Abstract
The formation of a complex between neuroglobin (Ngb) and cytochrome c (Cyt c) has an important biological role in preventing apoptosis. Binding of Ngb to Cyt c alone is sufficient to block the caspase 9 activation by ferric Cyt c that is released during ischemic insults. Therefore, a detailed information on the Ngb-Cyt c interactions is important for understanding apoptosis. However, the exact nature of the interactions between oxidized human neuroglobin (hNgb) and Cyt c is not well understood. In this work, we used a combination of computational modeling and surface plasmon resonance experiments to obtain and characterize the complex formation between oxidized hNgb and Cyt c. We identified important residues involved in the complex formation, including K72 in Cyt c, which is otherwise known to interact with the apoptotic protease-activation factor-1. Our computational results, together with an optimized structure of the hNgb-Cyt c complex, provide unique insights into how the hNgb-Cyt c complex can abate the apoptotic cascade without an hNgb-Cyt c redox reaction.
Introduction
Neuroglobin (Ngb) is a six-coordinated heme protein that is mainly expressed in nervous and endocrine tissues1 and the retina2. The structure of Ngb is similar to that of myoglobin3. Ngb has been proposed to be involved in the regulation of Alzheimer’s disease4. During hypoxic-ischemic insults, Ngb remains up-regulated and protects neurons5. However, the mechanism of Ngb neuroprotection is unclear6,7, and further studies investigating the neuroprotective role of Ngb are required8. Therefore, recent studies have increasingly explored the binding partners of Ngb9–12.
Cytochrome c (Cyt c), which is another heme protein13, is a known Ngb binding partner12,14. This mitochondrial heme protein triggers cell apoptosis during apoptotic cell signaling15. During ischemic insults, the cellular apoptotic pathway is activated, leading to the release of ferric Cyt c, which is an essential component of the apoptosome, into the cytoplasm16–18. Since a redox reaction between Ngb and Cyt c is not required to prevent the formation of the apoptosome, the binding of Ngb to Cyt c alone is sufficient to block the Cyt c-induced caspase 9 activation19. Thus, investigations targeting these two heme proteins in their ferric state are important to the scientific community.
According to studies using surface affinity-based surface plasmon resonance (SPR), nanopore-based nanopipettes, and solution-based isothermal titration calorimetry (ITC) experimental techniques, ferric human Ngb (referred to as hNgb hereafter) interacts with ferric Cyt c (referred to as Cyt c hereafter) with a micromolar affinity12,14,20. Electrochemical impedance spectroscopy measurements have also confirmed the formation of an hNgb-Cyt c complex12. Computational docking is useful to investigate the complex formation between proteins21. Several computational docking studies have been performed to predict Ngb-Cyt c complexes12,14,19. These computational techniques complement the limitations of the experimental methods used to study protein-protein interactions (PPIs)22. Molecular docking is useful in predicting the binding interfaces of interacting partners23,24. Since proteins are flexible biomolecules, a dynamic reorganization in the structures of the interacting partners occurs during PPIs12, and these conformational dynamics are the fundamental basis for their structure and function25. Molecular dynamics (MD) simulations are useful in investigations exploring the mechanisms by which the conformational flexibility of a protein is utilized in complex formation and stabilization26. Therefore, MD simulations are routinely used to study complex formation between a protein and its interacting partners21,27,28 and are crucial for an understanding of the physical basis of biomolecular structure and function25.
While experimental and docking-based computational investigations have been performed, the optimized structure of this important hNgb-Cyt c protein-protein complex has not been established. Therefore, the exact nature of the hNgb-Cyt c complex is not well understood. Here, we investigated the formation and stabilization of the complex between hNgb and Cyt c. We first modeled the hNgb-Cyt c complex using molecular docking and then performed MD simulations of the docked complex. The complex is stable (does not dissociate) for 700 ns of the simulation time, despite major structural changes. We observed an interesting structural rearrangement of the complex during stabilization. Based on our results, we predict that hNgb interacts with Cyt c via two salt bridges between D73 (hNgb) and K72 (Cyt c) and between E87 (hNgb) and K27 (Cyt c) as well as a hydrogen bond between T77 (hNgb) and K72 (Cyt c). The involvement of the amino acid residues predicted by the MD simulations was confirmed by SPR. To the best of our knowledge, this is the first computational investigation of the molecular mechanism underlying the hNgb-Cyt c complex formation, allowing conformational flexibility and structural rearrangements.
Results
Two salt bridges and a hydrogen bond are responsible for the formation and stabilization of the hNgb-Cyt c complex
The best predicted hNgb-Cyt c complex (see Materials and Methods section) structure obtained from docking studies was used in MD simulations. In this best predicted structure, several interfacial atomic contacts initiated the formation of the complex between hNgb and Cyt c. The formation of a complex between protein biomolecules is a dynamic process involving the structural reorganization of the interacting partners21. Additionally, proteins are flexible biomolecules, and their structure and function depend on their conformational flexibility25. The optimization of interfacial contacts via the structural adjustment of interacting protein molecules can be realized by dynamics21, providing the residues with enhanced flexibility. Therefore, to examine the stability and optimize the conformational integrity of the hNgb-Cyt c complex predicted from docking, we performed all-atom MD simulations in explicit solvent.
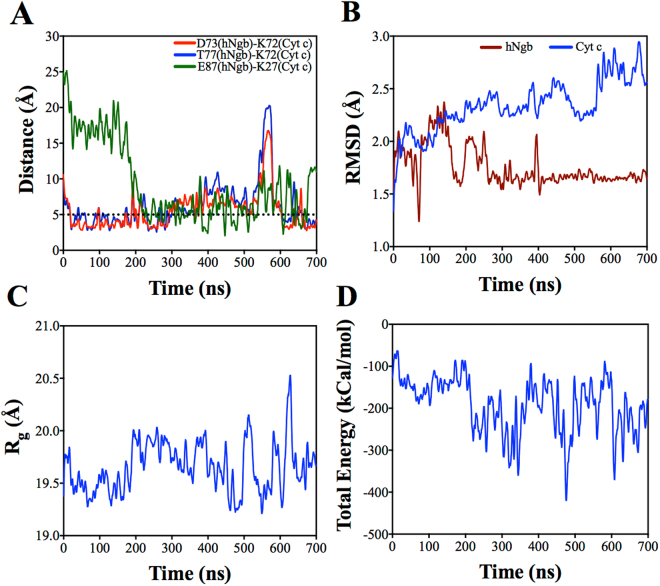
We investigated the amino acid residue contacts between hNgb and Cyt c during complex formation. As shown in Table 1, several residues exhibit interfacial contacts. We observed two salt bridges between D73 (hNgb) and K72 (Cyt c) and between E87 (hNgb) and K27 (Cyt c) as well as a hydrogen bond between T77 (hNgb) and K72 (Cyt c). The salt bridge formed between D73 (hNgb) and K72 (Cyt c) is stable as indicated by the stable distance <5 Å29 shown in Fig. 1A. However, compared to the salt bridge between D73 (hNgb) and K72 (Cyt c), the hydrogen bond between T77 (hNgb) and K72 (Cyt c) is not as stable. We compared the bond stability by the number of frames that showed interaction, based on contact analysis (Materials and Methods section). For the hydrogen bonding this number is ~1200. However, for the salt bridge the number of frames are ~1700. The residues D73 (hNgb) and T77 (hNgb) are both located in the E-helix of hNgb30. The repositioning of the E-helix, as an exogenous ligand binds to hNgb, has been proposed to alter the affinity of hNgb to Cyt c12.
Table 1.
Amino acid residues in both hNgb and Cyt c that showed interfacial contacts.
| hNgb residues | Cyt c residues |
|---|---|
| E87, Y88, S91 | Q16 |
| E87 | K27 |
| S84 | T28 |
| L70, D73, T77 | K72 |
| T77, N78 | K79 |
| K67, L70, V71, A74, L85, Y88, HEME | I81 |
| L70 | F82 |
| K67, L70 | A83 |
| L85, Y88 | HEME |
Several residues in one protein that establish interfacial contact with another protein are separated by coma on the same rows.
Figure 1.
(A) Distance between the predicted amino acid residues that form the complex between hNgb and Cyt c. The dotted line is drawn to show a reference line at 5 Å for salt bridge. (B) RMSD measurements for hNgb and Cyt c. (C) Radius of gyration (Rg) and (D) total interaction energy of hNgb-Cyt c complex as a function of simulation time.
As shown in Fig. 1B, as revealed by RMSD measurements, there are large rearrangements in Cyt c structure while hNgb structure remains mainly conserved. We have also measured radius of gyration (Rg) of the structure during the NVT simulations. As shown in Fig. 1C, Rg does not change significantly throughout the simulation, which suggests that the overall complex is thermally stable. Figure 1D shows the total interaction energy as a function of time. We did not observe significant fluctuation in the total energy of the system, which further assures the stability of the hNgb-Cyt c complex.
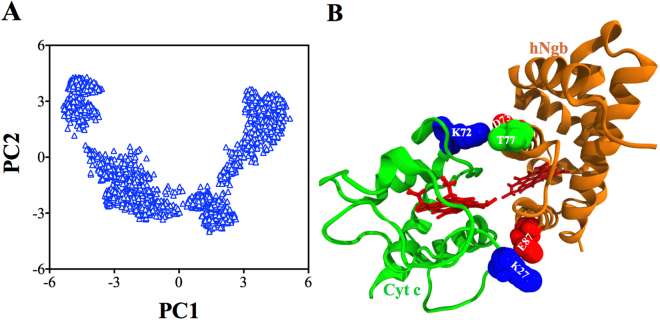
We performed principal component analysis (PCA) of the 700 ns MD simulation trajectories to evaluate the conformational differences between the hNgb-Cyt c complex structures. As shown in Fig. 2A, as compared to PC2, PC1 varies more broadly. An average hNgb-Cyt c complex structure obtained from the PCA-based cluster analysis is shown in Fig. 2B. The important residue pairs that are responsible to form specific salt bridges and hydrogen bonding are highlighted in Fig. 2B (VDW representation).
Figure 2.
(A) Projection of the hNgb-Cyt c complex structures along the direction of the first two principal components (PC1 and PC2). (B) An averaged hNgb-Cyt c complex structure obtained from PCA based cluster analysis.
SPR experiments confirm the involvement of the amino acid residues predicted by the MD simulation
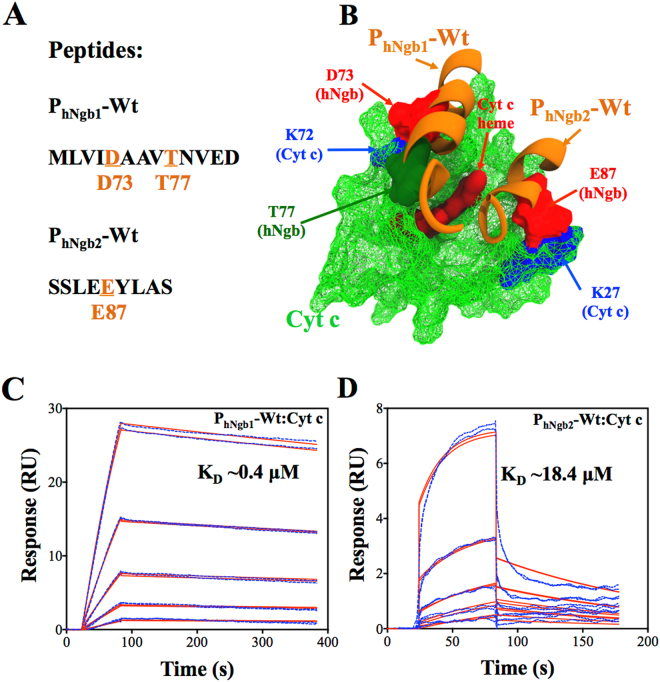
We performed SPR experiments to confirm the involvement of the amino acid residues predicted by the MD simulations. We used two peptides, PhNgb1-Wt and PhNgb2-Wt in the SPR experiments. The sequence of PhNgb1-Wt comprises of hNgb amino acids ranging from M69 to D81 and that of PhNgb2-Wt from S83 to S91. The peptides were dissolved in 100% DMSO and further diluted in the running buffer (10 mM HEPES pH 7.4, 150 mM NaCl, and 0.05% v/v surfactant P20 supplemented at 1% v/v DMSO) and injected over the Cyt c-immobilized surface at various concentrations. The name assignments and sequences of the peptides, used as analytes in the SPR experiments, are shown in Fig. 3A. Figure 3A displays the amino acid residues (orange color and underlined) predicted by the MD simulations to be involved in the formation of the hNgb-Cyt c complex. Figure 3B shows a pictorial view of sections of hNgb (orange) represented by the sequences of the wild-type peptides that are bound to Cyt c (green) and the amino acid residues responsible to form the complex are highlighted. Figures 3C,D show the SPR sensorgrams (colored dashed lines) of peptides PhNgb1-Wt and PhNgb2-Wt binding to Cyt c, respectively.
Figure 3.
(A) Names and sequences of the peptides used in the SPR experiments. The amino acid residues predicted by the MD simulations to be involved in the formation of the hNgb-Cyt c complex are shown in orange (with their position in hNgb) and are underlined. (B) Pictorial view of sections of hNgb (orange) represented by the sequences of the peptides bound to Cyt c (green). The SPR sensorgrams (blue dashed lines) of the binding of the peptides PhNgb1-Wt (C) and PhNgb2-Wt (D). Continuous red lines are fit to a 1:1 kinetics binding model. PhNgb1-Wt concentrations injected during SPR experiments were 0.625 µM, 1.25 µM, 2.5 µM, 5 µM, and 10 µM and PhNgb2-Wt concentrations were 6.25 µM, 12.5 µM, 25 µM, 50 µM, and 100 µM.
Qualitatively, the binding of the wild-type peptides to Cyt c confirm the prediction from the simulations that the amino acid residues mentioned above are essential for the hNgb-Cyt c complex formation. Quantitatively, we fit the SPR sensorgrams (red continuous lines), as shown in Fig. 3C, using a 1:1 kinetics model to derive the equilibrium dissociation constant (affinity or KD value). A KD value of 0.4 ± 0.2 μM (mean ± s.d. from three different experiments) was obtained for PhNgb1-Wt binding to Cyt c. We were interested in comparing affinities of the peptides PhNgb1-Wt and PhNgb2-Wt binding to Cyt c. Therefore, using the same fitting procedures, we fit the sensorgrams shown in Fig. 3D, and a KD value of 18.4 ± 4.3 μM (mean ± s.d. from three different experiments) was obtained for PhNgb2-Wt binding to Cyt c. PhNgb1-Wt had a 45-fold higher affinity (lower KD value) than PhNgb2-Wt, which is highly consistent with the stronger binding of the hNgb region represented by PhNgb1-Wt than that of the region represented by PhNgb2-Wt due to the formation of the bonds shown in Fig. 1A.
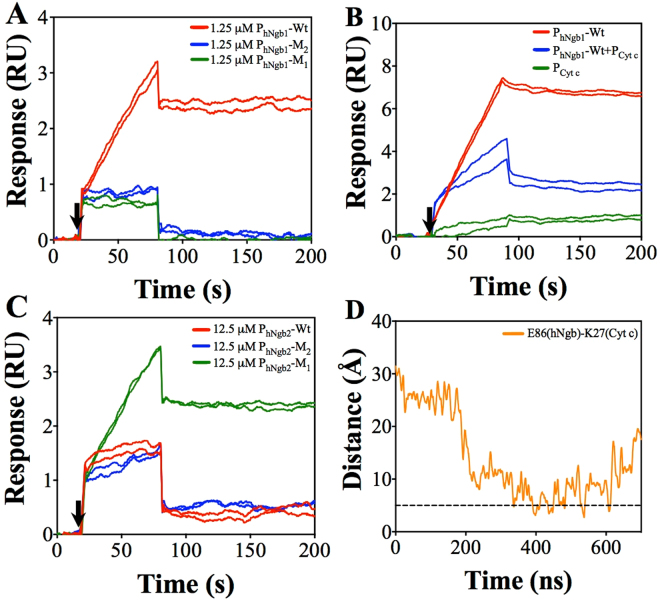
We conducted SPR experiments with mutated peptides. Qualitatively, as shown in Fig. 4A, the two mutant peptides PhNgb1-M1 (with D73A substitution) and PhNgb1-M2 (with T77A substitution) bind to Cyt c. Moreover, binding of these individual mutants showed weaker SPR signals as compared to SPR signals for PhNgb1-Wt, which is expected. These results also show the involvement of D73 (hNgb) and T77 (hNgb) together take part in binding to K72 (Cyt c), making a strong bonding as predicted by simulations. We could not perform quantitative investigations (via determination of KD values) for the mutated peptides due to solubility issues at higher concentrations. In addition, we also conducted an inhibition experiment with a peptide PCyt c (with amino acids L68 to P76 in Cyt c) and PhNgb1-Wt. As shown in Fig. 4B, a mixture of PhNgb1-Wt and PCyt c (in 1:2 molar ratio) showed diminished signal as compared to PhNgb1-Wt signal. This is because PCyt c with K72 (Cyt c) interacts with PhNgb1-Wt via D73 (hNgb)-K72 (Cyt c) or T77 (hNgb)-K72 (Cyt c) interactions in solution and reduces the available binding sites in PhNgb1-Wt to bind to immobilized Cyt c onto the ship surface. PCyt c, part of Cyt c sequence, does not bind to immobilized Cyt c. This experiment further confirms the involvement of the predicted amino acid residues (K72, D73 and T77) in the hNgb-Cyt c complex formation. We could not obtain purified peptide with double mutation (MLVIAAAVANVED) due to more hydrophobic in nature.
Figure 4.
(A) SPR sensorgrams for qualitative comparison of bindings for PhNgb1-Wt, PhNgb1-M1, and PhNgb1-M2. (B) SPR sensorgrams for inhibition of PhNgb1-Wt binding by PCyt c. (C) SPR sensorgrams for qualitative comparison of bindings for PhNgb2-Wt, PhNgb2-M1, and PhNgb2-M2. Black arrows in Fig. A to Fig. C show starting time of analyte injections. All peptides were injected for 60 s. (D) Distance between E86 (hNgb) and K27 (Cyt c) residues. The dotted line is drawn to show a reference line at 5 Å for salt bridge.
We also performed similar qualitative experiments for E87 (hNgb)-K27 (Cyt c) binding region. In addition to binding of PhNgb2-Wt (with E87 in hNgb), unlike weaker signals for PhNgb1-M1 and PhNgb1-M2 bindings as compared to PhNgb1-Wt bindings, a mutant peptide PhNgb2-M1 (with E87A substitution) also showed a stronger binding (Fig. 4C). Notably, there is E86 (hNgb) in the PhNgb2-Wt sequence. Based on this observation, in the absence of E87 (hNgb), we speculate that E86 (hNgb) also likely interacts with K27 (Cyt c). This possibility is also predicted in course of simulation after 300 ns, which is represented by bond distance as shown in Fig. 4D (occasionally less than 5 Å). However, the chance for this interaction is quite less as compared to the E87 (hNgb)-K27 (hNgb) interaction as shown in Fig. 1A. This observation is also supported by results from contact analysis. Based on our contact analysis, there were only ~450 frames showing E86 (hNgb)-K27 (Cyt c) interactions as compared to ~1700 frames showing E87 (hNgb)-K27 (Cyt c) interactions. We again conducted experiments with another mutant of PhNgb2-Wt, PhNgb2-M2 (with E86A and E87A substitutions). There is still a comparable binding between PhNgb2-M2 and PhNgb2-Wt. In simulation results, there is no indication of formation specific salt bridge and hydrogen bonding (except for E86 and E87) for this hNgb region. However, several interfacial contacts are observed as shown in Table 1, for this region, which might be non-specific. We speculate that this hNgb region also interacts with Cyt c via non-specific interactions like hydrophobic interactions. Therefore, we did not perform further inhibition experiments for this peptide sequence. Further extensive experiments are required for the interaction of this hNgb region with Cyt c.
Simulation results confirm that redox reaction is likely not required to abate the apoptotic cascade
Our simulation results show that K72 (Cyt c) is one of the amino acid residues that form complex with hNgb. Importantly, this K72 residue takes part in interactions of Cyt c with apoptotic protease-activation factor-131. Our simulation results show that the hNgb-Cyt c complex forms, involving K72 (Cyt c), even when both proteins in oxidized form; thus requires no redox reaction between hNgb and Cyt c for this process. Moreover, hNgb-Cyt c association is enough to suppress caspase-9 activation19. Hence, Cyt c likely competes with apoptosome formation by interacting with hNgb14,31. Therefore, our results confirm that the hNgb-Cyt c redox reaction is likely not required to abate the apoptotic cascade.
Alignment of heme groups and structural rearrangements were observed during the stabilization of the hNgb-Cyt c complex
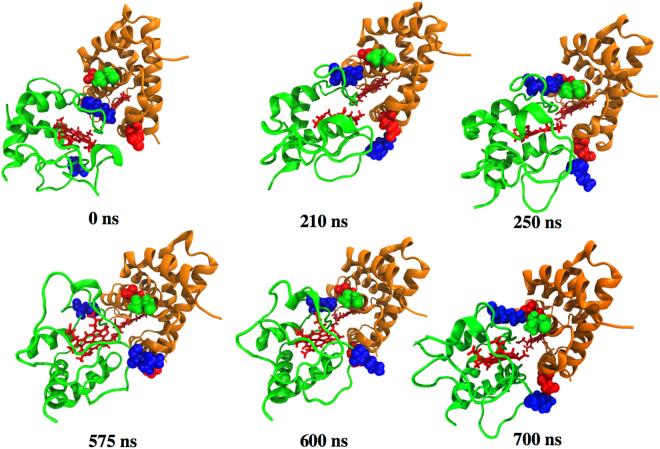
Figure 5 shows the hNgb-Cyt c complex structures at the following different simulation times: 0 ns (the start of the NVT simulations), 210 ns, 250 ns, 575 ns, 600 ns and 700 ns. In the starting complex (Fig. 5 at 0 ns), the heme groups of the two proteins are oriented in different directions or planes. Interestingly, both heme groups continuously align towards the same plane during the stabilization of the hNgb-Cyt c complex as shown in Fig. 5.
Figure 5.
Structures of the hNgb-Cyt c complex at different times during the simulation trajectory.
There is a large rearrangement in the Cyt c structure while hNgb structure is mainly conserved as revealed by the RMSD measurements. Interestingly, at 210 ns, Cyt c rotated around an axis passing through the Fe atoms (at the center of the heme groups) of both hNgb and Cyt c. This structural rearrangement at 210 ns during the complex stabilization initiates the formation of another salt bridge between E87 (hNgb) and K27 (Cyt c) (Fig. 1A). This structural rearrangement as well as new salt bridge formation is also shown in the simulation movie (Supporting file Movie-S1.mp4, after 150 ns of the NVT simulation). In course of simulation, at around 575 ns, another major structural rearrangement was observed, which temporarily increased bond distance between D73 (hNgb) and K72 (Cyt c) as well as between T77 (hNgb) and K72 (Cyt c), as shown in Fig. 1A. At this time no major destabilization in the heme groups was observed. This temporary destabilization is due to a structural modification in Cyt c while the structure of hNgb remains conserved, as supported by RMSD measurements (Fig. 1B). This structural rearrangement is also shown in the simulation movie (Supporting file Movie-S2.mp4, after 500 ns of the NVT simulation).
Discussion
We performed an MD simulations-based investigation of the formation and stabilization of the complex between hNgb and Cyt c. We obtained the best predicted hNgb-Cyt c complex from docking as described in the Materials and Methods section. In both models of the docked complex as proposed in a previous publication12, the protein interfaces involved in forming the complex are the same, with only a little difference in the initial amino acid contacts. Since MD incorporates conformational flexibility and structural adjustments (e.g. rotation of Cyt c at 210 ns as explained above) and allows optimization of the interfacial contacts of the complex, either model would give an optimized complex upon relaxation with MD. In the docking, we chose PDB ID 1OCD for Cyt c structure, with ~88% sequence homology with human Cyt c, as our template because this protein is easily available for purchase at lower cost for numerous experiments.
As supported by SPR results, our simulation results predict that the two residues D73 (hNgb) and T77 (hNgb), both E-helix residues, interact with K72 (Cyt c) and that E87 (hNgb) interacts with K27 (Cyt c) to establish the hNgb-Cyt c complex. The alignment of the heme groups as well as the interactions between the E-helix residues in hNgb and K72 in Cyt c are also crucial for the formation and stabilization of the hNgb-Cyt c complex. As previously mentioned, the repositioning of the E-helix alters the affinity of hNgb to Cyt c. This alignment in the heme groups also confirms the likely interaction between the heme groups for the formation (and stabilization) of the hNgb-Cyt c complex, which is consistent with a previous report based on docking results14. In addition, the involvement of K72 (Cyt c) in the formation of the complex between hNgb and Cyt c suggests that the interaction with hNgb likely competes with apoptosome formation, without requiring redox reaction between Ngb and Cyt c. Altogether, in the absence of a crystal structure, our results provide unique insight into the optimized structure of the hNgb-Cyt c complex, which has a biological role in preventing apoptosis. To the best of our knowledge, this is the first computational investigation of the molecular mechanism underlying the Ngb-Cyt c complex formation, allowing conformational flexibility and structural rearrangements. In future, following this investigation, more extensive computation-based investigations can be done, including much longer and repeated simulations for the wild type complex and all complexes with the mutation of all amino acid residues predicted in this report. Moreover, our current investigations explore avenues to conduct extensive experiments for the bindings (both specific and non-specific) of hNgb region represented by PhNgb2-Wt as well as calculation of quantitative binding energies for wild type and mutant proteins that could help further confirm the importance of each residue in the hNgb-Cyt c complex formation.
Materials and Methods
Materials
Equine heart Cyt c and dimethyl sulfoxide (DMSO) were purchased from Sigma Aldrich. The amine coupling kit, Series S sensor chip CM5, and HBS-P+ buffer (0.1 M HEPES, 1.5 M NaCl, and 0.5% v/v surfactant P20) were purchased from GE Healthcare. The peptides PhNgb1-Wt (sequence: MLVIDAAVTNVED), PhNgb1-M1 (sequence: MLVIDAAVANVED), PhNgb1-M2 (sequence: MLVIAAAVTNVED), PhNgb2-Wt (sequence: SSLEEYLAS), PhNgb2-M1 (sequence: SSLEAYLAS), PhNgb2-M2 (sequence: SSLAAYLAS), and PCyt c (sequence: LENPKKYIP), were purchased from GenScript.
Protein structures and docking
PDB ID 4MPM (Chain B)30 was used for hNgb, and PDB ID 1OCD32 was used for Cyt c. The ZDOCK protein docking server33 was used to predict the hNgb-Cyt c complexes. During the docking submission, SER84 (hNgb)-PRO76 (Cyt c), ASP73 (hNgb)-LYS73 (Cyt c), and heme propionate (hNgb)-LYS72 (Cyt c) group pairs were selected as the contacting residues. Of the top 10 predicted complexes obtained from docking, only one complex with the lowest Fe-Fe distance (16.85 Å), was selected as the best predicted complex. These contacting amino acid residues and Fe-Fe distance were selected based on a previous publication12.
Molecular dynamics (MD) simulations
The best predicted hNgb-Cyt c complex was used in the MD simulations. The input files (.psf and.pdb) for the complex were generated by Visual Molecular Dynamics (VMD)34 using the topology and parameters suggested by a previous publication35 for the oxidized Cyt c heme together with the CHARMM 36 force field. The complex was then solvated in a cubic box with TIP 3P (or TIP3) water and electrically neutralized by adding four Cl− ions, which resulted in a cubic solvation box with dimensions of 90 × 90 × 90 Å3 containing 68,749 atoms. All-atom MD simulations were performed using the NAMD simulation package36. The MD simulations of the complex began with 10,000 steps of energy minimization using the conjugate gradient and line search algorithm. The Particle Mesh Ewald (PME)37 method was used for the long-range interactions with a 12 Å non-bonded cut-off. The system was then equilibrated with a 1 fs integration time step at 300 K (27 °C) for 100 ps with the protein heavy atoms harmonically restrained. After equilibration, the simulation was continued for a 100 ps NPT run with the fully unrestrained protein using a 2 fs time step. The production simulation was then propagated for 700 ns using Langevin dynamics with a damping constant of 1 ps−1 under NVT conditions and a time step of 2 fs.
Surface plasmon resonance (SPR)
The SPR experiments were performed using a Biacore T200 SPR instrument (GE Healthcare). Cyt c, which was used as a ligand, was immobilized onto the CM5 chip up to a level of ~9000 Response Units (RU) in the presence of 10 mM sodium acetate buffer at pH 5.5. A standard amine coupling chemistry according to the manufacturer’s recommended protocol (GE Healthcare) was used to immobilize Cyt c. HBS-P+ buffer was diluted 10X to 10 mM HEPES pH 7.4, 150 mM NaCl, and 0.05% v/v surfactant P20 (HBS-P) in ddH2O and filtered through a 0.22 μM polystyrene membrane filter. HBS-P was used as the immobilization running buffer. The peptides were used as analytes to inject over the Cyt c-immobilized CM5 chip surface in various concentrations in an HBS-P+1% DMSO buffer. HBS-P+1% DMSO was used as the running buffer in the analyte-ligand binding experiments. A phosphoric acid solution (1:250 v/v ratio, H3PO4:ddH2O) was used to regenerate the CM5 chip sensor surface for PhNgb1-Wt and related mutants bindings to Cyt c, and 1 M NaCl was used to regenerate the chip for the bindings of PhNgb2-Wt and related mutants to Cyt c. All analytes were injected in duplicate in each cycle. The kinetics experiments were repeated for three times. All qualitative experiments, including inhibition experiments were repeated at least two times with the wild type peptides injected as positive controls in each experiment.
Data analysis
VMD34 was used to analyze and visualize the structure and simulation trajectories. The total interaction energy was calculated using NAMD energy plugin available in VMD. Each frame in the NVT simulation trajectory was saved at every 200 ps. An atomic distance of 3.5 Å and angle cutoff of 30 degrees were used to trace hydrogen bonding and the same atomic distance of 3.5 Å was used as the cut off to analyze interfacial contacts and salt bridges. For contacts analysis, only residues that show contacts for more than 1000 frames were considered. Carma38 was used to determine the radius of gyration (Rg) and to perform PCA as well as PCA-based cluster analyses. GraphPad prism (GraphPad Software, Inc.) was used to generate the graphs. The bonding distances between D73 (in hNgb, atom OD2) and K72 (in Cyt c, atom NZ), T77 (in hNgb, atom OG1) and K72 (in Cyt c, atom NZ), E87 (in hNgb, atom OE2) and K27 (in Cyt c, atom NZ) as well as between E86 (in hNgb, atom OE2) and K27 (in Cyt c, atom NZ) were plotted. All graphs obtained from analyses of MD simulation trajectories were averaged with 25 neighboring points and 4th order smoothing polynomial using the GraphPad prism software. Biacore T200 evaluation software version 1.0 (GE Healthcare) was used to analyze the SPR sensorgrams.
Electronic supplementary material
Acknowledgements
The experimental SPR sensorgrams were measured using a Biacore T200 instrument and were evaluated using the Biacore T200 evaluation software version 1.0 available in the Biacore Molecular Interaction Shared Resource (BMISR) facility at Georgetown University. The BMISR is supported by NIH grant P30CA51008. The authors declare no conflict of interest with the content of this article.
Author Contributions
P.B.T. conceived the project. P.B.T. performed the molecular docking, molecular dynamics simulations, and S.P.R. experiments. P.B.T. and P.P.C. analyzed the simulation data. P.B.T. and A.Ü. analyzed the SPR data. P.B.T. wrote the manuscript. P.P.C. and A.Ü. contributed to the manuscript editing.
Competing Interests
The authors declare no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-28836-6.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Burmester T, Weich B, Reinhardt S, Hankeln T. A vertebrate globin expressed in the brain. Nature. 2000;407:520–523. doi: 10.1038/35035093. [DOI] [PubMed] [Google Scholar]
- 2.Schmidt M, et al. How does the eye breathe?: Evidence for neuroglobin-mediated oxygen supply in the mammalian retina. J. Biol. Chem. 2003;278:1932–1935. doi: 10.1074/jbc.M209909200. [DOI] [PubMed] [Google Scholar]
- 3.Vallone B, Nienhaus K, Matthes A, Brunori M, Nienhaus GU. The structure of carbonmonoxy neuroglobin reveals a heme-sliding mechanism for control of ligand affinity. Proc. Natl. Acad. Sci. USA. 2004;101:17351–17356. doi: 10.1073/pnas.0407633101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Szymanski M, Wang R, Fallin MD, Bassett SS, Avramopoulos D. Neuroglobin and Alzheimer’s dementia: Genetic association and gene expression changes. Neurobiol. Aging. 2010;31:1835–1842. doi: 10.1016/j.neurobiolaging.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sun Y, Jin K, Mao XO, Zhu Y, Greenberg DA. Neuroglobin is up-regulated by and protects neurons from hypoxic-ischemic injury. Proc. Natl. Acad. Sci. USA. 2001;98:15306–15311. doi: 10.1073/pnas.251466698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Watanabe S, Wakasugi K. Neuroprotective function of human neuroglobin is correlated with its guanine nucleotide dissociation inhibitor activity. Biochem. Biophys. Res. Commun. 2008;369:695–700. doi: 10.1016/j.bbrc.2008.02.089. [DOI] [PubMed] [Google Scholar]
- 7.Di Pietro V, et al. Neuroglobin expression and oxidant/antioxidant balance after graded traumatic brain injury in the rat. Free Rad. Biol. Med. 2014;69:258–264. doi: 10.1016/j.freeradbiomed.2014.01.032. [DOI] [PubMed] [Google Scholar]
- 8.Baez, E. et al. Protection by neuroglobin expression in brain pathologies. Front. Neurol. 7, 10.3389/fneur.2016.00146 (2016). [DOI] [PMC free article] [PubMed]
- 9.Takahashi N, Wakasugi K. Identification of residues crucial for the interaction between human neuroglobin and the α-subunit of heterotrimeric Gi protein. Sci. Rep. 2016;6:24948. doi: 10.1038/srep24948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Azarov I, et al. Five-coordinate H64Q neuroglobin as a ligand-trap antidote for carbon monoxide poisoning. Sci. Transl. Med. 2016;8:368ra173. doi: 10.1126/scitranslmed.aah6571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu Z, et al. Roles of neuroglobin binding to mitochondrial complex III subunit cytochrome c1 in oxygen-glucose deprivation-induced neurotoxicity in primary neurons. Mol. Neurobiol. 2016;53:3249–3257. doi: 10.1007/s12035-015-9273-4. [DOI] [PubMed] [Google Scholar]
- 12.Tiwari PB, et al. Characterization of molecular mechanism of neuroglobin binding to cytochrome c: A surface plasmon resonance and isothermal titration calorimetry study. Ino. Chem. Commun. 2015;62:37–41. doi: 10.1016/j.inoche.2015.10.010. [DOI] [Google Scholar]
- 13.Fisher WR, Taniuchi H, Anfinsen CB. On the role of Heme in the formation of the structure of cytochrome c. J. Biol. Chem. 1973;248:3188–3195. [PubMed] [Google Scholar]
- 14.Bonding SH, Henty K, Dingley AJ, Brittain T. The binding of cytochrome c to neuroglobin: A docking and surface plasmon resonance study. Int. J. Biol. Macromol. 2008;43:295–299. doi: 10.1016/j.ijbiomac.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 15.Ow Y-LP, Green DR, Hao Z, Mak TW. Cytochrome c: functions beyond respiration. Nat. Rev. Mol. Cell Biol. 2008;9:532–542. doi: 10.1038/nrm2434. [DOI] [PubMed] [Google Scholar]
- 16.Brittain T, Skommer J, Raychaudhuri S, Birch N. An antiapoptotic neuroprotective role for neuroglobin. Int. J. Mol. Sci. 2010;11:2306. doi: 10.3390/ijms11062306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ascenzi P, Polticelli F, Marino M, Santucci R, Coletta M. Cardiolipin drives cytochrome c proapoptotic and antiapoptotic actions. IUBMB Life. 2011;63:160–165. doi: 10.1002/iub.440. [DOI] [PubMed] [Google Scholar]
- 18.De Marinis E, Fiocchetti M, Acconcia F, Ascenzi P, Marino M. Neuroglobin upregulation induced by 17 β-estradiol sequesters cytocrome c in the mitochondria preventing H2O2-induced apoptosis of neuroblastoma cells. Cell Death Dis. 2013;4:e508. doi: 10.1038/cddis.2013.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Raychaudhuri S, Skommer J, Henty K, Birch N, Brittain T. Neuroglobin protects nerve cells from apoptosis by inhibiting the intrinsic pathway of cell death. Apoptosis. 2010;15:401–411. doi: 10.1007/s10495-009-0436-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tiwari PB, et al. Quantitative study of protein-protein interactions by quartz nanopipettes. Nanoscale. 2014;6:10255–10263. doi: 10.1039/C4NR02964J. [DOI] [PubMed] [Google Scholar]
- 21.Tiwari PB, et al. Characterization of molecular interactions between Escherichia coli RNA polymerase and topoisomerase I by molecular simulations. FEBS Lett. 2016;590:2844–2851. doi: 10.1002/1873-3468.12321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li X, Wu M, Kwoh C-K, Ng S-K. Computational approaches for detecting protein complexes from protein interaction networks: a survey. BMC Genomics. 2010;11:S3. doi: 10.1186/1471-2164-11-S1-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xue LC, Dobbs D, Bonvin AMJJ, Honavar V. Computational prediction of protein interfaces: A review of data driven methods. FEBS Lett. 2015;589:3516–3526. doi: 10.1016/j.febslet.2015.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Singh AN, Baruah MM, Sharma N. Structure Based docking studies towards exploring potential anti-androgen activity of selected phytochemicals against prostate cancer. Sci. Rep. 2017;7:1955. doi: 10.1038/s41598-017-02023-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karplus M, McCammon JA. Molecular dynamics simulations of biomolecules. Nat. Struct. Mol. Biol. 2002;9:646–652. doi: 10.1038/nsb0902-646. [DOI] [PubMed] [Google Scholar]
- 26.Singh W, et al. Conformational dynamics, ligand binding and effects of mutations in NirE an S-Adenosyl-L-Methionine dependent methyltransferase. Sci. Rep. 2016;6:20107. doi: 10.1038/srep20107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shvetsov AV, et al. Structure of RecX protein complex with the presynaptic RecA filament: Molecular dynamics simulations and small angle neutron scattering. FEBS Lett. 2014;588:948–955. doi: 10.1016/j.febslet.2014.01.053. [DOI] [PubMed] [Google Scholar]
- 28.Vettoretti G, et al. Molecular dynamics dimulations reveal the mechanisms of allosteric activation of Hsp90 by aesigned aigands. Sci. Rep. 2016;6:23830. doi: 10.1038/srep23830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barlow DJ, Thornton JM. Ion-pairs in proteins. J. Mol. Biol. 1983;168:867–885. doi: 10.1016/S0022-2836(83)80079-5. [DOI] [PubMed] [Google Scholar]
- 30.Guimara BG, Hamdane D, Lechauve C, Marden MC, Golinelli-Pimpaneau B. The crystal structure of wild-type human brain neuroglobin reveals flexibility of the disulfide bond that regulates oxygen affinity. Acta Crystallogr. 2014;D70:1005–1014. doi: 10.1107/S1399004714000078. [DOI] [PubMed] [Google Scholar]
- 31.Fiocchetti, M., Cipolletti, M., Brandi, V., Polticelli, F. & Ascenzi, P. Neuroglobin and friends. J. Mol. Recog., e2654, 10.1002/jmr.2654. [DOI] [PubMed]
- 32.Qi PX, Beckman RA, Wand AJ. Solution Structure of horse heart ferricytochrome c and detection of redox-related structural changes by high-resolution 1H NMR. Biochemistry. 1996;35:12275–12286. doi: 10.1021/bi961042w. [DOI] [PubMed] [Google Scholar]
- 33.Pierce BG, Hourai Y, Weng Z. Accelerating protein docking in ZDOCK using an advanced 3D convolution library. PLoS ONE. 2011;6:e24657. doi: 10.1371/journal.pone.0024657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Humphrey W, Dalke A, Schulten K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 35.Autenrieth F, Tajkhorshid E, Baudry J, Luthey-Schulten Z. Classical force field parameters for the heme prosthetic group of cytochrome c. J. Comput. Chem. 2004;25:1613–1622. doi: 10.1002/jcc.20079. [DOI] [PubMed] [Google Scholar]
- 36.Phillips JC, et al. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Darden T, York D, Pedersen L. Particle mesh Ewald: An N.log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089–10092. doi: 10.1063/1.464397. [DOI] [Google Scholar]
- 38.Glykos N. Software news and updates carma: A molecular dynamics analysis program. J. Comput. Chem. 2006;27:1765–1768. doi: 10.1002/jcc.20482. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.