Abstract
Yellow fever (YF) remains a public health issue in endemic areas despite the availability of a safe and effective vaccine. In 2015–2016, urban outbreaks of YF were declared in Angola and the Democratic Republic of Congo, and a sylvatic outbreak has been ongoing in Brazil since December 2016. Of great concern is the risk of urban transmission cycles taking hold in Brazil and the possible spread to countries with susceptible populations and competent vectors. Vaccination remains the cornerstone of an outbreak response, but a low vaccine stockpile has forced a sparing-dose strategy, which has thus far been implemented in affected African countries and now in Brazil. Accurate laboratory confirmation of cases is critical for efficient outbreak control. A dearth of validated commercial assays for YF, however, and the shortcomings of serological methods make it challenging to implement YF diagnostics outside of reference laboratories. We examine the advantages and drawbacks of existing assays to identify the barriers to timely and efficient laboratory diagnosis. We stress the need to develop new diagnostic tools to meet current challenges in the fight against YF.
Introduction
The last two years have seen a re-emergence of yellow fever (YF) in countries in Africa and the Americas, which brings into acute focus the need for effective tools and protocols in medical practice and public health policy against this arboviral disease. Suitable YF diagnostics in humans, non-human primates (NHPs) and vectors constitute first-line defenses because timely laboratory confirmation of suspected YF cases is essential for effective outbreak control and the prevention of further spread. Meeting the current and future challenges of YF epidemics will require building up laboratory preparedness and proficiency, especially in the geographic areas of disease endemicity, and this build up should be informed by a thorough understanding of yellow fever virus (YFV) diagnostics. Here, we survey the field of YFV laboratory methodology in the context of the YF epidemiological situation in early 2018 as experts associated with the European Centre for Disease Prevention and Control (ECDC) Emerging Viral Diseases-Expert Laboratory Network (EVD-LabNet). We hope that this review of the strengths and limitations of the YF diagnostic toolkit, along with the included background information on the pathogen and disease, will assist diagnostics laboratories and public health officials in targeting areas of their practice for upgrade and research in the context of the ongoing fight against YF epidemics.
The YF epidemiological landscape, 2015–2018
Urban outbreaks of YF were declared in Angola in December 2015 and soon after in the Democratic Republic of the Congo (DRC). WHO declared the end of these outbreaks in January 2017 with a final register of 7334 suspected cases, 965 of which were laboratory-confirmed cases, including 137 fatalities1. In 2016, an unrelated outbreak was declared in Uganda2 and sporadic YF cases were also detected in Chad, Ghana, the Republic of Congo, and Guinea3. Nigeria is currently dealing with an active YFV outbreak4.
The cornerstone of the WHO coordinated international response to stop the transmission and anticipated spread of YF to other countries consisted of reactive and pre-emptive mass vaccination campaigns launched in Angola and the DRC5. A shortage of emergency vaccine supplies, however, led to a dose-sparing strategy implemented during the latest vaccination campaigns in Africa, which used one-fifth of the original dose6. Preliminary estimates of the seroconversion rate are not divergent from those achieved by full-dose vaccination7–9, but data are scarce on the duration of the immunity imparted by this approach.
In December 2016, a YF outbreak was declared in Brazil with over 3240 suspected (779 confirmed) human cases as of 13 December 201710. The number of cases declined from May 2017 onwards, but from July 2017 to 13 March 2018, 920 human cases (300 deaths) were reported in the states of Minas Gerais, São Paulo, Rio de Janeiro, and Espírito Santo and in the Federal District11. An alarming number of epizootics in NHPs have been reported from different Brazilian states during the considered time period, with the Sao Paulo metropolitan area accounting for 40% of them12. The presence of epizootics and confirmed cases near the urban areas of São Paulo and Rio de Janeiro and in municipalities that were previously considered not at risk of YF is a worrying trend because much of the population in these areas remain unvaccinated12. This outbreak, the most severe for several decades in Brazil, raises the concern that YF infections are no longer confined to jungle and remote areas as sylvatic transmission is now also occurring in the periphery of densely populated cities13.
In October 2017, the Brazilian public health authorities responded to the recorded epizootics with vaccination campaigns in the Northern areas of the city of São Paulo in an effort to prevent human cases in areas bordering epizootic prevalence and to control the risk of an urban outbreak14. A massive vaccination campaign took place in São Paulo in early 2018 using a fractionated dose of the vaccine15. Due to a limited vaccine stock, the Brazilian Ministry of Health adopted the WHO recommendation to administer a single dose of YF vaccine6; however, this strategy has generated some controversy regarding the duration of immunity16,17, and this decision is considered an emergency response to be re-evaluated in the short term18.
Colombia, Peru, Bolivia, Suriname, Ecuador, and French Guiana11 have also reported cases of epizootic and sylvatic YF in recent years. An aggravated risk of further disease spread was suggested by the increased incidence of sylvatic YF and the detection of human cases in Peru11. Likewise, Bolivia reported in February 2017 the first YF case in decades, which involved a non-vaccinated tourist, and four additional cases were confirmed in this country from May to July 201719.
Further international spread to areas with susceptible populations and competent mosquito vectors is a grave concern20. WHO considers the risk of YF spread at the regional level in the Americas to be low given the high vaccination coverage in Brazil’s neighboring countries, but the detection in August 2017 of a human case in French Guiana near the border with Brazil shows that the risk is real. At a global level, the risk remains restricted to non-vaccinated travelers12. Recently, the Evandro Chagas Institute reported the detection of YF in Ae. albopictus mosquitoes in 201712. The European Centre for Disease Prevention and Control (ECDC) stated a risk of uncertain magnitude for regions harboring endemic Ae. albopictus populations20,21. In non-endemic regions, such as Europe, preparedness and capability assessment activities for reference laboratories have to be undertaken to guarantee a timely diagnosis of suspected cases in travelers returning from areas with increased YFV circulation22.
This landscape of YF outbreaks has prompted WHO and partner organizations (UNICEF and GAVI) to revise their long-term YF strategy for the next 10-year period (2017–2026). The novel EYE (Eliminating Yellow fever Epidemics) strategy is a global and comprehensive long-term (2017–2026) scheme that builds on lessons learnt from recent outbreaks and aims to protect at-risk populations, prevent the international spread of the disease, and readily contain outbreaks23.
The pathogen
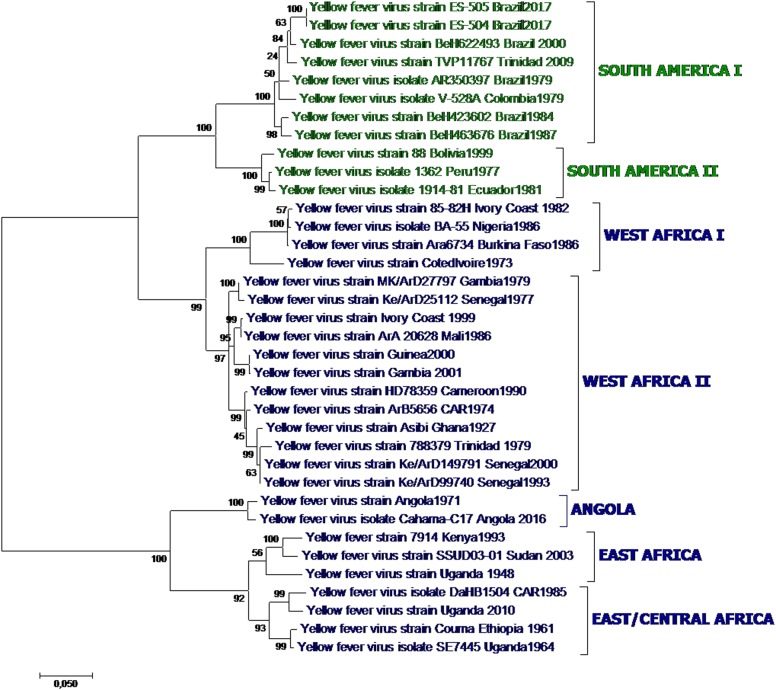
YFV is an enveloped virus with a single-stranded RNA genome and is a member of the genus Flavivirus, family Flaviviridae. Other flaviviruses of major importance to human health are dengue virus (DENV), West Nile virus (WNV), Zika virus (ZIKV), Japanese encephalitis virus (JEV), and tick-borne encephalitis virus (TBEV). YFV belongs to the YFV serogroup of mosquito-borne flaviviruses and is transmitted by Aedes mosquitoes. The circulating YFV strains constitute a single serotype, but seven major genotypes have been described (Fig. 1), five of which circulate in Africa and two in South America24.
Fig. 1.
Yellow fever phylogenetic analysis showing major YFV genotypes, based on alignment of a 1428 nt region of the prM-E junction region for 36 representative African and American YFV strains (Table 1) using the Maximum Likelihood method based on the general time reversible model (GTR). Individual strains are defined by name and country/year of isolation. Bootstrap values (500 replicates) for major branches are indicated
The evolutionary rates described for YFV are similar across the various genotypes and are estimated to be lower than those of other mosquito-borne flaviviruses, such as DENV25. Estimated genetic variance within the clade is 10–23% at the nucleotide level for the five African genotypes and 5% for the two American genotypes. The African genotypes are up to 16% dissimilar from the American genotypes26.
The strain of the recent Angola and DRC outbreaks is most closely related to that responsible for the 1971 Angola outbreak;27 likewise, preliminary sequencing of the 2016 Uganda strain showed that it is most closely related to a strain isolated in this country in 201028, which was of East/Central Africa genotype. Whole-genome sequence analysis of the current Brazil outbreak strain assigned it to South American genotype 1E29, which arose in Brazil during the 2008–2009 epidemics30, and identified eight mutations of possible functional importance that are still under investigation31. These changes, however, are not expected to affect the efficacy of currently available vaccines31.
Epidemiology and geographical spread
YFV is endemic and intermittently epidemic to tropical and subtropical areas of South America and Africa. Africa accounts for 90% of the global burden of YFV. The true incidence of YF is unknown because of insufficient reporting, ground surveillance, and limited access to specific diagnostics for YF and other common pathogens in differential diagnosis (i.e., malaria and viral hepatitis). Estimates based on African data sources from 2013 put the incidence at 84,000–170,000 severe cases per year, causing 29,000–60,000 deaths2. Autochthonous YFV transmission has not been detected in Asia yet, despite a large susceptible population and widespread competent mosquito vectors.
YFV is maintained in a sylvatic transmission cycle between NHPs and jungle mosquitoes (Aedes spp. in Africa, Haemagogus spp. and Sabethes spp. in South America), with humans getting infected when they enter forested areas for occupational, tourism, or leisure activities. The arrival of viremic persons in a densely populated urban environment could initiate a transmission cycle between humans and competent vectors present in the area, mainly Ae. aegypti. This so-called urban transmission cycle would have devastating consequences in Brazil, similar to those in the recent Angola and DRC outbreaks. Thus far, Ae. aegypti has not been involved in the ongoing Brazilian outbreak11,12, and the recent detection of YF in Ae. albopictus in rural areas in the state of Minas Gerais in Brazil deserves further investigation12. In Africa, the savannah transmission cycle connects the sylvatic and urban cycles by involving Aedes mosquitoes (Ae. furcifer, Ae. vittatus, Ae. luteocephalus and Ae. africanus in West Africa; Ae. africanus and Ae. simpsoni in East Africa) that feed on both humans and monkeys32.
NHPs involved in the sylvatic transmission of YFV in the Americas belong to the genera Aotus, Alouatta, Cebus, Ateles, Callithrix, and Saimiri. American NHPs exhibit different susceptibility to YFV. Alouatta (howler) monkeys are particularly susceptible, and they frequently die after YFV infection due to liver and renal failure and hemorrhage caused by the infection, whereas Callithrix and Cebus monkeys exhibit different grades of resistance. On the other hand, African NHPs experience inapparent infections while viremic33. The use of NHPs as sentinels for the early detection of the circulation of YFV is a proven useful surveillance tool to evidence the presence of sylvatic activity of the virus, leading to the activation of countermeasures (i.e., vector control and population vaccination) to control the spread of the virus and the occurrence of epidemics34,35. The collection of appropriate material for diagnosis is an essential part of epizootics investigation, and proper storage and transport are key to the reliability of the laboratory results. The prioritized samples for epizootics investigation are blood, serum, and tissues (liver, spleen, kidneys, heart, lung and brain, when possible). In the laboratory, viral isolation, genome detection, serology, histopathology, and immunohistochemistry (IHC) exams are attempted36.
During the recent Brazilian outbreak, a number of YFV-infected marmosets were detected in urban areas. Given the habitat versatility of marmosets, whose range includes forest edge areas, the question has been raised about their role not only as sentinels but also as a link in the transmission cycle of the virus and the spread of YF to urban areas37.
Except for one case of nosocomial transmission in the 1930s38, there are no reports of direct human-to-human YFV transmission outside the laboratory (see Biosafety below). However, transplacental-39,40, breastfeeding-41–43, and blood donation-based viral transmission44 has been described for the live attenuated YFV vaccines.
Lastly, the recent discovery of sexual transmission of Ebola virus (EBOV)45 has prompted investigations of this alternative, previously overlooked mode of transmission. Sexual transmission of ZIKV has been demonstrated46. Clinical and experimental investigations of YFV via sexual transmission are thus warranted.
YF disease
As in other flavivirus infections, most YFV-infected people are asymptomatic. When present, symptoms may include fever, headache, nausea, muscle pain, backache, vomiting, jaundice, and bleeding from the mouth, nose, eyes or stomach. In 25–50% of cases, the disease can progress into full hemorrhagic syndrome with multiorgan failure47. Treatment for YF is only supportive. The clinical course comprises three stages: infection, remission, and intoxication, often without clear stage demarcation. During the so-called period of remission, starting 3–4 days after the onset of symptoms, clinical signs subside and the patient may either go into remission or conversely deteriorate into the intoxication phase, which is characterized by high fever, nausea, vomiting, abdominal pain, and changes in consciousness48. Jaundice from excess bilirubin arises from liver cell damage (uniquely among hemorrhagic fevers), along with epistaxis, bleeding of the oral mucosa, hematemesis, and petechial hemorrhage. The patients may further deteriorate rapidly, and 20–50% will die 7–10 days after the onset of symptoms. Jaundice and increased liver enzymes, specifically serum aspartate aminotransferase (AST) levels over 1200 UI/l, have been correlated to disease severity and higher mortality49. Renal failure is also a manifestation of severe and fatal YF, and blood urea nitrogen (BUN) levels over 100 mg/mL were associated with an elevated risk of death49.
Vaccination against YF has been associated with rare cases of viscerotropic (yellow-fever vaccine-associated viscerotropic disease, YEL-AVD) and neurotropic disease (yellow-fever vaccine-associated neurotropic disease, YEL-AND)50. YEL-AVD clinical presentation is similar to wild-type YF disease with nonspecific initial symptoms, including fever, headache, malaise, myalgia, arthralgia, nausea, vomiting, and diarrhea, starting 2–8 days after vaccination. Jaundice can appear, along with thrombocytopenia and the elevation of hepatic transaminases, total bilirubin, and creatinine. Severe YEL-AVD is characterized by hypotension, hemorrhage, and acute renal and respiratory failure, leading to multiorgan system failure. Similarly, YEL-AND includes post-vaccinal encephalitis but also autoimmune disease with central or peripheral nervous system involvement, such as acute disseminated encephalomyelitis or Guillain-Barré syndrome. The clinical presentation varies but includes high fever and headache associated with confusion, lethargy, encephalitis, encephalopathy, and meningitis51.
Infection kinetics
Awareness of YFV infection kinetics is essential in designing optimal sampling strategies because the timing of sample-taking and the nature of the biological sample constrain diagnostic interpretation.
Viraemia
YFV infection by a mosquito bite typically has a 3–6-day incubation period (range: 2–9 days)52. It was traditionally assumed that YFV could be detected afterwards in the serum, plasma, or whole blood of symptomatic patients during the first 5 days of illness. Molecular diagnostics have now shown that viral RNA can be efficiently detected for longer periods in the blood and autopsy tissues of severe cases53–59 and up to 20 days after the onset of symptoms52,60,61.
Data are scarce on the detection of YFV in other body fluids, such as urine, saliva, or semen, following natural infection. It has been demonstrated that YFV can be detected for a longer time period in urine than in serum, and can be detected up to 25 days post-inoculation in cases of suspected adverse events after YF vaccination62. Recently, YFV RNA has been efficiently detected in urine samples from natural infection cases60,61,63 and in semen up to 20 days after disease onset61, which is also a substantially longer detection time than in serum. These observations strongly suggest urine as a valuable diagnostic sample for YF as observed previously for other flaviviruses, such as WNV and ZIKV, which deserves more attention. Likewise, the transmission of YF vaccine virus to babies born to vaccinated mothers suggests the presence of YFV in breast milk41–43.
Humoral immune response
Typically, anti-YFV IgM antibodies develop within a few days after the onset of illness with flaviviruses and can generally be detected for up to 3 months, whereas IgG antibodies develop within days subsequent to the IgM response and can be detected for years afterwards. The persistence of IgM antibodies for longer periods has been reported in a small percentage of vaccinees, which could interfere with diagnostic testing64. IgM production in response to secondary flavivirus infection (e.g., in YF cases with a prior history of infection by DENV) may be absent or small, hampering the serological identification of acute cases33,64.
YF diagnostics: state of the art
The clinical diagnosis of YF is problematic because the symptoms resemble those of a wide range of diseases, including dengue, other hemorrhagic viral diseases, leptospirosis, viral hepatitis, and malaria. All of these diseases have to be considered in differential diagnosis, and laboratory confirmation is essential. Detection of YFV-specific IgM in the absence of recent YF vaccination and negative diagnosis (including IgM antibodies) for other flaviviruses is considered confirmatory of YF. More robust corroboration of YFV infection, however, is provided by immunohistochemical detection of the YFV antigens, PCR amplification of YFV genomic sequences from blood or solid tissues, or by a test for viraemia involving the cultivation of YFV infectious particles. Generally, these assays are performed only in a few national or international reference laboratories.
YFV molecular diagnostics
Eleven quantitative real-time RT-PCR assays for molecular detection of the YFV genome have been described as of March 201865–75. In addition, four alternative assays oriented to field and point-of-care diagnosis have been reported in recent years based on isothermal amplification of the viral genome69,76–78.
In selecting an assay for the diagnosis of natural YFV infections, one should avoid those designed specifically for vaccine strains65,71 because their detection of wild strains would be less reliable79. In this work, we have reviewed assay specificity in the context of the differences between American and African strains and sequence diversity among the strains currently circulating in endemic areas. We matched the target sequences of published RT-PCR and isothermal amplification assays against an alignment of 61 complete YFV genomic sequences from GenBank (Table 1) to check the capability of each assay to detect all strains. Among the 14 assays described in international journals65–78, most were clearly unsuitable for many strains owing to excessive mismatches between the primers and/or probe and the target genome. Four real-time RT-PCR assays, namely, two TaqMan69,70, one LNA75 and one SYBR Green-based assay67, were studied in detail and are predicted to detect all considered strains (Fig. 2). Similar in silico analyses of published isothermal amplification protocols predicted functional sets of primers and probes in two assays (Fig. 3), whereas the number of mismatches in the other two assays77,78 made them unsuitable for at least some of the wild strains. The generic qRT-PCR protocol described in ref. 69 is currently recommended by PAHO as the benchmark method for YF diagnosis in reference laboratories. This robust assay performs equally well with a variety of commercial reagents, has been extensively validated and implemented in several laboratories, and provides a profile of sensitivity and specificity appropriate for reliable case detection69. Used as the assay of reference, it has enabled the homogenization and standardization of laboratory data between different settings in the current Brazilian outbreak.
Table 1.
Yellow fever virus strains (A: vaccine strains, B: wild type strains) included in the evaluation of suitability of published molecular detection assays
| A Yellow fever vaccine strains | |
|---|---|
| GenBank accession | Name |
| U21055.1 | YFV French neurotropic strain |
| U21056.1 | YFV French viscerotropic strain |
| X03700.1 | YFV 17D vaccine strain |
| U17067.1 | YFV vaccine strain 17D-213 |
| U17066.1 | YFV vaccine strain 17DD |
| DQ100292.1 | YFV strain 17DD-Brazil |
| DQ118157.1 | YFV isolate YF-AVD2791-93F/04 |
| JN628281.1 | YFV strain 17D Flavimun TVX |
| JN628280.1 | YFV strain 17D Flavimun WSL |
| JN628279.1 | YFV strain 17D RKI |
| JN811143.1 | YFV 17D YF-VAX Series C P11 |
| JN811142.1 | YFV 17D YF-VAX Series B P11 |
| JN811141.1 | YFV 17D YF-VAX Series A P11 |
| JN811140.1 | YFV 17D YF-VAX Series A P1 |
| KF769015.1 | YFV strain 17D-204 |
| GQ379163.1 | YFV strain case #2 |
| GQ379162.1 | YFV strain case #1 |
| JX503529.1 | YFV strain YF/Vaccine/USA/Sanofi-Pasteur-17D-204/UF795AA/YFVax |
| FJ654700.1 | YFV 17D/Tiantan |
| NC_002031.1 | YFV, NCBI reference sequence |
| B Yellow fever wild type strains | |||
|---|---|---|---|
| GenBank accession | Sequence name | Country/year | Genotype |
| KU921608.1 | YFV isolate CNYF01/2016 | China ex Angola/2016 | Angola |
| AY968064.1 | YFV strain Angola71 | Angola/1971 | Angola |
| KX027336.1 | YFV isolate CIC4 | China ex Angola/2016 | Angola |
| KX010996.1 | YFV isolate CIC3 | China ex Angola/2016 | Angola |
| KX010995.1 | YFV isolate CIC2 | China ex Angola/2016 | Angola |
| KX010994.1 | YFV isolate CIC1 | China ex Angola/2016 | Angola |
| KF907504.1 | YFV strain 88/1999 | Bolivia/1999 | Angola |
| AY968065.1 | YFV strain Uganda48a | Uganda/1948 | East Africa |
| DQ235229.1 | YFV strain Couma | Ethiopia/1961 | East/ Central Africa |
| JN620362.1 | YFV strain Uganda 2010 | Uganda/2010 | East/ Central Africa |
| JF912190.1 | YFV strain BeH655417 | Brazil/2002 | South America I |
| JF912189.1 | YFV strain BeAR646536 | Brazil/2001 | South America I |
| JF912188.1 | YFV strain BeH622493 | Brazil/2000 | South America I |
| JF912187.1 | YFV strain BeH622205 | Brazil/2000 | South America I |
| JF912186.1 | YFV strain BeH526722 | Brazil/1994 | South America I |
| JF912185.1 | YFV strain BeAR513008 | Brazil71992 | South America I |
| JF912184.1 | YFV strain BeH463676 | Brazil/1987 | South America I |
| JF912183.1 | YFV strain BeH423602 | Brazil /1984 | South America I |
| JF912182.1 | YFV strain BeH422973 | Brazil /1984 | South America I |
| JF912181.1 | YFV strain BeH413820 | Brazil /1983 | South America I |
| JF912180.1 | YFV strain BeH394880 | Brazil /1981 | South America I |
| JF912179.1 | YFV strain BeAR378600 | Brazil/1980 | South America I |
| KY885000 | YFV strain ES-504 | Brazil/2017 | South America I |
| KY885001 | YFV strain ES-505 | Brazil/2017 | South America I |
| JX898869.1 | YFV isolate DakArAmt7 | Cote d’Ivoire/1973 | West Africa I |
| AY603338.1 | YFV strain Ivory Coast 1999 | Cote d’Ivoire/1999 | West Africa I |
| U54798.1 | YFV strain 85-82H Ivory Coast | Cote d’Ivoire/1982 | West Africa I |
| AF094612.1 | YFV strain 79A/788379 | Trinidad/1979 | West Africa II |
| KF769016.1 | YFV strain Asibi | Ghana/1927 | West Africa II |
| JX898881.1 | YFV isolate ArD181439 | Senegal/2005 | West Africa II |
| JX898880.1 | YFV isolate ArD181564 | Senegal/2005 | West Africa II |
| JX898879.1 | YFV isolate ArD181676 | Senegal/2005 | West Africa II |
| JX898878.1 | YFV isolate ArD181250 | Senegal/2005 | West Africa II |
| JX898877.1 | YFV isolate ArD181464 | Senegal/2005 | West Africa II |
| JX898876.1 | YFV isolate ArD156468 | Senegal/2001 | West Africa II |
| JX898875.1 | YFV isolate ArD149815 | Senegal/2000 | West Africa II |
| JX898874.1 | YFV isolate ArD149194 | Senegal/2000 | West Africa II |
| JX898873.1 | YFV isolate ArD149214 | Senegal/2000 | West Africa II |
| JX898872.1 | YFV isolate ArD114972 | Senegal/1995 | West Africa II |
| JX898871.1 | YFV isolate ArD114896 | Senegal/1995 | West Africa II |
| JX898870.1 | YFV isolate ArD121040 | Senegal/1996 | West Africa II |
| JX898868.1 | YFV isolate HD117294 | Senegal/1995 | West Africa II |
| AY572535.1 | YFV strain Gambia 2001 | Gambia/2001 | West Africa II |
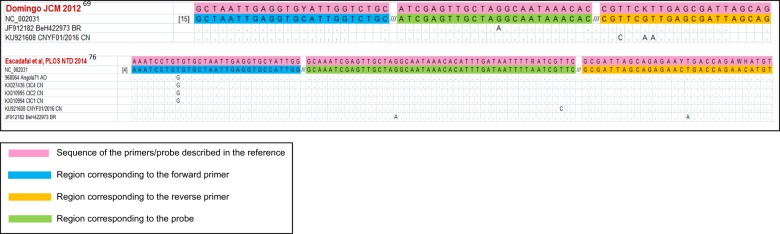
Fig. 2. Alignment of the primers and probes of shortlisted assays against relevant YFV target sequences.
The figure is restricted to the four assays described in references67,69,70,75, which generated the fewest mismatches overall in the comparison with the reference set of 61 YFV genomic sequences (Table 1). Perfectly matched YFV sequences are not shown. Primer and probe sequences are written 5′ to 3′ except for the reverse primers at the right edge of the figure, which are represented by the reverse-complement of the oligonucleotide sequence
Fig. 3. Alignment of the primers and probes of shortlisted assays against relevant YFV target sequences.
The figure is restricted to the two assays described in references69,76, which generated the fewest mismatches overall in the comparison with the reference set of 61 YFV genomic sequences (Table 1). Perfectly matched YFV sequences are not shown. Primer and probe sequences are written 5′ to 3′ except for the reverse primers at the right edge of the figure, which are represented by the reverse-complement of the oligonucleotide sequence
Two additional qRT-PCR methods have been recently reported for the detection and initial identification of YFV wild and vaccine strains by qRT-PCR as an alternative approach to sequencing. One method consists of a YFV American strain-specific qRT-PCR duplexed with a specific YF vaccine qRT-PCR74. The second approach consists of the use of a reference generic YF qRT-PCR method for case detection69, coupled to a generic YFV vaccine-strain qRT-PCR methodproviding a global approach covering all strains80. Since the specificity of these methods is based on single nucleotide differences between wild-type and vaccine strains, the identification of a natural infection versus a vaccine-related adverse event in a vaccinated patient in an area with no reported YF infections should be carefully characterized to exclude the possibility of mutations in the vaccine strain that could lead to case misclassification.
It was suggested recently that next-generation sequencing may be useful for the diagnosis of emerging infectious diseases in general, and hemorrhagic viral diseases specifically, as samples are analyzed in a non-biased manner with no assumptions regarding the pathogen involved. This approach may be valuable in identifying the causative agent at outbreak onset and in sporadic cases and characterizing novel pathogens or suspected new strains of known viruses, but its wider application for routine YF diagnostics has not been suggested81,82.
Commercial kits for YFV genome detection are provided by Genekam, Genesig, ViPrimePLUS, PCRmax, LifeRiver Bio-Tech, Altona, and Fast-Track Diagnostics (FTD). FTD combines a test for YFV with Brucella spp, Streptococcus pneumonia and Coxiella burnetii detection within a Tropical Fever Africa panel. The stated detection threshold is 100 copies/reaction for the ViPrimePLUS assay, and a more sensitive 1,000 copies/ml threshold for the FTD and LifeRiver assays; no information is provided for the other assays. The FTD Tropical Fever Africa and LifeRiver kits carry the conformity mark for European Economic Area regulations (CE). From Genekam, the dedicated YFV kit is CE-marked, but the three kits allowing YFV detection in combination with other pathogens (YFV + ZIKV + CHIKV; YFV + EBOV + Rift Valley fever virus; YFV + ZIKV) are not. Assays distributed by Genesig, VirPrimePLUS, Altona, and PCRmax are not CE-marked. To the best of our knowledge, no peer-reviewed reports on the evaluation of these kits using clinical samples from natural infections are available.
The recently sequenced YFV Brazilian strain displays eight non-silent base substitutions relative to previous isolates, seven in the NS3 and NS5 genes and one in the C protein gene31. Caution is advised about using any assays targeting these genes, but the new mutations should not affect the performance of the selected assays (Figs. 2 and 3), which all target the 5ʹ-noncoding region of the genome.
Special consideration is given to the use of paraffin-embedded or formalin-fixed samples for the molecular detection of YFV RNA. Even though qRT-PCR assays amplifying short regions of the YFV genome are very useful for these samples and support the results of histopathology or IHC, the detection of YFV RNA in these samples is, however, not entirely consistent, and false negatives might occur due to RNA degradation or damage during sample preparation and extraction, the generation of secondary structures during prolonged formalin fixation or the presence of inhibitors78.
Serology: an overview of current knowledge and methodologies
Limitations of the serological diagnosis of YFV infections
The serology criteria for YFV infection are the detection of either YFV-specific IgM species or a four-fold or greater increase in anti-YFV IgG antibody titers in acute and convalescent samples83. YF serological diagnosis, however, is complicated by cross-reactivity with other members of the genus Flavivirus (such as DENV, WNV, Saint Louis encephalitis virus (SLEV), or ZIKV), the phenomenon known as original antigenic sin, and the lack of extensively validated commercial assays.
Prior immunity to DENV is the most frequent confounder generating non-specificity in current serological tests84, which is relevant as DENV and YFV share distribution areas in the Americas and Africa, and a DENV vaccine has recently been introduced into some countries in the Americas.
The approach to YFV serological diagnosis recommended by WHO differs slightly depending on the epidemiological context, i.e., outbreaks versus endemicity or non-endemicity areas. The guidelines take into account the presence of antigenically related viruses but also the high prevalence of vaccination in the areas affected and the usual concomitant implementation of YFV vaccination campaigns85. Correct attribution of severe symptoms to either natural infection or the adverse effects of vaccination is particularly difficult in outbreak contexts, where the antigenic similarity between wild-type and vaccine strains precludes unambiguous serological identification. The distinction is only currently possible through molecular characterization of the causative agent by sequence analysis or alternatively by the molecular methods mentioned previously.
Assays for serological diagnosis of YFV infections
A variety of in-house methods have been described for YF serodiagnosis, surveillance purposes, or confirmation of the immune response to vaccination. Serodiagnosis is often requested from reference laboratories for individuals in which special circumstances may compromise the response to vaccination, such as pregnancy, immunosuppressive treatment, HIV infection, or other instances of inborn or acquired immunodeficiency.
The plaque reduction neutralization (PRNT) assay, or virus neutralization test (VNT), is the most specific method for the detection of YFV antibodies and the current “gold standard” for flavivirus differential diagnosis. A degree of cross-reactivity with other flaviviruses (e.g., DENV and ZIKV) has been observed in the PRNT assay during secondary flavivirus infections86, with the higher stringency PRNT 90% assay providing greater specificity than other assays (although sensitivity may decrease as a trade-off). PRNT assays, however, require specific cell culture facilities, standardized controls and well-trained personnel for reproducible results. This limitation confines PRNT to reference laboratories, which may create a diagnosis bottleneck in outbreak situations. Moreover, the time to final interpretation of results, which is usually 4–7 days, delays diagnosis and is not best-suited to decision-making during an outbreak response, i.e., regarding vaccination deployment.
Hemagglutination inhibition (HI) and complement fixation (CF) methods have been used in the past for the serodiagnosis of YF, but they have been used less frequently in recent years as they are non-discriminant of the IgM/IgG antibody class and perform poorly in comparison to alternative assays79,87.
Other tests that are currently used for the detection of IgM and IgG antibodies against YFV include in-house indirect immunofluorescence methods (IIF), which require well-trained personnel for correct interpretation, and ELISA79,87,88, MAC-ELISA89, and ELISA inhibition tests89,90. More recently, a multiplex microsphere immunoassay (MIA) test has been described for the detection of arboviral antibodies, including those against YFV91.
The United States Centers for Disease Control and Prevention (US CDC) have traditionally provided (via WHO) testing reagents for a MAC-ELISA assay to endemic countries92. This test uses whole-virus antigen propagated in mouse brain, it takes over two days to perform, and the reagents exhibit lot-to-lot variation (not all reagents are supplied by US CDC); storage conditions may also influence the quality of results. Prior standardization is therefore required in each practicing locale, which restricts the test to well-trained laboratories. Despite these limitations, the availability of these reagents has for years enabled IgM testing by laboratories in endemic regions. The results for an improved MAC-ELISA kit from the US CDC employing antigen produced in Vero cells with lyophilized and stabilized reagents have been published recently93. The test can be run in one day and is intended for standard laboratories. Because it uses whole-virus antigen, however, it inherits the cross-reactivity risk of earlier protocols. Data on field applications of this assay are urgently needed as it is currently one of the few reliable options for YF serology in many laboratories. An IgM capture ELISA using new monoclonal antibodies against YFV has been described recently and has a promising sensitivity and specificity profile;94 however, these reagents are not yet widely available.
We know of four commercial tests currently available for IgM- or IgG-based serological diagnosis of YF. An immunofluorescence assay is available from EUROIMMUN AG (Lübeck, Germany) in 5- or 10-sample packaging. Each serum sample is reacted in parallel against an antigenic substrate (whole virus in infected cells) and a non-antigenic control (non-infected cells), which favors better interpretation of the results and facilitates the identification of false positives. The test was validated by the manufacturer on 300 European serum samples (150 from Swiss YFV-17D vaccinees and 150 from German blood donors) with an overall specificity of 96% in the IgM version of the assay and 94.7% for IgG and an overall sensitivity of 94.4% for IgM and 94.7% for IgG95. A YFV seropositivity of 4% for IgM and 6% for IgG was reported in the negative control group. This result exceeds the proportion of YFV vaccinees among the general German population, and results from these tests must therefore be interpreted with caution. Low sensitivity was observed toward IgM antibodies in the sera of YF-17D vaccinees79. Nevertheless, this commercial assay is the only one with available validation data. The manufacturer also sells multiplex assays in which sera are tested against several antigens in parallel; the assays including YFV are as follows: Flavivirus Mosaic 1 (TBEV, WNV, JEV, and YFV), Flavivirus Profile 2 and Flavivirus Mosaic 3 (TBEV, WNV, JEV, YFV, and DENV 1–4) and Arbovirus Profile 3 (ZIKV, CHIKV, DENV, TBEV, WNV, JEV, and YFV). Performance varies among the different antigens, along with the reported specificity profile. The proportion of positive sera for anti-DENV antibodies that presented an anti-YFV positive result is 100% with respect to IgG detection and 22.2% with respect to IgM detection. Likewise, anti-JEV positive sera are anti-YFV-positive in 100% (IgG) and 33.3% (IgM) of cases; and the figures for anti-WNV positive samples are 91.7% (IgG) and 33.3% (IgM). No data are available for cross reactions on Zika-positive samples. Other issues to consider in using IIF assays are the requirement for experienced technical personnel at the interpretation stage, the small number of samples that can be assayed in parallel, and the higher cost per test relative to other routine methods.
ELISA tests for IgM or IgG antibodies in 96-well plates (human yellow fever virus IgM/IgG ELISA kit) are available from Abbexa Ltd. (Cambridge, UK). No data on the performance of this assay have been reported by the manufacturer. MyBiosource, Inc., (San Diego, CA, USA) manufactures sandwich ELISA kits (Qualitative Human Yellow Fever Antibody IgM (YFV-IgM) or IgG (YFV-IgG)) in 48- and 96-sample formats and provides figures for intra- and inter-assay precision. No data are available on assay validation in regard to sensitivity, specificity or cross-reactivity. This assay is labeled for in vitro research only and not for diagnostic use. The Tariki YF-ELISA (Tariki Fiebre Amarilla IgM) is an IgM capture ELISA produced and sold in Peru since 2013 by the National Institute of Health of that country. The reported overall sensitivity is 95% (95% confidence interval: 87–100%) with 98% specificity (95% confidence interval: 87–100%)96. However, few data are available on the validation procedure for this test or its wider application in laboratories of the region.
Other confirmatory assays for the diagnosis of YF
Histology and IHC
Even after the introduction of molecular methods, histological (hematoxylin-eosin staining) and immunohistochemical techniques continue to be valuable to reference laboratories as they provide supportive diagnoses in deceased cases and are useful for investigating epizootics. They provide a reliable diagnosis when antemortem serum or blood samples are not available, when specimens were not stored in conditions suitable for genome detection or viral isolation, or when hemolytic or autolytic processes are present.
The typical YF lesion is marked by lytic necrosis associated with hepatocyte apoptosis in the mid-zone of the liver lobule; cells bordering the central vein and portal triads are spared, and macro- and microvacuolar fatty changes can be observed in centrilobular cells. Eosinophilic degeneration of hepatocytes results in the formation of Councilman bodies and intranuclear eosinophilic granular inclusions. There is no disruption of the reticular architecture of the liver, and in nonfatal cases, healing is complete without postnecrotic fibrosis. Tubular necrosis in the kidneys is also observed frequently. The classical pathognomonic histological features of YFV infection are present only during the acute or late acute stages of the disease. Therefore, given a histological pattern of nonspecific hepatitis, ruling out a diagnosis of YF is contingent on additional evidence from serological and molecular tests.
The pathologic changes of YF-associated disease in NHPs are not fully resolved, and merit further study comparing the pathological findings in humans and other primates97. Divergences in the histopathological features of naturally and experimentally infected howler monkeys (Alouatta), where hepatic inflammatory mononuclear cell infiltration and hemorrhage are more pronounced than in humans or other primate species, must be carefully considered; otherwise, the diagnosis could be misleading as well as the identification of epizootics98.
Viral antigens can be detected in Kupffer cells and hepatocytes by IHC using YF-specific murine monoclonal antibodies or polyclonal rabbit sera. In addition, YFV antigens can also be detected in renal tubular epithelium and in groups of myocardial fibers68. YFV antigens and YFV RNA have been detected in the liver, kidney, spleen, lung, brain, and heart of deceased patients56,68, indicating that viral replication is not restricted to the liver and kidney, the major target organs.
Samples for IHC are preferably fixed using 10% neutral buffered formaldehyde99 and embedded in paraffin. Formaldehyde fixation prevents degradation and facilitates the manipulation and transport at room temperature of inactivated specimens to the reference laboratory; this effect is of great practical importance for field work and the investigation of epizootics in remote locations. The preferred protocol for YFV IHC uses specific antibodies against YF and the avidin-biotin complex technique. The quality, specificity, and careful validation of primary antibodies at laboratories are crucial for reliable results. The US CDC and the Evandro Chagas Institute (Brazil) produce and standardize qualified primary antibodies for this purpose. Different commercial reagents for detection are available, but the MACH-4 AP system (Biocare) is presently recommended as it provides increased sensitivity with minimal background by using a polymer-based detection system100.
Histopathological and IHC studies are laborious and require specific technical capability and expertise to provide reliable results; hence, they are practiced as reference techniques in expert laboratories.
Virus isolation
Laboratories embarking on YFV isolation must first establish appropriate biosafety practices (see Biosafety below). YFV can be isolated from blood collected during the initial febrile illness and from post-mortem tissues. The virus can be propagated in a variety of cell lines, including monkey epithelial and kidney fibroblasts (MA-104, Vero, LLC-MK2); rabbit- (MA-111) and baby hamster-derived lines (BHK); and Ae. pseudoscutellaris (AP-61) and Ae. albopictus (C6/36) mosquito cells. YFV may produce a cytopathic effect (CPE), and plaque formation is inconsistent and variable from strain to strain. While some strains produce detectable CPE or plaques within 1 or 2 days, many others require observation of the cells for 7–10 days. When CPE or plaques indicate that a virus has been isolated, the presence of viral RNA or antigens can be confirmed by RT-PCR or direct immunofluorescence using monoclonal antibodies.
YFV has been efficiently isolated by intrathoracic inoculation of mosquitoes and intracerebral inoculation of suckling mice or hamsters. Because of the requirement for laboratory animals and the availability of faster and simpler alternative protocols, this procedure is no longer recommended for routine diagnostic purposes. In addition, the efficiency of YFV isolation from clinical samples is greatly influenced by the presence of antibodies against the virus, sample storage conditions55, the isolation system implemented101, and the presence of metabolic products that can be detrimental to the growth of the virus on cell culture68.
Biosafety
YFV is a risk Group 3 pathogen in the WHO and European classification and should be handled in a Biosafety Level 3 (BSL3) laboratory102,103. Depending on the epidemiological context of the country of origin of the samples (i.e., the presence of other hemorrhagic fever viruses that could be included as a differential diagnosis), laboratory facilities and procedures appropriate to Laboratory Containment—BSL3 or higher should be in place for viral isolation. Work should be carried out only by staff vaccinated against YFV at least 10 days prior to any handling of the virus or samples from suspected cases. Over 40 instances of professionally acquired YFV infections were reported in the pre-vaccine era. These cases included a physician caring for a patient, laboratory staff handling biological samples from infected patients or laboratory animals, and one case of transmission from the bite of an infected mosquito38,104–107.
Standard inactivation measures for risk group 3 pathogens are applicable. YFV is inactivated by 2% glutaraldehyde108, β-propiolactone, 2–3% hydrogen peroxide, 70% ethanol, 500–5000 ppm chlorine, 3–8% formaldehyde, 1% iodine and phenol iodophors, or 0.5% phenol with detergent109. Furthermore, YFV may be inactivated by heat at >50°C for 30 min and by gamma irradiation109.
Concluding remarks
Astonishingly, for a well-known pathogen such as YFV, few diagnostic assays have been extensively validated using clinical samples from YF natural infections against different backgrounds of co-circulating flaviviruses. Performance statistics in terms of clinical-laboratory correlation are scarce for the available molecular methods. and only isolated cases have been reported, which have mainly occurred in travelers. Studies with sufficient statistical power are needed on the efficiency of YFV detection by molecular methods involving a follow-up of viraemia over time and examination of non-blood body fluids in parallel. Most reports on the persistence of viraemia have arisen from non-systematic observations in which YFV was generally identified by virus isolation, a technique with shortcomings in a diagnostic setting (as discussed above) and lower sensitivity than more recent molecular methods.
The analysis of body fluids other than serum or blood may widen the diagnostic window in natural infection cases, such as in ZIKV infection, where the pathogen has been detected in urine and semen. The possibility of detecting YFV in urine for longer periods than in serum60,62 warrants further investigation of urine as a useful diagnostic sample.
Detection of the NS1 antigen in the sera of acute YF cases holds promise for use as an alternative diagnostic target that affords high sensitivity and specificity in the early diagnosis of the disease33; however, available data evaluating this approach are currently limited to a recent publication110.
Current serological tests are unable to discriminate between cross-reactive flaviviral antibodies and between vaccine-acquired immunity and immunity from natural infection. The limited offering of commercial tests, scant data on their performance in the diagnosis of YF, and lack of well-defined validation panels hinder the rapid deployment of serological diagnostics during outbreaks.
Lastly, the implementation of YF diagnostic tools by regional laboratories in endemic countries remains challenging. Building up laboratory capacity and capability at the regional level would streamline case detection and foster the timely identification of new areas of transmission by removing bottlenecks at national reference laboratories that become overextended during epidemics and are forced to devote their resources to routine diagnosis. In this scenario, standardization of the assays used in reference laboratories (as currently recommended in the Americas), the establishment of regular quality control programs and interlaboratory comparisons using well-defined standards can provide insight into procedures or working protocols that need to be revised to improve detection capability and detect bias or uncertainties in test results related to the diagnostic laboratories. Furthermore, there is a strong need for standardization of the YFV case/laboratory definition across the Americas and Africa. A strong commitment would be required from authorities to invest heavily in laboratory equipment, logistics, staff training, quality assessment programs, and overall resource sustainability. Addressing the needs of remote laboratories in endemic regions entails developing affordable point-of-care YF diagnostics tests that must be easy to transport, run and interpret. Rigorous evaluation of new diagnostics tools before deployment will be essential. In addition, reference laboratories in non-endemic countries must be prepared and capabilities must be assessed to detect YF in returning travelers as an increasing number of cases have been exported related to the current outbreak in Brazil. For Europe, special attention is required in countries with endemic or intermittent presence of Ae. aegypti and Ae. albopictus.
Acknowledgements
The authors are grateful to José Enrique Mejía for critically revising the manuscript and to U. Erikli for copy editing.
Conflict of interest
This work was conducted within the EVD LabNet consortium under the auspices of the ECDC. The views expressed in this work are those of the authors and do not necessarily reflect the official position or policy of the ECDC. The authors declare no conflicts of interest.
References
- 1.WHO/AFRO. The yellow fever outbreak in Angola and Democratic Republic of the Congo ends (2017). http://www.who.int/csr/disease/yellowfev/en/.
- 2.WHO. Yellow Fever Fact Sheet (WHO, Geneva, Switzerland, 2016).
- 3.WHO. Yellow Fever Situation Report (WHO, Geneva, Switzerland, 2016).
- 4.Nigerian Centre for Disease Control (NCDC). Situation Report: Yellow Fever Outbreak in Nigeria (NCDC, Jabi Abuja, Nigeria, 2018).
- 5.WHO. Yellow Fever Strategic Response Plan (WHO, Geneva, Switzerland, 2016).
- 6.Vaccines and vaccination against yellow fever. WHO position paper—June 2013. Wkly Epidemiol. Rec. 88, 269–283 (2013). [PubMed]
- 7.Campi-Azevedo AC, et al. Subdoses of 17DD yellow fever vaccine elicit equivalent virological/immunological kinetics timeline. BMC Infect. Dis. 2014;14:391. doi: 10.1186/1471-2334-14-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martins RM, et al. 17DD yellow fever vaccine: a double blind, randomized clinical trial of immunogenicity and safety on a dose-response study. Hum. Vaccin Immunother. 2013;9:879–888. doi: 10.4161/hv.22982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu JT, Peak CM, Leung GM, Lipsitch M. Fractional dosing of yellow fever vaccine to extend supply: a modelling study. Lancet. 2016;388:2904–2911. doi: 10.1016/S0140-6736(16)31838-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.PAHO/WHO. Yellow Fever Epidemiological Update (PAHO/WHO, Washington D.C, USA, 2017).
- 11.PAHO/WHO. Yellow Fever Epidemiologicla Update (PAHO/WHO, Washington D.C, USA, 2018).
- 12.WHO. Emergencies Preparadness and Response: Yellow Fever, Brazil. Disease Outbreak News (WHO, Geneva, Switzerland, 2018).
- 13.Ministério da Saude (MS/SVS). COES- Febre Amarela INFORME—No 43/2017 (2017).
- 14.Prefectura de São Paulo. Secretaría de Salud. São Paulo vacuna 4,1 mil contra la fiebre amarilla en la Zona Norte. Noticias (2017).
- 15.Ministério da Saude (MS/SVS). Campanha de vacinação terá dose fracionada de febre amarela em três estados (2018).
- 16.Grobusch MP, et al. Yellow fever revaccination guidelines change - a decision too feverish? Clin. Microbiol Infect. 2013;19:885–886. doi: 10.1111/1469-0691.12332. [DOI] [PubMed] [Google Scholar]
- 17.Campi-Azevedo AC, et al. Booster dose after 10 years is recommended following 17DD-YF primary vaccination. Hum. Vaccin Immunother. 2016;12:491–502. doi: 10.1080/21645515.2015.1082693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Possas C, Martins RM, Oliveira RL, Homma A. Urgent call for action: avoiding spread and re-urbanisation of yellow fever in Brazil. Mem. Inst. Oswaldo Cruz. 2018;113:1–2. doi: 10.1590/0074-02760170361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.PAHO/WHO. Yellow Fever Epidemiological Update (PAHO/WHO, Washington D.C, USA, 2017).
- 20.European Centre for Disease Prevention and Control (ECDC). Outbreak of Yellow Fever in Angola (ECDC, Stockholm, Sweden, 2016).
- 21.Amraoui, F., Vazeille, M. & Failloux, A. B. French Aedes albopictus are able to transmit yellow fever virus. Euro Surveill21, 10.2807/1560-7917.ES.2016.21.39.30361 (2016). [DOI] [PMC free article] [PubMed]
- 22.Gossner CM, et al. Increased risk of yellow fever infections among unvaccinated European travellers due to ongoing outbreak in Brazil, July 2017 to March 2018. Eurosurveillance. 2018;23:18–00106. doi: 10.2807/1560-7917.ES.2018.23.11.18-00106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eliminate Yellow Fever Epidemics (EYE): a global strategy, 2017–2026. Wkly Epidemiol. Rec.92, 193-204 (2017). [PubMed]
- 24.Mutebi JP, et al. Genetic relationships and evolution of genotypes of yellow fever virus and other members of the yellow fever virus group within the Flavivirus genus based on the 3’ noncoding region. J. Virol. 2004;78:9652–9665. doi: 10.1128/JVI.78.18.9652-9665.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pugachev KV, et al. High fidelity of yellow fever virus RNA polymerase. J. Virol. 2004;78:1032–1038. doi: 10.1128/JVI.78.2.1032-1038.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mutebi JP, Wang H, Li L, Bryant JE, Barrett AD. Phylogenetic and evolutionary relationships among yellow fever virus isolates in Africa. J. Virol. 2001;75:6999–7008. doi: 10.1128/JVI.75.15.6999-7008.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grobbelaar AA, et al. Resurgence of yellow fever in Angola, 2015–2016. Emerg. Infect. Dis. 2016;22:1854–1855. doi: 10.3201/eid2210.160818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.WHO. Yellow Fever Outbreak in Angola and Democratic Republic of Congo (WHO, Geneva, Switzerland, 2016).
- 29.Mir D, et al. Phylodynamics of Yellow Fever Virus in the Americas: new insights into the origin of the 2017 Brazilian outbreak. Sci. Rep. 2017;7:7385. doi: 10.1038/s41598-017-07873-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de Souza RP, et al. Detection of a new yellow fever virus lineage within the South American genotype I in Brazil. J. Med Virol. 2010;82:175–185. doi: 10.1002/jmv.21606. [DOI] [PubMed] [Google Scholar]
- 31.Bonaldo MC, et al. Genome analysis of yellow fever virus of the ongoing outbreak in Brazil reveals polymorphisms. Mem. Inst. Oswaldo Cruz. 2017;112:447–451. doi: 10.1590/0074-02760170134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paules CI, Fauci AS. Yellow fever—once again on the radar screen in the Americas. N. Engl. J. Med. 2017;376:1397–1399. doi: 10.1056/NEJMp1702172. [DOI] [PubMed] [Google Scholar]
- 33.Monath TP, Vasconcelos PF. Yellow fever. J. Clin. Virol. 2015;64:160–173. doi: 10.1016/j.jcv.2014.08.030. [DOI] [PubMed] [Google Scholar]
- 34.Hamrick PN, et al. Geographic patterns and environmental factors associated with human yellow fever presence in the Americas. PLoS Negl. Trop. Dis. 2017;11:e0005897. doi: 10.1371/journal.pntd.0005897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Almeida MA, et al. Surveillance for yellow Fever virus in non-human primates in southern Brazil, 2001-2011: a tool for prioritizing human populations for vaccination. PLoS Negl. Trop. Dis. 2014;8:e2741. doi: 10.1371/journal.pntd.0002741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ministério da Saúde do Brasil. Serie A. Normas e Manuais Técnicos (2005).
- 37.Couto-Lima D, et al. Potential risk of re-emergence of urban transmission of Yellow Fever virus in Brazil facilitated by competent Aedes populations. Sci. Rep. 2017;7:4848. doi: 10.1038/s41598-017-05186-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Low GC, Fairley NH. Observations on laboratory and hospital infections with yellow fever in England. Br. Med J. 1931;1:125–128. doi: 10.1136/bmj.1.3655.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsai TF, Paul R, Lynberg MC, Letson GW. Congenital yellow fever virus infection after immunization in pregnancy. J. Infect. Dis. 1993;168:1520–1523. doi: 10.1093/infdis/168.6.1520. [DOI] [PubMed] [Google Scholar]
- 40.Bentlin MR, et al. Perinatal transmission of yellow fever, Brazil, 2009. Emerg. Infect. Dis. 2011;17:1779–1780. doi: 10.3201/eid1709.110242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Centers for Disease Control and Prevention (CDC Transmission of yellow fever vaccine virus through breast-feeding - Brazil, 2009. MMWR Morb. Mortal. Wkly Rep. 2010;59:130–132. [PubMed] [Google Scholar]
- 42.Kuhn S, Twele-Montecinos L, MacDonald J, Webster P, Law B. Case report: probable transmission of vaccine strain of yellow fever virus to an infant via breast milk. CMAJ. 2011;183:E243–E245. doi: 10.1503/cmaj.100619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Traiber C, Coelho-Amaral P, Ritter VR, Winge A. Infant meningoencephalitis caused by yellow fever vaccine virus transmitted via breastmilk. J. Pediatr. (Rio J.) 2011;87:269–272. doi: 10.2223/JPED.2067. [DOI] [PubMed] [Google Scholar]
- 44.Centers for Disease Control and Prevention (CDC Transfusion-related transmission of yellow fever vaccine virus--California, 2009. MMWR Morb. Mortal. Wkly Rep. 2010;59:34–37. [PubMed] [Google Scholar]
- 45.Sonnenberg P, Field N. Sexual and mother-to-child transmission of Ebola virus in the postconvalescent period. Clin. Infect. Dis. 2015;60:974–975. doi: 10.1093/cid/ciu981. [DOI] [PubMed] [Google Scholar]
- 46.D’Ortenzio E, et al. Evidence of Sexual Transmission of Zika Virus. N. Engl. J. Med. 2016;374:2195–2198. doi: 10.1056/NEJMc1604449. [DOI] [PubMed] [Google Scholar]
- 47.Quaresma JA, Pagliari C, Medeiros DB, Duarte MI, Vasconcelos PF. Immunity and immune response, pathology and pathologic changes: progress and challenges in the immunopathology of yellow fever. Rev. Med. Virol. 2013;23:305–318. doi: 10.1002/rmv.1752. [DOI] [PubMed] [Google Scholar]
- 48.Monath TP, Barrett AD. Pathogenesis and pathophysiology of yellow fever. Adv. Virus Res. 2003;60:343–395. doi: 10.1016/S0065-3527(03)60009-6. [DOI] [PubMed] [Google Scholar]
- 49.Tuboi SH, Costa ZG, da Costa Vasconcelos PF, Hatch D. Clinical and epidemiological characteristics of yellow fever in Brazil: analysis of reported cases 1998-2002. Trans. R. Soc. Trop. Med. Hyg. 2007;101:169–175. doi: 10.1016/j.trstmh.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 50.Barrett AD, Teuwen DE. Yellow fever vaccine—how does it work and why do rare cases of serious adverse events take place? Curr. Opin. Immunol. 2009;21:308–313. doi: 10.1016/j.coi.2009.05.018. [DOI] [PubMed] [Google Scholar]
- 51.Cottin P, Niedrig M, Domingo C. Safety profile of the yellow fever vaccine Stamaril(R): a 17-year review. Expert Rev. Vaccin. 2013;12:1351–1368. doi: 10.1586/14760584.2013.836320. [DOI] [PubMed] [Google Scholar]
- 52.Monath, T. P. (ed.) Arboviruses: Epidemiology and Ecology Vol. 5, 139-231 (CRC Press, Boca Raton, FL,1985).
- 53.Centers for Disease Control and Prevention (CDC Fatal yellow fever in a traveler returning from Amazonas, Brazil, 2002. MMWR Morb. Mortal. Wkly Rep. 2002;51:324–325. [PubMed] [Google Scholar]
- 54.Chen Z, et al. A fatal yellow fever virus infection in China: description and lessons. Emerg. Microbes Infect. 2016;5:e69. doi: 10.1038/emi.2016.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.de Filippis AM, et al. Outbreak of jaundice and hemorrhagic fever in the Southeast of Brazil in 2001: detection and molecular characterization of yellow fever virus. J. Med Virol. 2002;68:620–627. doi: 10.1002/jmv.10226. [DOI] [PubMed] [Google Scholar]
- 56.Filippis AM, et al. Isolation and characterization of wild type yellow fever virus in cases temporally associated with 17DD vaccination during an outbreak of yellow fever in Brazil. Vaccine. 2004;22:1073–1078. doi: 10.1016/j.vaccine.2003.11.002. [DOI] [PubMed] [Google Scholar]
- 57.McFarland JM, et al. Imported yellow fever in a United States citizen. Clin. Infect. Dis. 1997;25:1143–1147. doi: 10.1086/516111. [DOI] [PubMed] [Google Scholar]
- 58.Nassar Eda S, et al. Jungle yellow fever: clinical and laboratorial studies emphasizing viremia on a human case. Rev. Inst. Med Trop. Sao Paulo. 1995;37:337–341. doi: 10.1590/S0036-46651995000400009. [DOI] [PubMed] [Google Scholar]
- 59.Teichmann D, Gobels K, Niedrig M, Grobusch MP. Clinical and laboratory features of dengue virus-infected travellers previously vaccinated against yellow fever. Scand. J. Infect. Dis. 2003;35:427–429. doi: 10.1080/00365540310009888. [DOI] [PubMed] [Google Scholar]
- 60.Reusken C, et al. Urine as sample type for molecular diagnosis of natural yellow fever virus infections. J. Clin. Microbiol. 2017;55:3294–3296. doi: 10.1128/JCM.01113-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Barbosa, C. M. et al. Yellow fever virus RNA in urine and semen of convalescent patient, Brazil. Emerg. Infect. Dis.24, 176–178 (2018). [DOI] [PMC free article] [PubMed]
- 62.Domingo C, et al. Detection of yellow fever 17D genome in urine. J. Clin. Microbiol. 2011;49:760–762. doi: 10.1128/JCM.01775-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cui S, et al. Detection of yellow fever virus genomes from four imported cases in China. Int J. Infect. Dis. 2017;60:93–95. doi: 10.1016/j.ijid.2017.05.001. [DOI] [PubMed] [Google Scholar]
- 64.Gibney KB, et al. Detection of anti-yellow fever virus immunoglobulin m antibodies at 3-4 years following yellow fever vaccination. Am. J. Trop. Med Hyg. 2012;87:1112–1115. doi: 10.4269/ajtmh.2012.12-0182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bae HG, Nitsche A, Teichmann A, Biel SS, Niedrig M. Detection of yellow fever virus: a comparison of quantitative real-time PCR and plaque assay. J. Virol. Methods. 2003;110:185–191. doi: 10.1016/S0166-0934(03)00129-0. [DOI] [PubMed] [Google Scholar]
- 66.Chao DY, Davis BS, Chang GJ. Development of multiplex real-time reverse transcriptase PCR assays for detecting eight medically important flaviviruses in mosquitoes. J. Clin. Microbiol. 2007;45:584–589. doi: 10.1128/JCM.00842-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dash PK, Boutonnier A, Prina E, Sharma S, Reiter P. Development of a SYBR green I based RT-PCR assay for yellow fever virus: application in assessment of YFV infection in Aedes aegypti. Virol. J. 2012;9:27. doi: 10.1186/1743-422X-9-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Deubel V, et al. Molecular detection and characterization of yellow fever virus in blood and liver specimens of a non-vaccinated fatal human case. J. Med Virol. 1997;53:212–217. doi: 10.1002/(SICI)1096-9071(199711)53:3<212::AID-JMV5>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 69.Domingo C, et al. Advanced yellow fever virus genome detection in point-of-care facilities and reference laboratories. J. Clin. Microbiol. 2012;50:4054–4060. doi: 10.1128/JCM.01799-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Drosten C, et al. Rapid detection and quantification of RNA of Ebola and Marburg viruses, Lassa virus, Crimean-Congo hemorrhagic fever virus, Rift Valley fever virus, dengue virus, and yellow fever virus by real-time reverse transcription-PCR. J. Clin. Microbiol. 2002;40:2323–2330. doi: 10.1128/JCM.40.7.2323-2330.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mantel N, et al. Standardized quantitative RT-PCR assays for quantitation of yellow fever and chimeric yellow fever-dengue vaccines. J. Virol. Methods. 2008;151:40–46. doi: 10.1016/j.jviromet.2008.03.026. [DOI] [PubMed] [Google Scholar]
- 72.Mendez MC, et al. Development of a reverse transcription polymerase chain reaction method for yellow fever virus detection. Biomedica. 2013;33(Suppl 1):190–196. [PubMed] [Google Scholar]
- 73.Pang Z, et al. Comprehensive multiplex one-step real-time TaqMan qRT-PCR assays for detection and quantification of hemorrhagic fever viruses. PLoS One. 2014;9:e95635. doi: 10.1371/journal.pone.0095635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fischer, C. et al. Lineage-specific real-time RT-PCR for Yellow Fever Virus Outbreak Surveillance, Brazil. Emerg. Infect. Dis.23, 1867–1871 (2017). [DOI] [PMC free article] [PubMed]
- 75.Weidmann M, et al. Improved LNA probe-based assay for the detection of African and South American yellow fever virus strains. J. Clin. Virol. 2010;48:187–192. doi: 10.1016/j.jcv.2010.04.013. [DOI] [PubMed] [Google Scholar]
- 76.Escadafal C, et al. Rapid molecular assays for the detection of yellow fever virus in low-resource settings. PLoS Negl. Trop. Dis. 2014;8:e2730. doi: 10.1371/journal.pntd.0002730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kwallah A, et al. A real-time reverse transcription loop-mediated isothermal amplification assay for the rapid detection of yellow fever virus. J. Virol. Methods. 2013;193:23–27. doi: 10.1016/j.jviromet.2013.05.004. [DOI] [PubMed] [Google Scholar]
- 78.Nunes MR, et al. Analysis of a Reverse Transcription Loop-mediated Isothermal Amplification (RT-LAMP) for yellow fever diagnostic. J. Virol. Methods. 2015;226:40–51. doi: 10.1016/j.jviromet.2015.10.003. [DOI] [PubMed] [Google Scholar]
- 79.Domingo C, et al. First international external quality assessment study on molecular and serological methods for yellow fever diagnosis. PLoS One. 2012;7:e36291. doi: 10.1371/journal.pone.0036291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hughes, H. R. et al. Development of a real-time RT-PCR assay for the global differentiation of yellow fever virus vaccine adverse events from natural infections. J. Clin. Microbiol.10.1128/jcm.00323-18 (2018). [DOI] [PMC free article] [PubMed]
- 81.Brinkmann A, et al. Development and preliminary evaluation of a multiplexed amplification and next generation sequencing method for viral hemorrhagic fever diagnostics. PLoS Negl. Trop. Dis. 2017;11:e0006075. doi: 10.1371/journal.pntd.0006075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.McMullan LK, et al. Using next generation sequencing to identify yellow fever virus in Uganda. Virology. 2012;422:1–5. doi: 10.1016/j.virol.2011.08.024. [DOI] [PubMed] [Google Scholar]
- 83.PAHO/WHO. Laboratory Diagnosis of Yellow Fever Virus Infection (PAHO/WHO, 2018).
- 84.Houghton-Trivino N, Montana D, Castellanos J. Dengue-yellow fever sera cross-reactivity; challenges for diagnosis. Rev. Salud Publica (Bogota) 2008;10:299–307. doi: 10.1590/S0124-00642008000200010. [DOI] [PubMed] [Google Scholar]
- 85.WHO Interim Guidance. Yellow Fever Laboratory Diagnostic Testing in Africa (WHO, Geneva, Switzerland, 2016).
- 86.Lanciotti RS, et al. Genetic and serologic properties of Zika virus associated with an epidemic, Yap State, Micronesia, 2007. Emerg. Infect. Dis. 2008;14:1232–1239. doi: 10.3201/eid1408.080287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Deubel V, et al. Comparison of the enzyme-linked immunosorbent assay (ELISA) with standard tests used to detect yellow fever virus antibodies. Am. J. Trop. Med Hyg. 1983;32:565–568. doi: 10.4269/ajtmh.1983.32.565. [DOI] [PubMed] [Google Scholar]
- 88.Monath TP, Nystrom RR. Detection of yellow fever virus in serum by enzyme immunoassay. Am. J. Trop. Med. Hyg. 1984;33:151–157. doi: 10.4269/ajtmh.1984.33.151. [DOI] [PubMed] [Google Scholar]
- 89.Cammack N, Gould EA. Antigenic analysis of yellow fever virus glycoproteins: use of monoclonal antibodies in enzyme-linked immunosorbent assays. J. Virol. Methods. 1986;13:135–142. doi: 10.1016/0166-0934(86)90081-9. [DOI] [PubMed] [Google Scholar]
- 90.Vazquez S, et al. MAC-ELISA and ELISA inhibition methods for detection of antibodies after yellow fever vaccination. J. Virol. Methods. 2003;110:179–184. doi: 10.1016/S0166-0934(03)00128-9. [DOI] [PubMed] [Google Scholar]
- 91.Basile AJ, et al. Multiplex microsphere immunoassays for the detection of IgM and IgG to arboviral diseases. PLoS One. 2013;8:e75670. doi: 10.1371/journal.pone.0075670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Martin DA, et al. Standardization of immunoglobulin M capture enzyme-linked immunosorbent assays for routine diagnosis of arboviral infections. J. Clin. Microbiol. 2000;38:1823–1826. doi: 10.1128/jcm.38.5.1823-1826.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Basile AJ, et al. Development and validation of an ELISA kit (YF MAC-HD) to detect IgM to yellow fever virus. J. Virol. Methods. 2015;225:41–48. doi: 10.1016/j.jviromet.2015.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Adungo F, et al. Development and characterization of monoclonal antibodies to yellow fever virus and application in antigen detection and IgM capture enzyme-linked immunosorbent assay. Clin. Vaccin. Immunol. 2016;23:689–697. doi: 10.1128/CVI.00209-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Niedrig M, Kursteiner O, Herzog C, Sonnenberg K. Evaluation of an indirect immunofluorescence assay for detection of immunoglobulin M (IgM) and IgG antibodies against yellow fever virus. Clin. Vaccin. Immunol. 2008;15:177–181. doi: 10.1128/CVI.00078-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Berry GP. Yellow fever accidentally contracted in the laboratory: a study of seven cases. Am. J. Trop. Med. Hyg. 1931;11:365–434. doi: 10.4269/ajtmh.1931.s1-11.365. [DOI] [Google Scholar]
- 97.Fernandes NdA, et al. Outbreak of yellow fever among nonhuman primates, Espirito Santo, Brazil, 2017. Emerg. Infect. Dis. 2017;23:2038–2041. doi: 10.3201/eid2312.170685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Leal SG, et al. Frequency of histopathological changes in Howler monkeys (Alouatta sp.) naturally infected with yellow fever virus in Brazil. Rev. Soc. Bras. Med Trop. 2016;49:29–33. doi: 10.1590/0037-8682-0363-2015. [DOI] [PubMed] [Google Scholar]
- 99.Ministério da Saúde do Brasil. Normas e Manuais Técnicos. Guia de vigilância de epizootias em primatas não humanos e entomologia aplicada à vigilância da febre amarela(2005).
- 100.Parra, E. A. Critical requirements for processing and interpretating yellow fever immunohistochemistry. In Workshop UNASUR/CPLP on Scientific and Technological Actualization on Yellow Yever and other Emerging and Re-emerging Arbovirosis (Rio de Janeiro, Brazil, 2017).
- 101.Sariol CA, et al. Detection and genetic relationship of dengue virus sequences in seventeen-year-old paraffin-embedded samples from Cuba. Am. J. Trop. Med Hyg. 1999;61:994–1000. doi: 10.4269/ajtmh.1999.61.994. [DOI] [PubMed] [Google Scholar]
- 102.WHO. Laboratory Biosafety Manual (WHO, Geneva, Switzerland, 2004).
- 103.European Parliament and the Council of the European Union (EU). Directive 2000/54/ec of the European Parliament and of the Council of 18 September 2000 on the protection of workers from risks related to exposure to biological agents at work (seventh individual directive within the meaning of Article 16(1) of Directive 89/391/EEC). Offi. J. Eur. Commun. 21–45 (2000).
- 104.Hindle E. The transmission of yellow fever. Lancet. 1930;216:835–842. doi: 10.1016/S0140-6736(00)89729-9. [DOI] [Google Scholar]
- 105.Hanson RP, et al. Arbovirus infections of laboratory workers. Extent of problem emphasizes the need for more effective measures to reduce hazards. Science. 1967;158:1283–1286. doi: 10.1126/science.158.3806.1283. [DOI] [PubMed] [Google Scholar]
- 106.The Subcommittee on Arbovirus Laboratory Safety of the American Committee on Arthropod-Borne Viruses. Laboratory safety for arboviruses and certain other viruses of vertebrates. Am. J. Trop. Med. Hyg. 1980;29:1359–1381. doi: 10.4269/ajtmh.1980.29.1359. [DOI] [PubMed] [Google Scholar]
- 107.Graham JL, Jaeger RF. Inactivation of yellow fever virus by glutaraldehyde. Appl. Microbiol. 1968;16:177. doi: 10.1128/am.16.1.177-.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Graham, J. L. & Jaeger, R. F. Inactivation of yellow fever virus by glutaraldehyde. Appl. Microbiol. 16, 177 (1968). [DOI] [PMC free article] [PubMed]
- 109.Ricciardi-Jorge T, et al. Development of a quantitative NS1-capture enzyme-linked immunosorbent assay for early detection of yellow fever virus infection. Sci. Rep. 2017;7:16229. doi: 10.1038/s41598-017-16231-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ricciardi-Jorge, T. et al. Development of a quantitative NS1-capture enzyme-linked immunosorbent assay for early detection of yellow fever virus infection. Sci. Rep. 7, 16229 (2017). [DOI] [PMC free article] [PubMed]