Abstract
Reprogramming technology holds great promise for the study and treatment of Parkinson’s disease (PD) as patient-specific ventral midbrain dopamine (vmDA) neurons can be generated. This should facilitate the investigation of early changes occurring during PD pathogenesis, permitting the identification of new drug targets and providing a platform for drug screening. To date, most studies using reprogramming technology to study PD have employed induced pluripotent stem cells. Research into PD using direct reprogramming has been limited due to an inability to generate high yields of authentic human vmDA neurons. Nevertheless, direct reprogramming offers a number of advantages, and development of this technology is warranted. Previous reports have indicated that induced neural precursors (iNPs) derived from adult human fibroblasts by lineage factor-mediated direct reprogramming can give rise to dopamine neurons expressing tyrosine hydroxylase (TH+). Using normal adult human fibroblasts, the present study aimed to extend these findings and determine the capacity of iNPs for generating vmDA neurons, with the aim of utilising this technology for the future study of PD. While iNPs expressed late vmDA fate markers such as NURR1 and PITX3, critical early regional markers LMX1A, FOXA2 and EN1 were not expressed. Upon differentiation, iNPs gave rise to dopamine neuronal-like cells expressing TUJ1, TH, AADC, DAT, VMAT2 and GIRK2. To induce an authentic A9 phenotype, a series of experiments investigated temporal exposure to patterning factors. Exposure to SHH-C24II, purmorphamine, CHIR99021 and/or FGF8b during or after reprogramming was insufficient to induce expression of early vmDA regional markers. Addition of LMX1A/FOXA2 to the transfection cocktail did not induce a sustained vmDA iNP phenotype. This study reports for the first time that iNPs derived from healthy adult human cells by non-viral expression of lineage factors can give rise to dopamine neuronal-like cells. Direct-to-iNP reprogramming could be a suitable strategy for modelling PD in vitro using aged donor-derived cells.
Keywords: Induced neural precursor cells, Dopamine neurons, Direct reprogramming, Adult human fibroblasts, Parkinson’s disease
Introduction
Cell reprogramming technology offers exciting prospects for studying and treating Parkinson’s disease (PD)[1]. Human foetal mesencephalic tissue transplants have provided proof-of-principle that stem cell therapies hold promise for treating PD[2]. Cell reprogramming technology permits autologous or HLA-matched cell transplants, circumventing the requirement for immunosuppression and avoiding ethical issues related to using large amounts of foetal tissue. Another important application for cell reprogramming is disease modelling. The ability to generate patient-specific ventral midbrain dopamine (vmDA) neurons permits the investigation of early changes occurring during PD pathogenesis, the identification of new drug targets, and provides a platform for screening therapeutic compounds.
Reprogramming to an induced pluripotent stem cell (iPSC) fate is a popular method to derive human neurons. As iPSC technology was preceded by years of mouse and human embryonic stem cell work, existing protocols for deriving neuronal subtypes such as vmDA neurons are advanced[3, 4]. Induced pluripotent stem cells provide an indefinitely expandable cell source that is not restricted to a particular lineage. However, producing and validating iPSC clones is a lengthy process, taking 4 - 6 months to generate functional neurons from a starting population of fibroblasts[5]. Additionally, by reverting adult cells to an embryonic state, markers of age are erased, posing a problem for modelling age-related diseases[6]. For use in cell-based therapies, the potential presence of residual pluripotent cells in differentiated cultures poses a threat of tumour formation.
By contrast, direct-to-neuronal reprogramming technology is at an earlier stage of development. This technology is faster and more efficient than iPSC reprogramming, generating functional human induced neural (iN) cells within 1 – 3 weeks[7, 8]. As cells are not rejuvenated, age-related aspects of PD may be more readily studied in vitro. While mature iN cells are not suitable for transplantation therapy, the possibility of reprogramming in vivo[9, 10] may offer advantages to cell-based therapies, such as avoiding an immune response, a higher efficiency of neuronal generation, and a lower probability of tumour formation. However, for in vitro applications, direct-to-neuronal reprogramming generates a limited supply of post-mitotic neurons and for studies aiming to examine PD-related developmental changes in dopamine neurons, iN cells are not suitable. To date, the generation of authentic human vmDA iN cells from adult cells has not been described[1].
Direct reprogramming to induced neural stem cells/precursors (iNSC/iNPs) offers an intermediate strategy, where proliferative cells may be obtained in a faster process than iPSC reprogramming, generating iNSC/iNPs within 1 – 4 weeks, with additional time required for differentiation[5]. The three strategies for direct-to-iNSC/iNP reprogramming are to use pluripotency factors, lineage specific factors and/or chemicals[1]. Direct reprogramming using pluripotency factors may not be ‘direct’, but similar to iPSC reprogramming, inducing a transient state of pluripotency[11, 12]. Chromosomal abnormalities are more frequent in pluripotent factor- than lineage factor-reprogrammed iNSC/iNPs, which may increase the probability of tumour formation[13]. By using pluripotency factors, iNSC/iNPs may be rejuvenated, as observed with iPSC reprogramming. The mechanism underlying lineage factor-mediated direct-to-iNSC/iNP reprogramming has not been examined in detail, and as many different combinations of genes have been reported, mechanisms and stages of development may differ between protocols[1].
Reprogramming to an iNSC/iNP fate represents a promising strategy for generating neuronal lineages from somatic cells. A handful of studies have reported the generation of dopamine neurons from adult human pluripotent factor-reprogrammed iNSC/iNPs[14-17] (for a full review, see[1]). Several studies have reported generating dopamine neurons from human foetal or post-natal cells via lineage factor-mediated reprogramming[18-20], yet only two have generated tyrosine hydroxylase (TH+) cells from adult human donors[21, 22]. Further examination into the dopaminergic phenotype of TH+ cells generated from adult donors via this method has not been reported. Although reprogramming is less efficient in aged donor cells than younger cells[23], successful modelling of age-related diseases like PD will rely on using adult cells as a source for generating dopamine neurons. Finally, it is essential to confirm that directly reprogrammed dopamine neurons represent an authentic ventral midbrain A9 phenotype prior to PD modelling.
A method to reprogram adult human fibroblasts to an iNP fate by non-viral SOX2/PAX6 transfection has been developed in our laboratory[21]. Using non-viral methods of gene introduction reduces the chance of insertional mutagenesis which may be important for downstream applications. This method gives rise to TH+/NSE+ cells from adult human fibroblasts[21]. Based on the potential advantages of direct-to-iNSC/iNP reprogramming, the present study aimed to extend our previous work and determine the capacity of iNPs for generating dopamine neurons from adult human fibroblasts. Healthy donor cells were selected for optimising this protocol, with a view to utilising this technology for the study of PD in donor-derived cells.
Materials & Methods
Cell culture
Adult human fibroblast cells from healthy donors (n = 3), aged 39 – 56 years, were sourced from Cell Applications, Inc. No substantial differences were observed between cell lines throughout the study. Plasmid transfections were carried out using Lipofectamine LTX, Plus reagent (Invitrogen) and plasmid constructs at a ratio 3:1:1 (v/v/w) as per[24]. The plasmids contained cDNAs encoding human SOX2, human PAX6, mouse LMX1A and/or mouse FOXA2, and were used at 1.6 – 2.5 μg/plasmid/well (6-well format). The vectors pLV.PGK.mLmx1a and pLV.PGK.mFoxa2 were a gift from Malin Parmar, Lund University (Addgene plasmids #33013 and #33014)[25]. Transfection was confirmed by concurrent transfection with plasmids expressing fluorescent protein, and/or performing immunocytochemistry on cells fixed 1 – 3 days post-transfection.
Three days after transfection, cells were transferred to reprogramming medium containing Neurobasal-A medium (Gibco), 1 mM valproic acid (Sigma), 0.3% D-(+)-Glucose (Sigma), 1x Penicillin-Streptomycin-Glutamine (Gibco), 1x B27 supplement with retinoic acid (Gibco), 20 ng/ml EGF (Peprotech), 2 μg/ml Heparin (Sigma), with 25 ng/ml midkine (Peprotech) supplemented until day 17 – 21 post-transfection. Media was supplemented with 20 – 500 ng/ml SHH-C24II (R&D Systems), 0.7 – 3 μM CHIR99021 (Stemgent or Miltenyi Biotec), 2 μM purmorphamine (Stemgent) and/or 100 ng/ml FGF8b (Peprotech), as indicated.
To differentiate iNPs, cells were cultured in basal differentiation media containing Neurobasal-A medium (Gibco), 0.3% D-(+)-Glucose (Sigma), 1x Penicillin-Streptomycin-Glutamine (Gibco), 1x B27 supplement without retinoic acid (Gibco) and 1x N2 supplement (Gibco). Patterning media was supplemented with 200 ng/ml SHH-C24II (Peprotech) and 100 ng/ml FGF8b (Peprotech). Maturation media was supplemented with 20 ng/ml BDNF (Creative Biomart or Peprotech), 20 ng/ml GDNF (Merck Millipore or Peprotech), 1 ng/ml TGFβ3 (Peprotech), 500 μM dibutyryl cAMP (Sigma) and 200 μM L-ascorbic acid (Sigma). Cells were differentiated for up to one week in Patterning media and up to four weeks in Maturation media.
Quantitative real-time PCR
RNA was isolated using the Nucleospin RNA kit (Macherey-Nagel) and reverse transcription was conducted using SuperScript III First-Strand Synthesis (Invitrogen), with an equivalent amount of RNA per sample. Three independent duplex qPCR reactions were performed for each sample using the TaqMan system (Applied Biosystems) with ribosomal 18S rRNA as the internal standard and an equivalent of 4 ng mRNA per reaction. Analysis was performed using the comparative Ct (ΔΔCt) method[26].
Immunocytochemistry
The following primary antibodies were used: AADC (Millipore/AB1569), ASCL1 (Millipore/AB5696), DAT (Abcam/AB5990), FOXA2 (Santa Cruz/SC-6554), FOXG1 (Abcam/AB18259), GIRK2 (Abcam/AB30738), KI67 (DAKO/M7240), LMX1A (Millipore/AB10533), NESTIN (Abcam/AB22035), NGN2 (R&D Systems/MAB3314), NURR1 (Santa Cruz/SC-990), SOX1 (Millipore/AB15766), SOX2 (R&D Systems/MAB2018), TH (Millipore/MAB5280), TH (Millipore/AB152), TUJ1 (Biolegend/801202), VMAT2 (Millipore/AB1598P). Fluorescent imaging was performed on a Nikon Eclipse TE2000U microscope with an Optronics Microfire camera or Photometrics Evolve EMCCD camera. Phase contrast images were taken on the Nikon Eclipse TS100 microscope with a Nikon Digital Sight DS-U1 camera. For cell quantification experiments, the investigator was blinded to the condition during image acquisition and counting.
Results
SOX2/PAX6-iNPs show temporal changes in the expression of genes related to vmDA development
To give rise to dopamine neurons, it was reasoned that iNPs would need to express genes implicated in the development of vmDA neurons in vivo. To investigate this, a time course experiment examining the expression of a range of neural stem, progenitor and regional markers was performed during SOX2/PAX6-reprogramming, focussing on genes involved in vmDA development (Figure 1A; Study One). Over the course of reprogramming, and particularly after the first passage (day 31), cells transitioned from extended, polarised fibroblast morphologies to epithelial-like morphologies, with regular dimensions and cells growing in discrete patches (Figure 1B). In addition, floating and semi-adherent clusters of cells reminiscent of neurospheres were observed.
Figure 1:
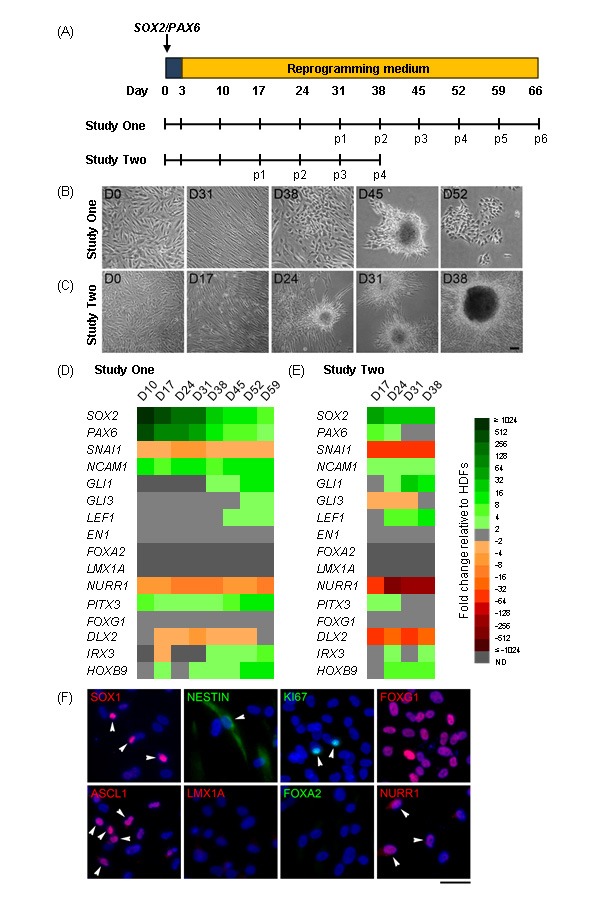
Characterisation of SOX2/PAX6-iNPs. (A) Schematic detailing experimental outline for Studies One and Two. Weekly passaging was commenced at day 31 (Study One) or day 17 (Study Two) post-transfection. (B) – (C) Phase contrast images of fibroblasts (Fib) reprogramming into iNPs over time. Scale bar: 100 μm. (D) – (E) Heat map depicting gene expression in iNPs over time as fold changes relative to fibroblasts. ND: not detected. (F) Expression of neural progenitor and regional markers in p4 SOX2/PAX6-iNPs. Arrowheads indicate some cells with positive staining. Scale bar: 50 μm.
Gene expression analysis (Figure 1D) found that SOX2 and PAX6 expression subsided over time, but remained elevated relative to fibroblasts. The fibroblast marker SNAI1 was downregulated while the neural marker NCAM1 was upregulated relative to fibroblasts. GLI1, GLI3 and LEF1, which are downstream targets and effectors of SHH and WNT signalling, were upregulated later during reprogramming. Importantly, the early vmDA regional markers EN1, FOXA2 and LMX1A were not upregulated by reprogramming, while the late vmDA marker NURR1 was downregulated and PITX3 was upregulated relative to fibroblasts.
In Study One, the expression of a number of genes was upregulated late during reprogramming after the commencement of weekly passaging. Study Two sought to determine if weekly passaging could be commenced earlier to reduce the duration of reprogramming (Figure 1A). Changes in cell morphologies were observed earlier (Figure 1C), and similar patterns of gene expression were seen at earlier time points (Figure IE).
Protein expression of neural precursor and regional markers in SOX2/PAX6-iNPs was examined (Figure 1F). Widespread expression of the anterior marker FOXG1 was found, with heterogeneous expression of SOX1, NESTIN, NURR1, KI67 and ASCL1. The vmDA progenitor markers LMX1A and FOXA2 were not observed.
SOX2/PAX6-iNPs show minimal response to timed exposure to patterning molecules
Studies One and Two found that SOX2/PAX6-iNPs upregulated GLI1 and LEF1 towards the end of reprogramming. These genes are involved in SHH and WNT signal transduction, and indicate that SOX2/PAX6-iNPs may respond to patterning molecules. To determine if SOX2/PAX6-iNPs would differentiate into dopamine neurons in response to commonly used patterning and maturation cues, cells were differentiated by applying BDNF, GDNF, TGFβ3, dcAMP and ascorbic acid, as a maturation medium, with or without a preceding period of patterning by SHH-C24II (SHH) and FGF8b (Figure 2A). A population of differentiated cells co-expressed TUJ1 with TH or AADC, and TH+ cells also expressed AADC, GIRK2, VMAT2 and DAT (Figure 2B). Quantification revealed that the addition of SHH/FGF8 had no effect on the yields of TUJ1+, TH+/TUJ1+ or AADC+/TUJ1+ cells (Figure 2C). Overall the yields of TUJ1+ cells co-expressing TH or AADC were low (≤ 2.1% of all cells), however the proportion of TUJ1+ cells that co-expressed TH or AADC was at least 24.9% (Figure 2D).
Figure 2:
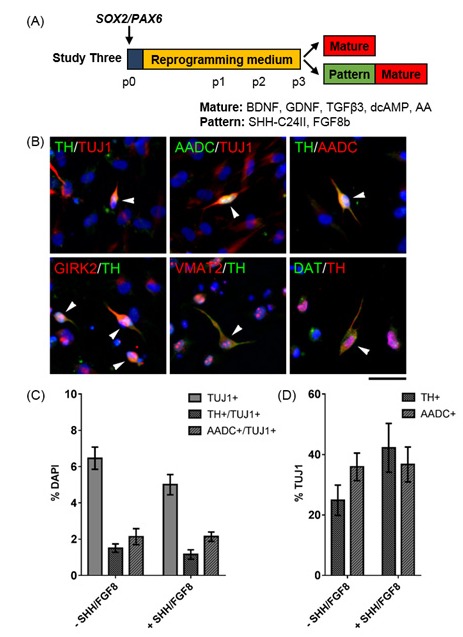
Determining the dopaminergic differentiation capacity of SOX2/PAX6-iNPs. (A) Schematic detailing experimental outline for Study Three. (B) Expression of dopaminergic markers in cultures differentiated with Patterning and Maturation media. Arrowheads indicate cells with positive staining. Scale bar: 50 μm. (C) Quantification of TUJ1+, TUJ1+/TH+ and TUJ1+/AADC+ cells in differentiated iNP cultures. Data are presented as mean ± SEM, n = 16 - 32 fields of view from two independent cell lines (number of cells counted per condition ≥ 2519). Unpaired t-tests revealed no effect of SHH/FGF8 patterning on yields of TUJ1+, TH+/TUJ1+ or AADC+/TUJ1+ cells. (D) Quantification of TH+ and AADC+ cells out of TUJ1 in differentiated iNP cultures. Data are presented as mean ± SEM, n = 16 fields of view from two independent cell lines (number of cells counted per condition ≥ 2519). Unpaired t-tests revealed no effect of SHH/FGF8 patterning on yields of TH+ or AADC+ cells.
Pluripotent stem cell studies have indicated that early exposure to potent SHH and WNT signalling is critical to pattern differentiating neural progenitors towards an authentic vmDA fate[3, 4, 27, 28]. Established SOX2/PAX6-iNPs may be too mature to respond to SHH/FGF8 patterning. We, therefore, investigated if exposure to SHH, or the Smoothened agonist purmorphamine (PUR), with WNT signalling by the GSK3β inhibitor CHIR99021 (CHIR) during reprogramming could induce expression of vmDA markers in iNPs (Figure 3A). Cells were exposed to these patterning conditions from the first passage onwards (Study Four), for the full duration of reprogramming (Study Five), or until the start of weekly passaging (Study Six). At p3, cells exposed to SHH/CHIR or PUR/CHIR for the last phase or the full duration of reprogramming showed upregulated GLI1 relative to unpatterned control iNPs (Figure 3B). Late patterning induced downregulation of NURR1 and PITX3, whereas patterning for the full duration of reprogramming upregulated both NURR1 and PITX3 (Figure 3B). Nevertheless, FOXA2, LMX1A and EN1 were unchanged by exposure to patterning molecules during reprogramming (Figure 3B). Importantly, exposure to patterning molecules for the full duration of reprogramming caused cellular toxicity, and this precluded further examination of the differentiation capacity of these cells. It was reasoned that early, restricted exposure to patterning factors might be sufficient to induce similar changes in vmDA gene expression without causing cellular toxicity (Study Six). However, few sustained changes in gene expression were observed when cells were exposed to SHH/CHIR or PUR/CHIR for the first period of reprogramming only. Differentiation of cells in Study Six found that PUR/CHIR-patterned iNPs died upon exposure to maturation medium (Figure 3C), while SHH/CHIR-patterning had no effect on TUJ1+ or TH+ cell yields (not shown).
Figure 3:
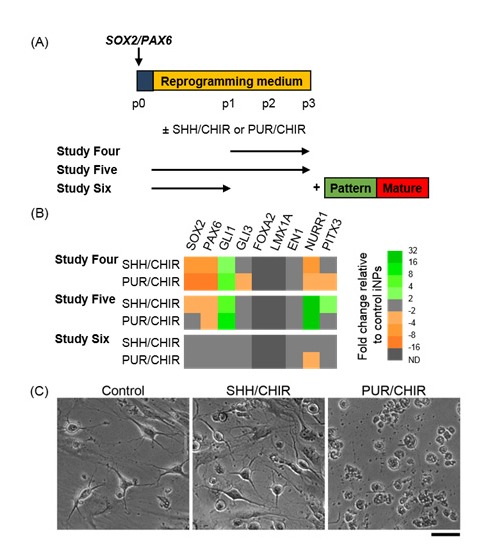
Investigating the effect of exposure to patterning molecules during SOX2/PAX6-mediated reprogramming. (A) Schematic detailing experimental outline for Studies Four – Six. SHH: 200 ng/ml SHH-C24II, PUR: 2 μM purmorphamine, CHIR: 0.7 μM CHIR99021. (B) Heat map depicting gene expression in iNPs at p3 as fold changes relative to control SOX2/PAX6-iNPs cultured under standard reprogramming conditions. (C) Phase contrast images of differentiated iNP cultures from Study Six after one week in Patterning media and one week in Maturation media .Scale bar: 50 μm.
SOX2 alone produces iNPs under standard reprogramming conditions
PAX6 is an anterior neuroectodermal marker that is not expressed in the ventral mesencephalon, the region where vmDA neurons arise[29,30]. Several studies have demonstrated that to pattern differentiating pluripotent stem cells to a floor plate fate, PAX6 induction must be avoided by early potent ventralisation[28, 31]. SOX2-iNPs have been generated from rodent and human foetal cells when combined with a feeder layer or a cocktail of small molecules[32, 33]. To determine if SOX2 alone could induce an iNP fate in adult fibroblasts under our direct reprogramming conditions, Study Seven investigated reprogramming fibroblasts by SOX2 transfection with standard culture conditions (Figure 4A). Similar changes in cellular morphologies during reprogramming were observed in SOX2-iNPs as in SOX2/PAX6-iNPs (Figure 4B). Gene expression analysis revealed similar profiles between SOX2/PAX6-iNPs and SOX2-iNPs (Figure 4C). At the protein level, SOX2-iNPs expressed SOX1, NESTIN, NGN2 and NURR1, but as observed in SOX2/PAX6-iNPs, there was no expression of either LMX1A or FOXA2 (Figure 4D).
Figure 4:
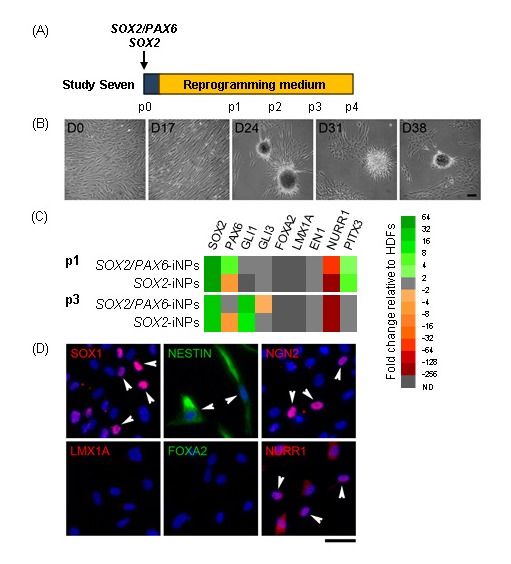
Characterisation of SOX2-iNPs. (A) Schematic detailing experimental outline for Study Seven. Weekly passaging was commenced at day 17 post-transfection. (B) Phase contrast images of SOX2-iNPs over the course of reprogramming. Scale bar: 100 μm. (C) Heat map depicting gene expression in SOX2/PAX6- and SOX2-iNPs at p1 and p3 as fold changes relative to fibroblasts. (D) Expression of neural progenitor and regional markers in p4 SOX2-iNPs. Arrowheads indicate some cells with positive staining. Scale bar: 50 μm.
SOX2-iNPs show minimal response to early exposure to SHH/CHIR
As SOX2-iNPs could be generated without PAX6 transfection, it was of interest to determine if these cells would respond to patterning cues. The concentration of 0.7 μM CHIR used to pattern differentiating pluripotent stem cells to a midbrain fate as described in a previous study[4] may require optimisation for this system, due to potentially varying degrees of endogenous WNT signalling[34]. To determine if SOX2-iNPs could respond to SHH/CHIR-patterning, cells were exposed to early SHH in conjunction with CHIR at 0.7, 1.5 or 3 μM for the first period of reprogramming (Figure 5A). Patterning SOX2/PAX6-iNPs at this point did not induce lasting changes in vmDA gene expression in Study Six, however the responsiveness of SOX2-iNPs to patterning cues may be altered in the absence of PAX6 expression. Additionally, pluripotent stem cell studies indicate that early patterning is essential[3, 4, 28, 31], and prolonged exposure to patterning molecules induced toxicity in Study Five. Transcriptional analysis found few changes in gene expression compared to unpatterned SOX2-iNPs, three weeks after withdrawal of patterning factors (Figure 5B). GLI1 and LEF1 were upregulated by higher concentrations of CHIR (1.5 or 3 μM). However no changes in vmDA gene expression or differentiation capacity was observed as a result of early SHH/CHIR-patterning (Figure 5B, C).
Figure 5:
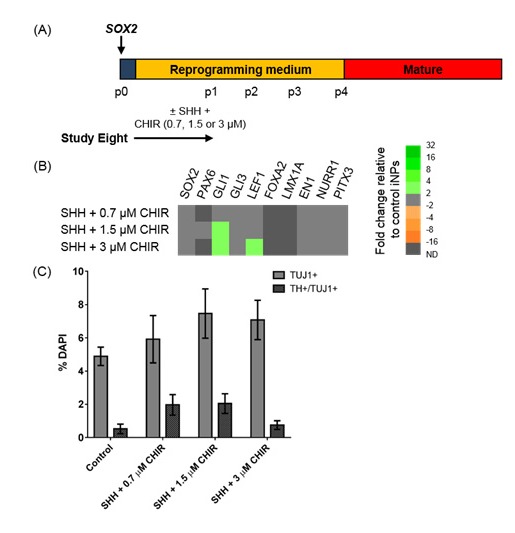
Investigating the effect of exposure to patterning molecules during SOX2-mediated reprogramming. (A) Schematic detailing experimental outline for Study Eight. (B) Heat map depicting gene expression in iNPs as fold changes relative to control SOX2-iNPs cultured under standard reprogramming conditions. (C) Quantification of TUJ1+ and TUJ1+/TH+ cells in differentiated iNP cultures. Data are presented as mean ± SEM, n = 8 fields of view from one cell line (number of cells counted per condition ≥ 819). An ordinary one-way ANOVA revealed no effect of SHH/CHIR patterning on yields of TUJ1+ or TH+/TUJ1+ cells.
Addition of LMX1A and FOXA2 has little effect on vmDA identity in iNPs
As media components were not sufficient to induce early vmDA genes in iNPs, we investigated whether the addition of LMX1A and FOXA2 was able to promote a vmDA phenotype. Fibroblasts were transfected with SOX2, SOX2/PAX6, SOX2/LMX1A or SOX2/LMX1A/FOXA2 and cultured under standard reprogramming conditions (Figure 6A). While expression of the transgenes was observed following transfection by immunocytochemistry (Figure 6B) and qPCR (not shown), at p3 iNPs did not show endogenous expression of either LMX1A or FOXA2 (Figure 6C). Furthermore, upon differentiation, there was no change in the yields of TUJ1+ or TH+/TUJ1+ cells compared to SOX2/PAX6- or SOX2-iNPs (Figure 6D).
Figure 6:
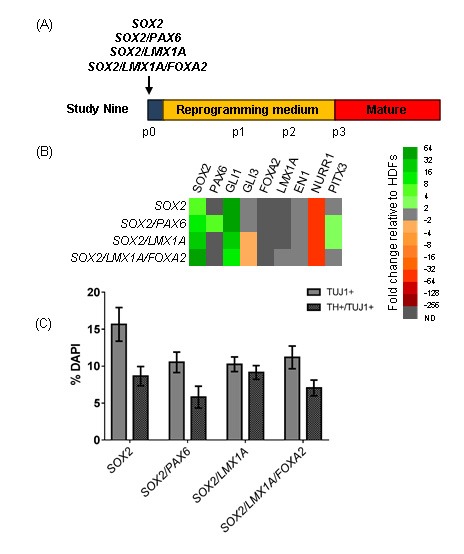
Investigating the effect of adding vmDA transgenes during reprogramming. (A) Schematic detailing experimental outline for Study Nine. Cells were cultured in standard reprogramming medium. (B) Immunocytochemistry at three days post-transfection confirms transfection of plasmids. Scale bar: 50 μm. (C) Heat map depicting gene expression in iNPs as fold changes relative to fibroblasts. (D) Quantification of TUJ1+ and TUJ1+/TH+ cells in differentiated iNP cultures. Data are presented as mean ± SEM, n = 6 – 13 fields of view from one cell line (number of cells counted per condition ≥ 738). An ordinary one-way ANOVA revealed no effect of the transfection factor combination on yields of TUJ1+ or TH+/TUJ1+ cells.
Previous studies have indicated that for exogenous LMX1A to exert its effect, SHH exposure is required[35, 36]. Therefore fibroblasts were transfected with SOX2/LMX1A or SOX2/LMX1A/FOXA2 and reprogrammed with early SHH/CHIR patterning, after which point CHIR was withdrawn and SHH concentration was dropped to 20 ng/ml (Figure 7A). The literature is mixed as to the requirement for FGF8 exposure to induce a vmDA fate[3, 4, 27, 37-39], however, it was reasoned that it was unlikely to be detrimental to the acquisition of a vmDA fate, so FGF8 was added for the full duration of reprogramming. Under these patterning conditions, we still did not observe the induction of endogenous LMX1A or FOXA2, and the expression of other vmDA genes such as EN1, NURR1 and PITX3 was unchanged relative to unpatterned iNPs (Figure 7B). Upon differentiation, SHH/CHIR/FGF8-patterned iNPs gave rise to higher yields of TUJ1+ cells than unpatterned iNPs (Figure 7C). The yields of AADC+/TUJ1+ cells were modestly increased in SOX2/LMX1A-iNPs by SHH/CHIR/FGF8-patterning, however, this was not observed in SOX2/LMX1A/FOXA2-iNPs, nor was a difference observed in TH+/TUJ1+ cell yields (Figure 7C).
Figure 7:
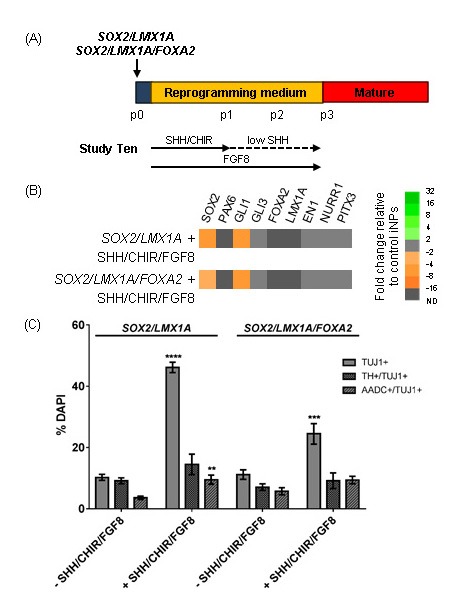
Investigating the effect of combining vmDA transgenes with patterning molecules during reprogramming.(A) Schematic detailing experimental outline for Study Ten. Standard SHH concentration was 200 ng/ml, low SHH was 20 ng/ml. (B) Heat map depicting gene expression in iNPs as fold changes relative to iNPs generated with the same transgene combinations but reprogrammed in the absence of patterning cues. (C) Quantification of TUJ1+, TUJ1+/TH+ and TUJ1+/AADC+ cells in differentiated iNP cultures. Data are presented as mean ± SEM, n = 6 – 13 fields of view from one cell line (number of cells counted per condition ≥ 1559). Unpaired t-tests were performed to compare the effect of patterning with SHH/CHIR/FGF8 on differentiation yields.
Finally, to determine if the increased TUJ1+ and AADC+/TUJ1+ cell yields were the result of FGF8 or the result of combining LMX1A with patterning conditions, cells were reprogrammed with SOX2/LMX1A and cultured under the same patterning conditions with or without FGF8 (Figure 8A). Additionally, to determine if stronger ventralisation could induce a vmDA fate, these conditions were examined with a high SHH concentration (high; 500 ng/ml) or the standard concentration (med; 200 ng/ml). Gene expression analysis showed no upregulation of endogenous vmDA markers in SOX2/LMX1A-iNPs (Figure 8B). Upon differentiation, both conditions cultured with FGF8 showed significantly increased yields of TUJ1+ cells compared to cells cultured in control reprogramming medium, or patterning media without FGF8, regardless of the concentration of SHH (Figure 8C). There was no significant effect of patterning on TH+/TUJ1+ and AADC+/TUJ1+ cell yields.
Figure 8:
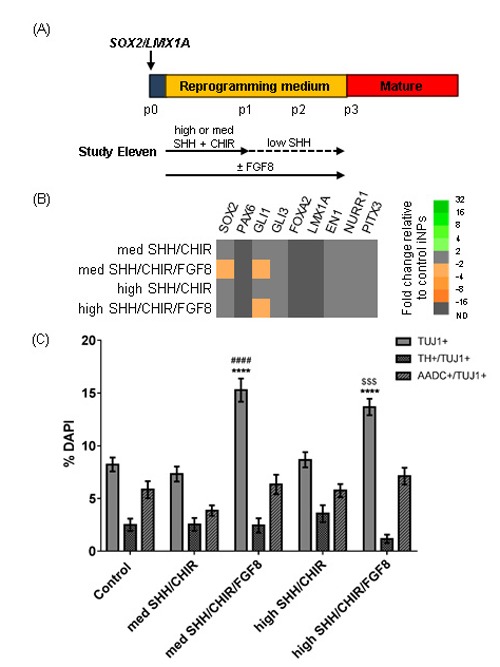
Investigating the effect of FGF8 and SHH concentration during SOX2/LMX1A-mediated reprogramming. (A) Schematic detailing experimental outline for Study Eleven. High SHH concentration was 500 ng/ml, med SHH was 200 ng/ml, low SHH was 20 ng/ml. (B) Heat map depicting gene expression in iNPs at p3 as fold changes relative to SOX2/LMX1A-iNPs reprogrammed standard reprogramming medium. (C) Quantification of TUJ1+, TUJ1+/TH+ and TUJ1+/AADC+ cells in differentiated iNP cultures. Data are presented as mean ± SEM, n = 16 – 33 fields of view from one cell line (number of cells counted per condition ≥ 3066). An ordinary one way ANOVA was performed for each stain, with Tukey’s multiple comparison test. Comparisons are as follows: to control iNPs (*), to med SHH/CHIR-patterned iNPs (#) or to high SHH/CHIR-patterned iNPs ($).
Discussion
This study aimed to generate vmDA neurons from adult human cells using lineage factor-mediated direct-to-iNP reprogramming. The expression of a range of vmDA markers was examined in SOX2/PAX6-iNPs, and it was found that iNPs expressed PITX3 and NURR1. Additionally, iNPs could be differentiated into neuronal-like cells that expressed a range of dopaminergic markers, including TH, AADC, DAT, VMAT2 and GIRK2. This is the first report to describe the potential for adult human somatic cells to be converted into dopamine neurons via non-viral lineage factor-mediated direct-to-iNSC/iNP reprogramming. Previous studies have reported the generation of TH+ cells from adult human cells without further characterisation of a vmDA identity[21, 22].
As authentic A9 dopamine neurons are required for downstream applications, further optimisation of the generation of vmDA neurons was examined in healthy cells (Figure 9). While expression of many late vmDA markers was observed (NURR1, PITX3, TH, AADC, DAT, VMAT2 and GIRK2), the acquisition of a correct vmDA identity would be expected to be accompanied by the presence of early, sustained regional markers such as FOXA2, LMX1A/B, EN1 and CORIN, as well as enhanced dopaminergic differentiation. Exposure to patterning molecules is important for differentiating neural stem/precursor cells to adopt desired regional fates. SHH/FGF8 exposure to established SOX2/PAX6-iNPs did not enhance dopaminergic differentiation, and exposure to patterning molecules during reprogramming was insufficient to induce a true vmDA iNP fate. As PAX6 is detrimental to the generation of an authentic vmDA identity, PAX6 was removed, and iNPs comparable to SOX2/PAX6-iNPs were generated from SOX2-transfected cells, consistent with previous studies[32, 33]. Nonetheless, exposure of patterning molecules to SOX2-iNPs still did not induce a true vmDA fate. Over expression of genes involved in the specification of a vmDA fate has been widely used in stem cell and fate conversion studies. However, the addition of exogenous LMX1A/FOXA2 with or without patterning molecules did not induce a vmDA fate in iNPs. Exposure to FGF8b for the full duration of reprogramming increased the yield of TUJ1+ cells following differentiation, however, this effect was not specific for dopaminergic neurons. Overall, while these strategies did not induce an authentic vmDA fate in iNPs derived from adult human cells, these results are consistent with other lineage factor-mediated direct reprogramming studies[18, 38, 40, 41].
Figure 9:
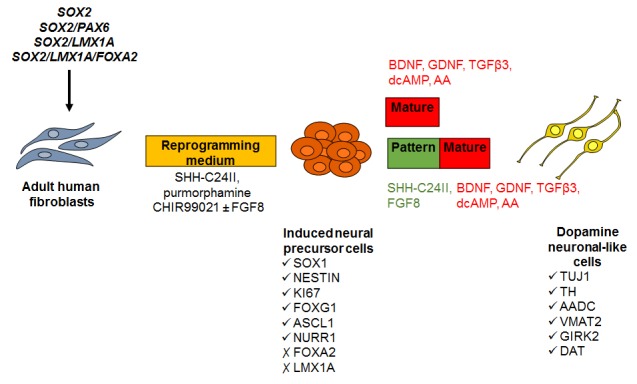
Schematic overview of the experimental processes trialled to optimise the generation of vmDA neurons from human iNPs. Transfection of adult human fibroblasts with SOX2 and PAX6 followed by culture in reprogramming medium generates iNP cells that express SOX1, NESTIN, FOXG1, ASCL1 and NURR1, and can be differentiated into dopamine-like neurons positive for TH, AADC, VMAT2, GIRK2 and DAT. In order to enhance the yield and induce an authentic vmDA identity of these cells, a range of neural and dopaminergic lineage transgenes were trialled coupled with exposure to the patterning factors SHH-C24II, purmorphamine, CHIR99021 and/or FGF8 during reprogramming. The effect of exposure to the patterning factors SHH-C24II and FGF8 during the early stage of differentiation to promote a vmDA fate was also investigated.
AA = ascorbic acid.
Using pluripotent factor-mediated reprogramming, rodent vmDA iNPs have been generated by adding CHIR99021, SHH, FGF8 and JI1 during reprogramming[42]. Another pluripotent factor-mediated reprogramming study derived vmDA iNSCs from adult human fibroblasts by exposure to a low concentration of SHH and FGF8[17], however the requirement for SHH/FGF8 was not examined. The success of this strategy probably lies in the fact that the iNSCs were at a very primitive stage of development. LIF-dependent neural stem cells have been reported to express mesencephalic genes[43], however, iNSCs showed pluripotent characteristics and could be converted into iPSCs[17]. The reliance on pluripotency factors for reprogramming raises the question as to whether these cells are directly reprogrammed without passing through a pluripotent stage [11, 12]. If iNSCs transition through a state of pluripotency, then they do not reflect true direct reprogramming, and may be more akin to pluripotent stem cells.
Amongst the lineage factor-mediated direct-to-iNSC/iNP reprogramming studies, the precise requirements for patterning factors have not been thoroughly examined. Correlating with the findings in Study Three, a mouse iNP study reported that established iNPs did not respond to SHH/FGF8, however, addition of these molecules to the reprogramming media was not examined, and overexpression of FOXA2 with or without LMX1A was required to induce vmDA genes[40]. Another rodent iNP study found that when lineage factor-reprogrammed iNPs were treated with FGF8 during reprogramming, very low numbers of TH+/TUJ1+ cells were generated, and NURR1/FOXA2 overexpression was required to improve TH yields and induce vmDA markers[38, 41]. Due to the differences in rodent and human development, methods for fate specification in rodent cells may not be directly applicable to human cells.
Only one other study has attempted to generate vmDA iNPs by lineage factor-mediated reprogramming in human cells[18]. This group took a similar approach to the present study, adding CHIR99021 and purmorphamine to the reprogramming medium during non-viral SOX2-mediated reprogramming of human post-natal fibroblasts[18, 33]. The requirement of these small molecules was not reported, only the effect of a cocktail of small molecules[33] or the effect of the addition of LMX1A[18]. Furthermore, the authenticity of the vmDA fate of iNPs generated by this method is unclear; while iNPs expressed SOX2, LMX1A and FOXA2, PAX6 was also expressed, suggesting that iNPs do not represent authentic vmDA progenitors[18]. Additionally, the long-term maintenance of a vmDA iNP fate in the absence of exogenous LMX1A was not shown, and the yields of TH+ neurons were not reported[18].
Currently, the derivation of authentic vmDA iNPs from adult human cells via lineage factor-mediated direct reprogramming remains elusive. In the present study, the addition of LMX1A/FOXA2 to the reprogramming cocktail, with or without exposure to patterning factors, did not induce a vmDA progenitor fate. The transient nature of plasmid transfection may mean that transgenes are not expressed for sufficient time to initiate endogenous vmDA-related gene expression programs, or that any changes induced by LMX1A/FOXA2 are not sustained in the long-term. While the use of transiently expressed, non-integrating vectors presents advantages for clinical translation, this strategy may be insufficient for generating vmDA progenitors from adult human fibroblasts, which are more difficult to reprogram than younger cells[23]. Future work could investigate the use of alternative gene expression systems, such as lentiviruses, stabilised RNA or Sendai viruses. Sustained, widespread transgene expression combined with patterning molecules may be effective at inducing a vmDA progenitor fate in directly reprogrammed adult human iNPs. Recent chemical reprogramming studies suggest that reprogramming efficiency could be improved by targeting certain pathways, such as the SHH, FGF2 pathways; inhibition of cellular senescence, autophagy, Rho/ROCK, TGFβ and GSK3β signalling pathways; the use of additional epigenetic modifiers; and hypoxia and/or antioxidant use[44-48].
The present study drew from patterning strategies employed in pluripotent stem cells to reprogram to a vmDA iNP fate. However, direct reprogramming, especially in adult cells, may require more powerful cues to induce desired fates. vmDA progenitors arise early in development, prior to the formation of the neuroectoderm. Future studies should, therefore, specifically aim to generate midbrain floor plate progenitors, rather than attempt to adapt methods for generating neuroepithelial-like precursors. This might be achieved through the use of potent SHH signalling and/or sustained GLI1 or FOXA2 overexpression[49], along with WNT1 signalling and/or LMX1A overexpression. However, signalling through these pathways was examined in the present study and found to be insufficient. Additional chemical components may be required to not only improve reprogramming efficiency, but also to induce the correct regional fate. Default anterior neuroectoderm specification might be targeted by blocking DKK1 expression[31]. Signalling through the p38 MAPK and JAK2/STAT3 pathways has recently been implicated in floor plate specification from human pluripotent stem cells[50].
Conclusion
Direct-to-iNSC/iNP reprogramming is a relatively new technology that offers a number of potential advantages over iPSCs and iN cells for the study and treatment of PD. This study has demonstrated for the first time that healthy adult human fibroblasts can be reprogrammed by non-viral lineage factor-mediated reprogramming into iNPs, capable of giving rise to dopamine neuronal-like cells. However, the expression of early vmDA regional markers was not seen in iNPs. Exposure to patterning molecules and/or vmDA transgenes was insufficient to induce an authentic vmDA fate in iNPs. Nevertheless, this study indicates that direct-to-iNSC/iNP reprogramming by lineage factors may be suitable for modelling PD in vitro using aged donor-derived cells. Future studies may improve the vmDA fate of derived cells by optimising gene expression systems and investigating the effect of chemical agents on reprogramming.
Acknowledgments
Neurological Foundation of New Zealand
Glossary
Abbreviations
- PD
Parkinson's disease
- vmDA
Ventral midbrain dopamine neuron
- iNP
Induced neural precursor cell
- TH
Tyrosine hydroxylase
- FGF8b
Fibroblast growth factor 8b
- iPSC
Induced pluripotent stem cell
- iN
Induced neuron
- iNSC
Induced neural stem cell
- NSE
Neuron specific enolase
- EGF
Epidermal growth factor
- SHH
Sonic hedgehog
- BDNF
Brain-derived neurotrophic factor
- GDNF
Glial-derived neurotrophic factor
- TGFP3
Transforming growth factor P3
- AADC
Aromatic L-amino acid decarboxylase
- VMAT2
Vesicular monoamine transporter 2
- DAT
Dopamine transporter
- TUJ1
Beta-3 tubulin
- PUR
Purmorphamine
- CHIR
CHIR99021
Potential Conflicts of Interests
None
References
- 1.Playne R, Connor B. Understanding Parkinson's Disease through the Use of Cell Reprogramming. Stem Cell Rev. 2017;13(2):151–69. doi: 10.1007/s12015-017-9717-5. [DOI] [PubMed] [Google Scholar]
- 2.Barker RA, Drouin-Ouellet J, Parmar M. Cell-based therapies for Parkinson disease—past insights and future potential. Nat Rev Neurol. 2015;11(9):492–503. doi: 10.1038/nrneurol.2015.123. [DOI] [PubMed] [Google Scholar]
- 3.Kriks S, Shim JW, Piao J, Ganat YM, Wakeman DR, Xie Z, Carrillo-Reid L, Auyeung G, Antonacci C, Buch A, Yang L, Beal MF, Surmeier DJ, Kordower JH, Tabar V, Studer L. Dopamine neurons derived from human ES cells efficiently engraft in animal models of Parkinson's disease. Nature. 2011;480(7378):547–51. doi: 10.1038/nature10648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kirkeby A, Grealish S, Wolf DA, Nelander J, Wood J, Lundblad M, Lindvall O, Parmar M. Generation of regionally specified neural progenitors and functional neurons from human embryonic stem cells under defined conditions. Cell Rep. 2012;1(6):703–14. doi: 10.1016/j.celrep.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 5.Mertens J, Marchetto MC, Bardy C, Gage FH. Evaluating cell reprogramming, differentiation and conversion technologies in neuroscience. Nat Rev Neurosci. 2016;17(7):424–37. doi: 10.1038/nrn.2016.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miller JD, Ganat YM, Kishinevsky S, Bowman RL, Liu B, Tu EY, Mandal PK, Vera E, Shim JW, Kriks S, Taldone T, Fusaki N, Tomishima MJ, Krainc D, Milner TA, Rossi DJ, Studer L. Human iPSC-based modeling of late-onset disease via progerin-induced aging. Cell Stem Cell. 2013;13(6):691–705. doi: 10.1016/j.stem.2013.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pang ZP, Yang N, Vierbuchen T, Ostermeier A, Fuentes DR, Yang TQ, Citri A, Sebastiano V, Marro S, Südhof TC, Wernig M. Induction of human neuronal cells by defined transcription factors. Nature. 2011;476(7359):220–3. doi: 10.1038/nature10202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu ML, Zang T, Zou Y, Chang JC, Gibson JR, Huber KM, Zhang CL. Small molecules enable neurogenin 2 to efficiently convert human fibroblasts into cholinergic neurons. Nat Commun. 2013;4:2183. doi: 10.1038/ncomms3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo Z, Zhang L, Wu Z, Chen Y, Wang F, Chen G. In vivo direct reprogramming of reactive glial cells into functional neurons after brain injury and in an Alzheimer's disease model. Cell Stem Cell. 2014;14(2):188–202. doi: 10.1016/j.stem.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Torper O, Ottosson DR, Pereira M, Lau S, Cardoso T, Grealish S, Parmar M. In Vivo Reprogramming of Striatal NG2 Glia into Functional Neurons that Integrate into Local Host Circuitry. Cell Rep. 2015;12(3):474–81. doi: 10.1016/j.celrep.2015.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maza, Caspi I, Zviran A, Chomsky E, Rais Y, Viukov S, Geula S, Buenrostro JD, Weinberger L, Krupalnik V, Hanna S, Zerbib M, Dutton JR, Greenleaf WJ, Massarwa R, Novershtern N, Hanna JH. Transient acquisition of pluripotency during somatic cell transdifferentiation with iPSC reprogramming factors. Nat Biotechnol. 2015;33(7):769–74. doi: 10.1038/nbt.3270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bar-Nur O, Verheul C, Sommer AG, Brumbaugh J, Schwarz BA, Lipchina, Huebner AJ, Mostoslavsky G, Hochedlinger K. Lineage conversion induced by pluripotency factors involves transient passage through an iPSC stage. Nat Biotechnol. 2015;33(7):761–8. doi: 10.1038/nbt.3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weissbein U, Ben-David U, Benvenisty N. Virtual karyotyping reveals greater chromosomal stability in neural cells derived by transdifferentiation than those from stem cells. Cell Stem Cell. 2014;15(6):687–91. doi: 10.1016/j.stem.2014.10.018. [DOI] [PubMed] [Google Scholar]
- 14.Wang L, Wang L, Huang W, Su H, Xue Y, Su Z, Liao B, Wang H, Bao X, Qin D, He J, Wu W, So KF, Pan G, Pei D. Generation of integration-free neural progenitor cells from cells in human urine. Nat Methods. 2013;10(1):84–9. doi: 10.1038/nmeth.2283. [DOI] [PubMed] [Google Scholar]
- 15.Lu J, Liu H, Huang CT, Chen H, Du Z, Liu Y, Sherafat MA, Zhang SC. Generation of integration-free and region-specific neural progenitors from primate fibroblasts. Cell Rep. 2013;3(5):1580–91. doi: 10.1016/j.celrep.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu S, Ambasudhan R, Sun W, Kim HJ, Talantova M, Wang X, Zhang M, Zhang Y, Laurent T, Parker J, Kim HS, Zaremba JD, Saleem S, Sanz-Blasco S, Masliah E, McKercher SR, Cho YS, Lipton SA, Kim J, Ding S. Small molecules enable OCT4-mediated direct reprogramming into expandable human neural stem cells. Cell Res. 2014;24(1):126–9. doi: 10.1038/cr.2013.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miura T, Sugawara T, Fukuda A, Tamoto R, Kawasaki T, Umezawa A, Akutsu H. Generation of primitive neural stem cells from human fibroblasts using a defined set of factors. Biol Open. 2015;4(11):1595–607. doi: 10.1242/bio.013151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mirakhori F, Zeynali B, Rassouli H, Salekdeh GH, Baharvand H. Direct conversion of human fibroblasts into dopaminergic neural progenitor-like cells using TAT-mediated protein transduction of recombinant factors. Biochem Biophys Res Commun. 2015;459(4):655–61. doi: 10.1016/j.bbrc.2015.02.166. [DOI] [PubMed] [Google Scholar]
- 19.Zou Q, Yan Q, Zhong J, Wang K, Sun H, Yi X, Lai L. Direct conversion of human fibroblasts into neuronal restricted progenitors. J Biol Chem. 2014;289(8):5250–60. doi: 10.1074/jbc.M113.516112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shahbazi E, Moradi S, Nemati S, Satarian L, Basiri M, Gourabi H, Zare Mehrjardi N, Günther P, Lampert A, Händler K, Hatay FF, Schmidt D, Molcanyi M, Hescheler J, Schultze JL, Saric T, Baharvand H. Conversion of Human Fibroblasts to Stably Self-Renewing Neural Stem Cells with a Single Zinc-Finger Transcription Factor. Stem Cell Reports. 2016;6(4):539–51. doi: 10.1016/j.stemcr.2016.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maucksch C, Firmin E, Butler-Munro C, Montgomery J, Dottori M, Connor B. Non-Viral Generation of Neural Precursor-like Cells from Adult Human Fibroblasts. J Stem Cells Regen Med. 2012;8(3):162–70. doi: 10.46582/jsrm.0803009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu KR, Shin JH, Kim JJ, Koog MG, Lee JY, Choi SW, Kim HS, Seo Y, Lee S, Shin TH, Jee MK, Kim DW, Jung SJ, Shin S, Han DW, Kang KS. Rapid and Efficient Direct Conversion of Human Adult Somatic Cells into Neural Stem Cells by HMGA2/let-7b. Cell Rep. 2015. p. pii: S2211-1247(14)01067-5. [DOI] [PubMed]
- 23.Trokovic R, Weltner J, Noisa P, Raivio T, Otonkoski T. Combined negative effect of donor age and time in culture on the reprogramming efficiency into induced pluripotent stem cells. Stem Cell Res. 2015;15(1):254–62. doi: 10.1016/j.scr.2015.06.001. [DOI] [PubMed] [Google Scholar]
- 24.Connor B, Firmin E, Maucksch C, Liu R, Playne R, Jones K, Dottori M. Direct Conversion of Adult Human Fibroblasts into Induced Neural Precursor Cells by Non-Viral Transfection. Protoc Exch. 2015. [DOI]
- 25.Pfisterer U, Kirkeby A, Torper O, Wood J, Nelander J, Dufour A, Bjorklund A, Lindvall O, Jakobsson J, Parmar M. Direct conversion of human fibroblasts to dopaminergic neurons. Proc Natl Acad Sci U S A. 2011;108(25):10343–8. doi: 10.1073/pnas.1105135108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-AACT method. Methods. 2001;25(4):p. 402–08. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 27.Xi J, Liu Y, Liu H, Chen H, Emborg ME, Zhang SC. Specification of midbrain dopamine neurons from primate pluripotent stem cells. Stem Cells. 2012;30(8):1655–63. doi: 10.1002/stem.1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Denham M, Bye C, Leung J, Conley BJ, Thompson LH, Dottori M. Glycogen synthase kinase 3p and activin/nodal inhibition in human embryonic stem cells induces a pre-neuroepithelial state that is required for specification to a floor plate cell lineage. Stem Cells. 2012;30(11):2400–11. doi: 10.1002/stem.1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vitalis T, Cases O, Engelkamp D, Verney C, Price DJ. Defect of tyrosine hydroxylase-immunoreactive neurons in the brains of mice lacking the transcription factor Pax6. J Neurosci. 2000;20(17):6501–16. doi: 10.1523/JNEUROSCI.20-17-06501.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kirkeby A, Parmar M. Building authentic midbrain dopaminergic neurons from stem cells - lessons from development. Transl Neurosci. 2012;3(4):p. 314–19. [Google Scholar]
- 31.Fasano CA, Chambers SM, Lee G, Tomishima MJ, Studer L. Efficient derivation of functional floor plate tissue from human embryonic stem cells. Cell Stem Cell. 2010;6(4):336–47. doi: 10.1016/j.stem.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 32.Ring KL, Tong LM, Balestra ME, Javier R, Andrews-Zwilling Y, Li G, Walker D, Zhang WR, Kreitzer AC, Huang Y. Direct reprogramming of mouse and human fibroblasts into multipotent neural stem cells with a single factor. Cell Stem Cell. 2012;11(1):100–9. doi: 10.1016/j.stem.2012.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mirakhori F, Zeynali B, Rassouli H, Shahbazi E, Hashemizadeh S, Kiani S, Salekdeh GH, Baharvand H. Induction of Neural Progenitor-Like Cells from Human Fibroblasts via a Genetic Material-Free Approach. PLoS One. 2015;10(8):e0135479. doi: 10.1371/journal.pone.0135479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moya N, Cutts J, Gaasterland T, Willert K, Brafman DA. Endogenous WNT signaling regulates hPSC-derived neural progenitor cell heterogeneity and specifies their regional identity. Stem Cell Reports. 2014;3(6):1015–28. doi: 10.1016/j.stemcr.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Andersson E, Tryggvason U, Deng Q, Friling S, Alekseenko Z, Robert B, Perlmann T, Ericson J. Identification of intrinsic determinants of midbrain dopamine neurons. Cell. 2006;124(2):393–405. doi: 10.1016/j.cell.2005.10.037. [DOI] [PubMed] [Google Scholar]
- 36.Panman L, Andersson E, Alekseenko Z, Hedlund E, Kee N, Mong J, Uhde CW, Deng Q, Sandberg R, Stanton LW, Ericson J, Perlmann T. Transcription factor-induced lineage selection of stem-cell-derived neural progenitor cells. Cell Stem Cell. 2011;8(6):663–75. doi: 10.1016/j.stem.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 37.Cooper O, Hargus G, Deleidi M, Blak A, Osborn T, Marlow E, Lee K, Levy A, Perez-Torres E, Yow A, Isacson O. Differentiation of human ES and Parkinson's disease iPS cells into ventral midbrain dopaminergic neurons requires a high activity form of SHH, FGF8a and specific regionalization by retinoic acid. Mol Cell Neurosci. 2010;45(3):258–66. doi: 10.1016/j.mcn.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lim MS, Lee SY, Park CH. FGF8 is Essential for Functionality of Induced Neural Precursor Cell-derived Dopaminergic Neurons. Int J Stem Cells. 2015;8(2):228–34. doi: 10.15283/ijsc.2015.8.2.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kirkeby A, Nolbrant S, Tiklova K, Heuer A, Kee N, Cardoso T, Ottosson DR, Lelos MJ, Rifes P, Dunnett SB, Grealish S, Perlmann T, Parmar M. Predictive Markers Guide Differentiation to Improve Graft Outcome in Clinical Translation of hESC-Based Therapy for Parkinson's Disease. Cell Stem Cell. 2017;20(1):135–48. doi: 10.1016/j.stem.2016.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tian C, Li Y, Huang Y, Wang Y, Chen D, Liu J, Deng X, Sun L, Anderson K, Qi X, Li Y, Mosley RL, Chen X, Huang J, Zheng JC. Selective Generation of Dopaminergic Precursors from Mouse Fibroblasts by Direct Lineage Conversion. Sci Rep. 2015;5:12622. doi: 10.1038/srep12622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lim MS, Chang MY, Kim SM, Yi SH, Suh-Kim H, Jung SJ, Kim MJ, Kim JH, Lee YS, Lee SY, Kim DW, Lee SH, Park CH. Generation of Dopamine Neurons from Rodent Fibroblasts through the Expandable Neural Precursor Cell Stage. J Biol Chem. 2015;290(28):17401–14. doi: 10.1074/jbc.M114.629808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim HS, Kim J, Jo Y, Jeon D, Cho YS. Direct lineage reprogramming of mouse fibroblasts to functional midbrain dopaminergic neuronal progenitors. Stem Cell Res. 2014;12(1):60–8. doi: 10.1016/j.scr.2013.09.007. [DOI] [PubMed] [Google Scholar]
- 43.Li W, Sun W, Zhang Y, Wei W, Ambasudhan R, Xia P, Talantova M, Lin T, Kim J, Wang X, Kim WR, Lipton SA, Zhang K, Ding S. Rapid induction and long-term self-renewal of primitive neural precursors from human embryonic stem cells by small molecule inhibitors. Proc Natl Acad Sci U S A. 2011;108(20):8299–304. doi: 10.1073/pnas.1014041108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li Y, Zhang Q, Yin X, Yang W, Du Y, Hou P, Ge J, Liu C, Zhang W, Zhang X, Wu Y, Li H, Liu K, Wu C, Song Z, Zhao Y, Shi Y, Deng H. Generation of iPSCs from mouse fibroblasts with a single gene, Oct4, and small molecules. Cell Res. 2011;21(1):196–204. doi: 10.1038/cr.2010.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cheng L, Hu W, Qiu B, Zhao J, Yu Y, Guan W, Wang M, Yang W, Pei G. Generation of neural progenitor cells by chemical cocktails and hypoxia. Cell Res. 2014;24(6):665–79. doi: 10.1038/cr.2014.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hu W, Qiu B, Guan W, Wang Q, Wang M, Li W, Gao L, Shen L, Huang Y, Xie G, Zhao H, Jin Y, Tang B, Yu Y, Zhao J, Pei G. Direct Conversion of Normal and Alzheimer's Disease Human Fibroblasts into Neuronal Cells by Small Molecules. Cell Stem Cell. 2015;17(2):204–12. doi: 10.1016/j.stem.2015.07.006. [DOI] [PubMed] [Google Scholar]
- 47.Zhang M, Lin YH, Sun YJ, Zhu S, Zheng J, Liu K, Cao N, Li K, Huang Y, Ding S. Pharmacological Reprogramming of Fibroblasts into Neural Stem Cells by Signaling-Directed Transcriptional Activation. Cell Stem Cell. 2016;18(5):653–67. doi: 10.1016/j.stem.2016.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zheng J, Choi KA, Kang PJ, Hyeon S, Kwon S, Moon JH, Hwang, Kim YI, Kim YS, Yoon BS, Park G, Lee J, Hong S, You S. A combination of small molecules directly reprograms mouse fibroblasts into neural stem cells. Biochem Biophys Res Commun. 2016;476(1):42–8. doi: 10.1016/j.bbrc.2016.05.080. [DOI] [PubMed] [Google Scholar]
- 49.Denham M, Thompson LH, Leung J, Pébay A, Björklund A, Dottori M. Gli 1 is an inducing factor in generating floor plate progenitor cells from human embryonic stem cells. Stem Cells. 2010;28(10):1805–15. doi: 10.1002/stem.510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chi L, Fan B, Zhang K, Du Y, Liu Z, Fang Y, Chen Z, Ren X, Xu X, Jiang C, Li S, Ma L, Gao L, Liu L, Zhang X. Targeted Differentiation of Regional Ventral Neuroprogenitors and Related Neuronal Subtypes from Human Pluripotent Stem Cells. Stem Cell Reports. 2016;7(5):941–54. doi: 10.1016/j.stemcr.2016.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]


