Abstract
Intercropping plays a vital role in greenhouse production, and affects soil physicochemical properties and soil microbial communities structure, but influences of intercropping on the relationship of microorganisms are reported in continuous cropping soil rarely. Here, we investigated the effects of seven intercropping systems [alfalfa (Medicago sativa L.)/cucumber, trifolium (Trifolium repens L.)/cucumber, wheat (Triticum aestivum L.)/cucumber, rye (Secale cereale L.)/cucumber, chrysanthemum (Chrysanthemum coronrium L.)/cucumber, rape (Brassica campestris L.)/cucumber, mustard (Brassica juncea L.)/cucumber] on soil bacterial and fungal communities compared to the cucumber continuous cropping system in the greenhouse. The results showed that intercropping increased microbial OTU richness and fungal communities diversity, soil bacterial communities diversity was abundant in the trifolium-cucumber and mustard-cucumber systems. Nevertheless, there was no significant differences of microbial communities structure between intercropping and monoculture systems. Redundancy analysis indicated that soil microbial communities composition was indirectly influenced by soil properties. In addition, network analysis demonstrated that simple inter-relationships of fungal taxa were observed in the intercropping soil, and trifolium, wheat, and mustard intercropping systems had a complex connection between bacterial taxa. Taken together, trifolium and mustard as the intercrops significantly increased cucumber continuous cropping soil bacterial and fungal communities diversity. Moreover, intercropping strongly changed the relationships of microbial taxa, though did not shape notably soil microbial communities structure.
Keywords: soil microbe, intercropping, bacteria, fungi, co-occurrence networks
Introduction
Agricultural management practices and cropping systems can evidently influence crop yield, soil physicochemical characteristics, and soil microbial activity and composition, and they have attracted attention in agricultural production so far (Acosta-Martínez et al., 2010; Li et al., 2010; Singh et al., 2016). Intercropping, the coinstantaneous cultivation of more than one crop species in the same location, is attributed to the efficiency and complementation of resources in temporal and spatial patterns, and to the enhancement of the resistance to diseases, pests, and hostile plants (Li et al., 1999; Banik et al., 2006; Hinsinger et al., 2011; Brooker et al., 2015). However, not all of the intercropping systems can tend to the better. Evidence illustrated that legume and cereal species intercropped had lower biomass and nitrate accumulation than sole crop (Li et al., 2001; Corre-Hellou et al., 2011). Understanding the roles of several intercropping systems can be conducive to explore new strategies to improve agricultural development in a sustainable way.
Soil microorganisms play an important role in soil biogeochemical processes such as nitrogen, phosphorus and other elements cycles (de Graaff et al., 2010; Mangan et al., 2010; Urbanová et al., 2015). Soil microbial communities composition and diversity are imperative to maintain the plant biodiversity, soil health and productivity (van der Heijden et al., 1998; Mangan et al., 2010). Furthermore, several reports showed that changes in soil microbial diversity and structure are confirmed to be connected with plant species, soil physicochemical characteristics as well as land-use types (Lauber et al., 2008; Mitchell et al., 2010; Bell et al., 2013). However, little is known about the influence of intercropping on continuous cropping soil microbial communities.
Soil microbes were not isolated in the microbial community, but existed in the complicated interaction systems that decided microbial community structure (Freilich et al., 2010). In previous study, most analytical methods of soil microbial communities were applied to illustrate microbial diversity, community composition and their variations with biotic or abiotic factors, but were rarely able to explain the relationships between soil microbial species (Deng et al., 2012). Co-occurrence network analysis gives new insight into the inter-species interactions of soil microbial communities, and promotes the understanding of the niche spaces among community members (Barberán et al., 2012). Nevertheless, the microbial interactions of intercropping systems were poorly understood in continuous cropping soil.
In the present study, we performed a metagenomic analysis of bacterial and fungal communities based on Miseq sequencing of 16S rRNA and ITS genes to illustrate their variation in the seven intercropping systems. The purpose of this study was to explore the responses of microbial communities to intercropping systems on the condition of continuous cropping soils. We hypothesized that intercropping influenced soil bacterial and fungal communities by variation of soil properties, and co-occurrence patterns of bacterial and fungal taxa distinctly responded to several intercropping systems in the continuous cropping soils.
Materials and methods
Field site description and experimental design
The experiment was located at Horticultural Experimental Station Northeast Agricultural University (45°41′N, 126°37′E), Harbin, Heilongjiang Province, China. The field soil was black soil (Mollisoil), that was under continuous cucumber cropping for 3 years. The soil chemical properties, measured by Bao (2005), were as follows: pH 6.64; EC, 0.88 mS cm−1; -N 13.32 mg kg−1; -N, 253.04 mg kg−1; available phosphorus (AP), 277.62 mg kg−1; available potassium (AK), 359.03 mg kg−1.
The experiment was a randomized block design with three replicates in the greenhouse, and the size of each plot was 6 × 0.5 m. 7 crops [alfalfa (A), trifolium (T), wheat (W), rye (Ry), Chrysanthemum (C), rape (Ra), and mustard (M)] were intercropped with cucumber as experimental group, and cucumber monoculture (CM) was the control. The spring experiment were ranged from 21 April 2015, cucumber seedlings 3-true-leaves, to 30 June 2015, cucumber harvest period. 12 cucumber plants were in one row as the test district, and two protective lines were on both sides. After 10 days of planting the cucumber, the seeds of the intercrops were sowed on the outside of cucumber about 10 cm. The number of intercrop seeds was as follows: (I) wheat and rye were 30 seedlings, respectively; (II) trifolium and alfalfa were 40 seedlings, respectively; (III) rape and mustard were 3 seedlings, respectively; (IV) chrysanthemum was 5 seedlings. When the intercrops grew to 20 cm high, it was pruned for 10 cm to avoid effects on growth of cucumber and their residue were left on soil surface. The compound NPK fertilizer (16:16:8) was performed with 300 kg ha−1 for each plot. The fall trial began on 29 July and ended on 6 October, and the specific operation was consistent with that in spring. A more detailed description of the experiment can be referred to (Chang et al., 2017).
Soil sampling and soil physicochemical properties analysis
The soil samples, totally 48, were obtained from 24 test district on 30 June and 6 October, and then were mixed and sieved through a 2 mm mesh to thoroughly homogenize and remove the roots, plant residues and stones. Afterwards the soils were transferred to the laboratory, every sample was divided into two parts: some archived at −80°C for DNA extraction, and the others for chemical analyze stored at −4°C.
Soil physicochemical characteristics were analyzed based on Bao (2005). Soil pH and EC were determined in a soil water suspension (1:2.5 w/v) using a glass electrode and a conductivity meter, respectively. For soil inorganic nitrogen concentration (-N, -N), available P and K were extracted with 2 M KCl, 0.5 M NH4+OAc (pH = 7) and 1 M NaHCO3 (pH = 8.5), respectively, and then soil filtrates were determined by Continuous Flow Analyser (SAN++, Skalar, Breda, Netherlands). Soil moisture contents were determined by drying at 105°C for 24 h.
DNA extraction, PCR and Miseq sequencing
Soil total DNA was extracted from 0.5 g soil using the MoBioPowerSoil™ DNA Isolation Kit (Mo Bio Laboratories Inc. Carlsbad, CA, USA) according to the manufacturer's instructions. DNA concentration and purity were determined with a NanoDrop 2000 Spectrophotometer (Thermo Scientific, USA).
The V3-V4 hypervariable region of the 16S rRNA gene and the ITS1 region of fungal ITS gene were used as the bacterial-specific fragment and the fungal-specific fragment with the 338F/806R (Xu et al., 2016) and ITS1F/ITS2 (Bellemain et al., 2010) primer sets for bacteria and fungi, respectively. These primer pairs were modified with a 6-bp unique barcode sequence at the 5′ end to identify samples. All amplification was performed in 25 μl reactions contained 0.5 μl of each primer, 1 μl of template, 2 μl 2.5 mM dNTPs, 0.5 μl of FastPfu Polymerase (Transgen Biotech, Beijing, China), 0.5 μl of × 5 FastPfu buffer, and 20 μl deionized H2O. The PCR conditions, performed in an ABI GeneAmp® 9700 PCR System (ABI, MA, USA), were described as follows: 5 min of initial denaturation step at 94°C, followed by 35 cycles of 94°C for 30 s, 50°C for 30 s and 72°C for 30 s, and a final extension step at 72°C for 10 min for the 16S V3-V4 rRNA gene; and 3 min of initial denaturation step at 95°C, followed by 35 cycles of 94°C at 30 s, 55°C for 30 s and 72°C for 45 s, and a final extension at 72°C for 10 min for ITS genes. The products from the three replicate amplifications of the bacterial 16S rRNA gene and fungal ITS gene were separately pooled and evaluated on 2% agarose gels (TBE buffer). Amplicons were purified with a DNA gel extraction kit (Axygen, China), quantified with a QuantiFluorTM-ST fluorometer (Promega, Madison, WI, USA), pooled at equimolar concentrations, and finally sequenced on an Illumina Miseq PE300 platform at Majorbio Bio-Pharm Technology Co., Ltd. (Shanghai, China).
Soil bacterial and fungal abundance analysis
Quantification of 16S rRNA and ITS genes were performed on a iQ5 Real-Time PCR Detection System (Bio-Rad, USA) with primers 338F/518R (Muyzer et al., 1993) and ITS1F/ITS4 (Gardes and Bruns, 1993), respectively. The PCR reaction mix contained 10 μl SYBR Green I PCR master mix (Applied Biosystems, USA), 0.2 μl each primer (10 μ mol l−1), 2.5 μl template DNA (sample DNA or plasmid DNA for standard curves), and finally fit it up with sterile deionized water to 20 μl. The qPCR conditions contained an initial denaturation at 95°C for 5 min, followed by 37 cycles of denaturation at 95°C for 45 s, annealing at 56°C or 58°C for 45 s for bacteria and fungi, respectively. The standard samples were diluted to yield a series of 10-fold concentrations and subsequently used for qPCR standard curves, The R2-value for each standard curve exceeded 0.99, indicating linear relationships over the concentration ranges used in this study. All of the amplifications were run in triplicate with the DNA extracted from each soil sample.
Sequence data analysis
The raw data yielded from Illumina sequencing were analyzed using QIIME software (v1.9.0) and the UPARSE pipeline as described before (Zhong et al., 2015). The UPARSE pipeline was performed for taxonomic assignment with similarities >97% (Edgar, 2013). Taxonomic classification was conducted with SILVA (version 119; http://www.arb-silva.de) and UNITE (version 7.0; http://unite.ut.ee/index.php) databases for bacteria and fungi, respectively. To preclude bias as a result of several sequencing depth, all samples were subsequently subsampled based on the minimum number of soil microbial sequencing depth of this study. The raw data have been deposited in the NCBI SRA database (SRP122874).
Statistical analysis
The bacterial and fungal diversity indices, Chao 1 richness, Shannon index, Simpson index and coverage were calculated by QIIME (Caporaso et al., 2010). Heatmap analysis was used to compare the top 50 classified genera in per sample of two growing seasons with the gplot package in R (R v.3.2.5) (R Development Core Team, 2006). Non-metric multidimensional scaling (NMDS) and Redundancy analysis (RDA) were carried out to reveal the microbial structure and the relationship between environmental factors and microbial abundance, and Anosim (analysis of similarity), adonis (non-parametric MANOVA), and MRPP (multi-response permutation procedure) were used to compare the microbial community differences of two cropping seasons (24 samples per season) with the Bray-Curtis distance and 999 permutations, they were performed in R using the vegan package. To demonstrate the relationship of different species among several samples, network analysis based on spearman's rank analysis in this study was performed using the 50 most abundant genera of bacterial and fungal communities. The co-occurrence patterns of soil microbial communities were explored based on strong (ρ > 0.6) and significant correlations (P < 0.01), and were visualized with the Gephi (Jacomy et al., 2009). The size of each node represented the number of connections, the node was colored by taxonomy. Soil properties, bacterial and fungal abundances, alpha diversity indices, and spearman's rank analysis were performed at 0.05 probability level in SPSS software (Version 17.0).
Results
Soil physicochemical properties
Soil physicochemical properties of all samples were summarized in Table 1. Compared with cucumber monoculture, soil AP content was significantly (P < 0.05) decreased under intercropping systems in the spring. rye-cucumber system had a significantly (P < 0.05) higher soil -N content and -N content in the spring, and alfalfa-cucumber and chrysanthemum-cucumber systems significantly (P < 0.05) increased soil -N content and -N content in the fall. Wheat-cucumber and rye-cucumber systems had a significantly (P < 0.05) higher EC in the two cropping seasons. No significant differences were found in soil moisture content, soil pH and AK content between intercropping and monoculture systems.
Table 1.
Effects of different crop modes on soil chemical properties at two sampling times in a black soil.
| Crop seasona | Treatmentsb | Moisture (%) | pH | EC (mS·cm−1) | -N (mg·kg−1) | -N (mg·kg−1) | APc (mg·kg−1) | AKc (mg·kg−1) |
|---|---|---|---|---|---|---|---|---|
| S | A | 24.94 ± 5.44a | 7.32 ± 0.05a | 0.14 ± 0.01cd | 27.66 ± 8.17b | 35.13 ± 9.37a | 467.14 ± 53.44b | 533.20 ± 110.66a |
| T | 23.44 ± 0.41a | 7.18 ± 0.05a | 0.12 ± 0.00d | 15.33 ± 6.96b | 63.61 ± 26.32a | 427.53 ± 51.22b | 561.11 ± 78.41a | |
| W | 22.47 ± 0.71a | 7.16 ± 0.09a | 0.31 ± 0.02a | 16.51 ± 2.76b | 62.78 ± 24.81a | 308.54 ± 83.92c | 566.91 ± 49.71a | |
| Ry | 22.54 ± 4.01a | 7.22 ± 0.15a | 0.25 ± 0.05b | 56.31 ± 28.18a | 54.58 ± 25.28a | 276.72 ± 66.06c | 460.27 ± 21.98a | |
| C | 19.51 ± 1.40a | 7.22 ± 0.17a | 0.15 ± 0.04cd | 30.37 ± 11.53b | 41.65 ± 14.32a | 237.93 ± 49.81c | 589.08 ± 49.52a | |
| Ra | 21.00 ± 4.73a | 7.17 ± 0.09a | 0.19 ± 0.00c | 20.39 ± 9.53b | 37.18 ± 15.57a | 430.65 ± 50.62b | 619.80 ± 160.82a | |
| M | 24.71 ± 2.66a | 7.20 ± 0.21a | 0.15 ± 0.00cd | 35.92 ± 6.91ab | 33.29 ± 14.66a | 505.27 ± 111.20b | 503.53 ± 54.84a | |
| CM | 23.41 ± 1.31a | 7.10 ± 0.06a | 0.13 ± 0.01cd | 26.77 ± 9.94b | 44.96 ± 21.57a | 621.85 ± 41.44a | 617.54 ± 36.36a | |
| F | A | 25.50 ± 2.86a | 6.81 ± 0.14a | 0.17 ± 0.01b | 13.15 ± 3.86a | 43.64 ± 2.99a | 388.36 ± 40.70a | 546.25 ± 37.31a |
| T | 26.68 ± 1.36a | 6.78 ± 0.05a | 0.22 ± 0.04b | 12.82 ± 2.92a | 36.51 ± 3.58ab | 386.53 ± 37.81a | 517.75 ± 26.07b | |
| W | 19.98 ± 8.27a | 6.91 ± 0.05a | 0.37 ± 0.07a | 22.25 ± 8.56a | 34.14 ± 3.03b | 390.63 ± 19.06a | 431.84 ± 38.31b | |
| Ry | 26.49 ± 2.73a | 6.85 ± 0.05a | 0.42 ± 0.05a | 17.63 ± 3.03a | 34.25 ± 7.82b | 427.94 ± 12.86a | 530.36 ± 70.57ab | |
| C | 25.82 ± 1.76a | 6.80 ± 0.06a | 0.38 ± 0.04a | 19.60 ± 7.38a | 43.91 ± 3.41a | 358.98 ± 26.13a | 525.23 ± 56.65ab | |
| Ra | 24.39 ± 0.88a | 6.84 ± 0.08a | 0.23 ± 0.02b | 19.36 ± 8.49a | 39.16 ± 8.27ab | 368.92 ± 59.10a | 506.17 ± 95.25ab | |
| M | 25.87 ± 2.33a | 6.86 ± 0.11a | 0.19 ± 0.01b | 21.33 ± 3.87a | 32.78 ± 1.70b | 393.76 ± 34.35a | 515.80 ± 40.82ab | |
| CM | 19.11 ± 9.03a | 6.75 ± 0.08a | 0.26 ± 0.06b | 18.81 ± 4.15a | 33.66 ± 1.72b | 425.29 ± 30.81a | 521.20 ± 22.51ab |
S and F indicated experiment conducted in spring and fall seasons, respectively.
A, Alfalfa; T, Trifolium; W, Wheat; Ry, Rye; C, Chrysanthemum; Ra, Rape; M, Mustard; CM, Cucumber monoculture.
AP, AK indicated soil available phosphorus and available potassium, respectively.
Data with different letters in each column indicate significantly different between treatments at 0.05 level.
Soil bacterial and fungal abundance
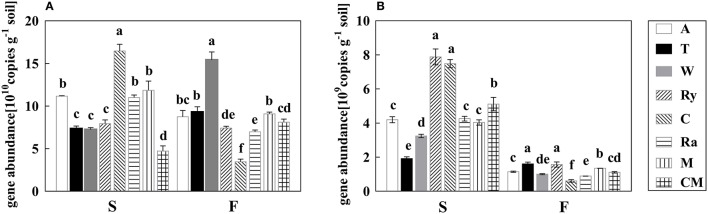
Soil bacterial abundance was significantly (P < 0.05) higher under all intercropping systems than cucumber monoculture in the spring, and wheat-cucumber, trifolium-cucumber, mustard-cucumber systems significantly (P < 0.05) increased bacterial abundance in the fall (Figure 1A). Besides, chrysanthemums-cucumber and rape-cucumber systems had significantly lower bacterial abundance in the fall (P < 0.05), and the highest bacterial abundance was existed in chrysanthemum-cucumber and wheat-cucumber systems in the two growing seasons, respectively.
Figure 1.
The abundances of bacterial 16S rRNA gene (A) and fungal ITS gene (B) under different treatments in the spring (S) and fall (F) cropping seasons. Data with different letters in each column indicate significantly different between treatments at 0.05 level.
Compared with cucumber monoculture, fungal abundance significantly (P < 0.05) decreased under intercropping systems except rye-cucumber and chrysanthemum-cucumber systems in the spring, and chrysanthemum-cucumber and rape-cucumber systems had a significantly (P < 0.05) lower fungal abundance in the fall (Figure 1B). Rye-cucumber and chrysanthemum-cucumber systems in the spring, and trifolium-cucumber, rye-cucumber, and mustard-cucumber systems in fall were significantly (P < 0.05) increased fungal abundance.
Soil microbial community diversity and composition
The process of raw sequencing reads generated about the total of 1788705 high quality V3-V4 sequences and 1798779 high quality ITS1 sequences, average read length of bacteria and fungi were 437 and 262 bp, and sequences of all samples were clustered to 134463 bacterial OTUs and 15541 fungal OTUs with a 97% identity threshold, respectively. The alpha-diversity indices of bacterial and fungal communities were observed for Table 2. Bacterial and fungal diversity represented by Shannon index and Simpson index. Bacterial Shannon index of trifolium-cucumber and mustard-cucumber systems were remarkably higher than monoculture in spring and fall (P < 0.05), However, for fungal Shannon index, every treatment was significantly higher than monoculture under two growing seasons except mustard-cucumber system in the spring. Simpson index of bacterial and fungal communities were opposite to Shannon index. Bacterial richness, estimated by Chao1 index, was higher under rape-cucumber, wheat-cucumber, and rye-cucumber systems than under monoculture in spring, and under intercropping systems except wheat-cucumber and chrysanthemum-cucumber systems in fall. For fungal richness, intercropping systems were significantly higher than cucumber monoculture in spring, but mustard-cucumber systems were significantly lower than monoculture in fall (P < 0.05). The coverage of soil bacterial and fungal communities was more than 95 and 99%, showing that the current sequencing depth in this study was enough to cover the soil bacterial and fungal communities diversity, respectively.
Table 2.
Diversity indices of soil microbial communities based on 16S rRNA and ITS genes analyzed from Illumina Miseq sequencing.
| Classified | Crop season | Treatments | Sequences | Number of OTUs | Shannon | Simpson | Chao 1 | Coverage (%) |
|---|---|---|---|---|---|---|---|---|
| Bacteria | S | A | 39, 665 ± 766 | 2, 857 ± 32cd | 6.69 ± 0.02de | 0.0053 ± 0.0003a | 3871.88 ± 10cd | 95.58 |
| T | 36, 371 ± 173 | 2, 977 ± 63ab | 6.80 ± 0.03c | 0.0042 ± 0.0003b | 3832.83 ± 39d | 95.67 | ||
| W | 39, 824 ± 3, 593 | 2, 934 ± 23ab | 6.82 ± 0.01bc | 0.0037 ± 0.0002c | 4112.14 ± 5ab | 95.44 | ||
| Ry | 40, 889 ± 1, 004 | 2, 954 ± 19ab | 6.89 ± 0.01a | 0.0031 ± 0.0000d | 4118.91 ± 12ab | 95.55 | ||
| C | 43, 255 ± 1, 576 | 2, 824 ± 68de | 6.82 ± 0.03bc | 0.0032 ± 0.0001d | 3887.53 ± 37c | 95.59 | ||
| Ra | 40, 746 ± 810 | 2, 985 ± 6a | 6.83 ± 0.01b | 0.0034 ± 0.0004cd | 4158.64 ± 47a | 95.41 | ||
| M | 31, 087 ± 104 | 2, 909 ± 31bc | 6.72 ± 0.01d | 0.0054 ± 0.0001a | 3858.57 ± 27cd | 95.62 | ||
| CM | 32, 749 ± 1, 751 | 2, 763 ± 13e | 6.68 ± 0.01e | 0.0055 ± 0.0001a | 4100.61 ± 20b | 95.56 | ||
| F | A | 32, 187 ± 841 | 2, 855 ± 9a | 6.62 ± 0.05b | 0.0060 ± 0.0000c | 3877.42 ± 12b | 95.56 | |
| T | 42, 883 ± 59 | 2, 754 ± 70a | 6.85 ± 0.13a | 0.0041 ± 0.0000e | 4035.48 ± 55a | 95.62 | ||
| W | 39, 120 ± 334 | 2, 623 ± 112b | 6.53 ± 0.02bc | 0.0073 ± 0.0002a | 3313.75 ± 64e | 96.12 | ||
| Ry | 31, 186 ± 700 | 2, 650 ± 26b | 6.55 ± 0.02bc | 0.0054 ± 0.0003d | 3683.83 ± 65c | 96.01 | ||
| C | 32, 438 ± 917 | 2, 621 ± 63b | 6.60 ± 0.11bc | 0.0056 ± 0.0002d | 3587.66 ± 2d | 96.12 | ||
| Ra | 40, 137 ± 1, 565 | 2, 783 ± 61a | 6.60 ± 0.02bc | 0.0066 ± 0.0002b | 4002.13 ± 16a | 95.54 | ||
| M | 37376 ± 310 | 2, 850 ± 21a | 6.75 ± 0.02a | 0.0037 ± 0.0001f | 3829.88 ± 18b | 95.65 | ||
| CM | 41, 353 ± 657 | 2, 582 ± 10b | 6.48 ± 0.06c | 0.0075 ± 0.0001a | 3637.16 ± 29cd | 95.90 | ||
| Fungi | S | A | 39, 257 ± 1, 609 | 323 ± 5ab | 3.36 ± 0.21a | 0.0761 ± 0.0005cd | 439.86 ± 1b | 99.68 |
| T | 33, 031 ± 122 | 313 ± 5bc | 3.34 ± 0.06a | 0.0656 ± 0.0023d | 393.95 ± 6d | 99.68 | ||
| W | 35, 871 ± 2, 329 | 330 ± 9a | 3.47 ± 0.16a | 0.0667 ± 0.0113d | 464.74 ± 14a | 99.61 | ||
| Ry | 31, 575 ± 1, 165 | 302 ± 8c | 3.40 ± 0.09a | 0.0641 ± 0.0116d | 417.72 ± 11c | 99.71 | ||
| C | 41, 051 ± 2, 684 | 285 ± 3d | 3.34 ± 0.05a | 0.0674 ± 0.0081d | 376.85 ± 8d | 99.72 | ||
| Ra | 41, 839 ± 687 | 313 ± 5bc | 3.39 ± 0.12a | 0.0908 ± 0.0155bc | 424.45 ± 8bc | 99.70 | ||
| M | 38, 081 ± 1, 727 | 268 ± 4e | 3.06 ± 0.05b | 0.1018 ± 0.0026ab | 349.53 ± 11e | 99.74 | ||
| CM | 32, 256 ± 1, 321 | 254 ± 13f | 2.93 ± 0.06b | 0.1113 ± 0.0073a | 329.54 ± 20f | 99.71 | ||
| F | A | 32, 917 ± 1, 138 | 358 ± 3b | 3.62 ± 0.04a | 0.0567 ± 0.0097cd | 481.98 ± 3a | 99.63 | |
| T | 37, 713 ± 731 | 387 ± 7a | 3.70 ± 0.07a | 0.0538 ± 0.0068d | 501.87 ± 20a | 99.64 | ||
| W | 31, 899 ± 393 | 350 ± 4b | 3.42 ± 0.05b | 0.0578 ± 0.0068cd | 489.01 ± 39a | 99.65 | ||
| Ry | 43, 318 ± 1, 204 | 345 ± 3b | 3.65 ± 0.04a | 0.0542 ± 0.0084d | 419.32 ± 6bc | 99.68 | ||
| C | 44, 516 ± 34 | 324 ± 3c | 3.46 ± 0.12b | 0.0689 ± 0.0016bc | 422.98 ± 1b | 99.66 | ||
| Ra | 37, 513 ± 534 | 353 ± 6b | 3.26 ± 0.01c | 0.0755 ± 0.0126b | 411.55 ± 5bc | 99.67 | ||
| M | 32, 617 ± 7 | 309 ± 10cd | 3.27 ± 0.10c | 0.0762 ± 0.0023b | 392.08 ± 7c | 99.70 | ||
| CM | 41, 255 ± 4, 568 | 292 ± 10d | 2.94 ± 0.00d | 0.1218 ± 0.0052a | 429.26 ± 3b | 99.68 |
Data with different letters in each column indicate significantly different between treatments at 0.05 level.
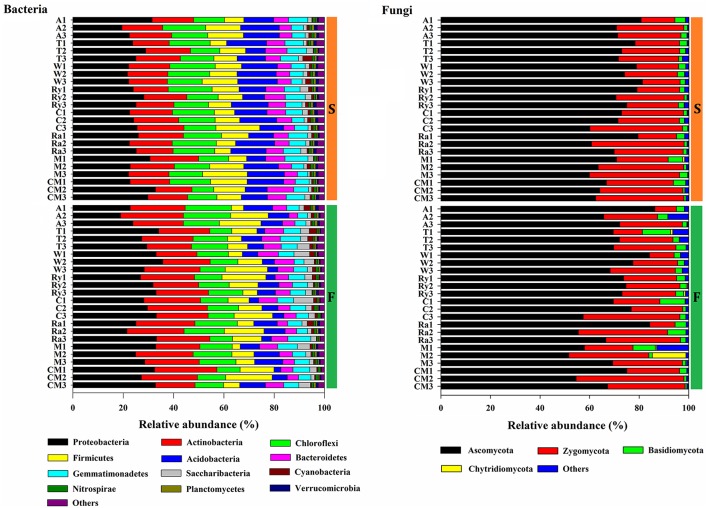
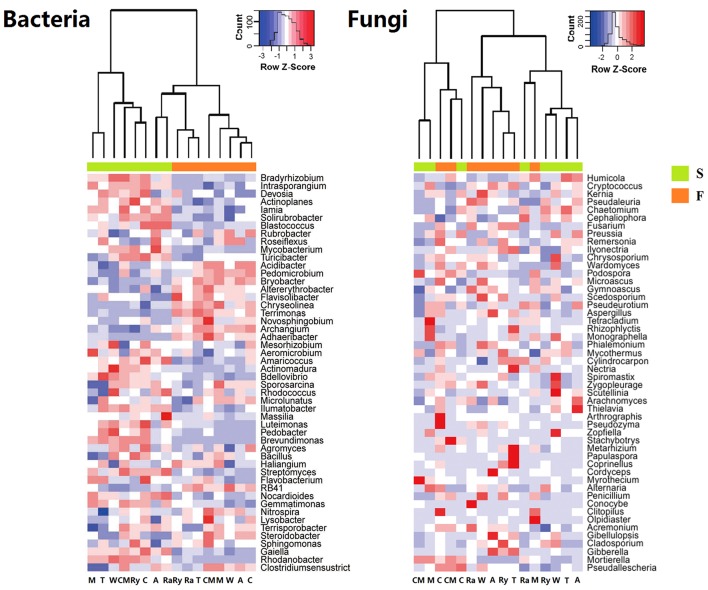
The predominant phyla of bacterial community were Proteobacteria, Actinobacteria, Chloroflexi, Firmicutes, Acidobacteria, Bacteroidetes, and Gemmatimonadetes, these phyla occupied more than 90% of the total sequences (Figure 2). In addition, Saccharibacteria, Cyanobacteria, Planctomycetes, Verrucomicrobia, Nitrospirae, Latescibacteria, and Parcubacteria were detected at relatively low abundances (relative abundance < 1%). Ascomycota, Zygomycota, and Basidiomycota were the dominant fungal phyla at all treatments, which accounted for more than 95% of the sequences, and Chytridiomycota was minor phyla, with a relatively lower abundance (Figure 2). Figure 3 demonstrated that the influence of intercropping systems on the 50 most abundant genera of soil bacterial and fungal communities in the two growing seasons. Relative abundance of soil microbial community from high to low is represented by red through white to blue. Of bacterial community, intercropping systems increased relative abundances of Aeromicrobium and Nocardioides, but Bradyrhizobium, Clostridium_sensu_stricto_1, Acidibacter, IIumatobacter, and Sterodobacter of intercropping systems had relative low abundances in the two growing seasons (Figure 3). With regard to soil fungal genera, Chaetomium, Gymnoascus, and Arthrographis had relative high abundances in the intercropping systems, and Mortierella was just the opposite, compared with monoculture in the two growing seasons (Figure 3).
Figure 2.
Changes in the relative abundances of bacterial and fungal phyla under different treatments in the spring (S) and fall (F) cropping seasons. Others includes phyla below 0.1% of relative abundance and the unclassified phyla.
Figure 3.
Heatmap of top 50 genera of soil bacterial and fungal communities in the spring (S) and fall (F) cropping seasons. Legends showed the Z-scores, demonstrating all samples were represented by the median-centered Z-scores as the relative abundance levels.
Soil bacterial and fungal communities structure
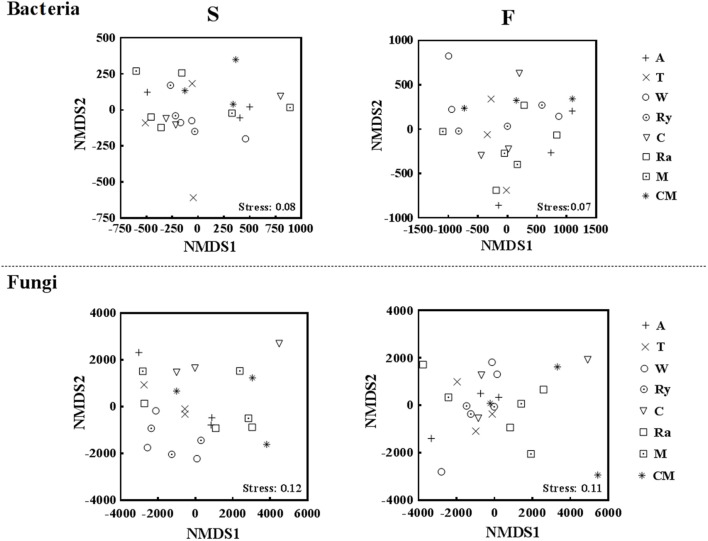
NMDS analysis at the OTU level showed that the differences of bacterial and fungal β-diversity based on the euclidean distance dissimilarity (Figure 4). The NMDS plot illustrated that triplicates of the same treatment were not situated closely, and soil bacterial and fungal communities of all samples had a distinct difference in the two growing seasons (Table 3). In addition, there was no obvious distinction between intercropping systems and monoculture in spring, but mustard-cucumber and alfalfa-cucumber systems were separated from monoculture in fall (Figure 4). For soil fungal community, alfalfa-cucumber, trifolium-cucumber, wheat-cucumber and rye-cucumber systems had a relatively distinguishing boundaries with monoculture in the two growing seasons (Figure 4).
Figure 4.
Nonmetric multidimensional scaling (NMDS) based on euclidean distance plot of all soil bacterial and fungal communities in the spring (S) and fall (F) cropping seasons.
Table 3.
Dissimilarity comparison of soil microbial communities structure between spring and fall cropping seasons.
| Spring vs. Fall | ANOSIM | Adonis | MRPP | |||||
|---|---|---|---|---|---|---|---|---|
| R | P | F | R2 | P | Delta (δ) | Effect size (A) | P | |
| Bacaterial community | 0.666 | 0.001 | 14.13 | 0.235 | 0.001 | 0.354 | 0.112 | 0.001 |
| Fungal community | 0.197 | 0.001 | 5.088 | 0.100 | 0.001 | 0.353 | 0.043 | 0.001 |
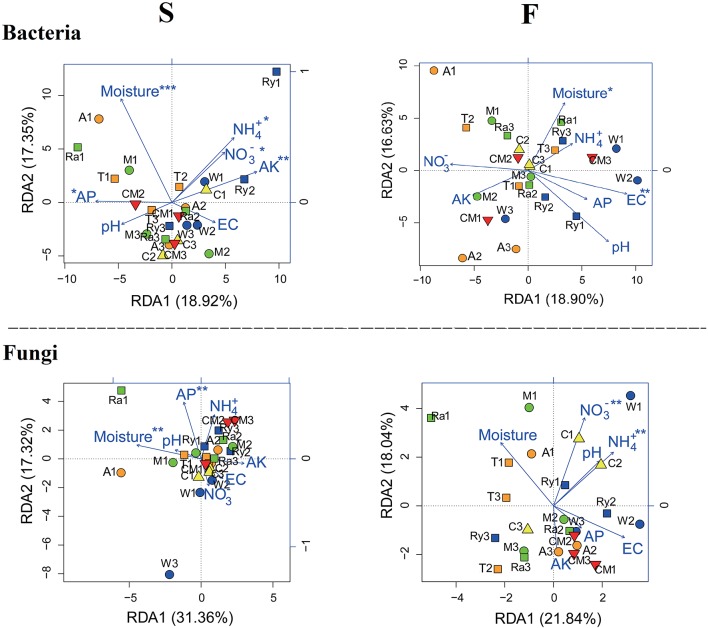
Redundancy analysis (RDA) (Figure 5) used to shed light on the influence of variation of soil physicochemical characteristics on the soil microbial community structure and compositions was showed that soil moisture and AP were the crucial environmental variations that correlated with the soil bacterial and fungal communities composition among soil samples in spring. In fall, EC and soil moisture were strongly correlated with soil bacterial community, however, soil fungal community was mainly influenced by soil -N and -N.
Figure 5.
Redundancy analysis (RDA) demonstrating the relationships between soil environmental factors and soil microbial communities during spring (S) and fall (F) cropping seasons in the control (CM), alfalfa (A), trifolium (T), wheat (W), chrysanthemum (C), rye (Ry), mustard (M), and rape (Ra). *P < 0.05, **P < 0.01, ***P < 0.001.
Co-occurrence network analysis
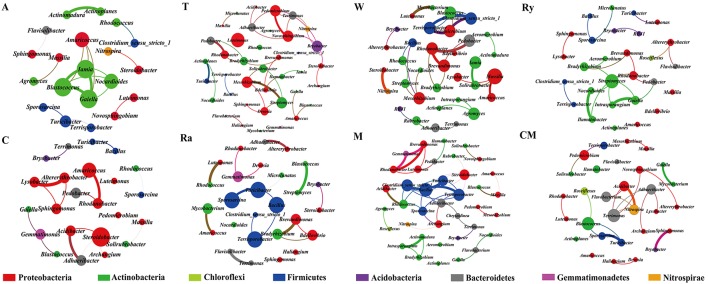
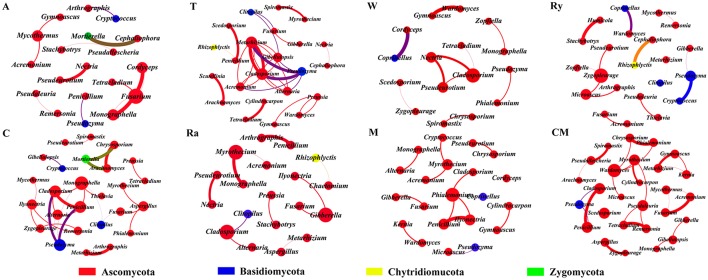
Most of the analytical methods based on metagenomic data focus on single properties of the studied communities, with little attention to the interactions between microbial species. Network analysis is widely performed to explore the interactions of microbial taxon in the complex microbial communities. In this study, network analysis was applied to illustrate the differences of soil bacterial and fungal communities between intercropping systems and monoculture in the two growing seasons (Figures 6, 7), and the network properties of soil bacterial and fungal communities were summarized in Table 4. The structural features, nodes and edges, of bacterial networks under trifolium-cucumber, mustard-cucumber, and wheat-cucumber systems were higher than under cucumber monoculture, showing that they had more connection and closer relationships of soil microbial taxa. For fungal community, the intercropping systems, especially for wheat-cucumber systems, had a less connection and more alienated relationships, compare with the monoculture. The hubs connecting mostly with members of others in intercropping systems were Blastococcus, Luteimonas, Massiliu, Streptomyces, Steroidobacter, Sporosarcina, and Bacillus, belonging to the phyla of Proteobacteria, Actinobacteria, and Firmicutes, and Terrimonas, the phylum of Bacteroidetes, was the major taxa in the monoculture (Figure 6). Of fungal networks, The keystone species of intercropping systems were Fusarium, Cladosporium, Zygopleurage, Monographella, Myrothecium, Gibberella, and Phialemonium; Wardomyces was hub connected chiefly with other species under the monoculture cultivation (Figure 7).
Figure 6.
Network of co-occurring bacterial genera based on correlation analysis for intercropping cultivations and cucumber monoculture. A connection stands for a strong (Spearman's ρ > 0.6) and significant (P < 0.01) correlation. The size of each node is proportional to the degree, the thickness of each edge is proportional to the value of Spearman's correlation coefficients. Nodes colored by taxonomy.
Figure 7.
Network of co-occurring fungal genera based on correlations analysis for intercropping cultivations and cucumber monoculture. A connection stands for a strong (Spearman's ρ > 0.6) and significant (P < 0.01) correlation. The size of each node is proportional to the degree, the thickness of each edge is proportional to the value of Spearman's correlation coefficients. Nodes colored by taxonomy.
Table 4.
Network properties of soil bacterial and fungal communities.
| Classified | Treatments | Average degree (AD) | Network diameter (ND) | Average path length (APL) | Clustering coefficient (CC) | Modularity (MD) | Nodes | Edges |
|---|---|---|---|---|---|---|---|---|
| Bacteria | A | 1.700 | 2 | 1.105 | 0.750 | 0.682 | 20 | 17 |
| T | 2.103 | 9 | 2.803 | 0.416 | 0.756 | 39 | 41 | |
| W | 1.941 | 4 | 1.797 | 0.483 | 0.829 | 34 | 33 | |
| Ry | 1.643 | 4 | 1.842 | 0.315 | 0.678 | 28 | 23 | |
| C | 1.565 | 2 | 1.308 | 0.750 | 0.821 | 23 | 18 | |
| Ra | 1.481 | 4 | 1.467 | 0.556 | 0.823 | 27 | 20 | |
| M | 2.368 | 6 | 2.368 | 0.448 | 0.645 | 38 | 45 | |
| CM | 1.733 | 5 | 2.169 | 0.487 | 0.762 | 30 | 26 | |
| Fungi | A | 1.263 | 2 | 1.200 | 0.583 | 0.820 | 19 | 12 |
| T | 2.696 | 5 | 1.978 | 0.460 | 0.691 | 23 | 31 | |
| W | 1.250 | 2 | 1.167 | 0.778 | 0.781 | 16 | 10 | |
| Ry | 1.182 | 3 | 1.450 | 0.482 | 0.842 | 22 | 13 | |
| C | 2.000 | 6 | 2.471 | 0.652 | 0.727 | 26 | 26 | |
| Ra | 1.158 | 2 | 1.214 | 0.576 | 0.860 | 19 | 11 | |
| M | 1.238 | 2 | 1.188 | 0.583 | 0.838 | 21 | 13 | |
| CM | 2.000 | 4 | 2.000 | 0.658 | 0.786 | 27 | 27 |
Discussion
Intercropping is thought to be an environmentally friendly method for disease management and nutrient complementary, and could also change the microclimatic conditions (Li et al., 1999, 2014; Cong et al., 2015). Previous papers showed that soil physicochemical properties, soil carbon and nitrogen, soil bulk density and pH, are improved by intercropping systems (Morris and Garrity, 1993; Fan et al., 2006; Bedoussac and Justes, 2009; Liu et al., 2014). We found that intercropping systems did not significantly change the continuous cropping soil physicochemical characteristics such as soil moisture, pH, EC, and AK in the two growing seasons (Table 1). These results were likely caused by short-term test and the complex environment of greenhouse. However, soil AP content was significantly decreased under intercropping systems, which may be caused by the competitive interaction among neighboring plants (Guadet and Keddy, 1995; Goldberg, 1996; Zhang and Lamb, 2011).
According to the qPCR results, wheat-, trifolium-, and mustard-cucumber systems increased the continuous cropping soil bacterial absolute abundance, and fungal abundance was increased under rye-cucumber systems, but decreased under wheat- and rape-cucumber systems in the two growing seasons (Figure 1). The results were in line with the previous findings that intercropping changed soil microbial abundance (Tjamos et al., 1992; Zhou et al., 2011). Soil microbial diversity is closely related to soil ecosystem stability and nutrient transformation, and intercropping could manage to various agroecosystem services by improving soil quality (Cong et al., 2015). Our results showed that soil microbial diversity indices were increased under intercropping systems (Table 2), indicating that intercropping improved continuous cropping soil quality and contribute to strengthen the stability of facility ecosystem.
Plant species, root exudates and soil types could affect soil microbial community (Marschner et al., 2001; Wieland et al., 2001; Broeckling et al., 2008; Lauber et al., 2008, 2009). Soil microbial taxonomic composition forcefully changed between intercropping and cucumber continuous cropping soils (Figures 2, 3). Proteobacteria was the most abundant phylum in the cucumber continuous cropping soil, but Actinobacteria and Chloroflexi were more abundant under the intercropping systems. This finding is in line with a previous study that Actinobacteria and Chloroflexi were present at a higher percentage in mulberry-soybean intercropping systems (Li et al., 2016). For fungal community, Ascomycota in the wheat-cucumber system was the most abundant phyla compared to other treatments, the result is also in accordance with a previous study in which Ascomycota were the main phyla in the wheat monoculture and intercropping systems (Granzow et al., 2017). Additionally, in this study we found that genera Aeromicrobium, Nocardioides and Gymnoascus were enriched under intercropping systems, but Clostridium_sensu_stricto_1, Acidibacter, IIumatobacter, Steroidobacter, and Mortierella were inverse. Aeromicrobium and Nocardioides belong to Actinobacteria, Actinobacteria were well-known for species that have beneficial association with plants (Kim et al., 2011; Palaniyandi et al., 2013). Mortierella could generate antagonistic substance, Arachidonic acid, which is an elicitor of phytoalexins in plants to suppress plant disease (Eroshin et al., 1996; Tagawa et al., 2010). It was decreased in the intercropping treatments, the soil is a complex environment, the concrete reasons need to be further studied in the future. High concentrations of soil available phosphorous and host species determined the arbuscular mycorrhiza (AM) community, that is the reason why Glomeromycota, concluded AM that can improve the nutrients supply and soil structure, was not detected in this experiments (Parniske, 2008; Gosling et al., 2013). Many studies indicated that Bradyrhizobium not only have a wide range of legumes, but also are abundant in non-legume plants (Trinick and Hadobas, 1988; VanInsberghe et al., 2015). We found that wheat-cucumber and mustard-cucumber intercropping systems had a higher relative abundances of Bradyrihizobium in spring and fall, respectively.
NMDS and RDA analyses demonstrated that soil microbial community structure did not have significant changes between intercropping and monoculture soils (Figures 4, 5). However, bacterial and fungal communities structure were significantly altered in the two growing seasons (Table 3). Previous studies showed that temperature regulated strongly soil microbial community structure and soil microbial communities varied broadly in their suitable temperature (Alster et al., 2016). Temperature of two growing seasons had a great difference in the northeast of China, thus, we speculate that the key factor was temperature led to variations of soil microbial communities structure in the two growing seasons. Soil moisture was related to soil microbial distribution, Banerjee et al. (2016) reported that soil moisture play an important role in soil microbial activities and composition. We found that soil moisture, AP, EC, -N, and -N were related to soil microbial community structure (Figure 5). These results illustrated that soil characteristics indirectly influenced soil bacterial and fungal community structure. In our study, intercropping did not significantly affect soil microbial community structure. These results need to carry out long-term test to confirm in the future.
A network analysis was performed to compare the complexity of operating in the intercropping soil and the continuous cropping soil. Network analysis was constructed using all positive correlations of top 50 most abundant genera of soil microbial community (Figure 6). The number of correlations of fungal community was lower in the intercropping systems than in the control, and bacterial community of trifolium, mustard, and wheat intercropping systems, especially wheat intercropping systems, had more positive correlations than the control (Table 4). In previous study, researchers found that rhizosphere of wild oat had the more complex networks compared to the bulk soils, it means that rhizosphere is contributed to the interaction between soil microbial taxon (Shi et al., 2016). In addition, plants have the same great effect on soil microbial communities as soil, intercropping systems could increase plant diversity, different plant species secrete diverse root exudates, which can affect species-specific shift in the microbial community as well as the soil microbial diversity (Grayston et al., 1998; Marchner et al., 2004; Broeckling et al., 2008; Zhou and Wu, 2012; Li et al., 2014). Plant residues can regulate the soil biochemical cycle, so that it generate strong microbial activities and interactions, and the amount and degradation rate of plant residues have great differences among intercropping treatments, which may also be the reason for the differences in the microbes interactions of all treatments (Martens, 2000; Philippot et al., 2013). Moreover, Soil microbial interactions can create intense positive feedbacks in the plant communities, which might contribute directly to decrease plant diversity (Bever et al., 2010). Now that we have determined the differences between intercropping and continuous cropping soils with respect to soil microbial communities and soil physicochemical properties, this will provide theoretical basis for microbial remediation of continuous cropping soil in the future.
Conclusions
In summary, our results illustrated that trifolium-cucumber and mustard-cucumber systems increased remarkably the bacterial diversity, and fungal diversity was more abundant in the intercropping systems. However, soil microbial community structure was not significantly altered by seven intercropping crops. We found that soil microbial community varied with soil characteristics, indicating the effect of intercropping on soil microbial community was indirectly affected by soil environmental factors. Moreover, the bacterial and fungal co-occurrence patterns were influenced by intercropping systems in essence. Intercropping systems had a complex relationships between soil bacterial community, and had less connection and relationships of fungal taxa.
Author contributions
FW designed this experiment. SL executed the experiment and finished the manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was funded by the China Agricultural Research System (CARS-23-C-10).
References
- Acosta-Martínez V., Burow G., Zobeck T. M., Allen V. G. (2010). Soil microbial communities and function in alternative systems to continuous cotton. Soil Sci. Soc. Am. J. 74, 1181–1192. 10.2136/sssaj2008.0065 [DOI] [Google Scholar]
- Alster C. J., Koyama A., Johnson N. G., Wallenstein M. D., von Fishcher J. C. (2016). Temperature sensitivity of soil microbial communities: an appicaiton of macromolecular rate theory to microbial respiration. J. Geophys. Res. Biogeosci. 121, 1420–1433. 10.1002/2016JG003343 [DOI] [Google Scholar]
- Banerjee S., Helgason B., Wang L. F., Winsley T., Ferrari B. C., Siciliano S. D. (2016). Legacy effects of soil moisture on microbial community structure and N2O emissions. Soil Biol. Biochem. 95, 40–50. 10.1016/j.soilbio.2015.12.004 [DOI] [Google Scholar]
- Banik P., Midya A., Sarkar B. K., Ghose S. S. (2006). Wheat and chickpea intercropping systems in an additive series experiment: advantages and weed smothering. Eur. J. Agron. 24, 325–332. 10.1016/j.eja.2005.10.010 [DOI] [Google Scholar]
- Bao S. D. (2005). Soil Analysis in Agricultural Chemistry (in Chinese), 3rd Edn. Beijing: China Agricultural Press. [Google Scholar]
- Barberán A., Bates S. T., Casamayor E. O., Fierer N. (2012). Using network analysis to explore co-occurrence patterns in soil microbial communities. ISME J. 6, 343–351. 10.1038/ismej.2011.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedoussac L., Justes E. (2009). The efficiency of a durum wheat-winter pea intercrop to improve yield and wheat grain protein concentration depends on N availability during early growth. Plant Soil 330, 19–35. 10.1007/s11104-009-0082-2 [DOI] [Google Scholar]
- Bell T. H., Yergeau E., Maynard C., Juck D., Whyte L. G., Greer C. W. (2013). Predictable bacterial composition and hydrocarbon degradation in Arctic soils following diesel and nutrient disturbance. ISME J. 7, 1200–1210. 10.1038/ismej.2013.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellemain E., Carlsen T., Brochmann C., Coissac E., Taberlet P., Kauserud H. (2010). ITS as an environmental DNA barcode for fungi: an in silico approach reveals potential PCR biases. BMC Microbiol. 10:189. 10.1186/1471-2180-10-189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bever J. D., Dickie I. A., Facelli E., Facelli J. M., Klironomos J., Moora M., et al. (2010). Rooting theories of plant community ecology in microbial interactions. Trends Ecol Evol. 25, 468–478. 10.1016/j.tree.2010.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broeckling C. D., Broz A. K., Bergelson J., Manter D. K., Vivanco J. M. (2008). Root exudates regulate soil fungal community composition and diversity. Appl. Environ. Microbiol. 74, 738–744. 10.1128/AEM.02188-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooker R. W., Bennett A. E., Cong W. F., Daniell T. J., George T. S., Hallett P. D., et al. (2015). Improving intercropping: a synthesis of research in agronomy, plant physiology and ecology. New Phytol. 206, 107–117. 10.1111/nph.13132 [DOI] [PubMed] [Google Scholar]
- Caporaso J. G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F. D., Costello E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. 10.1038/nmeth.f.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C. L., Fu X. P., Zhou X. G., Guo M. Y., Wu F. Z. (2017). Effects of seven different companion plants on cucumber productivity, soil chemical characteristics and Pseudominas community. J. Integr. Agric. 16, 2206–2214. 10.1016/S2095-3119(17)61698-8 [DOI] [Google Scholar]
- Cong W. F., Hoffland E., Li L., Six J., Sun J. H., Bao X. G., et al. (2015). Intercropping enhances soil carbon and nitrogen. Global Change Biol. 21, 1715–1726. 10.1111/gcb.12738 [DOI] [PubMed] [Google Scholar]
- Corre-Hellou G., Dibet A., Hauggaard-Nielsen H., Crozat Y., Gooding M., Ambus P., et al. (2011). The competitive abiliy of pea-barley intercrops against weeds and the interactions with crop productivity and soil N availability. Field Crops Res. 122, 264–272. 10.1016/j.fcr.2011.04.004 [DOI] [Google Scholar]
- de Graaff M. A., Classen A. T., Castro H. F., Schadt C. W. (2010). Labile soil carbon inputs mediate the soil microbial community composition and plant residue decomposition rates. New Phytol. 188, 1055–1064. 10.1111/j.1469-8137.2010.03427.x [DOI] [PubMed] [Google Scholar]
- Deng Y., Jiang Y. H., Yang Y., He Z., Luo F., Zhou J. (2012). Molecular ecological network analyses. BMC Bioinformatics 13:113. 10.1186/1471-2105-13-113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998. 10.1038/nmeth.2604 [DOI] [PubMed] [Google Scholar]
- Eroshin V. K., Dedyukhina E. G., Chistyakova T. I., Zhelifonova V. P., Kurtzman C. P., Bothast R. J. (1996). Arachidonic-acid production by species of Mortierella. World J. Microbiol. Biotechnol. 12, 91–96. 10.1007/BF00327809 [DOI] [PubMed] [Google Scholar]
- Fan A., Chen X., Li Z. (2006). Effects of intercropping systems of trees with soybean on soil physicochemical properties in juvenile plantations. J. For. Res. 17, 226–230. 10.1007/s11676-006-0052-y [DOI] [Google Scholar]
- Freilich S., Kreimer A., Meilijson I., Gophma U., Sharan R., Ruppin E. (2010). The large-scale organizaiton of the bacterial network of ecological co-occurrence interactions. Nucleic Acids Res. 38, 3857–3868. 10.1093/nar/gkq118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardes M., Bruns T. D. (1993). ITS primers with enhanced specificity for basidiomycetees-application to the identification of mycorrhizae and rusts. Mol. Ecol. 2, 113–118. 10.1111/j.1365-294X.1993.tb00005.x [DOI] [PubMed] [Google Scholar]
- Goldberg D. E. (1996). Competitive ability: definitions, contingency and correlated traits. Philos. Trans. R. Soc. B Biol. Sci. 351, 1377–1385. 10.1098/rstb.1996.0121 [DOI] [Google Scholar]
- Gosling P., Mead A., Proctor M., Hammond J. P., Bending G. D. (2013). Contrasting arbuscular mycorrhizal communities colonizing different host plants show a similar response to a soil phosphorus concentration gradient. New Phytol. 198, 546–556. 10.1111/nph.12169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granzow S., Kaiser K., Wemheuer B., Birgit P., Rolf D., Stefan V., et al. (2017). The effects of cropping regimes on fungal and bacterial communities of wheat and faba bean in a greenhouse pot experiment differ between plant species and compartment. Front. Microbiol. 8:902. 10.3389/fmicb.2017.00902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grayston S. J., Wang S. Q., Campbell C. D., Edwards A. C. (1998). Selective influence of plant species on microbial diversity in the rhizosphere. Soil Biol. Biochem. 30, 369–378. 10.1016/S0038-0717(97)00124-7 [DOI] [Google Scholar]
- Guadet C. L., Keddy P. A. (1995). Competitive performance and species distribution in shortline plant communities: a comparative approach. Ecology 76, 280–291. 10.2307/1940649 [DOI] [Google Scholar]
- Hinsinger P., Betencourt E., Bernard L., Brauman A., Plassard C., Shen J., et al. (2011). P for two, sharing a scarce resource: soil phosphorus acquisition in the rhizosphere of intercropped species. Plant Physiol. 156, 1078–1086. 10.1104/pp.111.175331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacomy M., Bastian M., Heymann S. (2009). Gephi: an open source software for exploring and manipulating networks, in International AAAI Conference on Weblogs and Social Media; Third International AAAI Conference on Weblogs and Social Media (Paris: ). [Google Scholar]
- Kim Y. C., Leveau J., McSpadden Gardener B. B., Pierson E. A., Pierson L. S., III., Ryu C. M. (2011). The multifactorial basis for plant health promotion by plant-associated bacteria. Appl. Environ. Microbiol. 77, 1548–1555. 10.1128/AEM.01867-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauber C. L., Hamady M., Knight R., Fierer N. (2009). Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl. Environ. Microbiol. 75, 5111–5120. 10.1128/AEM.00335-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauber C. L., Strickland M. S., Bradford M. A., Fierer N. (2008). The influence of soil properties on the structure of bacterial and fungal communities across land-use types. Soil Biol. Biochem. 40, 2407–2415. 10.1016/j.soilbio.2008.05.021 [DOI] [Google Scholar]
- Li C., Li X., Kong W., Wu Y., Wang J. (2010). Effect of monoculture soybean on soil microbial community in the Northeast China. Plant Soil 330, 423–433. 10.1007/s11104-009-0216-6 [DOI] [Google Scholar]
- Li L., Sun J. H., Zhang F. S., Li X. L., Yang S. C., Rengel Z. (2001). Wheat/maize or wheat/soybean strip intercropping. I. Yield advantages and interspecific interactions on nutrients. Field Crops Res. 71, 123–137. 10.1016/S0378-4290(01)00156-3 [DOI] [Google Scholar]
- Li L., Tilman D., Lambers H., Zhang F. S. (2014). Plant diversity and overyielding: insights from belowground facilitation of intercropping in agriculture. New Phytol. 203, 63–69. 10.1111/nph.12778 [DOI] [PubMed] [Google Scholar]
- Li L., Yang S., Li X., Zhang F., Christie P. (1999). Interspecific complementary and competitive interactions between intercropped maize and faba bean. Plant Soil 212, 105–114. 10.1023/A:1004656205144 [DOI] [Google Scholar]
- Li X., Sun M., Zhang H., Xu N., Sun G. (2016). Use of mulberry-soybean intercropping in salt-alkali soil impacts the diversity of the soil bacterial community. Microb. Biotechnol. 9, 293–304. 10.1111/1751-7915.12342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T., Cheng Z., Meng H., Ahmad I., Zhao H. (2014). Growth, yield and quality of spring tomato and physicochemical properties of medium in a tomato/garlic intercropping system under plastic tunnel organic medium cultivation. Sci. Hortic. 170, 159–168. 10.1016/j.scienta.2014.02.039 [DOI] [Google Scholar]
- Mangan S. A., Schnitzer S. A., Herre E. A., Mack K. M., Valencia M. C., Sanchez E. I., et al. (2010). Negative plant-soil feedback predicts tree-species relative abundance in a tropical forest. Nature 466, 752–755. 10.1038/nature09273 [DOI] [PubMed] [Google Scholar]
- Marchner P., Growley D., Yang C. H. (2004). Development of specific rhizosphere of bacterial communities in relation to plant species, nutrition and soil type. Plant Soil 261, 199–208. 10.1023/B:PLSO.0000035569.80747.c5 [DOI] [Google Scholar]
- Marschner P., Yang C. H., Lieberei R., Crowley D. E. (2001). Soil and plant specific effects on bacterial community composition in the rhyzosphere. Soil Biol. Biochem. 33, 1437–1445. 10.1016/S0038-0717(01)00052-9 [DOI] [Google Scholar]
- Martens D. A. (2000). Plant residue biochemistry regulates soil carbon cycling and carbon sequestration. Soil Biol. Biochem. 32, 361–369. 10.1016/S0038-0717(99)00162-5 [DOI] [Google Scholar]
- Mitchell R. J., Campbell C. D., Chapman S. J., Cameron C. M. (2010). The ecological engineering impact of a single tree species on the soil microbial community. J. Ecol. 98, 50–61. 10.1111/j.1365-2745.2009.01601.x [DOI] [Google Scholar]
- Morris R. A., Garrity D. P. (1993). Resource capture and utilization in intercropping: water. Field Crops Res. 34, 303–317. 10.1016/0378-4290(93)90119-8 [DOI] [Google Scholar]
- Muyzer G., De Waal E. C., Uitterlinden A. G. (1993). Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59, 695–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palaniyandi S. A., Yang S. H., Zhang L., Suh J. W. (2013). Effects of actinobacteria on plant disease suppression and growth promotion. Appl. Microbiol. Biotechnol. 97, 9621–9636. 10.1007/s00253-013-5206-1 [DOI] [PubMed] [Google Scholar]
- Parniske M. (2008). Arbuscular mycorrhiza: the mother of plant root endosymbioses. Nat Rev Microbiol. 6, 763–775. 10.1038/nrmicro1987 [DOI] [PubMed] [Google Scholar]
- Philippot L., Raaijmakers J. M., Lemanceau P., Van der Putten W. H. (2013). Going back to the roots: the microbial ecology of the rhizosphere. Nat. Rev. Microbiol. 11, 789–799. 10.1038/nrmicro3109 [DOI] [PubMed] [Google Scholar]
- R Development Core Team (2006). R, a Language and Environment for Statistic Computing. Vienna: R Foundation for Statistical Computing; Available online at: http://www.R-project.org [Google Scholar]
- Shi S. J., Nuccio E. E., Shi Z. J., He Z. L., Zhou J. Z., Firestone M. K. (2016). The interconnected rhizosphere: high network complexity dominates rhizosphere assemblages. Ecol. Lett. 19, 926–936. 10.1111/ele.12630 [DOI] [PubMed] [Google Scholar]
- Singh K., Mish A. K., Singh B., Singh R. P., Patra D. D. (2016). Tillage effects on crop yield and physicochemical properties of sodic soils. Land Degrad. Dev. 27, 223–230. 10.1002/ldr.2266 [DOI] [Google Scholar]
- Tagawa M., Tamaki H., Manome A., Koyama O., Kamagata Y. (2010). Isolation and characterization of antagonistic fungi against potato scab pathogens from potato field soils. FEMS Microbiol. Lett. 305, 136–142. 10.1111/j.1574-6968.2010.01928.x [DOI] [PubMed] [Google Scholar]
- Tjamos E. C., Papavizas G. C., Cook R. J. (1992). Biological Control of Plant Diseases. Athens: Plenum Press. [Google Scholar]
- Trinick M. J., Hadobas P. A. (1988). Biology of the Parasponia-Bradyrhizobium symbiosis. Plant Soil. 110, 177–185. [Google Scholar]
- Urbanová M., Šnajdr J., Baldrian P. (2015). Composition of fungal and bacterial communities in forest litter and soil is largely determined by dominant trees. Soil Biol. Biochem. 84, 53–64. 10.1016/j.soilbio.2015.02.011 [DOI] [Google Scholar]
- van der Heijden M. G. A., Klironomos J. N., Ursic M., Moutoglis P., Streitwolf-Engel R., Boller T., et al. (1998). Mycorrhizal fungal diversity determines plant biodiversity, ecosystem variability and productivity. Nature 396, 69–72. 10.1038/23932 [DOI] [Google Scholar]
- VanInsberghe D., Maas K. R., Cardenas E., Strachan C. R., Hallam S. J., Mohn W. W. (2015). Non-symbiotic Bradyrhizobium ecotypes dominate north American forest soils. ISME J. 9, 2435–2441. 10.1038/ismej.2015.54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieland G., Neumann R., Backhaus H. (2001). Variation of microbial communities in soil, rhizosphere, and rhizoplane in response to crop species, soil type, and crop development. Appl. Environ. Microbiol. 67, 5849–5854. 10.1128/AEM.67.12.5849-5854.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu N., Tan G., Wang H., Gai X. (2016). Effect of biochar additions to soil on nitrogen leaching, microbial biomass and bacterial community structure. Eur. J. Soil Biol. 74, 1–8. 10.1016/j.ejsobi.2016.02.004 [DOI] [Google Scholar]
- Zhang S., Lamb E. G. (2011). Plant competitive ability and the transitivity of competitive hierarchies change with plant age. Plant Ecol. 213, 15–23. 10.1007/s11258-011-0002-4 [DOI] [Google Scholar]
- Zhong Y., Yan W., Shangguan Z. (2015). Impact of long-term N additions upon coupling between soil microbial community structure and activity, and nutrient-use efficiencies. Soil Biol. Biochem. 91, 151–159. 10.1016/j.soilbio.2015.08.030 [DOI] [Google Scholar]
- Zhou X., Wu F. (2012). Dynamics of the diversity of fungal and Fusarium communities during continuous cropping of cucumber in the greenhouse. FEMS Microbiol. Ecol. 80, 469–478. 10.1111/j.1574-6941.2012.01312.x [DOI] [PubMed] [Google Scholar]
- Zhou X., Yu G., Wu F. (2011). Effects of intercropping cucumber with onion or garlic on soil enzyme activities, microbial communities and cucumber yield. Euro. J. Soil Biol. 47, 279–287. 10.1016/j.ejsobi.2011.07.001 [DOI] [Google Scholar]