanisms. Physiol Rev. 2012;92:1235–1316. doi: 10.1152/physrev.00037.2010. [DOI] [PubMed] [Google Scholar]
- 49.Faiman C, Winter JSD. Gonadotropins and sex hormone patterns in puberty: Clinical data. In: Grumbach MM, Grave GD, Mayer FE, editors. Control of the Onset of Puberty. New York: John Wiley & Sons; 1974. pp. 32–55. [Google Scholar]
- 50.Reiter EO, Grumbach MM. Neuroendocrine control mechanisms and the onset of puberty. Ann Rev Physiol. 1982;44:595–613. doi: 10.1146/annurev.ph.44.030182.003115. [DOI] [PubMed] [Google Scholar]
- 51.Terasawa E, Bridson WE, Nass TE, Noonan JJ, Dierschke DJ. Developmental changes in the luteinizing hormone secretory pattern in peripubertal female rhesus monkeys: Comparisons between gonadally intact and ovariectomized animals. Endocrinology. 1984;115:2233–2240. doi: 10.1210/endo-115-6-2233. [DOI] [PubMed] [Google Scholar]
- 52.Chongthammakun S, Terasawa E. Negative feedback effects of estrogen on luteinizing hormone-releasing hormone release occur in pubertal, but not prepubertal, ovariectomized female rhesus monkeys. Endocrinology. 1993;132:735–743. doi: 10.1210/endo.132.2.8425492. [DOI] [PubMed] [Google Scholar]
- 53.Ojeda SR. The mystery of mammalian puberty: how much more do we know? Perspect Biol Med. 1991;34:365–383. doi: 10.1353/pbm.1991.0017. [DOI] [PubMed] [Google Scholar]
- 54.Thind KK, Goldsmith PC. Infundibular gonadotropin-releasing hormone neurons are inhibited by direct opioid and autoregulatory synapses in juvenile monkeys. Neuroendocrinology. 1988;47:203–216. doi: 10.1159/000124914. [DOI] [PubMed] [Google Scholar]
- 55.Blank MS, Panerai AE, Friesen HG. Opioid peptides modulate hormone secretion during sexual development. Science. 1979;203:1129–1131. doi: 10.1126/science.424743. [DOI] [PubMed] [Google Scholar]
- 56.Blank MS, Murphy JR. Luteinizing hormone sensitivity to naloxone in maturing male chimpanzees. Brain Res Bull. 1991;27:241–245. doi: 10.1016/0361-9230(91)90075-u. [DOI] [PubMed] [Google Scholar]
- 57.Mahesh VB, Nazian SJ. Role of sex steroids in the initiation of puberty. J Steroid Biochem. 1979;11:587–591. doi: 10.1016/0022-4731(79)90086-4. [DOI] [PubMed] [Google Scholar]
- 58.Sirinathsinghji DJS, Motta M, Martini L. Induction of precocious puberty in the female rat after chronic naloxone administration during the neonatal period: The opiate “brake” on prepubertal gonadotrophin secretion. J Endocrinol. 1985;104:299–307. doi: 10.1677/joe.0.1040299. [DOI] [PubMed] [Google Scholar]
- 59.Masotto C, Negro-Vilar A. Activation of gamma-amino butyric acid B-receptors abolishes naloxone-stimulated luteinizing hormone release. Endocrinology. 1987;121:2251–2255. doi: 10.1210/endo-121-6-2251. [DOI] [PubMed] [Google Scholar]
- 60.Moguilevsky JA, Carbone S, Szwarcfarb B, Rondina D. Sexual maturation modifies the GABAergic control of gonadotrophin secretion in female rats. Brain Res. 1991;563:12–16. doi: 10.1016/0006-8993(91)91508-x. [DOI] [PubMed] [Google Scholar]
- 61.Bourguignon J-P, Gerard A, Mathieu J, Mathieu A, Franchimont P. Maturation of the hypothalamic control of pulsatile gonadotropin-releasing hormone secretion at onset of puberty. I. Increased activation of N-methyl-D-aspartate receptors. Endocrinology. 1990;127:873–881. doi: 10.1210/endo-127-2-873. [DOI] [PubMed] [Google Scholar]
- 62.Bourguignon J-P, Gérard A, Alvarez-Gonzalez M-L, Fawe L, Franchimont P. Gonadal-independent developmental changes in activation of N-methyl-D-aspartate receptors involved in gonadotropin-releasing hormone secretion. Neuroendocrinology. 1992;55:634–641. doi: 10.1159/000126182. [DOI] [PubMed] [Google Scholar]
- 63.Donoso AO, López FJ, Negro-Vilar A. Glutamate receptors of the non-N-methyl-D-aspartic acid type mediate the increase in luteinizing hormone releasing hormone release by excitatory amino acid in vitro. Endocrinology. 1990;126:414–420. doi: 10.1210/endo-126-1-414. [DOI] [PubMed] [Google Scholar]
- 64.Petersen SL, McCrone S, Keller M, Gardner E. Rapid increase in LHRH mRNA levels following NMDA. Endocrinology. 1991;129:1679–1681. doi: 10.1210/endo-129-3-1679. [DOI] [PubMed] [Google Scholar]
- 65.Weesner GD, Bergen HT, Pfaff DW. Alpha-1 adrenergic regulation of luteinizing hormone-releasing hormone (LHRH) gene expression in the rat. Prog 73rd Ann Mtg Endocrine Soc; 1991; p. 285. [Google Scholar]
- 66.Ojeda SR, Urbanski HF, Rogers LC, Hill DF, Moholt-Siebert M, Costa ME. Developmental regulation of GnRH secretion. In: Delemarre-van de Waal HA, Schoemaker J, Plant TM, editors. Control of the Onset of Puberty III. The Netherlands: Elsevier Science Publishers B.V; 1989. pp. 55–62. [Google Scholar]
- 67.Kottler ML, Hamel A, Malville E, Richard N. GnRH deficiency: new insights from genetics. J Soc Biol. 2004;198:80–87. [PubMed] [Google Scholar]
- 68.Mastronardi C, Smiley GG, Raber J, Kusakabe T, Kawaguchi A, Matagne V, Dietzel A, Heger S, Mungenast AE, Cabrera R, Kimura S, Ojeda SR. Deletion of the Ttf1 gene in differentiated neurons disrupts female reproduction without impairing basal ganglia function. J Neurosci. 2006;26:13167–13179. doi: 10.1523/JNEUROSCI.4238-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cukier P, Wright H, Rulfs T, Silveira LF, Teles MG, Mendonca BB, Arnhold IJ, Heger S, Latronico AC, Ojeda SR, Brito VN. Molecular and gene network analysis of thyroid transcription factor 1 (TTF1) and enhanced at puberty (EAP1) genes in patients with GnRH-dependent pubertal disorders. Horm Res Paediatr. 2013;80:257–266. doi: 10.1159/000354643. [DOI] [PubMed] [Google Scholar]
- 70.Topaloglu AK, Reimann F, Guclu M, Yalin AS, Kotan LD, Porter KM, Serin A, Mungan NO, Cook JR, Ozbek MN, Imamoglu S, Akalin NS, Yuksel B, O’Rahilly S, Semple RK. TAC3 and TACR3 mutations in familial hypogonadotropic hypogonadism reveal a key role for Neurokinin B in the central control of reproduction. Nat Genet. 2008;41:354–358. doi: 10.1038/ng.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Grachev P, Li XF, Kinsey-Jones JS, di Domenico AL, Millar RP, Lightman SL, O’Byrne KT. Suppression of the GnRH pulse generator by neurokinin B involves a kappa-opioid receptor-dependent mechanism. Endocrinology. 2012;153:4894–4904. doi: 10.1210/en.2012-1574. [DOI] [PubMed] [Google Scholar]
- 72.Kinsey-Jones JS, Grachev P, Li XF, Lin YS, Milligan SR, Lightman SL, O’Byrne KT. The inhibitory effects of neurokinin B on GnRH pulse generator frequency in the female rat. Endocrinology. 2012;153:307–315. doi: 10.1210/en.2011-1641. [DOI] [PubMed] [Google Scholar]
- 73.Heger S, Mastronardi C, Dissen GA, Lomniczi A, Cabrera R, Roth CL, Jung H, Galimi F, Sippell W, Ojeda SR. Enhanced at puberty 1 (EAP1) is a new transcriptional regulator of the female neuroendocrine reproductive axis. J Clin Invest. 2007;117:2145–2154. doi: 10.1172/JCI31752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dissen GA, Lomniczi A, Heger S, Neff TL, Ojeda SR. Hypothalamic enhanced at puberty 1 (EAP1) is required for menstrual cyclicity in non-human primates. Endocrinology. 2012;153:350–361. doi: 10.1210/en.2011-1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Roth CL, Mastronardi C, Lomniczi A, Wright H, Cabrera R, Mungenast AE, Heger S, Jung H, Dubay C, Ojeda SR. Expression of a tumor-related gene network increases in the mammalian hypothalamus at the time of female puberty. Endocrinology. 2007;148:5147–5161. doi: 10.1210/en.2007-0634. [DOI] [PubMed] [Google Scholar]
- 76.Ojeda SR, Hill J, Hill DF, Costa ME, Tapia V, Cornea A, Ma YJ. The Oct-2 POU-domain gene in the neuroendocrine brain: A transcriptional regulator of mammalian puberty. Endocrinology. 1999;140:3774–3789. doi: 10.1210/endo.140.8.6941. [DOI] [PubMed] [Google Scholar]
- 77.Mueller JK, Dietzel A, Lomniczi A, Loche A, Tefs K, Kiess W, Danne T, Ojeda SR, Heger S. Transcriptional regulation of the human KiSS1 gene. Mol Cell Endocrinol. 2011;342:8–19. doi: 10.1016/j.mce.2011.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yeung KT, Das S, Zhang J, Lomniczi A, Ojeda S, Xu CF, Neubert TA, Samuels HH. A novel transcription complex that selectively modulates apoptosis of breast cancer cells through regulation of FASTKD2. Mol Cell Biol. 2011;31:2287–2298. doi: 10.1128/MCB.01381-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mueller JK, Koch I, Lomniczi A, Loche A, Rulfs T, Castellano JM, Kiess W, Ojeda S, Heger S. Transcription of the human EAP1 gene is regulated by upstream components of a puberty-controlling Tumor Suppressor Gene network. Mol Cell Endocrinol. 2012;351:184–198. doi: 10.1016/j.mce.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Steeg PS, Ouatas T, Halverson D, Palmieri D, Salerno M. Metastasis suppressor genes: Basic biology and potential clinical use. Clin Breast Cancer. 2003;4:51–62. doi: 10.3816/cbc.2003.n.012. [DOI] [PubMed] [Google Scholar]
- 81.Sandau US, Mungenast AE, McCarthy J, Biederer T, Corfas G, Ojeda SR. The Synaptic Cell Adhesion Molecule, SynCAM1, Mediates Astrocyte-to-Astrocyte and Astrocyte-to-GnRH Neuron Adhesiveness in the Mouse Hypothalamus. Endocrinology. 2011;152:2353–2363. doi: 10.1210/en.2010-1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sandau US, Mungenast AE, Alderman Z, Sardi SP, Fogel AI, Taylor B, Parent AS, Biederer T, Corfas G, Ojeda SR. SynCAM1, a Synaptic Adhesion Molecule, is Expressed in Astrocytes and Contributes to erbB4 Receptor-Mediated Control of Female Sexual Development. Endocrinology. 2011;152:2364–2376. doi: 10.1210/en.2010-1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fortes MR, Reverter A, Nagaraj SH, Zhang Y, Jonsson NN, Barris W, Lehnert S, Boe-Hansen GB, Hawken RJ. A single nucleotide polymorphism-derived regulatory gene network underlying puberty in 2 tropical breeds of beef cattle. J Anim Sci. 2011;89:1669–1683. doi: 10.2527/jas.2010-3681. [DOI] [PubMed] [Google Scholar]
- 84.Tanikawa C, Okada Y, Takahashi A, Oda K, Kamatani N, Kubo M, Nakamura Y, Matsuda K. Genome wide association study of age at menarche in the Japanese population. PLoS ONE. 2013;8:e63821. doi: 10.1371/journal.pone.0063821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Perry JR, Stolk L, Franceschini N, Lunetta KL, Zhai G, McArdle PF, Smith AV, Aspelund T, Bandinelli S, Boerwinkle E, Cherkas L, Eiriksdottir G, Estrada K, Ferrucci L, Folsom AR, Garcia M, Gudnason V, Hofman A, Karasik D, Kiel DP, Launer LJ, van MJ, Nalls MA, Rivadeneira F, Shuldiner AR, Singleton A, Soranzo N, Tanaka T, Visser JA, Weedon MN, Wilson SG, Zhuang V, Streeten EA, Harris TB, Murray A, Spector TD, Demerath EW, Uitterlinden AG, Murabito JM. Meta-analysis of genome-wide association data identifies two loci influencing age at menarche. Nat Genet. 2009;41:648–650. doi: 10.1038/ng.386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Elks CE, Perry JR, Sulem P, Chasman DI, Franceschini N, He C, Lunetta KL, Visser JA, Byrne EM, Cousminer DL, Gudbjartsson DF, Esko T, Feenstra B, Hottenga JJ, Koller DL, Kutalik Z, Lin P, Mangino M, Marongiu M, McArdle PF, Smith AV, Stolk L, van Wingerden SH, Zhao JH, Albrecht E, Corre T, Ingelsson E, Hayward C, Magnusson PK, Smith EN, Ulivi S, Warrington NM, Zgaga L, Alavere H, Amin N, Aspelund T, Bandinelli S, Barroso I, Berenson GS, Bergmann S, Blackburn H, Boerwinkle E, Buring JE, Busonero F, Campbell H, Chanock SJ, Chen W, Cornelis MC, Couper D, Coviello AD, d’Adamo P, de FU, de Geus EJ, Deloukas P, Doring A, Smith GD, Easton DF, Eiriksdottir G, Emilsson V, Eriksson J, Ferrucci L, Folsom AR, Foroud T, Garcia M, Gasparini P, Geller F, Gieger C, Gudnason V, Hall P, Hankinson SE, Ferreli L, Heath AC, Hernandez DG, Hofman A, Hu FB, Illig T, Jarvelin MR, Johnson AD, Karasik D, Khaw KT, Kiel DP, Kilpelainen TO, Kolcic I, Kraft P, Launer LJ, Laven JS, Li S, Liu J, Levy D, Martin NG, McArdle WL, Melbye M, Mooser V, Murray JC, Murray SS, Nalls MA, Navarro P, Nelis M, Ness AR, Northstone K, Oostra BA, Peacock M, Palmer LJ, Palotie A, Pare G, Parker AN, Pedersen NL, Peltonen L, Pennell CE, Pharoah P, Polasek O, Plump AS, Pouta A, Porcu E, Rafnar T, Rice JP, Ring SM, Rivadeneira F, Rudan I, Sala C, Salomaa V, Sanna S, Schlessinger D, Schork NJ, Scuteri A, Segre AV, Shuldiner AR, Soranzo N, Sovio U, Srinivasan SR, Strachan DP, Tammesoo ML, Tikkanen E, Toniolo D, Tsui K, Tryggvadottir L, Tyrer J, Uda M, van Dam RM, van Meurs JB, Vollenweider P, Waeber G, Wareham NJ, Waterworth DM, Weedon MN, Wichmann HE, Willemsen G, Wilson JF, Wright AF, Young L, Zhai G, Zhuang WV, Bierut LJ, Boomsma DI, Boyd HA, Crisponi L, Demerath EW, van Duijn CM, Econs MJ, Harris TB, Hunter DJ, Loos RJ, Metspalu A, Montgomery GW, Ridker PM, Spector TD, Streeten EA, Stefansson K, Thorsteinsdottir U, Uitterlinden AG, Widen E, Murabito JM, Ong KK, Murray A. Thirty new loci for age at menarche identified by a meta-analysis of genome-wide association studies. Nat Genet. 2010;42:1077–1085. doi: 10.1038/ng.714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.He C, Kraft P, Chen C, Buring JE, Pare G, Hankinson SE, Chanock SJ, Ridker PM, Hunter DJ, Chasman DI. Genome-wide association studies identify loci associated with age at menarche and age at natural menopause. Nat Genet. 2009;41:724–728. doi: 10.1038/ng.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Warde-Farley D, Donaldson SL, Comes O, Zuberi K, Badrawi R, Chao P, Franz M, Grouios C, Kazi F, Lopes CT, Maitland A, Mostafavi S, Montojo J, Shao Q, Wright G, Bader GD, Morris Q. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010;38:W214–W220. doi: 10.1093/nar/gkq537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kim JY, Kim KB, Eom GH, Choe N, Kee HJ, Son HJ, Oh ST, Kim DW, Pak JH, Baek HJ, Kook H, Hahn Y, Kook H, Chakravarti D, Seo SB. KDM3B is the H3K9 demethylase involved in transcriptional activation of lmo2 in leukemia. Mol Cell Biol. 2012;32:2917–2933. doi: 10.1128/MCB.00133-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Krishnan S, Horowitz S, Trievel RC. Structure and function of histone H3 lysine 9 methyltransferases and demethylases. Chembiochem. 2011;12:254–263. doi: 10.1002/cbic.201000545. [DOI] [PubMed] [Google Scholar]
- 91.Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33(Suppl):245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 92.Bjornsson HT, Fallin MD, Feinberg AP. An integrated epigenetic and genetic approach to common human disease. Trends Genet. 2004;20:350–358. doi: 10.1016/j.tig.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 93.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, Rao A. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Koh KP, Yabuuchi A, Rao S, Huang Y, Cunniff K, Nardone J, Laiho A, Tahiliani M, Sommer CA, Mostoslavsky G, Lahesmaa R, Orkin SH, Rodig SJ, Daley GQ, Rao A. Tet1 and Tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell Stem Cell. 2011;8:200–213. doi: 10.1016/j.stem.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ficz G, Branco MR, Seisenberger S, Santos F, Krueger F, Hore TA, Marques CJ, Andrews S, Reik W. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature. 2011;473:398–402. doi: 10.1038/nature10008. [DOI] [PubMed] [Google Scholar]
- 96.Guo JU, Su Y, Zhong C, Ming GL, Song H. Emerging roles of TET proteins and 5-hydroxymethylcytosines in active DNA demethylation and beyond. Cell Cycle. 2011;10:2662–2668. doi: 10.4161/cc.10.16.17093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 98.Khorasanizadeh S. The nucleosome: from genomic organization to genomic regulation. Cell. 2004;116:259–272. doi: 10.1016/s0092-8674(04)00044-3. [DOI] [PubMed] [Google Scholar]
- 99.Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21:381–395. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Falkenberg KJ, Johnstone RW. Histone deacetylases and their inhibitors in cancer, neurological diseases and immune disorders. Nat Rev Drug Discov. 2014;13:673–691. doi: 10.1038/nrd4360. [DOI] [PubMed] [Google Scholar]
- 101.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 102.Yun M, Wu J, Workman JL, Li B. Readers of histone modifications. Cell Res. 2011;21:564–578. doi: 10.1038/cr.2011.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ruthenburg AJ, Li H, Patel DJ, Allis CD. Multivalent engagement of chromatin modifications by linked binding modules. Nat Rev Mol Cell Biol. 2007;8:983–994. doi: 10.1038/nrm2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Pekowska A, Benoukraf T, Zacarias-Cabeza J, Belhocine M, Koch F, Holota H, Imbert J, Andrau JC, Ferrier P, Spicuglia S. H3K4 tri-methylation provides an epigenetic signature of active enhancers. EMBO J. 2011;30:4198–4210. doi: 10.1038/emboj.2011.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Huang B, Jiang C, Zhang R. Epigenetics: the language of the cell? Epigenomics. 2014;6:73–88. doi: 10.2217/epi.13.72. [DOI] [PubMed] [Google Scholar]
- 106.Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152:1298–1307. doi: 10.1016/j.cell.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Cech TR, Steitz JA. The noncoding RNA revolution-trashing old rules to forge new ones. Cell. 2014;157:77–94. doi: 10.1016/j.cell.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 108.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 109.Okamura K, Chung WJ, Ruby JG, Guo H, Bartel DP, Lai EC. The Drosophila hairpin RNA pathway generates endogenous short interfering RNAs. Nature. 2008;453:803–806. doi: 10.1038/nature07015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kim VN. Small RNAs just got bigger: Piwi-interacting RNAs (piRNAs) in mammalian testes. Genes Dev. 2006;20:1993–1997. doi: 10.1101/gad.1456106. [DOI] [PubMed] [Google Scholar]
- 111.Girard A, Sachidanandam R, Hannon GJ, Carmell MA. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442:199–202. doi: 10.1038/nature04917. [DOI] [PubMed] [Google Scholar]
- 112.Gruber AJ, Zavolan M. Modulation of epigenetic regulators and cell fate decisions by miRNAs. Epigenomics. 2013;5:671–683. doi: 10.2217/epi.13.65. [DOI] [PubMed] [Google Scholar]
- 113.Spitale RC, Tsai MC, Chang HY. RNA templating the epigenome: long noncoding RNAs as molecular scaffolds. Epigenetics. 2011;6:539–543. doi: 10.4161/epi.6.5.15221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, Cabili MN, Jaenisch R, Mikkelsen TS, Jacks T, Hacohen N, Bernstein BE, Kellis M, Regev A, Rinn JL, Lander ES. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.McCarthy MM, Nugent BM. Epigenetic contributions to hormonally-mediated sexual differentiation of the brain. J Neuroendocrinol. 2013;25:1133–1140. doi: 10.1111/jne.12072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Schwarz JM, Nugent BM, McCarthy MM. Developmental and hormone-induced epigenetic changes to estrogen and progesterone receptor genes in brain are dynamic across the life span. Endocrinology. 2010;151:4871–4881. doi: 10.1210/en.2010-0142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kurian JR, Olesen KM, Auger AP. Sex differences in epigenetic regulation of the estrogen receptor-alpha promoter within the developing preoptic area. Endocrinology. 2010;151:2297–2305. doi: 10.1210/en.2009-0649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Matsuda KI, Mori H, Nugent BM, Pfaff DW, McCarthy MM, Kawata M. Histone deacetylation during brain development is essential for permanent masculinization of sexual behavior. Endocrinology. 2011;152:2760–2767. doi: 10.1210/en.2011-0193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kauffman AS, Navarro VM, Kim J, Clifton DK, Steiner RA. Sex differences in the regulation of Kiss1/NKB neurons in juvenile mice: implications for the timing of puberty. Am J Physiol Endocrinol Metab. 2009;297:E1212–E1221. doi: 10.1152/ajpendo.00461.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Yang C, Ye J, Li X, Gao X, Zhang K, Luo L, Ding J, Zhang Y, Li Y, Cao H, Ling Y, Zhang X, Liu Y, Fang F. DNA Methylation Patterns in the Hypothalamus of Female Pubertal Goats. PLoS ONE. 2016;11:e0165327. doi: 10.1371/journal.pone.0165327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kurian JR, Keen KL, Terasawa E. Epigenetic changes coincide with in vitro primate GnRH neuronal maturation. Endocrinology. 2010;151:5359–5368. doi: 10.1210/en.2010-0555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Messina A, Langlet F, Chachlaki K, Roa J, Rasika S, Jouy N, Gallet S, Gaytan F, Parkash J, Tena-Sempere M, Giacobini P, Prevot V. A microRNA switch regulates the rise in hypothalamic GnRH production before puberty. Nat Neurosci. 2016;19:835–844. doi: 10.1038/nn.4298. [DOI] [PubMed] [Google Scholar]
- 123.Lomniczi A, Loche A, Castellano JM, Ronnekleiv OK, Bosh M, Kaidar G, Knoll JG, Wright H, Pfeifer GP, Ojeda SR. Epigenetic control of female puberty. Nature Neuroscience. 2013;16:281–289. doi: 10.1038/nn.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Urbanski HF, Ojeda SR. Gonadal-independent activation of enhanced afternoon luteinizing hormone release during pubertal development in the female rat. Endocrinology. 1987;121:907–913. doi: 10.1210/endo-121-3-907. [DOI] [PubMed] [Google Scholar]
- 125.Urbanski HF, Ojeda SR. The juvenile-peripubertal transition period in the female rat: Establishment of a diurnal pattern of pulsatile luteinizing hormone secretion. Endocrinology. 1985;117:644–649. doi: 10.1210/endo-117-2-644. [DOI] [PubMed] [Google Scholar]
- 126.Ojeda SR, Urbanski HF. Puberty in the rat. In: Knobil E, Neill JD, editors. The Physiology of Reproduction. 2. Vol. 2. New York: Raven Press; 1994. pp. 363–409. [Google Scholar]
- 127.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, Jaenisch R, Wagschal A, Feil R, Schreiber SL, Lander ES. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 128.Toro CA, Wright H, Aylwin CF, Ojeda SR, Lomniczi A. Trithorax dependent changes in chromatin landscape at enhancer and promoter regions drive female puberty. Nat Commun. 2018;9:57. doi: 10.1038/s41467-017-02512-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Shilatifard A. The COMPASS family of histone H3K4 methylases: mechanisms of regulation in development and disease pathogenesis. Annu Rev Biochem. 2012;81:65–95. doi: 10.1146/annurev-biochem-051710-134100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Simon JA, Kingston RE. Mechanisms of polycomb gene silencing: knowns and unknowns. Nat Rev Mol Cell Biol. 2009;10:697–708. doi: 10.1038/nrm2763. [DOI] [PubMed] [Google Scholar]
- 131.Herz HM, Mohan M, Garruss AS, Liang K, Takahashi YH, Mickey K, Voets O, Verrijzer CP, Shilatifard A. Enhancer-associated H3K4 monomethylation by Trithorax-related, the Drosophila homolog of mammalian Mll3/Mll4. Genes Dev. 2012;26:2604–2620. doi: 10.1101/gad.201327.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Schuettengruber B, Martinez AM, Iovino N, Cavalli G. Trithorax group proteins: switching genes on and keeping them active. Nat Rev Mol Cell Biol. 2011;12:799–814. doi: 10.1038/nrm3230. [DOI] [PubMed] [Google Scholar]
- 133.Kim HG, Kurth I, Lan F, Meliciani I, Wenzel W, Eom SH, Kang GB, Rosenberger G, Tekin M, Ozata M, Bick DP, Sherins RJ, Walker SL, Shi Y, Gusella JF, Layman LC. Mutations in CHD7, encoding a chromatin-remodeling protein, cause idiopathic hypogonadotropic hypogonadism and Kallmann syndrome. Am J Hum Genet. 2008;83:511–519. doi: 10.1016/j.ajhg.2008.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Lomniczi A, Wright H, Castellano JM, Matagne V, Toro CA, Ramaswamy S, Plant TM, Ojeda SR. Epigenetic regulation of puberty via Zinc finger protein-mediated transcriptional repression. Nat Commun. 2015;6:10195. doi: 10.1038/ncomms10195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 136.Maisonpierre PC, Le Beau MM, Espinosa R, III, Ip NY, Belluscio L, de la Monte SM, Squinto S, Furth ME, Yancopoulos GD. Human and rat brain-derived neurotrophic factor and neurotrophin-3: Gene structures, distributions, and chromosomal localizations. Genomics. 1991;10:558–568. doi: 10.1016/0888-7543(91)90436-i. [DOI] [PubMed] [Google Scholar]
- 137.Tomikawa J, Uenoyama Y, Ozawa M, Fukanuma T, Takase K, Goto T, Abe H, Ieda N, Minabe S, Deura C, Inoue N, Sanbo M, Tomita K, Hirabayashi M, Tanaka S, Imamura T, Okamura H, Maeda K, Tsukamura H. Epigenetic regulation of Kiss1 gene expression mediating estrogen-positive feedback action in the mouse brain. Proc Natl Acad Sci U S A. 2012;109:E1294–E1301. doi: 10.1073/pnas.1114245109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Abreu AP, Dauber A, Macedo DB, Noel SD, Brito VN, Gill JC, Cukier P, Thompson IR, Navarro VM, Gagliardi PC, Rodrigues T, Kochi C, Longui CA, Beckers D, de ZF, Montenegro LR, Mendonca BB, Carroll RS, Hirschhorn JN, Latronico AC, Kaiser UB. Central precocious puberty caused by mutations in the imprinted gene MKRN3. N Engl J Med. 2013;368:2467–2475. doi: 10.1056/NEJMoa1302160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Settas N, Dacou-Voutetakis C, Karantza M, Kanaka-Gantenbein C, Chrousos GP, Voutetakis A. Central precocious puberty in a girl and early puberty in her brother caused by a novel mutation in the MKRN3 gene. J Clin Endocrinol Metab. 2014;99:E647–E651. doi: 10.1210/jc.2013-4084. [DOI] [PubMed] [Google Scholar]
- 140.Jeong HR, Lee HS, Hwang JS. Makorin ring finger 3 gene analysis in Koreans with familial precocious puberty. J Pediatr Endocrinol Metab. 2017;30:1197–1201. doi: 10.1515/jpem-2016-0471. [DOI] [PubMed] [Google Scholar]
- 141.Grandone A, Capristo C, Cirillo G, Sasso M, Umano GR, Mariani M, Miraglia Del GE, Perrone L. Molecular Screening of MKRN3, DLK1, and KCNK9 Genes in Girls with Idiopathic Central Precocious Puberty. Horm Res Paediatr. 2017;88:194–200. doi: 10.1159/000477441. [DOI] [PubMed] [Google Scholar]
- 142.Grandone A, Cirillo G, Sasso M, Capristo C, Tornese G, Marzuillo P, Luongo C, Rosaria UG, Festa A, Coppola R, Miraglia Del GE, Perrone L. MKRN3 levels in girls with central precocious puberty and correlation with sexual hormone levels: a pilot study. Endocrine. 2017 doi: 10.1007/s12020-017-1281-x. [DOI] [PubMed] [Google Scholar]
- 143.Christoforidis A, Skordis N, Fanis P, Dimitriadou M, Sevastidou M, Phelan MM, Neocleous V, Phylactou LA. A novel MKRN3 nonsense mutation causing familial central precocious puberty. Endocrine. 2017;56:446–449. doi: 10.1007/s12020-017-1232-6. [DOI] [PubMed] [Google Scholar]
- 144.Brito VN, Latronico AC. Underdiagnosis of central precocious puberty in boys with loss-of-function mutations of MKRN3. J Pediatr. 2017;183:202–203. doi: 10.1016/j.jpeds.2016.12.023. [DOI] [PubMed] [Google Scholar]
- 145.Simsek E, Demiral M, Ceylaner S, Kirel B. Two Frameshift Mutations in MKRN3 in Turkish Patients with Familial Central Precocious Puberty. Horm Res Paediatr. 2017;87:405–411. doi: 10.1159/000450923. [DOI] [PubMed] [Google Scholar]
- 146.Bessa DS, Macedo DB, Brito VN, Franca MM, Montenegro LR, Cunha-Silva M, Silveira LG, Hummel T, Bergada I, Braslavsky D, Abreu AP, Dauber A, Mendonca BB, Kaiser UB, Latronico AC. High Frequency of MKRN3 Mutations in Male Central Precocious Puberty Previously Classified as Idiopathic. Neuroendocrinology. 2017;105:17–25. doi: 10.1159/000446963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Kansakoski J, Raivio T, Juul A, Tommiska J. A missense mutation in MKRN3 in a Danish girl with central precocious puberty and her brother with early puberty. Pediatr Res. 2015;78:709–711. doi: 10.1038/pr.2015.159. [DOI] [PubMed] [Google Scholar]
- 148.de VL, Gat-Yablonski G, Dror N, Singer A, Phillip M. A novel MKRN3 missense mutation causing familial precocious puberty. Hum Reprod. 2014;29:2838–2843. doi: 10.1093/humrep/deu256. [DOI] [PubMed] [Google Scholar]
- 149.Perry JR, Day F, Elks CE, Sulem P, Thompson DJ, Ferreira T, He C, Chasman DI, Esko T, Thorleifsson G, Albrecht E, Ang WQ, Corre T, Cousminer DL, Feenstra B, Franceschini N, Ganna A, Johnson AD, Kjellqvist S, Lunetta KL, McMahon G, Nolte IM, Paternoster L, Porcu E, Smith AV, Stolk L, Teumer A, Tsernikova N, Tikkanen E, Ulivi S, Wagner EK, Amin N, Bierut LJ, Byrne EM, Hottenga JJ, Koller DL, Mangino M, Pers TH, Yerges-Armstrong LM, Hua ZJ, Andrulis IL, Anton-Culver H, Atsma F, Bandinelli S, Beckmann MW, Benitez J, Blomqvist C, Bojesen SE, Bolla MK, Bonanni B, Brauch H, Brenner H, Buring JE, Chang-Claude J, Chanock S, Chen J, Chenevix-Trench G, Collee JM, Couch FJ, Couper D, Coviello AD, Cox A, Czene K, D’adamo AP, Davey SG, De VI, Demerath EW, Dennis J, Devilee P, Dieffenbach AK, Dunning AM, Eiriksdottir G, Eriksson JG, Fasching PA, Ferrucci L, Flesch-Janys D, Flyger H, Foroud T, Franke L, Garcia ME, Garcia-Closas M, Geller F, de Geus EE, Giles GG, Gudbjartsson DF, Gudnason V, Guenel P, Guo S, Hall P, Hamann U, Haring R, Hartman CA, Heath AC, Hofman A, Hooning MJ, Hopper JL, Hu FB, Hunter DJ, Karasik D, Kiel DP, Knight JA, Kosma VM, Kutalik Z, Lai S, Lambrechts D, Lindblom A, Magi R, Magnusson PK, Mannermaa A, Martin NG, Masson G, McArdle PF, McArdle WL, Melbye M, Michailidou K, Mihailov E, Milani L, Milne RL, Nevanlinna H, Neven P, Nohr EA, Oldehinkel AJ, Oostra BA, Palotie A, Peacock M, Pedersen NL, Peterlongo P, Peto J, Pharoah PD, Postma DS, Pouta A, Pylkas K, Radice P, Ring S, Rivadeneira F, Robino A, Rose LM, Rudolph A, Salomaa V, Sanna S, Schlessinger D, Schmidt MK, Southey MC, Sovio U, Stampfer MJ, Stockl D, Storniolo AM, Timpson NJ, Tyrer J, Visser JA, Vollenweider P, Volzke H, Waeber G, Waldenberger M, Wallaschofski H, Wang Q, Willemsen G, Winqvist R, Wolffenbuttel BH, Wright MJ, Boomsma DI, Econs MJ, Khaw KT, Loos RJ, McCarthy MI, Montgomery GW, Rice JP, Streeten EA, Thorsteinsdottir U, van Duijn CM, Alizadeh BZ, Bergmann S, Boerwinkle E, Boyd HA, Crisponi L, Gasparini P, Gieger C, Harris TB, Ingelsson E, Jarvelin MR, Kraft P, Lawlor D, Metspalu A, Pennell CE, Ridker PM, Snieder H, Sorensen TI, Spector TD, Strachan DP, Uitterlinden AG, Wareham NJ, Widen E, Zygmunt M, Murray A, Easton DF, Stefansson K, Murabito JM, Ong KK. Parent-of-origin-specific allelic associations among 106 genomic loci for age at menarche. Nature. 2014;514:92–96. doi: 10.1038/nature13545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Day FR, Thompson DJ, Helgason H, Chasman DI, Finucane H, Sulem P, Ruth KS, Whalen S, Sarkar AK, Albrecht E, Altmaier E, Amini M, Barbieri CM, Boutin T, Campbell A, Demerath E, Giri A, He C, Hottenga JJ, Karlsson R, Kolcic I, Loh PR, Lunetta KL, Mangino M, Marco B, McMahon G, Medland SE, Nolte IM, Noordam R, Nutile T, Paternoster L, Perjakova N, Porcu E, Rose LM, Schraut KE, Segre AV, Smith AV, Stolk L, Teumer A, Andrulis IL, Bandinelli S, Beckmann MW, Benitez J, Bergmann S, Bochud M, Boerwinkle E, Bojesen SE, Bolla MK, Brand JS, Brauch H, Brenner H, Broer L, Bruning T, Buring JE, Campbell H, Catamo E, Chanock S, Chenevix-Trench G, Corre T, Couch FJ, Cousminer DL, Cox A, Crisponi L, Czene K, Davey SG, de Geus EJCN, de MR, De VI, Dennis J, Devilee P, Dos-Santos-Silva I, Dunning AM, Eriksson JG, Fasching PA, Fernandez-Rhodes L, Ferrucci L, Flesch-Janys D, Franke L, Gabrielson M, Gandin I, Giles GG, Grallert H, Gudbjartsson DF, Guenel P, Hall P, Hallberg E, Hamann U, Harris TB, Hartman CA, Heiss G, Hooning MJ, Hopper JL, Hu F, Hunter DJ, Ikram MA, Im HK, Jarvelin MR, Joshi PK, Karasik D, Kellis M, Kutalik Z, LaChance G, Lambrechts D, Langenberg C, Launer LJ, Laven JSE, Lenarduzzi S, Li J, Lind PA, Lindstrom S, Liu Y, Luan J, Magi R, Mannermaa A, Mbarek H, McCarthy MI, Meisinger C, Meitinger T, Menni C, Metspalu A, Michailidou K, Milani L, Milne RL, Montgomery GW, Mulligan AM, Nalls MA, Navarro P, Nevanlinna H, Nyholt DR, Oldehinkel AJ, O’Mara TA, Padmanabhan S, Palotie A, Pedersen N, Peters A, Peto J, Pharoah PDP, Pouta A, Radice P, Rahman I, Ring SM, Robino A, Rosendaal FR, Rudan I, Rueedi R, Ruggiero D, Sala CF, Schmidt MK, Scott RA, Shah M, Sorice R, Southey MC, Sovio U, Stampfer M, Steri M, Strauch K, Tanaka T, Tikkanen E, Timpson NJ, Traglia M, Truong T, Tyrer JP, Uitterlinden AG, Edwards DRV, Vitart V, Volker U, Vollenweider P, Wang Q, Widen E, van Dijk KW, Willemsen G, Winqvist R, Wolffenbuttel BHR, Zhao JH, Zoledziewska M, Zygmunt M, Alizadeh BZ, Boomsma DI, Ciullo M, Cucca F, Esko T, Franceschini N, Gieger C, Gudnason V, Hayward C, Kraft P, Lawlor DA, Magnusson PKE, Martin NG, Mook-Kanamori DO, Nohr EA, Polasek O, Porteous D, Price AL, Ridker PM, Snieder H, Spector TD, Stockl D, Toniolo D, Ulivi S, Visser JA, Volzke H, Wareham NJ, Wilson JF, Spurdle AB, Thorsteindottir U, Pollard KS, Easton DF, Tung JY, Chang-Claude J, Hinds D, Murray A, Murabito JM, Stefansson K, Ong KK, Perry JRB. Genomic analyses identify hundreds of variants associated with age at menarche and support a role for puberty timing in cancer risk. Nat Genet. 2017;49:834–841. doi: 10.1038/ng.3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Bohne A, Darras A, D’Cotta H, Baroiller JF, Galiana-Arnoux D, Volff JN. The vertebrate makorin ubiquitin ligase gene family has been shaped by large-scale duplication and retroposition from an ancestral gonad-specific, maternal-effect gene. BMC Genomics. 2010;11:721. doi: 10.1186/1471-2164-11-721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Omwancha J, Zhou XF, Chen SY, Baslan T, Fisher CJ, Zheng Z, Cai C, Shemshedini L. Makorin RING finger protein 1 (MKRN1) has negative and positive effects on RNA polymerase II-dependent transcription. Endocrine. 2006;29:363–373. doi: 10.1385/ENDO:29:2:363. [DOI] [PubMed] [Google Scholar]
- 153.Li W, Notani D, Rosenfeld MG. Enhancers as non-coding RNA transcription units: recent insights and future perspectives. Nat Rev Genet. 2016;17:207–223. doi: 10.1038/nrg.2016.4. [DOI] [PubMed] [Google Scholar]