Abstract
Evolving strategies to counter cancer initiation and progression rely on the identification of novel therapeutic targets that exploit the aberrant genetic changes driving oncogenesis. Several chromatin associated enzymes have been shown to influence post-translational modification (PTM) in DNA, histones, and non-histone proteins. Any deregulation of this core group of enzymes often leads to cancer development. Ubiquitylation of histone H2B in mammalian cells was identified over three decades ago. An exciting really interesting new gene (RING) family of E3 ubiquitin ligases, known as RNF20 and RNF40, monoubiquitinates histone H2A at K119 or H2B at K120, is known to function in transcriptional elongation, DNA double-strand break (DSB) repair processes, maintenance of chromatin differentiation, and exerting tumor suppressor activity. RNF20 is somatically altered in breast, lung, prostate cancer, clear cell renal cell carcinoma (ccRCC), and mixed lineage leukemia, and its reduced expression is a key factor in initiating genome instability; and it also functions as one of the significant driving factors of oncogenesis. Loss of RNF20/40 and H2B monoubiquitination (H2Bub1) is found in several cancers and is linked to an aggressive phenotype, and is also an indicator of poor prognosis. In this review, we summarized the current knowledge of RNF20 in chronic inflammation-driven cancers, DNA DSBs, and apoptosis, and its impact on chromatin structure beyond the single nucleosome level.
Keywords: cancer, nuclear factor kappaB, RNF20, ubiquitins
Introduction
Cancer is a disease that afflicts men and women, young and old, without any specific etiology. Remarkably, cancer has been studied for over more than several decades; however, the development of cancer is yet a mystery. Several risk factors have been identified as potential initiators of cancer. Development of cancer was often associated with high consumption of red meat, heavy smoking, chronic alcohol intake, viral infections, lifestyle, and environmental factors. Recent advances in research and technology have identified additional inherent risk factors that may or may not be heritable, which range from cellular allelic mutations, somatic mutations, accumulating mutations such as hot spot mutation, homozygous gene deletion, or gene amplification, non-synonymous single nucleotide polymorphisms, inflammatory tumor microenvironment, angiogenesis, and epigenetic alterations in the genome of normal cells that transforms them into cancer cells with characteristic properties such as uncontrolled cell proliferation, and are associated with invasive and metastatic potential [1–5]. These mutations are often associated with poor prognosis. The epigenetic impact in cancer development is yet a largely unexplored area and it is potentially an evolving strategy to counter the development and progression of cancer [6–12].
Several studies have indicated that cancer cells are often associated with modifications or alterations in their chromatin landscape and are associated with DNA replication and repair [13–15]. Five diverse types of DNA repair systems have been recognized based on the type of DNA damage: (i) direct reversal, (ii) base excision repair (BER), (iii) nucleotide excision repair (NER), (iv) DNA mismatch repair (MMR), and (v) double-strand break (DSB) repair [15–17]. In DNA DSB repair, the most important marker is the histone H2A variant H2AX, which is also a surrogate marker for DSB repair. Ring finger protein 20 is an E3 ligase that ubiquitinates histone H2B [18,19]. Ubiquitination of histone H2B (H2Bub1) has been demonstrated to be implicated in chromatin dynamics during transcription regulation, and previous studies have indicated that it is also involved in homologous recombination by altering chromatin structure [20]. Interestingly, it was also observed that ubiquitination of H2B promotes the accumulation of chromatin remodeling factor SNF2H in DNA repair and sustains euchromatin structure [20,21].
A RING (really interesting new gene) family of E3 ubiquitin ligases enables homo- or heterodimeric complex formations such as RNF20–RNF40, BRCA1–BARD1, BIRC7, CHIP, cIAP, IDOL, RNF4, Prp19, Mdm2–MdmX, and RING1B–Bmi [22]. In general, it is believed that the RING family of E3 ligases co-operates with E2 enzyme through the RING domain [19,22]. Both RNF20 and RNF40 contain a RING domain in their C-terminal and are orthologs of yeast BRE1 [23–25]. The RING domain is absolutely essential for ubiquitin ligase activity, formation of homo/heterodimeric complexes, and stability of E3 ubiquitin ligases [23–26]. Knockdown of either RNF20 or RNF40 leads to degradation of both the proteins [26]. In contrast, the RING domain of RNF20/40 and BRE1 is not required for interacting with RAD6 enzyme [27–29]. The present review is designed to collate the existing literature, and critically analyze our current understanding and the recent advances in RNF20/40 mediated processes and their implication in cancer development.
RNF20 in chromatin and histones remodeling – implication in cancer phenotype gene expression
All eukaryotic cells contain identical DNA that has a unique ability to differentiate and maintain specific phenotypes and functions. The key regulators of this unique and important specific capability are the chromatin associated histones on the DNA. Several studies have demonstrated that modifications such as addition or deletion on DNA and/or histones by methylation, acetylation, SUMOylation, ADP ribosylation, ubiquitination, phosphorylation, and several other modifications on histone serine, threonine, and lysine residues or the DNA itself by specific enzymes regulating several processes such as maintaining cell identity, cell cycle regulation, proliferation, and genome integrity [6,10–12,30]. These changes in the genome of the cells are known as epigenetic changes that regulate activation or repression of gene expression [30–33]. In eukaryotic cells, 147 bps of DNA are wrapped around the core octamural globular histone proteins known as the histone octamer containing pairs of H2A, H2B, H3, and H4. The histone octamer forms the fundamental unit known as the nucleosome, which is the building block for chromatin and chromosomes [12,30,33]. Thus, the state of chromatin (i.e. euchromatin or heterochromatin formation) determines the gene expression pattern, resulting in profound changes in the cellular phenotypes and critical biological functions [9,34,35]. Deregulated epigenetic changes have been associated with the development of several diseases including chronic inflammation-driven cancers [32,33,36–39].
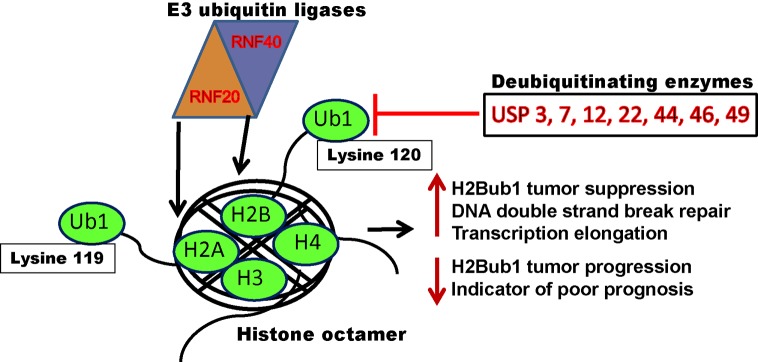
In cancer cells, numerous epigenetic alterations are observed in genes regulating cell cycle, oncogenes, tumor suppressor genes, and apoptosis related genes, such as aberrant methylation or acetylation of the histones and/or DNA [12,32,34,39]. Histones have been shown to be ubiquitinated and are associated with transcription regulation. Addition of ubiquitin molecules to histones can directly modulate transcription factors and their cofactors. The equilibrium between ubiquitination and deubiquitination is vital for normal cell function, and any disparity frequently leads to malignant transformation. Ubiquitin is a 76-aa polypeptide responsible for the addition of ~8.5 kDa to the overall mass of a histone (~11–15 kDa). Ubiquitin contains seven lysine (K) residues. The process of ubiquitination is an ATP-driven process involving the E1 and E2 enzymes, permitting E3 ubiquitin ligase to ubiquitinate histone directly or indirectly [12,40–42]. In general, histone ubiquitination is a natural physiological cellular process and participates in DNA DSB repair, regulation of transcription, and genome stability. Histone polyubiquitination is often not processed for proteosomal degradation; instead it regulates gene transcription [43]. Remarkably, it has been reported that histones H2A and H2B are more prone to ubiquitination [11,12,18,44,45]. Ubiquitylation of histone H2B in mammalian cells was identified in 1980 [46] (Figure 1).
Figure 1. Reversible post-translational modification (PTM) ubiquitination of H2A (K119) and H2B (K120) on specific amino acid residues on core histone tails regulates various biological processes, including transcription elongation, inflammation, DNA replication, and DNA DSB repair processes, and is highly dynamic in nature.
These modifications are added on to histones by RNF20/40 E3 ubiquitin ligases. These PTM marks are identified and removed by active enzymes known as deubiquitinating enzymes. Any deregulation of this process often leads to malignant cellular transformation.
H2A monoubiquitination on K119 plays a role in transcriptional silencing of polycomb proteins and in genome maintenance compared with H2B, which is ubiquitinated at K120. These ubiquitin marks can be deubiquitinated by MYSM1 (2A-DUB) [47]. In yeast, histone H2B is monoubiquitinated on lysine 123 (K123) by ubiquitin-conjugating enzyme (E2) RAD6 and ubiquitin ligase (E3) BRE1 [23–25]. H2B is ubiquitinated on its C-terminal tail and significantly increases transcript length [20,48,49]. In eukaryotic cells, RNF20/RNF40 can exclusively monoubiquitinate K120 on histone H2B [21,26]. Moreover, H2B ubiquitinations are reversible and are promptly removed by deubiquitinating enzymes (DUBs). Several DUBs have been identified that have been shown to deubiquitinate mammalian H2B, such as USP3, USP7, USP12, USP22, USP44, USP46, and USP49 [50–55]. H2B monoubiquitination (H2Bub1) plays a pivotal role in both activation of transcription and tumor suppression [56,57]. Aberrant H2Bub1 is the key to initiation of malignant transformation and directly influences chromatin structure beyond the level of the single nucleosome [58,59]. Ubiquitin-conjugating enzyme E2A (UBE 2A or RAD6A) and the RNF20/40 E3 ligase complex are responsible for catalysis in H2B ubiquitination. Deregulation in any of these process leads to the development of tumors, as evidenced with hypermethylation of RNF20 promoters in breast tumor samples [60]. Mutations in cell division cycle 73 (CDC73) lead to loss of maintenance of H2Bub1 PTM both in vitro and in vivo.
Abnormally regulated or mutated CDC73 has been reported in several tumors such as breast, colorectal, gastric, parathyroid, renal, and in patients with familial disorder-hyperparathyroidism jaw tumor syndrome [61]. Interestingly, in malignant breast cancer samples, low levels of H2Bub1 were correlated with tumor suppression compared with normal and benign samples [60]. Deregulated expression and hyperactivation of deubiquitination enzymes also upset the overall expression of H2Bub1 [50,59,62]. In addition, USP22 was found to carry homozygous gene deletions, gene amplifications, and non-synonymous single nucleotide polymorphisms in a variety of tumor types [43]. In a study by Zhang et al. [63] (2011), USP22 was found to be up-regulated in breast cancer patient samples and was associated with aggressive phenotypes and decreased levels of H2Bub1 compared with benign tumors. Furthermore, USP22 overexpression was associated with breast cancer lymph node metastasis and recurrence, and was a pointer of poor prognosis [63,64]. An in vitro reconstituted transcription assay demonstrated that H2B ubiquitination regulates the transcription elongation regulator PAF complex and the RNF20/40 heterodimer, indicating that RNF20 transcription regulation is complex in cells [20].
Role of RNF20 in inflammation and inflammation-driven cancers
Chronic inflammation has been widely associated with diseases such as asthma, Alzheimer’s disease, rheumatoid arthritis, atherosclerosis, multiple sclerosis, and a variety of cancers, and is a key driver of cancer progression [4,11,65–73]. Rudolf Virchow in 1858 proposed chronic inflammation in cancer [66,69,74]. Virchow postulated that it is the tumor microenvironment that sustains persistent inflammation, which drives the initiation and development of oncogenesis [68,70,75]. Acute inflammation is mainly a self-limiting process and can be treated therapeutically; however, prolonged chronic inflammation is mostly detrimental [2,65,76,77].
Chronic inflammation is associated with the production of pro-inflammatory cytokines and chemokines that constitutively activate pro-survival transcriptional factors that act as key regulators of tumor promotion and progression [11,66,70,78]. Cancer development in the presence of chronic inflammation involves the constant presence of activated oncogenes and transcriptional factors such as nuclear factor-κB (NF-κB), signal transducer and activator of transcription 3, activator protein 1, hypoxia-inducible factor 1α, forkhead box protein M1, peroxisome proliferator associated receptor γ, Wnt/β-catenin, c-Met (hepatocyte growth factor receptor), and hedgehog (HH/GLI) [2,4,70,71,79–81]. The transcriptional factor NF-κB was discovered in 1986 by David Baltimore [82–84]. The mammalian NF-κB family of transcription factors is composed of RelA (p65), c-Rel, RelB, p50 (NF-κB1), and p52 (NF-κB2). They all share a conserved REL homology domain of ∼300 amino acids that play a pivotal role in their functions such as DNA dimerization, binding, and heterodimerization interaction with inhibitory κBs (IκBs), which are the intracellular inhibitors of NF-κB [3,4,11,71,81,85–88]. In resting cells, the majority of NF-κB complexes are primarily cytoplasmic and present in an inactive form due to their binding to the IκB family of proteins that prevent DNA binding and, as a consequence, prevent nuclear accumulation [71,77,81,85,88–90].
Ubiquitination of histone H2B is predominantly through E3 ligase RNF20 and it is found to be down-regulated in a majority of cancers [24,43,60,61,73,91–95]. Inflammation is a securely regulated process that can be very effectively turned on or off in normal physiological conditions [87,96,97]. Factors linking chronic inflammation and cancer are of great interest, and several lines of evidence suggest that the constitutive activation of pro-inflammatory transcription factors plays a pivotal role in the sustained cell proliferation observed in cancers [1,3,66,70,81]. Notably, inflammatory bowel disease patients are more prone to developing colorectal cancer [98,99]. H2Bub1 is considered to play a crucial role during transcription, and any change in levels of H2Bub1 affects the transcriptional response to epidermal growth factor [100], estrogen [60,95], interferon γ [101], and androgens [102].
A recent report by Tarcic et al. (2016) [73] showed that RNF20 depletion with a concomitant reduction in H2Bub1 augments tumor necrosis factor-induced activation of NF-κB and its subsequent pro-inflammatory cytokine and chemokine genes. They showed that mice with reduced RNF20 and H2Bub1 are more susceptible to chronic colon inflammation and colorectal cancer, which is associated with activation of NF-κBp65 and reduced H3K9 trimethylation on a subset of NF-κB target genes. In vivo RNF20+/− mice were shown to be predisposed to acute and chronic colonic inflammation and development of colorectal cancer. Surprisingly, RNF20/40 and H2Bub1 were markedly reduced in the epithelium and stroma of ulcerative colitis patients and in human colorectal tumor samples [73]. Genetic instability has been identified to play a critical role in the development of colorectal cancer [103]. Barber et al. (2008) [103] for the first time identified five genes SMC1L1 (two independent mutations), CSPG6, NIPBL, STAG3, and RNF20 involved in sister chromatid cohesion and mutations in these genes can lead to chromosome instability in colorectal cancers.
Breast cancer
Breast cancer is the second most common cancer that afflicts women, with an estimated 1.67 million women diagnosed with breast cancer in 2012 [5,104,105]. Breast cancer was ranked fifth in cancer-associated deaths amongst all cancers globally in 2012 [5,104]. Despite outstanding progress in the identification of specific genes involved in the breast cancer progression, our current knowledge of the complex machinations of the epigenetic landscape is still limited. The RNF20/40 heterodimeric complex is a known major E3 ligase that is responsible for H2Bub1 on K120 and also facilitates H3 methylation on K4 and K79, thereby regulating transcription [106–108]. Interestingly, in human breast cancer MCF7, MDA-MB-231, and T47D cell lines, RNF20/40 was found to be highly expressed compared with the normal human breast epithelial cell line MCF-10A [109]. In this particular study by Duan et al. (2015) [109] they showed that RNF20/40 is physically associated with motor protein Eg5 and is functionally involved in spindle assembly in breast cancer cells. Proteins that are involved in mitosis are often found to be overexpressed in a variety of tumor cells, primarily due to an elevated mitotic index [109–111]. Furthermore, athymic nude mice receiving MCF-7 cells infected by lentiviruses carrying empty vectors or MCF-7 cells with lentivirus-delivered Eg5, RNF20, or RNF40 knockdown, the tumor growth was significantly suppressed compared with control mice, suggesting that an RNF20/40-Eg5 axis is involved in breast carcinogenesis [109].
Protein interaction networks data show relationship between disease-causing genes is often greater than expected by chance and serves as a powerful means for the identification of new genes in diseases such as breast cancer [112–116]. RNF20 was found to be associated with four other genes (SNAI1, WHSC1L1, BCAS3, and MTA3) that share common domains with DNA damage repair (DDR) enzymes. However, it is RNF20 that has been implicated as a modulator of DDR in breast cancer cells [116]. Monoubiquitylation of histone H2B (H2Bub1) is catalyzed primarily by the RNF20/RNF40 complex and removed by multiple DUBs.
RNF20 knockdown significantly reduces H2Bub1 expression and promotes migration in both breast cancer cells and in non-transformed mammary epithelial cells. The Let-7 family of miRNAs is a tumor suppressor that was shown to regulate H2B ubiquitination and reduced breast cancer cell migration, suggesting additional mechanisms of action may be involved in its tumor-suppressor effects [117]. Silencing of RNF20 in breast cancer cells can function as a tumor promoter [100]. It was found that depletion of RNF20 increased breast cancer cell proliferation and migration potential. It is of note that RNF20 promotes CpG island hypermethylation in several breast cancers and that down-regulation of H2B ubiquitination promotes tumorigenesis [100]. In NIH3T3 mouse cells, RNF20 silencing up-regulated formation of colonies in soft agar, indicating neoplastic transformation of cells [100]. In conclusion, up-regulation of RNF20/40-mediated up-regulation of H2Bub1 may down-regulate cancer progression and may be a therapeutic target for cancer prevention and treatment.
Mixed-lineage leukemia-rearranged leukemia
The mixed-lineage leukemia (MLL) proto-oncogene MLL1 was found to be involved in chromosomal translocations occurring frequently in acute myeloid leukemia (AML), acute lymphoblastic leukemia (ALL), infant acute leukemia, and in patients treated with topoisomerase II inhibitors [118]. MLL rearrangements initiate aggressive forms of acute leukemia and are associated with poor outcome [118]. In a study by Wang et al. (2013) [93] they reported the role of RNF20 in the pathogenesis of MLL-fusion leukemia. They found that RNF20 is an additional chromatin regulator that is necessary for MLL-fusion-mediated leukemogenesis and that suppression of RNF20 in leukemia cells leads to inhibition of cell proliferation in vitro. Suppression of RNF20 slowed down leukemia progression in an in vivo animal model and was associated with down-regulation of the MLL-AF9 target gene [93].
In this context, another class of chromatin remodelers, histone deacetylases, has recently emerged as a promising target in MLL-rearranged ALL. Histone deacetylase inhibitors such as LBH589 (panobinostat) have shown promise as an antileukemic against MLL-rearranged infant ALL cells in vitro, with a promising therapeutic index and was effective at nanomolar concentrations [119]. In a recent study by Garrido et al. (2018) [120] they showed that panobinostat could inhibit tumor growth in an ALL xenograft mouse model and prolonged survival of mice. It was also reported that the antileukemic effect was targetted by the suppression of H2B ubiquitination by inhibiting the RNF20/RNF40/WAC E3 ligase complex and inducing apoptosis [120]. This finding also implicates RNF20 as a potential target including other classes of E3 ligases with existing or other new compounds that may potentially act specifically against ubiquitin ligases [121,122].
Prostate cancer
Prostate cancer is the fourth most commonly occurring cancer globally and the second most common cancer in men [123]. Histone modification by methylation, acetylation, or ubiquitination has been reported to be deregulated in a variety of cancer cells [100]. The concomitant activation of polycomb ubiquitin ligases RNF2 and deubiquitinase USP22 is significant during cancer progression because USP22 activation allows transcriptional up-regulation of cell cycle related genes [50]. For instance, H2A ubiquitin ligase RNF2/RING1b and H2B deubiquitinase USP22 are associated with poor prognosis in numerous cancers [50]. Interestingly, genes encoding polycomb group protein BMI-1 and EZH2 are found to be amplified in metastatic prostate cancer, with a concomitant increase in levels of H2Aub1 and H3K27me3 [124]. The Oncomine database reveals that metastatic prostate cancer cells have decreased the levels of RNF20 [102]. In a previous study, it was shown that RNF20 and RNF40 interact with androgen receptor and modulate its transcritpional activity in androgen-dependent LNCaP prostate cancer cells, and depletion of RNF20 or RNF40 is strongly correlated with inhibition of LNCaP cell proliferation and a reduction in H2Bub1 levels [102].
Lung cancer
Lung cancer is the primary cause of cancer-related deaths worldwide. Approximately 85% of lung cancers are non-small-cell lung cancer (NSCLC), while lung adenocarcinoma accounts for approximately 50% of NSCLC [125,126]. Using human lung cancer A549, H1299, and H460 cell lines, and normal lung epithelial cells, suppression of H2Bub1 by RNF20 knockdown was associated with significant decrease in H3K4 and H3K79 trimethylation. It was also observed that RNF20 knockdown and down-regulation of H2Bub1 affect several cellular signaling pathways and enhanced proliferation, migration, invasion, and cisplatin resistance of these cells [127]. Furthermore, lung cancer patients with H2Bub1-negative cancers have shorter survival outcomes compared with H2Bub1-positive patients [127]. The present study, has for the first time demonstrated that loss of H2Bub1 is associated with enhanced malignancy and poor differentiation of lung adenocarcinoma.
Clear cell renal cell carcinoma
Clear cell renal cell carcinoma (ccRCC) is the foremost subtype of kidney cancer. Excessive lipid accumulation in the kidney is one of the characteristics of the aggressive form of ccRCC [128,129]. In general, lipid accumulation and lipogenesis is increased in several cancers [130,131], and lipid metabolites are produced by activation of lipogenesis [132,133]. Increased lipid synthesis is another hallmark of cancer [134]. Sterol regulatory element-binding protein (SREBP) plays a central role in lipid metabolism and membrane biology. Incidentally, Brown and Goldstein identified a nuclear protein that is bound to sterol regulatory element of the low-density lipoprotein receptor and controls transcription [135–137]. Subsequently, SREBP1c was demonstrated as a major determinant of adipocyte determination and differentiation [138]. SREBP targets such as fatty acid synthase and LDL-receptor are often found to be elevated in cancer cells, thus implicating SREBP in deregulated lipogenesis in cancer cells and targetting lipid supply serves as a potential target for anticancer therapy [139,140]. RNF20 has been suggested to act as tumor suppressor in chronic inflammation-driven cancer [73]. RNF20 has also been implicated in polyubiquitinating and degrading SREBP1c upon protein kinase A activation, thus down-regulating lipid metabolism [137,141].
In a recent study, RNF20 was demonstrated to have tumor suppressor activity in ccRCC. RNF20 overexpression inhibited lipogenesis and ccRCC cell proliferation by down-regulating SREBP1c [142]. Furthermore, RNF20 overexpression greatly reduced tumor growth and lipid storage in a xenograft mouse model [142]. In the clinical setting, ccRCC patients with low levels of RNF20 and SREBP1 activation have been reported to be linked with poor prognosis [142]. In addition, tumor-associated mutant p53 has been shown to bind and transcriptionally activate SREBP2 and activate the mevalonate pathway [143]; it is highly possible that p53 and SREBPs may potentially regulate each other. Furthermore, p53 has been shown to have well-known roles in oncogenesis and RNF20 has a role in DNA damage response, it would be critical to determine whether RNF20-mediated suppression of SREBP1c will impact on these pathways, and further strengthen the link between SREBPs and cancer [144].
Other cancers
There was a loss of global H2Bub1 in 77% (313 of 407) of high-grade serous ovarian cancers and it was observed at all the stages (I–IV) of tumor development [145]. Numerous studies have implicated RNF20 in oncogenesis and it was found to be somatically mutated or deleted in various cancers including breast, colon, lung, and prostate cancer (Table 1) [7,100,146–148].
Table 1. RNF20 somatic alterations for a given cancer type.
| Cancer type | Number of new cases diagnosed in U.S.A. and Canada | Overall alterations (%) (deletions, mutations, amplification, multiple) | References |
|---|---|---|---|
| Breast | 276, 989 | 1.1 | [148,171] |
| Lung | 252, 826 | 1.6 | [148,183] |
| Prostate | 202, 499 | 3.3 | [148,184] |
| Colorectal | 160, 640 | 3.3 | [148,175] |
| Uterine | 79, 607 | 5.8 | [148,176] |
| Pancreatic | 58, 230 | 3.7 | [148,185] |
Role of RNF20 in DNA DSB repair
Chromatin dynamics is highly regulated by numerous intracellular signaling networks. These networks often control the extent of extracellular signal activation that helps in normal cellular homeostasis. However, deregulated signaling directly threatens genomic stability, which may result in malignant transformation or cell death [44]. Several risk factors have been identified, such as ionizing radiation, radiomimetic chemicals, environmental factors, and replication fork stalling, which potentially cause cellular genomic DNA damage and DNA DSBs [149–153]. DNA damage is often countered in normal cells by DNA damage response signaling processes. The major DSB repair pathway is the error-prone non-homologous end-joining (NHEJ) and homologous recombination between sister chromatids [154–156]. The initial response to DSB is characterized by extensive PTMs such as acetylation, phosphorylation, ubiquitination, and SUMOylation [44,157].
Dynamic alterations in chromatin and associated histone marks have been identified as important events in DSB and DNA repair mechanisms [158–160]. H2Bub1 has been demonstrated to be a critical event in DDR in eukaryotic cells [89,90,91]. H2Bub1 was first found to be induced in DSB and DDR in yeast. In yeast, the BRE1 mutant showed higher sensitivity to ionizing radiation and was associated with RAD51, a key molecule in homologous recombination repair [160]. Preliminary studies have indicated that BRE1 functions in an RAD51-dependent fashion; however, the molecular mechanisms need to be further elucidated [160]. Upon DSB, protein kinase ataxia telangiectasia mutated (ATM) catalyzes phosphorylation of HNF20 and HNF40 on serine residues. p-HNF20/40 is then recruited to the sites of DSB where it ubiquitinates DNA damage associated H2Bub1 [26]. In another study by Nakamura et al. (2011) [21] they showed that RNF20 functions with the MRE11, RAD50, and NBS1 repair complex (termed as the MRN complex) at DSB sites and augments the repair process through SNF2H-mediated chromatin reorganization [20,21]. This process of H2B ubiquitination is required for both NHEJ (XRCC4 and Ku80) and homologous recombination repair (RAD51, BRCA1, and BRCA2) mechanisms [21,26,44].
Mutated or defective NBS1 potentiates cell death upon ionizing radiation due to impaired homologous recombination and NHEJ repair mechanisms [161,162]. RNF20 was shown to interact with the several critical domains for protein–protein interaction in the C-terminal of NBS1; however, ATM interaction with RNF20/40 is yet to be reported [20,21]. Cancer cells treated with doxorubicin (a DNA damaging agent) have been linked to global loss of H2Bub1, especially those cells encoding proteins that play a pivotal role in DDR by either maintaining or increasing the levels of H2Bub1 [163]. Using a p53 overexpression model, H2Bub1 was found to be at the transcribed region of the p53 target CDKN1A and was associated with recruitment of RNA Pol II and a concomitant increase in CDKN1A [163]. Silencing of RNF20 by si/shRNA in cells augmented ionizing radiation and DNA damaging agents such as camptothecin, neocarzinostatin, and mitomycin C, with severe impairment of DNA repair mechanisms [21,26,91]. Moreover, overexpression of mutant H2B and silencing of RNF20 did not have any additional effect on cells, indicating that RNF20 functions by ubiquitinating H2B in DSB repair. In addition, RNF20 in DSB repair is correlated with euchromatin structure, strongly supporting the evidence that defects in DSB repair protein accumulation at the DSB sites were released by compounds that induce chromatin relaxation [21,164].
A study by Fierz et al. (2011) [58] showed that H2B ubiquitination obstructs chromatin compaction, resulting in an open and biochemically accessible fiber conformation. Furthermore, H2B ubiquitination by RNF20 is followed by accumulation of chromatin remodeling factor SNF2H. Indeed, several studies have shown that depletion of SNF2H sensitized cells to ionizing radiation and DNA damaging agents, and the effect was comparable with RNF20 silencing [20,21,165,166]. Recently, Klement et al. (2014) [167] reported that RNF20-SNF2H is involved in DSB repair and induces euchromatin structure in a SNF2H-dependent manner. Heterochromatin acts as a barrier to DNA repair, with a strong correlation with increased somatic mutations in cancer [167]. In cancer cells, low levels of H2Bub1 contribute to cancer progression and influence several aspects of chromatin function, including transcription regulation and DNA repair [117]. Ionizing radiation induces an overabundance of diverse types of DNA damage, of which DSB accounts for less than 5% compared with DNA single-strand breaks and DNA base damage that is more commonly observed. HeLa and oropharyngeal squamous cell carcinoma (UMSCC74A and UMSCC6) cells that were subjected to radiation demonstrated ionizing radiation-induced complex DNA damage (CDD).
It was demonstrated that H2Bub1 is specifically induced for several hours after irradiation [168]. RNF20/40 has been previously reported to be involved in DSB repair [21]. Carter et al. (2018) [168] showed that abrogation of RNF20 is strongly associated with suppression of H2Bub1 and DNA transcription. In a recent study it was shown that PARP1 inhibitors (Olaparib and BMN673) could preferentially inhibit the proliferation of RNF20-deficient cells by inducing severe DNA DSB (γ-H2AX) and apoptosis (cleaved Caspase-3) [148]. In a recent report by Guppy et al. (2017) [148] they have introduced a new approach known as synthetic lethal targetting to deliver highly specific tumor cell killing. BRCA1 and BRCA2 ubiquitin ligases are involved in DSB homologous recombination repair, also known as the error-free DNA DSB repair pathway [148,169,170]. BRCA1 and BRCA2 are mutated in breast, ovarian [171–173], prostate, lung, and colorectal cancer, and are potential biomarkers for a synthetic lethal targetting strategy [174–177].
Conclusion
Deregulated epigenetic changes have been implicated in the development of several inflammation-driven diseases, including cancer. Numerous epigenetic alterations have been identified in histones, which determine the euchromatin or heterochromatin state that impedes critical physiological functions and may lead to malignant transformation. Ubiquitination of histones is one of the critical histone PTMs occurring on histone H2B and is regulated by the E3 ligase RNF20/40. The balance between H2Bub1 and deubiquitinating enzyme USP22 is critical and disruption leads to tumorigenesis, as H2Bub1 has been implicated in both transcription and DNA DSB repair pathways. Several lines of evidence suggest that RNF20’s role in DSB is mediated by ubiquitination of H2B. Another possibility that has been suggested is histone methylation concomitant with ubiquitination. However, there are technical difficulties in achieving this as it has been shown that the human genome has at least 17 RNF20 genes and would require recently developed techniques such as CRISPR-Cas9 and TALEN to overcome these problems [178–182]. RNF20 has been suggested to act as a tumor suppressor in chronic inflammation-driven cancer.
Interestingly, RNF20 depletion has been shown to enhance NF-κB-dependent gene transcription, and TNF-mediated H2Bub1 down-regulation augments NF-κB’s response in the up-regulation of proinflammatory cytokines or chemokines that may act in an autocrine or paracrine fashion to sustain the prosurvival gene expression in cancer cells. RNF20 now adds another element by which histone polyubiquitination is often not processed for proteosomal degradation; instead it regulates gene transcription, thereby impacts the lifespan of NF-κB-p65 regulated proinflammatory genes and may regulate SREBP1c degradation. The exact molecular role of RNF20 varies in different types of cancers; therefore it is valuable to determine the role for DUBs, protein kinases, and/or pharmacological drugs in modulating RNF20 levels that may provide clinical benefits. Targetting these pathways may ultimately be exploited for cancer treatment. However, several additional in vitro, in vivo, and transgenic rodent studies will determine if RNF20/40 and H2Bub1 can be used as a promising target to be exploited for the prevention and treatment of cancer. Therefore, addressing these questions will hopefully advance our understanding on the role of RNF20/40 and H2Bub1 in chromatin remodeling during transcription and DNA DSB repair, and the potential of epigenetics based therapies for cancer.
Abbreviations
- ALL
acute lymphoblastic leukemia
- ATM
ataxia telangiectasia mutated
- BARD
BRCA1 associated RING domain
- BIRC
baculoviral IAP repeat containing
- BMI
BMI proto-oncogene, polycomb ring finger
- BRCA
breast cancer gene
- ccRCC
clear cell renal cell carcinoma
- CDC73
cell division cycle 73
- CHIP
carboxyl terminus of HSC70-interacting protein
- cIAP
cellular inhibitor of apoptosis
- CRISPR-Cas9
clustered regularly interspaced short palindromic repeats, associated protein-9 nuclease
- DSB
double-strand break
- DUB
deubiquitinating enzyme
- H2Bub1
ubiquitination of histone H2B
- IκB
inhibitory κB
- IDOL (MYLIP)
myosin regulatory light chain interacting protein
- LDL
low-denisty lipoprotein
- MLL
mixed-lineage leukemia
- NF-κB
nuclear factor-κB
- NHEJ
non-homologous end-joining
- NSCLC
non-small-cell lung cancer
- POL II
RNA polymerase II
- PRP19
E3 ubiquitin protein ligase PRP19
- PTM
post-translational modification
- RING
really interesting new gene
- RFN20
E3 ubiquitin ligases
- SREBP
sterol regulatory element-binding protein
- TALEN
transcription activator-like effector nuclease
Competing interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
This work was supported by grants from the National Medical Research Council of Singapore [713-000-177-511 and 713-000-214-511]; the Medical Science Cluster, and the National Research Foundation Singapore; and the Singapore Ministry of Education under its Research Centers of Excellence initiative to Cancer Science Institute of Singapore, National University of Singapore (to A.P.K.).
Author contribution
M.K.S., A.P.K., F.A., and G.S. designed and wrote the manuscript.
References
- 1.Hanahan D., et al. (2011) Hallmarks of cancer: the next generation. Cell 144, 646–674 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- 2.Grivennikov S.I., et al. (2010) Immunity, inflammation, and cancer. Cell 140, 883–899 10.1016/j.cell.2010.01.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Colotta F., et al. (2009) Cancer-related inflammation, the seventh hallmark of cancer: links to genetic instability. Carcinogenesis 30, 1073–1081 10.1093/carcin/bgp127 [DOI] [PubMed] [Google Scholar]
- 4.Chai E.Z., et al. (2015) Analysis of the intricate relationship between chronic inflammation and cancer. Biochem. J. 468, 1–15 10.1042/BJ20141337 [DOI] [PubMed] [Google Scholar]
- 5.Bray F., et al. (2012) Global cancer transitions according to the Human Development Index (2008-2030): a population-based study. Lancet Oncol. 13, 790–801 10.1016/S1470-2045(12)70211-5 [DOI] [PubMed] [Google Scholar]
- 6.Baylin S.B., et al. (2011) A decade of exploring the cancer epigenome - biological and translational implications. Nat. Rev. Cancer 11, 726–734 10.1038/nrc3130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gao J., et al. (2013) Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 6, 11 10.1126/scisignal.2004088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jones P.A., et al. (2002) The fundamental role of epigenetic events in cancer. Nat. Rev. Genet. 3, 415–428 10.1038/nrg816 [DOI] [PubMed] [Google Scholar]
- 9.Juo Y.Y., et al. (2015) Epigenetic therapy for solid tumors: from bench science to clinical trials. Epigenomics 7, 215–235 10.2217/epi.14.73 [DOI] [PubMed] [Google Scholar]
- 10.Shanmugam M.K., et al. (2016) Epigenetic Modifications and Their Potential Role in Tumorigenesis (Dincer Y., ed.), pp. 31–82, Nova Science Publishers, Inc, New York [Google Scholar]
- 11.Shanmugam M.K., et al. (2013) Role of epigenetics in inflammation-associated diseases. Subcell. Biochem. 61, 627–657 10.1007/978-94-007-4525-4_27 [DOI] [PubMed] [Google Scholar]
- 12.Shanmugam M.K., et al. (2018) Role of novel histone modifications in cancer. Oncotarget 9, 11414–11426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mehta A., et al. (2014) Sources of DNA double-strand breaks and models of recombinational DNA repair. Cold Spring Harb. Perspect. Biol. 6, a016428 10.1101/cshperspect.a016428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Turgeon M.O., et al. (2018) DNA damage, repair, and cancer metabolism. Front. Oncol. 8, 15 10.3389/fonc.2018.00015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aparicio T., et al. (2014) DNA double-strand break repair pathway choice and cancer. DNA Repair (Amst.) 19, 169–175 10.1016/j.dnarep.2014.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martin L.P., et al. (2008) Platinum resistance: the role of DNA repair pathways. Clin. Cancer Res. 14, 1291–1295 10.1158/1078-0432.CCR-07-2238 [DOI] [PubMed] [Google Scholar]
- 17.Sharma R.A., et al. (2007) Targeting base excision repair to improve cancer therapies. Mol. Aspects Med. 28, 345–374 10.1016/j.mam.2007.06.002 [DOI] [PubMed] [Google Scholar]
- 18.Cao J., et al. (2012) Histone ubiquitination and deubiquitination in transcription, DNA damage response, and cancer. Front. Oncol. 2, 26 10.3389/fonc.2012.00026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deshaies R.J., et al. (2009) RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 78, 399–434 10.1146/annurev.biochem.78.101807.093809 [DOI] [PubMed] [Google Scholar]
- 20.Kato A., et al. (2015) RNF20-SNF2H pathway of chromatin relaxation in DNA double-strand break repair. Genes (Basel) 6, 592–606 10.3390/genes6030592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakamura K., et al. (2011) Regulation of homologous recombination by RNF20-dependent H2B ubiquitination. Mol. Cell 41, 515–528 10.1016/j.molcel.2011.02.002 [DOI] [PubMed] [Google Scholar]
- 22.Metzger M.B., et al. (2014) RING-type E3 ligases: master manipulators of E2 ubiquitin-conjugating enzymes and ubiquitination. Biochim. Biophys. Acta 1843, 47–60 10.1016/j.bbamcr.2013.05.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wood A., et al. (2003) Bre1, an E3 ubiquitin ligase required for recruitment and substrate selection of Rad6 at a promoter. Mol. Cell 11, 267–274 10.1016/S1097-2765(02)00802-X [DOI] [PubMed] [Google Scholar]
- 24.Hwang W.W., et al. (2003) A conserved RING finger protein required for histone H2B monoubiquitination and cell size control. Mol. Cell 11, 261–266 10.1016/S1097-2765(02)00826-2 [DOI] [PubMed] [Google Scholar]
- 25.Kao C.F., et al. (2004) Rad6 plays a role in transcriptional activation through ubiquitylation of histone H2B. Genes Dev. 18, 184–195 10.1101/gad.1149604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moyal L., et al. (2011) Requirement of ATM-dependent monoubiquitylation of histone H2B for timely repair of DNA double-strand breaks. Mol. Cell 41, 529–542 10.1016/j.molcel.2011.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim J., et al. (2009) RAD6-m ediated transcription-coupled H2B ubiquitylation directly stimulates H3K4 methylation in human cells. Cell 137, 459–471 10.1016/j.cell.2009.02.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim J., et al. (2009) Direct Bre1-Paf1 complex interactions and RING finger-independent Bre1-Rad6 interactions mediate histone H2B ubiquitylation in yeast. J. Biol. Chem. 284, 20582–20592 10.1074/jbc.M109.017442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tanny J.C., et al. (2007) Ubiquitylation of histone H2B controls RNA polymerase II transcription elongation independently of histone H3 methylation. Genes Dev. 21, 835–847 10.1101/gad.1516207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tessarz P., et al. (2014) Histone core modifications regulating nucleosome structure and dynamics. Nat. Rev. Mol. Cell Biol. 15, 703–708 10.1038/nrm3890 [DOI] [PubMed] [Google Scholar]
- 31.Van Speybroeck L. (2002) From epigenesis to epigenetics: the case of C. H. Waddington. Ann. N.Y. Acad. Sci. 981, 61–81 10.1111/j.1749-6632.2002.tb04912.x [DOI] [PubMed] [Google Scholar]
- 32.Simo-Riudalbas L., et al. (2015) Targeting the histone orthography of cancer: drugs for writers, erasers and readers. Br. J. Pharmacol. 172, 2716–2732 10.1111/bph.12844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baylin S.B., et al. (2016) Epigenetic determinants of cancer. Cold Spring Harb. Perspect. Biol. 8, pii: a019505 10.1101/cshperspect.a019505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jones P.A., et al. (2016) Targeting the cancer epigenome for therapy. Nat. Rev. Genet. 17, 630–641 10.1038/nrg.2016.93 [DOI] [PubMed] [Google Scholar]
- 35.Venkatesh S., et al. (2015) Histone exchange, chromatin structure and the regulation of transcription. Nat. Rev. Mol. Cell Biol. 16, 178–189 10.1038/nrm3941 [DOI] [PubMed] [Google Scholar]
- 36.Easwaran H., et al. (2014) Cancer epigenetics: tumor heterogeneity, plasticity of stem-like states, and drug resistance. Mol. Cell 54, 716–727 10.1016/j.molcel.2014.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Egger G., et al. (2004) Epigenetics in human disease and prospects for epigenetic therapy. Nature 429, 457–463 10.1038/nature02625 [DOI] [PubMed] [Google Scholar]
- 38.Esteller M. (2008) Epigenetics in cancer. N. Engl. J. Med. 358, 1148–1159 10.1056/NEJMra072067 [DOI] [PubMed] [Google Scholar]
- 39.Simo-Riudalbas L., et al. (2014) Cancer genomics identifies disrupted epigenetic genes. Hum. Genet. 133, 713–725 10.1007/s00439-013-1373-5 [DOI] [PubMed] [Google Scholar]
- 40.Villamil M.A., et al. (2013) The WD40-repeat protein-containing deubiquitinase complex: catalysis, regulation, and potential for therapeutic intervention. Cell Biochem. Biophys. 67, 111–126 10.1007/s12013-013-9637-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Blumenfeld N., et al. (1994) Purification and characterization of a novel species of ubiquitin-carrier protein, E2, that is involved in degradation of non-“N-end rule” protein substrates. J. Biol. Chem. 269, 9574–9581 [PubMed] [Google Scholar]
- 42.Ciechanover A., et al. (1980) ATP-dependent conjugation of reticulocyte proteins with the polypeptide required for protein degradation. Proc. Natl. Acad. Sci. U.S.A. 77, 1365–1368 10.1073/pnas.77.3.1365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thompson L.L., et al. (2013) Regulation of chromatin structure via histone post-translational modification and the link to carcinogenesis. Cancer Metastasis Rev. 32, 363–376 10.1007/s10555-013-9434-8 [DOI] [PubMed] [Google Scholar]
- 44.Shiloh Y., et al. (2011) RNF20-RNF40: a ubiquitin-driven link between gene expression and the DNA damage response. FEBS Lett. 585, 2795–2802 10.1016/j.febslet.2011.07.034 [DOI] [PubMed] [Google Scholar]
- 45.Fuchs G., et al. (2014) Writing and reading H2B monoubiquitylation. Biochim. Biophys. Acta 1839, 694–701 10.1016/j.bbagrm.2014.01.002 [DOI] [PubMed] [Google Scholar]
- 46.West M.H., et al. (1980) Histone 2B can be modified by the attachment of ubiquitin. Nucleic Acids Res. 8, 4671–4680 10.1093/nar/8.20.4671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhu P., et al. (2007) A histone H2A deubiquitinase complex coordinating histone acetylation and H1 dissociation in transcriptional regulation. Mol. Cell 27, 609–621 10.1016/j.molcel.2007.07.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li B., et al. (2007) The role of chromatin during transcription. Cell 128, 707–719 10.1016/j.cell.2007.01.015 [DOI] [PubMed] [Google Scholar]
- 49.Belotserkovskaya R., et al. (2003) FACT facilitates transcription-dependent nucleosome alteration. Science 301, 1090–1093 10.1126/science.1085703 [DOI] [PubMed] [Google Scholar]
- 50.Zhang X.Y., et al. (2008) The putative cancer stem cell marker USP22 is a subunit of the human SAGA complex required for activated transcription and cell-cycle progression. Mol. Cell 29, 102–111 10.1016/j.molcel.2007.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang Z., et al. (2013) USP49 deubiquitinates histone H2B and regulates cotranscriptional pre-mRNA splicing. Genes Dev. 27, 1581–1595 10.1101/gad.211037.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nicassio F., et al. (2007) Human USP3 is a chromatin modifier required for S phase progression and genome stability. Curr. Biol. 17, 1972–1977 10.1016/j.cub.2007.10.034 [DOI] [PubMed] [Google Scholar]
- 53.van der Knaap J.A., et al. (2005) GMP synthetase stimulates histone H2B deubiquitylation by the epigenetic silencer USP7. Mol. Cell 17, 695–707 10.1016/j.molcel.2005.02.013 [DOI] [PubMed] [Google Scholar]
- 54.Joo H.Y., et al. (2011) Regulation of histone H2A and H2B deubiquitination and Xenopus development by USP12 and USP46. J. Biol. Chem. 286, 7190–7201 10.1074/jbc.M110.158311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fuchs G., et al. (2012) RNF20 and USP44 regulate stem cell differentiation by modulating H2B monoubiquitylation. Mol. Cell 46, 662–673 10.1016/j.molcel.2012.05.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Davie J.R., et al. (1990) Level of ubiquitinated histone H2B in chromatin is coupled to ongoing transcription. Biochemistry 29, 4752–4757 10.1021/bi00472a002 [DOI] [PubMed] [Google Scholar]
- 57.Davie J.R., et al. (1994) Inhibition of transcription selectively reduces the level of ubiquitinated histone H2B in chromatin. Biochem. Biophys. Res. Commun. 203, 344–350 10.1006/bbrc.1994.2188 [DOI] [PubMed] [Google Scholar]
- 58.Fierz B., et al. (2011) Histone H2B ubiquitylation disrupts local and higher-order chromatin compaction. Nat. Chem. Biol. 7, 113–119 10.1038/nchembio.501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Johnsen S.A. (2012) The enigmatic role of H2Bub1 in cancer. FEBS Lett. 586, 1592–1601 10.1016/j.febslet.2012.04.002 [DOI] [PubMed] [Google Scholar]
- 60.Prenzel T., et al. (2011) Estrogen-dependent gene transcription in human breast cancer cells relies upon proteasome-dependent monoubiquitination of histone H2B. Cancer Res. 71, 5739–5753 10.1158/0008-5472.CAN-11-1896 [DOI] [PubMed] [Google Scholar]
- 61.Hahn M.A., et al. (2012) The tumor suppressor CDC73 interacts with the ring finger proteins RNF20 and RNF40 and is required for the maintenance of histone 2B monoubiquitination. Hum. Mol. Genet. 21, 559–568 10.1093/hmg/ddr490 [DOI] [PubMed] [Google Scholar]
- 62.Zhao Y., et al. (2008) A TFTC/STAGA module mediates histone H2A and H2B deubiquitination, coactivates nuclear receptors, and counteracts heterochromatin silencing. Mol. Cell 29, 92–101 10.1016/j.molcel.2007.12.011 [DOI] [PubMed] [Google Scholar]
- 63.Zhang Y., et al. (2011) Elevated expression of USP22 in correlation with poor prognosis in patients with invasive breast cancer. J. Cancer Res. Clin. Oncol. 137, 1245–1253 10.1007/s00432-011-0998-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu Y.L., et al. (2010) Increased expression of ubiquitin-specific protease 22 can promote cancer progression and predict therapy failure in human colorectal cancer. J. Gastroenterol. Hepatol. 25, 1800–1805 10.1111/j.1440-1746.2010.06352.x [DOI] [PubMed] [Google Scholar]
- 65.Aggarwal B.B., et al. (2009) Inflammation and cancer: how friendly is the relationship for cancer patients? Curr. Opin. Pharmacol. 9, 351–369 10.1016/j.coph.2009.06.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Aggarwal B.B., et al. (2006) Inflammation and cancer: how hot is the link. Biochem. Pharmacol. 72, 1605–1621 10.1016/j.bcp.2006.06.029 [DOI] [PubMed] [Google Scholar]
- 67.Balkwill F., et al. (2001) Inflammation and cancer: back to Virchow? Lancet 357, 539–545 10.1016/S0140-6736(00)04046-0 [DOI] [PubMed] [Google Scholar]
- 68.Balkwill F., et al. (2010) Cancer and inflammation: implications for pharmacology and therapeutics. Clin. Pharmacol. Ther. 87, 401–406 10.1038/clpt.2009.312 [DOI] [PubMed] [Google Scholar]
- 69.Mantovani A., et al. (2008) Cancer-related inflammation. Nature 454, 436–444 10.1038/nature07205 [DOI] [PubMed] [Google Scholar]
- 70.Sethi G., et al. (2012) Multifaceted link between cancer and inflammation. Biosci. Rep. 32, 1–15 10.1042/BSR20100136 [DOI] [PubMed] [Google Scholar]
- 71.Sethi G., et al. (2009) Potential pharmacological control of the NF-kappaB pathway. Trends Pharmacol. Sci. 30, 313–321 10.1016/j.tips.2009.03.004 [DOI] [PubMed] [Google Scholar]
- 72.Shanmugam M.K., et al. (2016) Cancer prevention and therapy through the modulation of transcription factors by bioactive natural compounds Semin. Cancer Biol., 40–41 10.1016/j.semcancer.2016.03.005 [DOI] [PubMed] [Google Scholar]
- 73.Tarcic O., et al. (2016) RNF20 links histone h2b ubiquitylation with inflammation and inflammation-associated. Cell Rep. 14, 1462–1476 10.1016/j.celrep.2016.01.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mantovani A. (2009) Cancer: inflaming metastasis. Nature 457, 36–37 10.1038/457036b [DOI] [PubMed] [Google Scholar]
- 75.Demaria S., et al. (2010) Cancer and inflammation: promise for biologic therapy. J. Immunother. 33, 335–351 10.1097/CJI.0b013e3181d32e74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Aggarwal B.B., et al. (2009) Targeting inflammatory pathways for prevention and therapy of cancer: short-term friend, long-term foe. Clin. Cancer Res. 15, 425–430 10.1158/1078-0432.CCR-08-0149 [DOI] [PubMed] [Google Scholar]
- 77.Tong L., et al. (2014) Rho protein GTPases and their interactions with NFkappaB: crossroads of inflammation and matrix biology. Biosci. Rep. 34, pii: e00115 10.1042/BSR20140021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Liu Y., et al. (1999) Human papillomavirus type 16 E6-enhanced susceptibility of L929 cells to tumor necrosis factor alpha correlates with increased accumulation of reactive oxygen species. J. Biol. Chem. 274, 24819–24827 10.1074/jbc.274.35.24819 [DOI] [PubMed] [Google Scholar]
- 79.Karin M. (2006) Nuclear factor-kappaB in cancer development and progression. Nature 441, 431–436 10.1038/nature04870 [DOI] [PubMed] [Google Scholar]
- 80.Karin M. (2006) NF-kappaB and cancer: mechanisms and targets. Mol. Carcinog. 45, 355–361 10.1002/mc.20217 [DOI] [PubMed] [Google Scholar]
- 81.Shanmugam M.K., et al. (2013) Role of NF-κB in tumorigenesis. Forum Immun. Dis. Ther. 4, 187–203 10.1615/ForumImmunDisTher.2013008382 [DOI] [Google Scholar]
- 82.Sen R., et al. (2006) Multiple nuclear factors interact with the immunoglobulin enhancer sequences. Cell 1986. 46: 705-716. J. Immunol. 177, 7485–7496 [PubMed] [Google Scholar]
- 83.Sen R., et al. (1986) Multiple nuclear factors interact with the immunoglobulin enhancer sequences. Cell 46, 705–716 10.1016/0092-8674(86)90346-6 [DOI] [PubMed] [Google Scholar]
- 84.Singh H., et al. (1986) A nuclear factor that binds to a conserved sequence motif in transcriptional control elements of immunoglobulin genes. Nature 319, 154–158 10.1038/319154a0 [DOI] [PubMed] [Google Scholar]
- 85.Sethi G., et al. (2008) Nuclear factor-kappaB activation: from bench to bedside. Exp. Biol. Med. (Maywood) 233, 21–31 10.3181/0707-MR-196 [DOI] [PubMed] [Google Scholar]
- 86.Sethi G., et al. (2008) TNF: a master switch for inflammation to cancer. Front. Biosci. 13, 5094–5107 10.2741/3066 [DOI] [PubMed] [Google Scholar]
- 87.Lawrence T., et al. (2001) Possible new role for NF-kappaB in the resolution of inflammation. Nat. Med. 7, 1291–1297 10.1038/nm1201-1291 [DOI] [PubMed] [Google Scholar]
- 88.Li F., et al. (2015) NF-kappaB in cancer therapy. Arch. Toxicol. 89, 711–731 10.1007/s00204-015-1470-4 [DOI] [PubMed] [Google Scholar]
- 89.Dey A., et al. (2008) Hexamethylene bisacetamide (HMBA) simultaneously targets AKT and MAPK pathway and represses NF kappaB activity: implications for cancer therapy. Cell Cycle 7, 3759–3767 10.4161/cc.7.23.7213 [DOI] [PubMed] [Google Scholar]
- 90.Tergaonkar V., et al. (2005) Distinct roles of IkappaB proteins in regulating constitutive NF-kappaB activity. Nat. Cell Biol. 7, 921–923 10.1038/ncb1296 [DOI] [PubMed] [Google Scholar]
- 91.Chernikova S.B., et al. (2012) Deficiency in mammalian histone H2B ubiquitin ligase Bre1 (Rnf20/Rnf40) leads to replication stress and chromosomal instability. Cancer Res. 72, 2111–2119 10.1158/0008-5472.CAN-11-2209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Urasaki Y., et al. (2012) Coupling of glucose deprivation with impaired histone H2B monoubiquitination in tumors. PLoS ONE 7, e36775 10.1371/journal.pone.0036775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wang E., et al. (2013) Histone H2B ubiquitin ligase RNF20 is required for MLL-rearranged leukemia. Proc. Natl. Acad. Sci. U.S.A. 110, 3901–3906 10.1073/pnas.1301045110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wang Z.J., et al. (2013) Decreased histone H2B monoubiquitination in malignant gastric carcinoma. World J. Gastroenterol. 19, 8099–8107 10.3748/wjg.v19.i44.8099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bedi U., et al. (2015) SUPT6H controls estrogen receptor activity and cellular differentiation by multiple epigenomic mechanisms. Oncogene 34, 465–473 10.1038/onc.2013.558 [DOI] [PubMed] [Google Scholar]
- 96.Medzhitov R. (2010) Inflammation 2010: new adventures of an old flame. Cell 140, 771–776 10.1016/j.cell.2010.03.006 [DOI] [PubMed] [Google Scholar]
- 97.O’Dea E., et al. (2010) The regulatory logic of the NF-kappaB signaling system. Cold Spring Harb. Perspect. Biol. 2, a000216 10.1101/cshperspect.a000216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Terzic J., et al. (2010) Inflammation and colon cancer. Gastroenterology 138, 2101e2105–2114e2105 10.1053/j.gastro.2010.01.058 [DOI] [PubMed] [Google Scholar]
- 99.Ullman T.A., et al. (2011) Intestinal inflammation and cancer. Gastroenterology 140, 1807–1816 10.1053/j.gastro.2011.01.057 [DOI] [PubMed] [Google Scholar]
- 100.Shema E., et al. (2008) The histone H2B-specific ubiquitin ligase RNF20/hBRE1 acts as a putative tumor suppressor through selective regulation of gene expression. Genes Dev. 22, 2664–2676 10.1101/gad.1703008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Buro L.J., et al. (2010) Menin and RNF20 recruitment is associated with dynamic histone modifications that regulate signal transducer and activator of transcription 1 (STAT1)-activated transcription of the interferon regulatory factor 1 gene (IRF1). Epigenetics Chromatin 3, 16 10.1186/1756-8935-3-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Jaaskelainen T., et al. (2012) Histone H2B ubiquitin ligases RNF20 and RNF40 in androgen signaling and prostate cancer cell growth. Mol. Cell. Endocrinol. 350, 87–98 10.1016/j.mce.2011.11.025 [DOI] [PubMed] [Google Scholar]
- 103.Barber T.D., et al. (2008) Chromatid cohesion defects may underlie chromosome instability in human colorectal cancers. Proc. Natl. Acad. Sci. U.S.A. 105, 3443–3448 10.1073/pnas.0712384105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Torre L.A., et al. (2015) Global cancer statistics, 2012. CA Cancer J. Clin. 65, 87–108 10.3322/caac.21262 [DOI] [PubMed] [Google Scholar]
- 105.Shin E.M., et al. (2014) DEAD-box helicase DP103 defines metastatic potential of human breast cancers. J. Clin. Invest. 124, 3807–3824 10.1172/JCI73451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Schulze J.M., et al. (2009) Linking cell cycle to histone modifications: SBF and H2B monoubiquitination machinery and cell-cycle regulation of H3K79 dimethylation. Mol. Cell 35, 626–641 10.1016/j.molcel.2009.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sun Z.W., et al. (2002) Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature 418, 104–108 10.1038/nature00883 [DOI] [PubMed] [Google Scholar]
- 108.Dover J., et al. (2002) Methylation of histone H3 by COMPASS requires ubiquitination of histone H2B by Rad6. J. Biol. Chem. 277, 28368–28371 10.1074/jbc.C200348200 [DOI] [PubMed] [Google Scholar]
- 109.Duan Y., et al. (2016) Ubiquitin ligase RNF20/40 facilitates spindle assembly and promotes breast carcinogenesis through stabilizing motor protein Eg5. Nat. Commun. 7, 12648 10.1038/ncomms12648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hicke L. (2001) Protein regulation by monoubiquitin. Nat. Rev. Mol. Cell Biol. 2, 195–201 10.1038/35056583 [DOI] [PubMed] [Google Scholar]
- 111.Rodriguez J.E., et al. (2015) The ubiquitin ligase MuRF1 regulates PPARalpha activity in the heart by enhancing nuclear export via monoubiquitination. Mol. Cell. Endocrinol. 413, 36–48 10.1016/j.mce.2015.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Pujol A., et al. (2010) Unveiling the role of network and systems biology in drug discovery. Trends Pharmacol. Sci. 31, 115–123 10.1016/j.tips.2009.11.006 [DOI] [PubMed] [Google Scholar]
- 113.Pujana M.A., et al. (2007) Network modeling links breast cancer susceptibility and centrosome dysfunction. Nat. Genet. 39, 1338–1349 10.1038/ng.2007.2 [DOI] [PubMed] [Google Scholar]
- 114.Stratton M.R., et al. (2009) The cancer genome. Nature 458, 719–724 10.1038/nature07943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Schadt E.E. (2009) Molecular networks as sensors and drivers of common human diseases. Nature 461, 218–223 10.1038/nature08454 [DOI] [PubMed] [Google Scholar]
- 116.Arroyo R., et al. (2015) Systematic identification of molecular links between core and candidate genes in breast cancer. J. Mol. Biol. 427, 1436–1450 10.1016/j.jmb.2015.01.014 [DOI] [PubMed] [Google Scholar]
- 117.Spolverini A., et al. (2017) let-7b and let-7c microRNAs promote histone H2B ubiquitylation and inhibit cell migration by targeting multiple components of the H2B deubiquitylation machinery. Oncogene 36, 5819–5828 10.1038/onc.2017.187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Krivtsov A.V., et al. (2007) MLL translocations, histone modifications and leukaemia stem-cell development. Nat. Rev. Cancer 7, 823–833 10.1038/nrc2253 [DOI] [PubMed] [Google Scholar]
- 119.van der Linden M.H., et al. (2016) Clinical and molecular genetic characterization of wild-type MLL infant acute lymphoblastic leukemia identifies few recurrent abnormalities. Haematologica 101, e95–e98 10.3324/haematol.2014.122119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Garrido Castro P., et al. (2018) The HDAC inhibitor panobinostat (LBH589) exerts in vivo anti-leukaemic activity against MLL-rearranged acute lymphoblastic leukaemia and involves the RNF20/RNF40/WAC-H2B ubiquitination axis. Leukemia 32, 323–331 10.1038/leu.2017.216 [DOI] [PubMed] [Google Scholar]
- 121.Ito T., et al. (2010) Identification of a primary target of thalidomide teratogenicity. Science 327, 1345–1350 10.1126/science.1177319 [DOI] [PubMed] [Google Scholar]
- 122.Vassilev L.T., et al. (2004) In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science 303, 844–848 10.1126/science.1092472 [DOI] [PubMed] [Google Scholar]
- 123.Attard G., et al. (2016) Prostate cancer. Lancet 387, 70–82 10.1016/S0140-6736(14)61947-4 [DOI] [PubMed] [Google Scholar]
- 124.Berezovska O.P., et al. (2006) Essential role for activation of the Polycomb group (PcG) protein chromatin silencing pathway in metastatic prostate cancer. Cell Cycle 5, 1886–1901 10.4161/cc.5.16.3222 [DOI] [PubMed] [Google Scholar]
- 125.Ferlay J., et al. (2010) Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int. J. Cancer 127, 2893–2917 10.1002/ijc.25516 [DOI] [PubMed] [Google Scholar]
- 126.Siegel R., et al. (2014) Cancer statistics, 2014. CA Cancer J. Clin. 64, 9–29 10.3322/caac.21208 [DOI] [PubMed] [Google Scholar]
- 127.Zhang K., et al. (2017) Loss of H2B monoubiquitination is associated with poor-differentiation and enhanced malignancy of lung adenocarcinoma. Int. J. Cancer 141, 766–777 10.1002/ijc.30769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Rezende R.B., et al. (1999) Differential diagnosis between monomorphic clear cell adenocarcinoma of salivary glands and renal (clear) cell carcinoma. Am. J. Surg. Pathol. 23, 1532–1538 10.1097/00000478-199912000-00011 [DOI] [PubMed] [Google Scholar]
- 129.Valera V.A., et al. (2011) Misdiagnosis of clear cell renal cell carcinoma. Nat. Rev. Urol. 8, 321–333 10.1038/nrurol.2011.64 [DOI] [PubMed] [Google Scholar]
- 130.Menendez J.A., et al. (2007) Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nat. Rev. Cancer 7, 763–777 10.1038/nrc2222 [DOI] [PubMed] [Google Scholar]
- 131.Kuhajda F.P. (2000) Fatty-acid synthase and human cancer: new perspectives on its role in tumor biology. Nutrition 16, 202–208 10.1016/S0899-9007(99)00266-X [DOI] [PubMed] [Google Scholar]
- 132.Medes G., et al. (1953) Metabolism of neoplastic tissue. IV. A study of lipid synthesis in neoplastic tissue slices in vitro. Cancer Res. 13, 27–29 [PubMed] [Google Scholar]
- 133.Ookhtens M., et al. (1984) Liver and adipose tissue contributions to newly formed fatty acids in an ascites tumor. Am. J. Physiol. 247, R146–R153 [DOI] [PubMed] [Google Scholar]
- 134.Krycer J.R., et al. (2010) The Akt-SREBP nexus: cell signaling meets lipid metabolism. Trends Endocrinol. Metab. 21, 268–276 10.1016/j.tem.2010.01.001 [DOI] [PubMed] [Google Scholar]
- 135.Wang X., et al. (1993) Nuclear protein that binds sterol regulatory element of low density lipoprotein receptor promoter. II. Purification and characterization. J. Biol. Chem. 268, 14497–14504 [PubMed] [Google Scholar]
- 136.Briggs M.R., et al. (1993) Nuclear protein that binds sterol regulatory element of low density lipoprotein receptor promoter. I. Identification of the protein and delineation of its target nucleotide sequence. J. Biol. Chem. 268, 14490–14496 [PubMed] [Google Scholar]
- 137.Beaven S.W. (2014) La mort de la lipogenese: RNF20 lashes ubiquitin to SREBP-1c. Hepatology 60, 776–778 10.1002/hep.27112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Tontonoz P., et al. (1993) ADD1: a novel helix-loop-helix transcription factor associated with adipocyte determination and differentiation. Mol. Cell. Biol. 13, 4753–4759 10.1128/MCB.13.8.4753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Guo D., et al. (2011) An LXR agonist promotes glioblastoma cell death through inhibition of an EGFR/AKT/SREBP-1/LDLR-dependent pathway. Cancer Discov. 1, 442–456 10.1158/2159-8290.CD-11-0102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Shao W., et al. (2012) Expanding roles for SREBP in metabolism. Cell Metab. 16, 414–419 10.1016/j.cmet.2012.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Lee J.H., et al. (2014) Ring finger protein20 regulates hepatic lipid metabolism through protein kinase A-dependent sterol regulatory element binding protein1c degradation. Hepatology 60, 844–857 10.1002/hep.27011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Lee J.H., et al. (2017) RNF20 suppresses tumorigenesis by inhibiting SREBP1c-PTTG1 axis in kidney cancer. Mol. Cell. Biol., 10.1128/MCB.00265-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Freed-Pastor W.A., et al. (2012) Mutant p53 disrupts mammary tissue architecture via the mevalonate pathway. Cell 148, 244–258 10.1016/j.cell.2011.12.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Berkers C.R., et al. (2013) Metabolic regulation by p53 family members. Cell Metab. 18, 617–633 10.1016/j.cmet.2013.06.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Dickson K.A., et al. (2016) The RING finger domain E3 ubiquitin ligases BRCA1 and the RNF20/RNF40 complex in global loss of the chromatin mark histone H2B monoubiquitination (H2Bub1) in cell line models and primary high-grade serous ovarian cancer. Hum. Mol. Genet. 25, 5460–5471 [DOI] [PubMed] [Google Scholar]
- 146.Shema E., et al. (2011) RNF20 inhibits TFIIS-facilitated transcriptional elongation to suppress pro-oncogenic gene expression. Mol. Cell 42, 477–488 10.1016/j.molcel.2011.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Cerami E., et al. (2012) The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401–404 10.1158/2159-8290.CD-12-0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Guppy B.J., et al. (2017) Synthetic lethal targeting of RNF20 through PARP1 silencing and inhibition. Cell Oncol. (Dordr.) 40, 281–292 10.1007/s13402-017-0323-y [DOI] [PubMed] [Google Scholar]
- 149.Jackson S.P., et al. (2009) The DNA-damage response in human biology and disease. Nature 461, 1071–1078 10.1038/nature08467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Ciccia A., et al. (2010) The DNA damage response: making it safe to play with knives. Mol. Cell 40, 179–204 10.1016/j.molcel.2010.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Giglia-Mari G., et al. (2011) DNA damage response. Cold Spring Harb. Perspect. Biol. 3, a000745 10.1101/cshperspect.a000745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Hartlerode A.J., et al. (2009) Mechanisms of double-strand break repair in somatic mammalian cells. Biochem. J. 423, 157–168 10.1042/BJ20090942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Hiom K. (2010) Coping with DNA double strand breaks. DNA Repair (Amst.) 9, 1256–1263 10.1016/j.dnarep.2010.09.018 [DOI] [PubMed] [Google Scholar]
- 154.Lieber M.R. (2010) The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu. Rev. Biochem. 79, 181–211 10.1146/annurev.biochem.052308.093131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Holthausen J.T., et al. (2010) Regulation of DNA strand exchange in homologous recombination. DNA Repair (Amst.) 9, 1264–1272 10.1016/j.dnarep.2010.09.014 [DOI] [PubMed] [Google Scholar]
- 156.Wyman C., et al. (2006) DNA double-strand break repair: all’s well that ends well. Annu. Rev. Genet. 40, 363–383 10.1146/annurev.genet.40.110405.090451 [DOI] [PubMed] [Google Scholar]
- 157.Bekker-Jensen S., et al. (2011) The ubiquitin- and SUMO-dependent signaling response to DNA double-strand breaks. FEBS Lett. 585, 2914–2919 10.1016/j.febslet.2011.05.056 [DOI] [PubMed] [Google Scholar]
- 158.van Attikum H., et al. (2009) Crosstalk between histone modifications during the DNA damage response. Trends Cell Biol. 19, 207–217 10.1016/j.tcb.2009.03.001 [DOI] [PubMed] [Google Scholar]
- 159.Corpet A., et al. (2009) A histone code for the DNA damage response in mammalian cells? EMBO J. 28, 1828–1830 10.1038/emboj.2009.180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Falk M., et al. (2010) Higher-order chromatin structure in DSB induction, repair and misrepair. Mutat. Res. 704, 88–100 10.1016/j.mrrev.2010.01.013 [DOI] [PubMed] [Google Scholar]
- 161.Tauchi H., et al. (2002) Nbs1 is essential for DNA repair by homologous recombination in higher vertebrate cells. Nature 420, 93–98 10.1038/nature01125 [DOI] [PubMed] [Google Scholar]
- 162.Xie A., et al. (2009) Role of mammalian Mre11 in classical and alternative nonhomologous end joining. Nat. Struct. Mol. Biol. 16, 814–818 10.1038/nsmb.1640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Minsky N., et al. (2008) Monoubiquitinated H2B is associated with the transcribed region of highly expressed genes in human cells. Nat. Cell Biol. 10, 483–488 10.1038/ncb1712 [DOI] [PubMed] [Google Scholar]
- 164.Oliveira D.V., et al. (2014) Histone chaperone FACT regulates homologous recombination by chromatin remodeling through interaction with RNF20. J. Cell Sci. 127, 763–772 10.1242/jcs.135855 [DOI] [PubMed] [Google Scholar]
- 165.Smeenk G., et al. (2013) Poly(ADP-ribosyl)ation links the chromatin remodeler SMARCA5/SNF2H to RNF168-dependent DNA damage signaling. J. Cell Sci. 126, 889–903 10.1242/jcs.109413 [DOI] [PubMed] [Google Scholar]
- 166.Toiber D., et al. (2013) SIRT6 recruits SNF2H to DNA break sites, preventing genomic instability through chromatin remodeling. Mol. Cell 51, 454–468 10.1016/j.molcel.2013.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Klement K., et al. (2014) Opposing ISWI- and CHD-class chromatin remodeling activities orchestrate heterochromatic DNA repair. J. Cell Biol. 207, 717–733 10.1083/jcb.201405077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Carter R.J., et al. (2018) Complex DNA damage induced by high linear energy transfer alpha-particles and protons triggers a specific cellular DNA damage response. Int. J. Radiat. Oncol. Biol. Phys. 100, 776–784 10.1016/j.ijrobp.2017.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Ohta T., et al. (2011) The BRCA1 ubiquitin ligase and homologous recombination repair. FEBS Lett. 585, 2836–2844 10.1016/j.febslet.2011.05.005 [DOI] [PubMed] [Google Scholar]
- 170.Sajesh B.V., et al. (2013) Synthetic genetic targeting of genome instability in cancer. Cancers (Basel) 5, 739–761 10.3390/cancers5030739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Ciriello G., et al. (2015) Comprehensive molecular portraits of invasive lobular breast cancer. Cell 163, 506–519 10.1016/j.cell.2015.09.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Cancer Genome Atlas Network (2012) Comprehensive molecular portraits of human breast tumours. Nature 490, 61–70 10.1038/nature11412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Cancer Genome Atlas Research Network (2011) Integrated genomic analyses of ovarian carcinoma. Nature 474, 609–615 10.1038/nature10166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Cancer Genome Atlas Research Network (2014) Comprehensive molecular profiling of lung adenocarcinoma. Nature 511, 543–550 10.1038/nature13385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Cancer Genome Atlas Network (2012) Comprehensive molecular characterization of human colon and rectal cancer. Nature 487, 330–337 10.1038/nature11252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Cancer Genome Atlas Research Network (2013) Integrated genomic characterization of endometrial carcinoma. Nature 497, 67–73 10.1038/nature12113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Taylor B.S., et al. (2010) Integrative genomic profiling of human prostate cancer. Cancer Cell 18, 11–22 10.1016/j.ccr.2010.05.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Marzluff W.F., et al. (2002) The human and mouse replication-dependent histone genes. Genomics 80, 487–498 10.1006/geno.2002.6850 [DOI] [PubMed] [Google Scholar]
- 179.Hsu P.D., et al. (2014) Development and applications of CRISPR-Cas9 for genome engineering. Cell 157, 1262–1278 10.1016/j.cell.2014.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Wright D.A., et al. (2014) TALEN-mediated genome editing: prospects and perspectives. Biochem. J. 462, 15–24 10.1042/BJ20140295 [DOI] [PubMed] [Google Scholar]
- 181.Gaj T., et al. (2013) ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 31, 397–405 10.1016/j.tibtech.2013.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Akincilar S.C., et al. (2016) Long-range chromatin interactions drive mutant TERT promoter activation. Cancer Discov. 6, 1276–1291 10.1158/2159-8290.CD-16-0177 [DOI] [PubMed] [Google Scholar]
- 183.Imielinski M., et al. (2012) Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell 150, 1107–1120 10.1016/j.cell.2012.08.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Robinson D., et al. (2015) Integrative clinical genomics of advanced prostate cancer. Cell 161, 1215–1228 10.1016/j.cell.2015.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Witkiewicz A.K., et al. (2015) Whole-exome sequencing of pancreatic cancer defines genetic diversity and therapeutic targets. Nat. Commun. 6, 6744 10.1038/ncomms7744 [DOI] [PMC free article] [PubMed] [Google Scholar]