Abstract
Direct oral anticoagulants (DOAC) are substrates for the ABCB1 transporter (also called P-glycoprotein), an active efflux pump. ABCB1 polymorphisms have been previously reported to influence the pharmacokinetics of several drugs such as immunosuppressants and tyrosine kinase inhibitors. Recently, in vivo studies have suggested that genetic variants might contribute to the inter-individual variability in DOAC plasma concentrations. Therefore, we evaluated the in vitro effect of the most common coding ABCB1 single nucleotide polymorphisms (SNP), 1236 C > T-2677G > T-3435C > T, and the coding ABCB1 1199 G > A SNP on the transport activity towards rivaroxaban. HEK293 cells were transfected to overexpress the ABCB1 wild-type (1236C-2677G-3435C, 1199 G) or variant proteins (1236C-2677G-3435T, 1236T-2677T-3435T or 1199 A). ABCB1 expression decreased the intracellular accumulation of rivaroxaban, when compared to control cells. This confirms the involvement of ABCB1 in the active transport of rivaroxaban. However, the ABCB1 1236 C > T-2677G > T-3435C > T and 1199 G > A SNPs had no significant influence on the intracellular accumulation of rivaroxaban when compared to the wild-type protein. These results suggest that the ABCB1 coding SNPs investigated in the present study are unlikely to contribute to the inter-individual variability in rivaroxaban plasma concentrations.
Introduction
The landscape of oral anticoagulant therapy has significantly changed in the last years. Direct oral anticoagulants (DOAC) have been approved for stroke prevention in non-valvular atrial fibrillation and the treatment as well as the secondary prevention of venous thromboembolism. They are increasingly used in clinical practice, partly due to their higher convenience for clinicians and patients (fixed-dose regimen, fewer interactions with drugs and food) compared with vitamin K antagonists (VKA)1. Four DOACs are currently available: three direct factor Xa inhibitors (apixaban, edoxaban, rivaroxaban) and one direct thrombin inhibitor (dabigatran etexilate). In 2016, guidelines issued by the European Society of Cardiology or the American College of Chest Physicians have recommended DOACs over VKAs in eligible patients2,3.
All DOACs are substrates for the ABCB1 transporter, also called P-glycoprotein (P-gp) or formerly designated as the multidrug resistance 1 (MDR1) protein4. This active efflux pump belongs to the ATP-binding cassette transporter superfamily and is involved in the disposition of multiple drugs from diverse classes such as anticancer agents, immunosuppressants or antibiotics5. ABCB1 expression at the apical membrane of enterocytes limits absorption, while its localization on the luminal membrane of hepatocytes and renal tubular cells enhances biliary and renal excretion respectively. In patients taking rivaroxaban, the ABCB1 protein expression in renal tubular cells is particularly important given that more than one third of the dose is eliminated unchanged in the urine6. Many medications that are commonly prescribed in patients with atrial fibrillation (AF) can inhibit or induce ABCB1 activity and thereby influence DOAC pharmacokinetics7. For instance, it has been demonstrated that the concurrent use of moderate inhibitors of ABCB1 such as amiodarone or verapamil led to a nearly 40% increase in rivaroxaban exposure8,9. Combining this with renal impairment or older age, two common characteristics in AF patients, produced even stronger effect on rivaroxaban pharmacokinetics.
Genetic polymorphisms have also been reported to influence ABCB1 activity and/or expression5. The ABCB1 gene, located on chromosome 7, is composed of 29 exons in a 251.3-kb genomic region. More than 60 coding single nucleotide polymorphisms (SNP) have been described for ABCB110. The three most common SNPs in the coding region are rs1128503 (1236 C > T, Gly412Gly), rs2032582 (2677 G > T, Ala893Ser) and rs1045642 (3435 C > T, Ile1145Ile). They are in strong linkage disequilibrium and present a minor allelic frequency around 50% in the Caucasian population. They have been widely investigated, with inconsistent effects on drug disposition, drug response and toxicity5. Another SNP of interest is the 1199 G > A non-synonymous coding SNP (rs2229109), with an allelic frequency around 6% in Caucasians. The amino acid change (Ser400Asn) caused by this SNP is located in a cytoplasmic loop involved in substrate recognition. In previous in vitro studies, it has been shown that 1199 G > A affects the transport of tacrolimus, vinblastine or tyrosine kinase inhibitors but that cyclosporine A, Rh123 and doxorubicin are transported in a similar extent by the wild-type and variant ABCB1 proteins11,12.
Despite the fixed-dose regimen of DOACs, significant inter-individual variability in peak and trough plasma concentrations has been described13,14. In a prospective cohort study, 40% of patients with atrial fibrillation had rivaroxaban plasma measurements outside the 5th–95th percentile interval observed in phase 3 trials15. Moreover, in an analysis of the ROCKET-AF trial, rivaroxaban exposure predicted the risk of major bleeding16. Inversely, it was recently highlighted that DOAC patients experiencing thromboembolic events had lower plasma levels in the first month of treatment17. Genetic polymorphisms have been proposed to explain why patients taking the same DOAC dose present highly variable plasma concentrations18. Therefore, this study aimed to evaluate the in vitro effect of the ABCB1 1236 C > T-2677G > T-3435C > T and 1199 G > A SNPs on the transport activity towards rivaroxaban.
Results
Generation of ABCB1 recombinant cell lines
HEK293 cells overexpressing ABCB1 wild-type and variant proteins have been previously generated and characterized11,19. For ABCB1 1236 C > T-2677G > T-3435C > T, the recombinant models used consisted of stably transfected cell lines with the pcDNA3.1 empty vector, HEKpcDNA3.1, the wild-type vector, HEK1236C-2677G-3435C or two different combinations of the variant cDNA, HEK1236C-2677G-3435T and HEK1236T-2677T-3435T, hereafter referred to as HEKcontrol, HEKCGC, HEKCGT and HEKTTT respectively. For ABCB1 1199 G > A, three recombinant cell lines were used: HEKcontrol, HEK1199G (wild-type protein) and HEK1199A (variant protein). To ensure similar ABCB1 surface expression among the different cell lines, cells were sorted using fluorescence activated cell sorting (FACS) with fluorescence parameters gated on the same level of intensity. As shown in Fig. 1A, similar ABCB1 surface expression was observed by analytical flow cytometry among recombinant cell lines, both for the ABCB1 1236 C > T-2677G > T-3435C > T and 1199 G > A SNPs. In contrast, there was no fluorescence detected in HEKcontrol ensuring no or at least very low basal expression in our negative controls.
Figure 1.
ABCB1 cell surface expression. Flow cytometry histograms of HEK293 cells transfected with (A) the empty pcDNA3.1 vector, ABCB1CGC, ABCB1CGT or ABCB1TTT and (B) the empty pcDNA3.1 vector, ABCB11199G, ABCB11199A. Cells were incubated with a FITC anti-ABCB1 antibody (blue) or a matched isotypic control (red).
Impact of ABCB1 1236 C > T-2677G > T-3435C > T polymorphisms on the intracellular accumulation of rivaroxaban
First, we assessed the impact of ABCB1 1236 C > T-2677G > T-3435C > T on the ABCB1 transport activity towards rivaroxaban. HEKcontrol, HEKCGC, HEKCGT and HEKTTT were incubated with different concentrations of rivaroxaban ranging from 50 to 1000 ng/ml. These concentrations were chosen to cover rivaroxaban plasma levels encountered in clinical practice (concentrations up to 600 ng/ml were described in real-life patients with AF)14,15. As shown in Fig. 2, the intracellular accumulation of rivaroxaban was significantly decreased in recombinant cell lines overexpressing ABCB1 when compared to control cells (Fig. 2, [50, 250 and 500 ng/ml], p < 0.01; [100 and 1000 ng/ml], p < 0.001). However, we observed that the intracellular accumulation of rivaroxaban was similar between HEKCGC, HEKCGT and HEKTTT (Fig. 2, p > 0.05 at all concentrations).
Figure 2.
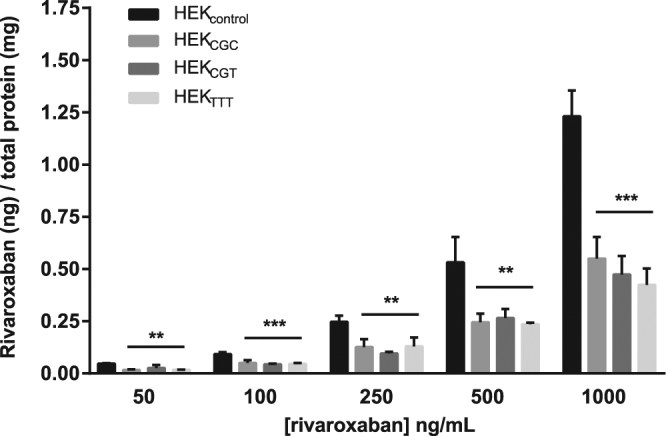
Impact of ABCB1 1236 C > T-2677G > T-3435C > T on the intracellular accumulation of rivaroxaban. Intracellular accumulation of rivaroxaban after 120 min of incubation (N = 3) at different concentrations in HEKcontrol, HEKCGC, HEKCGT and HEKTTT. The absolute amount of rivaroxaban (in ng) was divided by the total amount of proteins in cell extracts (in mg). *Compared to control cells: *p < 0.05, **p < 0.01, ***p < 0.001.
Impact of ABCB1 1199 G > A polymorphism on the intracellular accumulation of rivaroxaban
Same experiments were conducted to investigate the impact of ABCB1 1199 G > A on the ABCB1 activity towards rivaroxaban. Again, the intracellular accumulation of rivaroxaban was lower in cell lines overexpressing ABCB1, when compared with control cells whatever their genotype (Fig. 3, [from 50 to 1000 ng/ml], p < 0.001). As for the three other variants, we did not find any statistical difference in the intracellular accumulation of rivaroxaban between cells overexpressing the ABCB1 1199 G and 1199 A proteins (Fig. 3, p > 0.05 at all concentrations).
Figure 3.
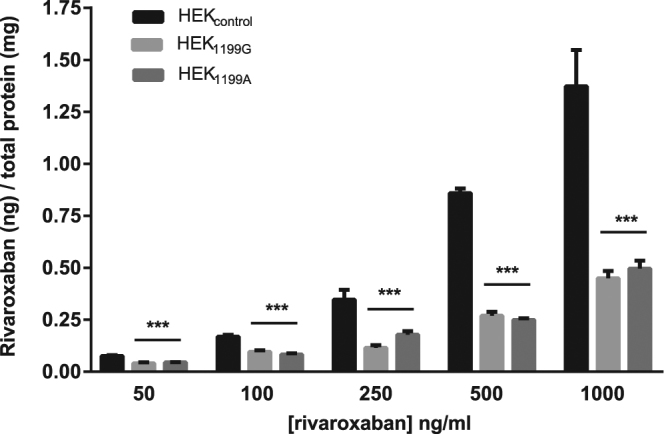
Impact of ABCB1 1199 G > A on the intracellular accumulation of rivaroxaban. Intracellular accumulation of rivaroxaban after 120 min of incubation (N = 3) at different concentrations in HEKcontrol, HEK1199G and HEK1199A. The absolute amount of rivaroxaban (in ng) was divided by the total amount of proteins in cell extracts (in mg). *Compared to control cells: *p < 0.05, **p < 0.01, ***p < 0.001.
Discussion
In this study, we investigated the in vitro impact of ABCB1 genetic polymorphisms on the transport activity towards rivaroxaban. Previously validated HEK293 recombinant cell models were used11,19. We confirmed the involvement of ABCB1 in the active transport of rivaroxaban6. Indeed, we found that ABCB1 expression decreases the intracellular accumulation of the drug. However, we failed to show a significant effect of the different SNPs investigated. The ABCB1 1236 C > T-2677G > T-3435C > T and 1199 G > A SNPs had no significant influence on the intracellular accumulation of rivaroxaban when compared to the wild-type protein.
To our knowledge, this is the first in vitro study to explore the role of ABCB1 genetic determinants in the disposition of DOACs. Few in vivo studies have been published so far. Recently, a case has been described reporting a rivaroxaban-treated patient admitted at the hospital for severe anemia related to gastrointestinal bleeding20. This patient presented abnormally high levels of anti-Xa activity and rivaroxaban plasma concentrations as well as an unexpected delayed clearance. The authors suggested that the homozygous presence of ABCB1 variant alleles 2677–3435TT might have contributed to the altered drug clearance. This observation seems in contradiction with our results showing no influence of those SNPs using in vitro models. However, it must be stressed that this observation relates to an isolated clinical case and, given the relatively high frequency of the homozygous genotype (up to 25% of the population of Caucasian origin is expected to be 2677–3435TT), it might simply reflect a spurious association that is only due to chance. Furthermore, this patient received concomitant simvastatin (40 mg q.d) which is known to inhibit CYP3A4 enzyme but also the ABCB1 and ABCG2 transporters and might have explained the findings. Finally, the patient presented a moderate renal impairment that could have worsened the renal elimination of the drug.
More recently, a randomized, two-center, crossover study was performed in healthy volunteers to study the influence of ABCB1 polymorphisms on the pharmacokinetics of dabigatran and rivaroxaban. Peak plasma concentration and area under the curve (AUC) were compared across three different ABCB1 genotypes i.e. 2677GG-3435CC, 2677GT-3435CT and 2677TT-3435TT and the effect of clarithromycin co-administration on the pharmacokinetics was also assessed. In agreement with the fact that rivaroxaban and dabigatran etexilate are both substrates for ABCB1, clarithromycin co-administration led to a two-fold increase in both drugs’ AUC, irrespective of ABCB1 genotype. Their conclusion meets the observations made in the present investigation as they inferred that ABCB1 genotype is not a significant determinant of inter-individual variability in dabigatran and rivaroxaban pharmacokinetics but that co-administration of a P-gp/CYP3A4 inhibitor with dabigatran etexilate or rivaroxaban may warrant caution.
ABCB1 is involved in the transport of many endogenous, dietary and drug compounds that do not share obvious common structural features. The ABCB1 1236 C > T-2677G > T-3435C > T SNPs have been inconstantly described to influence the clinical pharmacokinetics of immunosuppressants, anticancer agents, antiepileptic drugs or antidepressants5. A trend towards higher concentrations in patients carrying the variant haplotype seems to emerge, but therapy adjustments are currently not required or even recommended. The ABCB1 1199 G > A SNP has previously shown a substrate-dependent impact on transport activity. For instance, the variant allele (1199 G) decreased ABCB1 activity towards tacrolimus in HEK293 and K562 recombinant cell lines, but did not affect the transport of cyclosporine11. Similarly, the in vitro effect of 1199 G > A was lower for nilotinib compared to imatinib, two tyrosine kinase inhibitors12. Structural flexibility and multiple binding sites were suggested as explanations for the polyspecificity of ABCB121. In that way the 1199 G > A SNP, which is located in a cytoplasmic loop involved in substrate recognition, may affect the binding of some ABCB1 substrates while the transport of the others remains unchanged as this seems to be the case for rivaroxaban11.
Another explanation for our negative findings may rely in the fact that rivaroxaban is described as a weak to moderate substrate for ABCB14,22. In Caco-2 cells for instance, ABCB1 inhibition reduced rivaroxaban efflux by only 23% whereas the efflux of dabigatran was inhibited by 87%4. Moreover, rivaroxaban is also substrate for the ABCG2 transporter, known as breast cancer resistance protein (BCRP), and the cytochrome P450 (CYP) 3A4/3A5 enzyme. The active renal secretion by ABCB1 and ABCG2 and hepatic transformation by CYP3A4/3A5 account for 30% and 18% of total drug elimination respectively6. Although combined ABCB1, ABCG2 and CYP3A4 inhibitors were shown to increase rivaroxaban plasma concentrations by up to 150%, the impact of ABCB1 inhibition alone seems marginal23,24. Gong and colleagues demonstrated that rivaroxaban clearance was significantly reduced in mice lacking both ABCB1 and ABCG2, but did not differ for individual transporter knockout25. This suggests a compensatory role of one transporter when the other is inhibited. It is important to note that, in HEK293 cells, the endogenous expression of ABCB1 and ABCG2 is reported to be very low26.
Other genetic determinants could contribute to the inter-individual variability in DOAC plasma concentrations. Future studies are needed to assess the impact of ABCG2 polymorphisms on the transport of rivaroxaban. The ABCG2 rs2231142 (421 C > A, Gln141Lys) SNP, with an allelic frequency around 5–10% in Caucasians and 30–60% in East Asians, is of particular interest as it significantly affected plasma trough concentrations in patients treated with apixaban27,28. In the same study the CYP3A5*3 allele, which is highly prevalent in Caucasians, also contributed to higher apixaban levels. Finally, it could be relevant to investigate the CYP3A4*22 allele, which was associated with a lower CYP3A4 expression and/or activity29.
In summary, the ABCB1 1236 C > T-2677G > T-3435C > T and 1199 G > A SNPs did not affect the efflux of rivaroxaban in HEK293 recombinant cell lines. Therefore, we believe that these SNPs are unlikely to contribute to the inter-individual variability in rivaroxaban plasma concentrations. Future studies should focus on the role of ABCG2, CYP3A4 and CYP3A5 genetic polymorphisms in the transport of rivaroxaban.
Materials and Methods
Materials
Rivaroxaban and [13C6]-rivaroxaban-d5 were purchased from Alsachim (Strasbourg, France).
G418 was purchased from Roche Applied Science (Vilvoorde, Belgium).
Cell culture
Human Embryonic Kidney (HEK293) cells were grown in Dulbecco’s Modified Eagle medium (DMEM) Glutamax 4.5 g/l glucose (Gibco, Invitrogen) supplemented with 10% (v/v) of Fetal Bovin Serum (Gibco, Invitrogen) and 1% (v/v) of Penicillin-Streptomycin solution (Gibco, Invitrogen) at a temperature of 37 °C in the presence of 5% of CO2.
Generation of stable recombinant cell lines
The generation of stable recombinant cell lines has been described in previous studies11,19. Briefly, the expression vectors pcDNA3.1 with cDNA encoding ABCB1 (ABCB11199G and ABCB11236T-2677T-3435T) were used. Mutated plasmids ABCB11199A, ABCB11236C-2677G-3435C and ABCB11236C-2677G-3435T were obtained by site-directed mutagenesis. Plasmids were sequenced to confirm the presence of mutations and HEK293 cells were then transfected with pcDNA3.1 vectors. G418 (concentration 1 mg/ml) was added to select stable recombinant cell lines, which were characterized by flow cytometry, western-blot and immunofluorescence.
Characterization of ABCB1 cell surface expression
After one week of culture in the presence of G418, ABCB1 cell surface protein expression was characterized by flow cytometry as previously described with minor changes11. For each recombinant cell line, 106 cells were harvested by centrifugation and washed twice with ice-cold buffer solution (phosphate buffer saline (PBS), FBS 1%, EDTA 1 mM). Cells were then incubated 45 min on ice in the dark in buffer solution containing FITC Mouse anti-P-glycoprotein antibody diluted 1:10 (clone17F9 557002, BD Pharmingen) or its isotypic control diluted 1:10 (FITC Mouse IgG 2bk, clone27-35 555742, BD Pharmingen). Cells were finally washed with ice-cold buffer solution, centrifuged and resuspended in buffer solution. Samples were analyzed using Fluorescence-activated cell sorting (FACS) Canto II (BD).
Rivaroxaban accumulation
Rivaroxaban accumulation experiments were performed as previously described for tacrolimus or cyclosporin A11. One day before the experiment, 350.000 cells were seeded in poly-L-lysine-coated 24-well plates in 500 µl of complete medium. Rivaroxaban was added at 5 different concentrations: 50, 100, 250, 500 and 1000 ng/ml. Recombinant cells were incubated for 120 min at 37 °C, 5% of CO222. Cells were then washed twice with ice-cold PBS. Cold conditions were applied to block enzymatic activity and to avoid active transport out of the cells, thereby preventing any drug loss during washing steps. After centrifugation and removal of the supernatant, cells were detached with ice-cold PBS containing 0.2% EDTA. Cell pellets were conserved at −80 °C until quantification by LC-MS/MS analysis.
Rivaroxaban quantification
The amount of rivaroxaban extracted from the cell pellet was quantified by LC-MS/MS analysis according to a previously described method, with minor adaptive changes in the sample preparation30. The chromatography system was made up of an UPLC Acquity H-Class system (Waters) coupled with a tandem-quadrupole mass spectrometer Xevo TQ-S (Waters). Separation of the analytes was achieved on a Waters Cortecs UPLC C18 column (2.1 × 100 mm, 1.6 µm). Rivaroxaban was extracted with 100 µl of methanol and water (volume ratio 7:3) containing the internal standard ([13C6]-rivaroxaban-d5) at 2 ng/ml. Blank cell pellets were prepared similarly for the calibration curve, with extraction solutions containing rivaroxaban at concentrations ranging from 0.125 ng/ml to 50 ng/ml. Samples were vortexed and then sonicated for 10 minutes in an ultrasound bath. After centrifugation at 20,000 g for 10 minutes, the supernatant was transferred to a HPLC vial (Macheray-Nagel, Düren, Germany). An aliquot (2 µL) of the final extract was injected into the LC-MS/MS system. For each experiment, the absolute amount of rivaroxaban recovered from the cell extracts was normalized to the amount of proteins measured with the BCA kit (Thermoscientific, Ghent, Belgium).
Statistical analysis
All experiments were repeated twice. Statistical analyses were performed using GraphPad Prism 7.0 for Windows (San Diego California, USA). Analyses of variance were used under the null hypothesis that the means of the different groups were equal. When differences among means were significant, post-hoc Student-Newman-Keuls tests were carried out. In all cases, a p-value of less than 0.05 was considered statistically significant.
Data availability
All data generated or analyzed during the current study are available from the corresponding author on reasonable request.
Acknowledgements
The authors would like to thank S. Ibouraadaten and F. Uwambayinema for their assistance in analytical flow cytometry, Dr N. Dauguet for his help in cell sorting and R. Siriez for his contribution to LC-MS/MS analysis. AL Sennesael is a Research Fellow of the Fonds National de la Recherche Scientifique (FNRS).
Author Contributions
A.L.S., L.E., V.H., A.S. designed the research study. A.L.S. and N.P. performed the experiments and analysed the results. C.V. and L.P. supervised the LC-MS/MS analysis. A.L.S. wrote the first draft of the manuscript and the final version. All the co-authors were responsible for the review of the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Czuprynska, J., Patel, J. P. & Arya, R. Current challenges and future prospects in oral anticoagulant therapy. Br. J. Haematol., 10.1111/bjh.14714 (2017). [DOI] [PubMed]
- 2.Kirchhof P, et al. 2016 ESC Guidelines for the management of atrial fibrillation developed in collaboration with EACTS. Europace. 2016;18:1609–1678. doi: 10.1093/europace/euw295. [DOI] [PubMed] [Google Scholar]
- 3.Jain A, Cifu AS. Antithrombotic therapy for venous thromboembolic disease. JAMA. 2017;317:2008–2009. doi: 10.1001/jama.2017.1928. [DOI] [PubMed] [Google Scholar]
- 4.Hodin, S. et al. In Vitro Comparison of the Role of P-Glycoprotein and Breast Cancer Resistance Protein on Direct Oral Anticoagulants Disposition. Eur. J. Drug Metab. Pharmacokinet., 10.1007/s13318-017-0434-x (2017). [DOI] [PubMed]
- 5.Wolking S, Schaeffeler E, Lerche H, Schwab M, Nies AT. Impact of Genetic Polymorphisms of ABCB1 (MDR1, P-Glycoprotein) on Drug Disposition and Potential Clinical Implications: Update of the Literature. Clin. Pharmacokinet. 2015;54:709–735. doi: 10.1007/s40262-015-0267-1. [DOI] [PubMed] [Google Scholar]
- 6.Mueck W, Stampfuss J, Kubitza D, Becka M. Clinical pharmacokinetic and pharmacodynamic profile of rivaroxaban. Clin. Pharmacokinet. 2014;53:1–16. doi: 10.1007/s40262-013-0100-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jungbauer L, Dobias C, Stollberger C, Weidinger F. The frequency of prescription of P-glycoprotein-affecting drugs in atrial fibrillation. J. Thromb. Haemost. 2010;8:2069–2070. doi: 10.1111/j.1538-7836.2010.03943.x. [DOI] [PubMed] [Google Scholar]
- 8.Greenblatt DJ, et al. Impaired Rivaroxaban Clearance in Mild Renal Insufficiency With Verapamil Coadministration: Potential Implications for Bleeding Risk and Dose Selection. J. Clin. Pharmacol. 2018;58:533–540. doi: 10.1002/jcph.1040. [DOI] [PubMed] [Google Scholar]
- 9.Cheong EJY, Goh JJN, Hong Y, Kojodjojo P, Chan ECY. Rivaroxaban With and Without Amiodarone in Renal Impairment. J. Am. Coll. Cardiol. 2018;71:1395–1397. doi: 10.1016/j.jacc.2018.01.044. [DOI] [PubMed] [Google Scholar]
- 10.Hodges LM, et al. Very important pharmacogene summary: ABCB1 (MDR1, P-glycoprotein) Pharmacogenet. Genomics. 2011;21:152–161. doi: 10.1097/FPC.0b013e3283385a1c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dessilly G, et al. ABCB1 1199G > A genetic polymorphism (Rs2229109) influences the intracellular accumulation of tacrolimus in HEK293 and K562 recombinant cell lines. PLoS One. 2014;9:e91555. doi: 10.1371/journal.pone.0091555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dessilly G, et al. ABCB1 1199G > A polymorphism (rs2229109) affects the transport of imatinib, nilotinib and dasatinib. Pharmacogenomics. 2016;17:883–890. doi: 10.2217/pgs-2016-0012. [DOI] [PubMed] [Google Scholar]
- 13.Chaussade, E. et al. Real-life peak and trough dabigatran plasma measurements over time in hospitalized geriatric patients with atrial fibrillation. J Nutr Helath Aging, 10.1007/s12603-017-0982-4 (2017). [DOI] [PubMed]
- 14.Testa S, et al. Plasma levels of direct oral anticoagulants in real life patients with atrial fibrillation: Results observed in four anticoagulation clinics. Thromb. Res. 2016;137:178–183. doi: 10.1016/j.thromres.2015.12.001. [DOI] [PubMed] [Google Scholar]
- 15.Gulilat M, et al. Interpatient Variation in Rivaroxaban and Apixaban Plasma Concentrations in Routine Care. Can. J. Cardiol. 2017;33:1036–1043. doi: 10.1016/j.cjca.2017.04.008. [DOI] [PubMed] [Google Scholar]
- 16.Eikelboom JW, Quinlan DJ, Hirsh J, Connolly SJ, Weitz JI. Laboratory Monitoring of Non-Vitamin K Antagonist Oral Anticoagulant Use in Patients With Atrial Fibrillation: A Review. JAMA cardiology. 2017;2:566–574. doi: 10.1001/jamacardio.2017.0364. [DOI] [PubMed] [Google Scholar]
- 17.Testa, S. et al. Low drug levels and thrombotic complications in high risk atrial fibrillation patients treated with direct oral anticoagulants. J. Thromb. Haemost., 10.1111/jth.14001 (2018). [DOI] [PubMed]
- 18.Pare G, et al. Genetic determinants of dabigatran plasma levels and their relation to bleeding. Circulation. 2013;127:1404–1412. doi: 10.1161/CIRCULATIONAHA.112.001233. [DOI] [PubMed] [Google Scholar]
- 19.Dessilly G, Panin N, Elens L, Haufroid V, Demoulin JB. Impact of ABCB1 1236C > T-2677G > T-3435C > T polymorphisms on the anti-proliferative activity of imatinib, nilotinib, dasatinib and ponatinib. Sci. Rep. 2016;6:29559. doi: 10.1038/srep29559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ing Lorenzini K, Daali Y, Fontana P, Desmeules J, Samer C. Rivaroxaban-Induced Hemorrhage Associated with ABCB1 Genetic Defect. Front. Pharmacol. 2016;7:494. doi: 10.3389/fphar.2016.00494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chufan EE, Sim HM, Ambudkar SV. Molecular basis of the polyspecificity of P-glycoprotein (ABCB1): recent biochemical and structural studies. Adv. Cancer Res. 2015;125:71–96. doi: 10.1016/bs.acr.2014.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gnoth MJ, Buetehorn U, Muenster U, Schwarz T, Sandmann S. In vitro and in vivo P-glycoprotein transport characteristics of rivaroxaban. J. Pharmacol. Exp. Ther. 2011;338:372–380. doi: 10.1124/jpet.111.180240. [DOI] [PubMed] [Google Scholar]
- 23.Mueck W, Kubitza D, Becka M. Co-administration of rivaroxaban with drugs that share its elimination pathways: pharmacokinetic effects in healthy subjects. Br. J. Clin. Pharmacol. 2013;76:455–466. doi: 10.1111/bcp.12075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hellwig T, Gulseth M. Pharmacokinetic and pharmacodynamic drug interactions with new oral anticoagulants: what do they mean for patients with atrial fibrillation? Ann. Pharmacother. 2013;47:1478–1487. doi: 10.1177/1060028013504741. [DOI] [PubMed] [Google Scholar]
- 25.Gong IY, Mansell SE, Kim RB. Absence of both MDR1 (ABCB1) and breast cancer resistance protein (ABCG2) transporters significantly alters rivaroxaban disposition and central nervous system entry. Basic Clin. Pharmacol. Toxicol. 2013;112:164–170. doi: 10.1111/bcpt.12005. [DOI] [PubMed] [Google Scholar]
- 26.Ahlin G, et al. Endogenous gene and protein expression of drug-transporting proteins in cell lines routinely used in drug discovery programs. Drug Metab. Dispos. 2009;37:2275–2283. doi: 10.1124/dmd.109.028654. [DOI] [PubMed] [Google Scholar]
- 27.Ueshima S, et al. Impact of ABCB1, ABCG2, and CYP3A5 polymorphisms on plasma trough concentrations of apixaban in Japanese patients with atrial fibrillation. Pharmacogenet. Genomics. 2017;27:329–336. doi: 10.1097/FPC.0000000000000294. [DOI] [PubMed] [Google Scholar]
- 28.Hira, D. & Terada, T. BCRP/ABCG2 and high-alert medications: Biochemical, pharmacokinetic, pharmacogenetic, and clinical implications. Biochem. Pharmacol., 10.1016/j.bcp.2017.10.004 (2017). [DOI] [PubMed]
- 29.Elens L, van Gelder T, Hesselink DA, Haufroid V, van Schaik RH. CYP3A4*22: promising newly identified CYP3A4 variant allele for personalizing pharmacotherapy. Pharmacogenomics. 2013;14:47–62. doi: 10.2217/pgs.12.187. [DOI] [PubMed] [Google Scholar]
- 30.Sennesael AL, et al. An optimized dRVVT-based assay to estimate the intensity of anticoagulation in patients treated with direct oral anticoagulants. Thromb. Res. 2017;157:29–37. doi: 10.1016/j.thromres.2017.06.034. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during the current study are available from the corresponding author on reasonable request.



