Abstract
Lim mineralization protein-3 (LMP3) induces osteoblast differentiation by regulating the expression and activity of certain molecules involved in the osteogenic cascade, including those belonging to the bone morphogenetic protein (BMP) family. The complete network of molecular events involved in LMP3-mediated osteogenesis is still unknown. The aim of this study was to analyze the genome-wide gene expression profiles in human mesenchymal stem cells (hMSC) induced by exogenous LMP3 to mediate osteogenesis. For this purpose hMSC were transduced with a defective adenoviral vector expressing the human LMP3 gene and microarray analysis was performed 1 day post-adenoviral transduction. Cells transduced with the vector backbone and untransduced cells were used as independent controls in the experiments. Microarray data were independently validated by means of real-time PCR on selected transcripts. The statistical analysis of microarray data produced a list of 263 significantly (p < 0.01) differentially expressed transcripts. The biological interpretation of the results indicated, among the most noteworthy effects, the modulation of genes involved in the TGF-β1 pathway: 88 genes coding for key regulators of the cell cycle regulatory machinery and 28 genes implicated in the regulation of cell proliferation along with the development of connective, muscular, and skeletal tissues. These results suggested that LMP3 could affect the fine balance between cell proliferation/differentiation of mesenchymal cells mostly by modulating the TGF-β1 signaling pathway.
Key words: Lim mineralization protein-3 (LMP3), Microarray, Osteogenesis, Mesechymal cells, Gene expression
INTRODUCTION
Lim mineralization protein (LMP) is an intracellular osteoinductive molecule originally demonstrated to be expressed in calvarial cells upon glucocorticoid-induced osteoblastogenesis (2). Three different splice variants are transcribed from the human LMP gene, termed LMP1, LMP2, and LMP3 (18). All the LMP splice variants were detected among the osteoinductive growth factors expressed in an age-related fashion in the iliac crest bone, and LMP1 expression increased during dental pulp cells’ differentiation and nodule mineralization (5,48). LMP1 is the longest isoform (457 amino acid protein) and contains highly conserved LIM and PDZ protein interaction domains plus a nonconserved, unique region. LMP3 is the shortest splice variant, originating from a 17-bp insertion, producing a reading frame shift that introduces a stop codon. The resulting protein contains 153 amino acids, with an intact PDZ domain, only part of the unique region, and no LIM domains (28,42). Both LMP1 and LMP3 are known to play a critical role in BMP-mediated osteogenic cascade and have been largely employed in gene therapy approaches (16,18, 28). Both the isoforms were able to induce osteogenic differentiation of mesenchymal cells, expression of osteoblast markers gene and bone formation/regeneration in vivo (16,18,21,28,35,36,42,45).
Human mesenchymal stem cells (hMSC) isolated from bone marrow are the main source of osteoblast progenitors in vivo and have been widely used for cell-based therapy as a convenient cell culture model for the study of the osteogenic process as, under appropriate conditions, they differentiate in vitro into osteoblasts (7,9,26). The overexpression of exogenous human LMP1 and LMP3 genes induced osteogenic differentiation of hMSC in vitro and bone formation in different animal models (3,28,46). In addition, LMP3 induces successful osteogenic differentiation of rat and murine dermal fibroblasts, which proved to be effective in forming bone when implanted in a clinically relevant model of bone defects (16,25). The mechanism through which LMP3 induces osteogenesis is still poorly defined, but appeared to be mediated through the activation of BMPs and other key osteogenic marker genes. These include runt-related transcription factor 2 (RUNX2) and osterix (OSX), which were upregulated not earlier than 48 h upon LMP3 expression in hMCSs (21,28,45). In addition, a region in LMP1 and LMP3 has been demonstrated to interact with the Smad ubiquitination regulatory factor 1 (Smurf1), preventing ubiquitin-mediated degradation of the RunX2 and Smad proteins and mediating the initial steps of the osteogenic cascade (30). These results suggest that LMP3 could act as an upstream osteogenic factor, possibly acting on diverse aspects of cell homeostasis. To understand the molecular events associated with the initial steps of osteoblast differentiation induced by LMP, microarrays analysis was performed on hMSC transduced with an adenoviral vector carrying LMP3. The results suggest that LMP3 regulates the expression of genes important for proliferation and differentiation of mesenchymal cells, in particular, genes encoding members of the TGF-β1 signaling pathway, which are essential mediators of the musculoskeletal developmental process.
MATERIALS AND METHODS
hMSC Isolation and Culture
Human bone marrow stromal cells (human mesenchymal stem cells, hMSC) were purchased from Lonza (Basel, Switzerland) and had the following characteristics. They were obtained by human bone marrow specimens withdrawn from the posterior iliac crest of the pelvic bone of normal volunteers (healthy males and nonpregnant females between the ages of 18 and 45 years old); the cell population was tested as positive for CD105, CD166, CD29, and CD44, negative for CD14, CD34 and CD45, by flow cytometry. All cell culture media, serum, and supplements were purchased from Lonza unless otherwise specified. Cells were seeded using a seeding density of 5,000–6,000 cells/cm2 and then expanded in culture up to four culture passages for the following molecular analyses, as suggested by the manufacturer. A longer culture period was then used to assess the growth kinetics characteristics of the cells up to 20 passages.
Growth Kinetics
The proliferation rate of hMSC was assessed using the trypan blue exclusion assay to check cell viability as previously described (31). Both stained (dead) and unstained (viable) cells were counted in a counting chamber and the cell viability was calculated according to the following formula: % cell viability = number of unstained cells/total number of cells (unstained plus stained) × 100. Assays were performed in three independent experiments.
Adenoviral-Mediated Transduction
E1- and E3-deleted adenoviral vectors expressing the human LMP3 gene (AdLMP3) was constructed as previously described (28). In particular, LMP3 transgene was constructed using a full-length LMP3 cDNA, optimized for efficient translation, being 80% identical at the genomic level and 100% identical at the amino acid level (28). AdEGFP was used as a vector control for testing transduction efficiency whereas an empty ψ5 helper virus (Adψ5), the vector backbone, was used as an additional negative control for transduction. hMSC at the fourth culture passage were plated at 5 × 104 cells/cm2 density and grown until they reached 80% confluence. Cells were then infected with either AdLMP3 or AdEGFP or Adψ5 using an MOI of 100 plaque forming units (pfu) per cell. Untransduced cells served as additional negative controls. The efficiency of adenovirus-mediated transduction was then calculated by both evaluating the number of AdEGFP-transduced cells under fluorescence microscopy and quantifying transgenic LMP3 expression in AdLMP3-transduced cells by means of real-time PCR (see following paragraph). Moreover, the growth kinetics of hMSC was monitored up to 7 days postinfection in order to assess the effects of the osteogenic molecule on the cell viability and cycling.
RNA Isolation
The expression level of both endogenous LMP and transgenic LMP3 was assessed in nontransduced and adenoviral-transduced cells, respectively. For this purpose, total RNA was isolated from AdLMP3- and Adψ5-transduced and untransduced cells using RNeasy Mini Kit (Qiagen Inc., Valencia, CA) according to the manufacturer’s instructions. In order to avoid genomic DNA in samples, RNA was digested with amplification grade DNase I (Qiagen). The yield of RNA isolation was determined using spectrophotometry (Beckman DU800, Beckman Coulter, Inc., Fullerton, CA). The quality and integrity of total RNA were assessed using the Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA). Purified RNA (1 μg) was then reverse-transcribed using SuperScript™ First-Strand Synthesis System (Invitrogen, Carlsbad, CA) with random hexamers according to the manufacturer’s suggested procedures. The cDNA served as a template for PCRs in the following experimental steps. All the oligonucleotide primers used in this study were synthesized by PRIMM Srl (Milano, Italy).
Endogenous LMP Gene Expression
The endogenous LMP expression pattern in hMSC was assessed under basal and osteogenic conditions. For this purpose, untransduced cells were cultured for 0, 2, and 4 days in osteogenic medium (see following paragraphs for method details) and the LMP coding sequence was amplified by means of RT-PCR, using 2 μl of the template cDNA. The sequence-specific oligonucleotide primers were designed to amplify the three splicing variants of the gene, as previously described (5). The PCR products were separated on 3% agarose gel (Bio-Rad, Hercules, CA) stained with ethidium bromide (Sigma-Aldrich, St. Louis, MO). The glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene was amplified using the following oligonucleotide primers, and served as housekeeping gene in the PCR reaction: GAPDH_F 5′-TGGAAGATGGTGA TGGGATT-3′ and GAPDH_R 5′-GAGTCAACGGA TTTGGTCGT-3′.
Transgenic LMP3 Expresssion
The expression of the transgenic hLMP3 was assessed by qPCR in AdLMP3- and Adψ5-transduced cells 24 and 48 h after infection. For this purpose, 20 ng of the single-stranded cDNA, synthesized as previously described, was used for real-time PCR, performed in a reaction volume of 20 μl using the SYBR green PCR master mix (Applied Biosystem, Foster City, CA) and 1 μM of LMP sequence-specific primers. Target genes were normalized to the reference housekeeping 18S ribosomal RNA gene. The sequences of the oligonucleotide primers were designed, using Primer 3 software (http://primer3.sourceforge.net/), to selectively amplify the transgenic codon-optimized LMP3 delivered by the adenoviral vector, based on the base substitutions introduced in the genomic sequence during the codon optimization (28). The primer sequences were as follows: forward primer 5′-CTGTCCCTCGGTCTTAG TCG-3′ and reverse primer 5′-GCCTTCTGAGGTT TGGACTG-3′. The analysis was performed on an ABI Prism 7900 Sequence Detection System (Applied Biosystems, Foster City, CA). The PCR conditions were as follows: an initial incubation of 50°C for 2 min and 95°C for 10 min followed by 40 cycles of 94°C for 15 s and 60°C for 1 min. Standard curves were generated for all the assays to verify PCR efficiency. The threshold cycle (CT), which correlates inversely with the levels of target mRNA, was measured as the cycle number at which the reporter fluorescence emission exceeded a preset threshold level. The amplified transcripts were quantified using the comparative CT method as previously described (7), with the formula for relative fold change = 2−ΔΔCT, where ΔCT = [ΔCT gene of interest (treated sample) −ΔCT 18S rRNA (treated sample)] − [ΔCT gene of interest (control sample) − ΔCT 18S rRNA (control sample)]. ΔCT represents the mean CT value of each sample.
In Vitro Mineralization Assay
The osteogenic capability of LMP3 was measured by evaluating in vitro the mineralization occurring in hMSC 1 and 2 weeks following AdLMP3 transduction by means of alizarin red staining. For this purpose cells were plated in six-well plates and infected with 100 MOI of either AdLMP3 or Adψ5. Culture medium was changed 48 h after infection and cells were then incubated at 37°C in a humidified incubator with 5% CO2 up to 14 days. Confluent cells treated with an osteogenic medium, consisting of Dulbecco’s modified Eagle’s medium supplemented with 10% FBS, 1% penicillin/streptomycin, 2 mM l-glutamine, 100 nM dexamethasone, 50 μM ascorbic acid-2-phosphate, and 10 mM β-glycerophosphate (all purchased from Sigma, St. Louis, MO), served as positive control for osteogenic induction. Cells were fixed at the specified time points in 10% neutral buffered formalin and stained with alizarin red in order to assess the presence of calcium deposits. The experiment was performed in triplicate in order to demonstrate reproducibility.
Microarray Analysis
For microarray analysis fourth passage hMSC were plated in T75 flasks and transduced with 100 MOI of either AdLMP3 or Adψ5, while untransduced cells served as negative controls. All the experiments were carried out in three independent replicates; thus, an overall number of nine samples were analyzed. Thereafter, total RNA was isolated, as described above, from all samples 24 h following adenoviral-mediated transduction and analyzed using the Gene-Chip microarray technology (Affymetrix, Santa Clara, CA, USA), as already described elsewhere (15). Briefly, the syntheses of cDNA and biotinylated cRNA were performed according to the protocols provided by the manufacturer. Biotinylated fragmented cRNA probes were hybridized to the Human Genome Focus Arrays (Affymetrix), which contained probe sets for over 8,700 known transcripts and expressed sequence tags. Hybridization was performed at 45°C for 16 h in a hybridization oven (Affymetrix). The GeneChips were then automatically washed and stained with streptavidin–phycoerythrin conjugate using the GeneChip Fluidics Station (Affymetrix). Fluorescence intensities were scanned with the GeneChip Scanner 3000 (Affymetrix). Hybridizations were carried out independently for each condition using three biological replicates, according to the “minimum information about a microarray experiment” (MIAME) guidelines (4).
Data Analysis
Data analysis were performed using BRB Array-Tools (33). Probe level summaries were generated using Robust Multi-array Average (RMA) analysis. The normalized probe intensity values were then summarized within each probe set, using the median polish technique, to generate a single measure of expression (12). These expression measures were then log transformed, base 2. The complete raw data set obtained by microarray analysis was submitted to the “Gene Expression Omnibus” (GEO) database (http://www.ncbi.nlm.nih.gov/geo/, accession number GSE16614). Quality control on data set was performed using multidimensional scaling (MDS) on all the genes in order to test the segregation efficiency. MDS is a group of methods for representing high-dimensional data graphically in few (usually 2 or 3) dimensions. The objective in MDS is to preserve the pair-wise similarities or distances between objects in the low-dimensional graphical representation. MDS analysis is therefore aimed at examining the relations among samples providing a graphical representation of the pair-wise similarities or distances among samples. The similarity between gene expression patterns was measured by computing a centered correlation and samples were positioned in a three-dimensional space in which samples with gene expression profiles more similar to each other laid closer and formed an aggregation (cluster). In order to compute the probability of genes being differentially expressed between two classes the class comparison procedure was performed. For this purpose, a paired t-test with random variance model (significance threshold of unvariate test, p = 0.01) was used. Cells infected with Adψ5 were defined as control in the comparison procedure as to obtain the gene expression profiles specifically correlated to LMP3 effects. The paired t-test was chosen as appropriate as the comparison was between samples belonging to the same independent experiment (technical replicate), in order to improve the statistical power of the analysis. Moreover, in order to categorize genes modulated as a result of general effects related to the adenoviral transduction procedure, we also compared data from the two control groups (i.e., untransduced cells vs. Adψ-transduced cells). For this purpose this dataset was filtered as previously described and class comparison was performed using a two sample t-test. The resulting gene lists were annotated according to functional roles or biological processes according to the Gene Ontology (GO) Consortium directions (1).
Microarray Functional Analysis and Biological Interpretation
Global functional analysis of the gene list was performed using Ingenuity Pathway Analysis (IPA, http://www.ingenuity.com; Ingenuity® Systems, Redwood City, CA) software to determine whether the list of selected genes were significantly enriched with known gene ontologies. The biological meanings of the gene list were interpreted using Core Analysis. Both up- and downregulated genes were analyzed. The Functional Analysis served to identify the biological functions that were most significant to the data set. Fischer’s exact test was used to calculate a p-value determining the probability that each biological function assigned to that data set is due to chance alone. Smaller p-values indicate a higher level of enrichment in the input gene list of genes involved in the corresponding category/function/function annotation. Custom pathways were then generated using the path designer feature that allowed a graphical representation of the molecular relationships between genes/gene products.
Gene Expression Validation
The gene expression results obtained by means of microarray analysis were validated using quantitative real-time PCR to amplify selected transcripts. For this purpose RNA was isolated and reverse transcribed, as already described, from cells obtained from new experiments using three independent replicate samples for each tested conditions. The following genes were selected for validation, as either being indicative of the main functional groups of modulated transcripts or exerting the highest levels of modulation: cysteine rich transmembrane BMP regulator 1 (CRIM1), calponin 3 (CNN3), LIM and cysteine-rich domains 1 (LMCD1), transforming growth factor-β (TGF-β1), activin A receptor, type 1 (ACVR1), pleckstrin homology-like domain, family A, member 1 (PHLDA1), SMAD family member 3 (SMAD3), and growth arrest-specific 1 (GAS1). The GAPDH gene was amplified by real-time PCR from the same RNA samples and used as a housekeeping control gene. Real time PCR was then carried out following the procedures already specified in previous paragraphs, using the following sequence-specific primers, designed using Primer 3 software (http://primer3.sourceforge.net/): CRIM1_F, 5′-ttggatttgctttcagttggt-3′ and CRTM1_R, 5′-aactgccatcaatgtctctgg-3′; CNN3_F, 5′-atggcgagtaccaggatgac-3′and CNN3_R, 5′-atcaatgc cttggtcgctat-3′; LMCD1_F, 5′-gagaaggagagccaggtgtg-3′ and LMCD1_R, 5′-acttgcttgctgtcacatcg-3′; SMAD3_F, 5′-tgagactgacccaagtgcag-3′ and SMAD3_R, 5′-ggaag gtgctgaagacaagg-3′; TGF-β1_F, 5′-agctccacggagaa gaactg-3′ and TGF-β1_R, 5′-gtccttgcggaagtcaatgt-3′; ACVR1_F, 5′-gctgcgtactccactggtct-3′ and ACVR1_R, 5′-cattttgttttgccaaattga-3′; PHLDA1_F, 5′-ggtcaagct caaggaactgc-3′ and PHLDA1_R, 5′-cacacagtccacggtct tca-3′; GAS1_F, 5′-cggagcttgacttcttggac-3′ and GASl_R, 5′-tttctggagcttggaattgg-3′; GAPDH_F, 5′-tggaaggact catgaccaca-3′ and GAPDH_R, 5′-gtcttctgggtggcagt gat-3′.
RESULTS
Biological Features and Basal LMP Gene Expression
The hMSC showed an average doubling time of 38 ± 5 h for up to six culture passages and a progressive slowdown (e.g., 47 ± 3 h after the 10th passage) up to a drastic arrest after the 15th culture passage. The basal expression of endogenous LMP was then evaluated in hMSC, prior to perform the adenoviral-mediated transduction, using RT-PCR. The oligonucleotide primers employed in the reaction were designed to span the sequence that varies among the three splice variants of the gene (5). LMP1, the more abundant isoform, was stably expressed in undifferentiated cells and following 2 and 4 days of osteogenic induction (Fig. 1). Conversely, the expression of the minor isoform, LMP3, was extremely low in undifferentiated cells and increased in a time-related fashion during osteogenic induction (Fig. 1).
Figure 1.
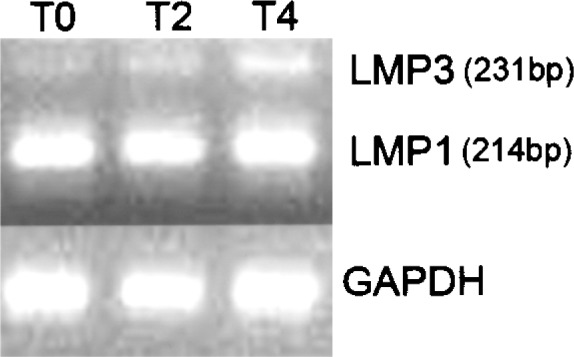
Endogenous LMP gene expression in untransduced cells. The expression of the endogenous LMP gene was assessed in untransduced hMSC, cultured for 0 (TO), 2 (T2), and 4(T4) days in osteogenic medium, by means of RT-PCR. LMP1 is expressed at higher levels in undifferentiated hMSC and during osteogenic induction. LMP3 expression in undifferentiated cells (TO) is barely detectable.
Efficient Adenoviral-Mediated Transduction of hMSC
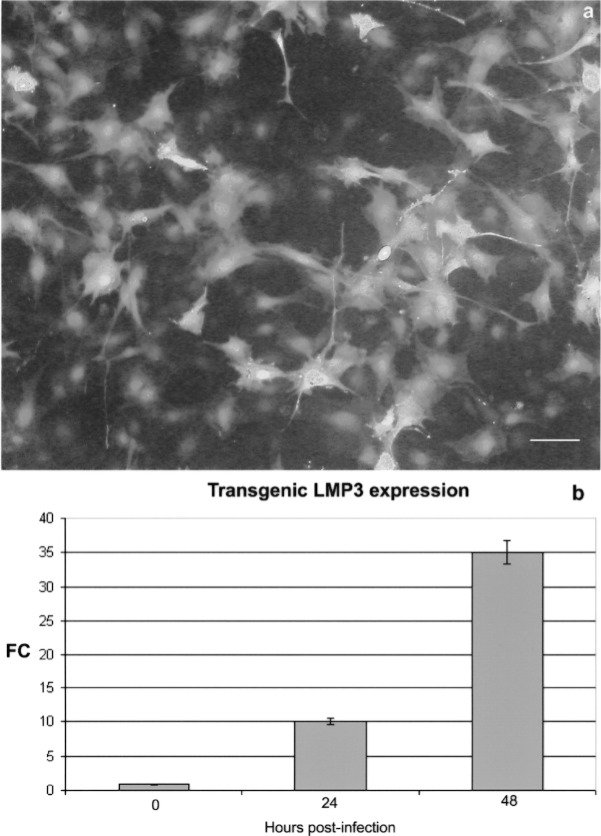
In order to evaluate the biological effects of LMP3 overexpression, the hMSC were transduced using a replication-defective adenoviral vector carrying the sequence-optimized cDNA of the human LMP3 gene (AdLMP3) (28). hMSC were transfected with 100 MOI of AdLMP3 with either Adψ5 or AdEGFP used as transduction controls, and mock-transduced cells used as a negative control. The efficiency of adenoviral-mediated transduction was assessed by counting eGFP-positive fluorescent cells following transduction with AdEGFP. As shown in Figure 2a, 60% of the cells were fluorescent 24 h following AdEGFP transduction and 90% 48 h posttransduction. Moreover, transgenic LMP3 expression was assessed in hMSC upon AdLMP3 transduction by means of quantitative RT-PCR. The expression of transgenic LMP3 significantly increased in a time-related fashion in LMP3-transduced cells compared to control Adψ5-transduced and untransduced cells (Fig. 2b).
Figure 2.
Adenoviral-mediated transduction efficiency. hMSC were infected using a replication-defective adenoviral vectors carrying either the human LMP3 or the EGFP gene; the adenoviral vector backbone (Adψ5) served as control. (a) EGFP served as a reporter gene, which allowed to assess the transgene expression in vitro by counting EGFP-positive cells under fluorescent invertoscope 24 h after transduction. Scale bar: 10 μm. (b) Transgenic LMP3 expression was assessed in AdLMP3-transduced cells by means of real-time PCR in time course. The GAPDH gene was used as a housekeeping control gene. Expression levels are expressed as mean fold change (FC) over the control, which was calculated according to the ΔΔCt method as reported in the Materials and Methods section.
Inhibition of Growth of hMSC Following LMP3 Overexpression
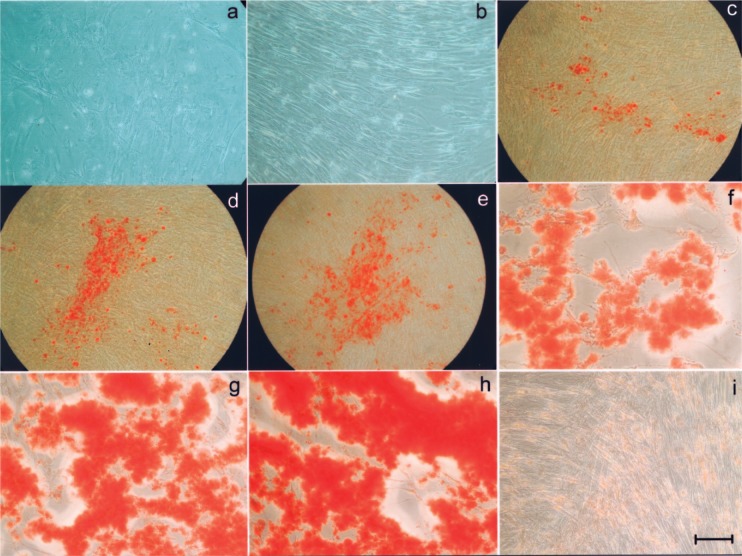
hMSC growth kinetics was monitored for 1 week following adenoviral-mediated transduction. Cell growth was arrested as early as 2 days following AdLMP3 transduction, suggesting that the progression of osteogenic differentiation (see below) induced by LMP3 correlated with cell growth arrest. Regions of cell detachment also were observed 7 days postinfection (Fig. 3a). Similar morphological signs were not observed in Adψ5-transduced cells (Fig. 3b).
Figure 3.
Osteogenic differentiation of LMP3-expressing hMSC. Cells were transduced using either AdLMP3 or Adψ5 and osteogenic differentiation was assessed in vitro by microscopic analysis and alizarin red staining in time course. Four days after AdLMP3 transduction slight morphological changes were observed in cell culture, with the presence of a reticular web overlapping on the cell layer (a); at the same time point Adψ5-transduced cells retained the bipolar spindle-like morphology of wild-type hMSC in an homogeneous monolayer culture (b). Alizarin red staining demonstrated the progressive matrix mineralization occurring as early as 2 days after AdLMP3 transduction: 48 h (c), 72 h (d), and 4 days (e). The mineralized matrix noticeably increased at later time points: 10 (f) and 14 days (g–h). No evidence of mineralization was evidenced in Adψ5-transduced cells up to the end of the observation (i). Scale bar: 10 μm (a, b, i); 50 μm (c–e); 5 μm (f–h).
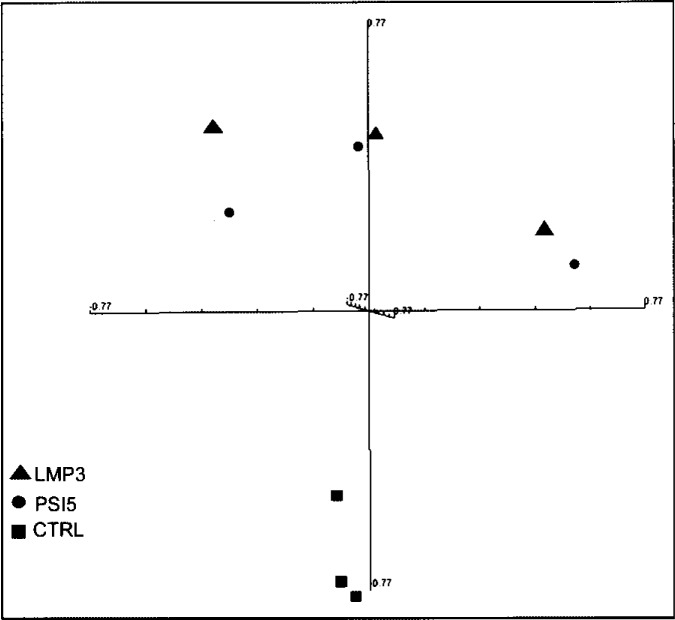
Figure 4.
Multidimensional scaling of microarray data. Unsupervised multidimensional scaling (MDS) of the expression profiles from LMP3-transduced and control cells was performed in order to assess the segregation efficiency of samples. The high dimensionality of the gene expression data has been reduced to the three dimensions that comprise the greatest variation across the data set. All samples are represented by spheres and positioned in the three-dimensional space, so that the distance between each pair of samples very closely approximates the centered correlation distance for the corresponding sample pair. A clear separation is achieved between adenoviral-transduced and control untransduced cells along one of the three components of the plot. Triangles: AdLMP3-transduced cells; circles: cells transduced with the viral backbone Adψ5; squares: control untransduced cells.
Osteogenic Differentiation and Matrix Mineralization of HMSC Induced by LMP3
To evaluate the osteogenic potential of LMP3, the morphological characteristics of hMSC following AdLMP3-mediated transduction were analyzed over time. Adψ5-transduced cells served as controls. Transduction of hMSC with AdLMP3 resulted in slight morphological changes in cells that could be observed as early as 4 days after infection in LMP3-expressing cells compared to control (Fig. 3a, b). In particular, the cell layer lost its homogeneity and a web-like structure overlapped the cell culture. Alizarin staining revealed that a mineralized matrix began to appear in culture, as a sign of cell differentiation, as early as 2 days after AdLMP3 transduction, and increased progressively thereafter (Fig. 3c–g). No evidence of matrix mineralization was observed in control cells (Fig. 3h).
Gene Expression Profiling
To examine the initial osteoinductive effects of human LMP3 on bone marrow-derived hMSC, microarray analysis was performed on RNA isolated from hMSC transduced with either AdLMP3 or Adψ5, using untransduced cells as an additional negative control. Three technical replicates per each experimental condition were performed, giving a total number of nine arrays analyzed.
The resulting data set was analyzed by unsupervised multidimensional scaling (MDS), in order to examine the samples’ segregation. MDS analysis showed that adenoviral-transduced cells belonging to the same replicated experiment tended to cluster in closer areas of the tridimensional space, while control untransduced samples were located in a distinct section of the plot (Fig. 5). This result indicated that at early time points after cell transduction, the main transcriptional modulation was due to adenoviral-related effects (see following paragraphs). Therefore, we further analyzed the LMP3 treatment group using the Adψ5-transduced cells as controls, in order to rule out any viral vector-related effect from the AdLMP3-induced expression profile.
Figure 5.
Top biological functions. The functional analysis of the gene list allowed the identification of the biological functions that were most significant to the data set. Genes from the gene list were grouped into functional categories displayed along the x-axis; the y-axis displays the significance (−log p-value). The threshold line is set as default at p = 0.05. Distinct bar graphs are used to list the top Molecular and Cellular Functions (a) and the top Physiological System Development and Functions (b).
Specific Gene Expression Profiles Induced in hMSC by LMP3
All data from the AdLMP3- and Adψ5-transduced hMSC was analyzed using a paired t-test with random variance model (significance threshold of univariate test = 0.01). On the basis of MDS results, we paired samples (AdLMP3-treated cells and corresponding Adψ5 controls) originating from the same replicated experiment. This allowed for segregation of gene expression profiles specifically due to the transgenic LMP3 expression in treated cells. As a result of the paired t-test, 263 transcripts were observed to be differentially expressed between the two classes, with a p-value ranging from 3.4 × 10−6 to 9.9 × 10−3 (see the complete gene list published on GEO database; http://www.ncbi.nlm.nih.gov/geo/, accession number GSE16614). A restricted gene list of most significantly modulated genes (p < 0.001) is shown in Table 1.
TABLE 1.
GENES MODULATED BY LMP3 IN hMSC
| FC* | p-Value† | Probe Set ID | Gene Symbol‡ | GenBank ID | Description | GO Biological Process§ |
|---|---|---|---|---|---|---|
| ↑ | 0.0001322 | 203935_at | ACVR1 | NM_001105 | activin A receptor, type I | G1/S transition of mitotic cell cycle |
| ↓ | 0.0005144 | 201924_at | AFF1 | NM_005935 | AF4/FMR2 family, member 1 | positive regulation of transcription |
| ↓ | 0.0006217 | 219723_x_at | AGPAT3 | NM_020132 | l-acylglycerol-3-phosphate O-acyl-transferase 3 | metabolic process |
| ↑ | 0.0009348 | 20415l_x_at | AKR1C1 | NM_001353 | aldo-keto reductase family 1, member C1 | lipid metabolic process |
| ↑ | 0.0005113 | 209699_x_at | AKR1C2 | U05598 | aldo-keto reductase family 1, member C2 | lipid metabolic process |
| ↑ | 7.97E-05 | 209160_at | AKR1C3 | AB018580 | aldo-keto reductase family 1, member C3 | prostaglandin metabolic process |
| ↓ | 0.0004019 | 209645_s_at | ALDH1B1 | NM_000692 | aldehyde dehydrogenase 1 family, member B1 | carbohydrate metabolic process |
| ↓ | 0.000398 | 202686_s_at | AXL | NM_021913 | AXL receptor tyrosine kinase | protein amino acid phosphorylation |
| ↑ | 0.0003068 | 202705_at | CCNB2 | NM_004701 | cyclin B2 | cell cycle |
| ↑ | 8.62E-05 | 209714_s_at | CDKN3 | AF213033 | cyclin-dependent kinase inhibitor 3 | cell cycle |
| ↓ | 5.32E-05 | 201445_at | CNN3 | NM_001839 | calponin 3, acidic | smooth muscle contraction |
| ↓ | 7.71E-05 | 203477_at | COL15A1 | NM_001855 | collagen, type XV, alpha 1 | cell adhesion |
| ↓ | 0.0002589 | 20255l_s_at | CRIM1 | BG546884 | cysteine rich transmembrane BMP regulator 1 | regulation of cell growth |
| ↓ | 0.0009697 | 214377_s_at | CTRL | BF508685 | chymotrypsin-like | proteolysis |
| ↓ | 0.000185 | 214446_at | ELL2 | NM_012081 | elongation factor, RNA polymerase II, 2 | transcription |
| ↑ | 0.0004684 | 204163_at | EMILIN1 | NM_007046 | elastin microfibril interfacer 1 | cell adhesion |
| ↑ | 3.40E-06 | 202017_at | EPHX1 | NM_000120 | epoxide hydrolase 1, microsomal (xenobiotic) | response to toxin |
| ↑ | 0.0003472 | 219117_s_at | FKBP11 | NM_016594 | FK506 binding protein 11, 19 kDa | protein folding |
| ↑ | 0.0006456 | 213524_s_at | G0S2 | NM_015714 | G0/G1 switch 2 | cell cycle |
| ↑ | 0.0005706 | 204457_s_at | GAS1 | NM_002048 | growth arrest-specific 1 | cell cycle |
| ↓ | 0.0006808 | 206432_at | HAS2 | NM_005328 | hyaluronan synthase 2 | — |
| ↑ | 0.0009572 | 214022_s_at | IFITM1 | AA749101 | interferon induced transmembrane protein 1 (9–27) | cell surface receptor linked signal transduction |
| ↑ | 0.0001397 | 202409_at | IGF2 | X07868 | insulin-like growth factor 2 (soma-tomedin A) | skeletal system development |
| ↓ | 0.0003656 | 201393_s_at | IGF2R | NM_000876 | insulin-like growth factor 2 receptor | transport |
| ↓ | 0.0009478 | 201596_x_at | KRT18 | NM_000224 | keratin 18 | apoptosis/cell cycle/morphogenesis |
| ↓ | 0.0002906 | 218574_s_at | LMCD1 | NM_014583 | LIM and cysteine-rich domains 1 | negative regulation of transcription |
| ↓ | 0.0007359 | 211926_s_at | MYH9 | AI827941 | myosin, heavy chain 9, non-muscle | cell morphogenesis involved in differentiation |
| ↑ | 0.0006921 | 219295_s_at | PCOLCE2 | NM_013363 | procollagen C-endopeptidase enhancer 2 | — |
| ↓ | 0.000457 | 20273 l_at | PDCD4 | NM_014456 | programmed cell death 4 | apoptosis/cell cycle/cell aging |
| ↑ | 0.0004643 | 217996_at | PHLDA1 | NM_007350 | pleckstrin homology-like domain, family A, member 1 | apoptosis |
| ↓ | 0.000827 | 205048_s_at | PSPH | NM_003832 | phosphoserine phosphatase | L-serine metabolic process |
| ↑ | 5.00E-04 | 218045_x_at | PTMS | NM_002824 | parathymosin | DNA replication |
| ↑ | 0.0003426 | 203455_s_at | SAT1 | NM_002970 | spermidine/spermine Nl-acetyltransferase 1 | polyamine metabolic process |
| ↓ | 0.0003915 | 205396_at | SMAD3 | BF971416 | SMAD family member 3 | negative regulation of transcription |
| ↓ | 0.0002897 | 221499_s_at | STX16 | AK026970 | syntaxin 16 | transport |
| ↑ | 0.0003462 | 203085_s_at | TGFB1 | BC000125 | transforming growth factor, beta 1 | skeletal system development |
FC: fold change, arrows indicate the direction of gene modulation.
p-Value indicating the significance of differential expression calculated using a t-test with random variance model.
Genes are listed in alphabetical order according to the official gene symbol.
GO: gene ontology, primary GO biological process terms are indicated, a — sign stands for missing GO annotations for selected transcripts.
The functional analysis of the gene list aimed at the biological interpretation of the results was accomplished using IPA software. This allowed to identify the biological functions that were most significant to the data set (p < 0.05; named “Top Biological Functions”), based on IPA Knowledge Base (www.ingenuity.com). Among the “Top Biological Functions,” the IPA software categorized genes alternatively in two functional groups, based on their involvement in “Molecular and Cellular Functions” and in “Physiological System Development and Functions.” In particular, among the Molecular and Cellular Functions category an overall number of 88 genes, coding for key regulators of the cell cycle regulatory machinery, were listed as being involved in “cell death” and “cell growth and proliferation” (Fig. 5a). Also, among the Physiological System Development and Function categories 28 genes were categorized as molecules implicated in the development and function of the Connective Tissue and of the Skeletal Muscular System (Fig. 5b).
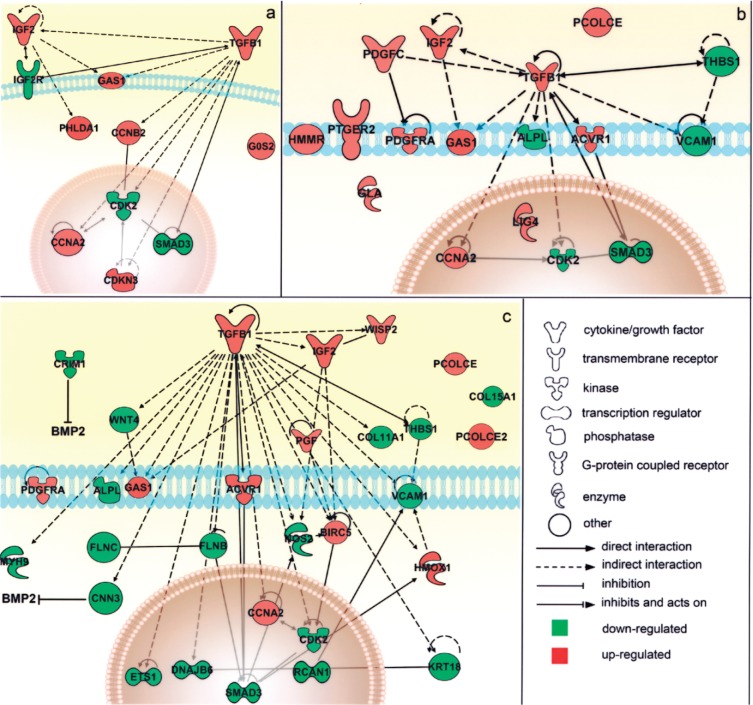
The reciprocal interactions of all the molecules involved in the above-mentioned functions were then schematized using the Path Designer function of IPA that enabled the reconstruction of the molecular pathways involved in the experimental model, based on the information provided by the IPA Knowledge Base. This allowed defining the active role of the TGF-β1 signaling pathway in both the cell proliferation and tissue development functions (Fig. 6). In fact, among the most significant molecular events within the LMP3-associated gene expression profile in hMSC (see Table 1) was the upregulation of genes in the TGF-β family, including ACVR1, encoding the BMP type I receptor ALK2, and IGF-2, along with the downregulation of the BMP inhibitors CRIM1 and CNN3. In contrast, the SMAD3 gene, coding for an important TGF-β1 intracellular mediator, was downregulated (Fig. 6).
Figure 6.
Schematic representation of the main modulated genes and their reciprocal interactions in the cell. Genes significantly associated to the main biological functions resulted from the functional analysis were associated in the graphical representation of their reciprocal interaction, represented as nodes, and the biological relationship between two nodes is represented as an edge (line). All edges are supported by at least 1 reference from the literature, from a textbook, or from canonical information stored in the Ingenuity Pathways Knowledge Base (www.ingenuity.com). The node color indicates the direction of gene modulations: upregulation (red) or downregulation (green). Nodes are displayed using various shapes that represent the functional class of the gene product. Separate graphs show distinct functional categories significantly associated to the gene list. (a) Cell death/cell growth and proliferation: the transcriptional modulation of the genes in this group pointed toward cell cycle arrest and the inhibition of cell proliferation, which usually precede cell differentiation. (b) Connective tissue development and function, mainly involving genes that interact reciprocally within the TGF-β1 signaling pathway. (c) Muscular and skeletal tissue development and function, which includes genes that interact either directly (such as the ACVR1 receptor on the cell membrance and the SMAD3 regulator in the nucleus) or indirectly (such as IGF2) with the TGF-β1 pathway and genes involved in extracellular matrix deposition (including PCOLCE, PCOLCE2, and the collagen isoforms COL11A1 and COL15A1), suggesting the initial osteoblastic commitment of cells (see text for additional details).
Gene Expression Validation
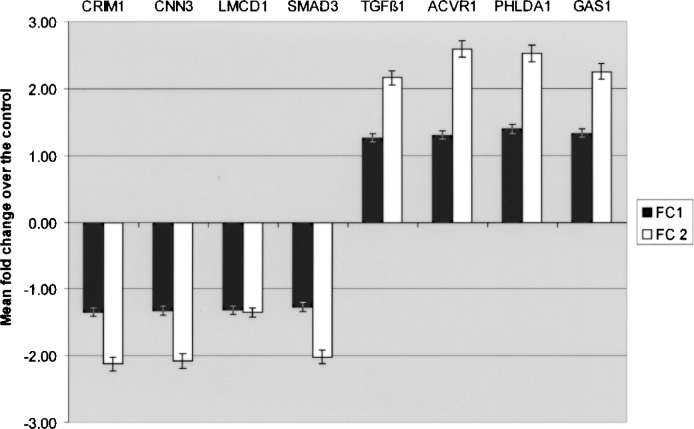
Gene expression results obtained by microarray analysis were also validated by real-time PCR performed on eight selected transcripts. The assay confirmed the modulated expression of the target genes tested, showing consistent alignment with the trends shown in the microarray analysis (Fig. 7).
Figure 7.
Real-time PCR validation. The expression of selected transcript from the dataset was analyzed by means of real-time PCR using sequence-specific primers and GAPDH as a housekeeping control gene. Expression levels are expressed as mean fold change over the control, which was calculated according to the ΔΔCt method as reported in the Materials and Methods section. FC1: fold change obtained by means of microarray analysis; FC2: fold change obtained in real-time PCR.
Transcriptional Modulation Related to Adenoviral Infection
MDS analysis suggested that a significant proportion of the gene modulation could be brought back to the defective adenoviral entry in cells. Therefore, we also focused on genes modulated coherently in hMSC transduced with Adψ5, using untransduced cells as controls. For this purpose data were first filtered as mentioned above; 1,526 out of 8,793 transcripts passed the 1.2-fold criteria. The class comparison on this dataset was carried out using a two-sample t -test (significance threshold of unvariate test = 0.001), which allowed to categorize 68 genes with a p-value ranging from 2.3 × 10−6 to 0.97 × 10−3 (data not shown). The resulting gene list was annotated according to GOA classification of biological processes. This allowed to identify mainly upregulated genes, whose function is involved the regulation of cell cycling, DNA remodeling, and recombination (Table 2).
TABLE 2.
GENES MODULATED BY THE ADENOVIRAL BACKBONE IN hMSC
| FC* | Gene Symbol | Gene Title | GenBank ID | Functional Category† |
|---|---|---|---|---|
| ↑ | CCNA2 | cyclin A2 | AI346350 | cell cycle/proliferation/mitosis |
| ↑ | GMNN | geminin, DNA replication inhibitor | NM_015895 | |
| ↑ | CDK2 | cyclin-dependent kinase 2 | M68520 | |
| ↑ | CKS2 | CDC28 protein kinase regulatory subunit 2 | NM_001827 | |
| ↑ | CDC20 | cell division cycle 20 homolog (S. cerevisiae) | NM_001255 | |
| ↑ | CCNB2 | cyclin B2 | NM_004701 | |
| ↑ | MSH2 | mutS homolog 2, colon cancer, nonpolyposis type 1 (E. coli) | U04045 | |
| ↑ | TPX2 | TPX2, microtubule-associated, homolog (Xenopus laevis) | AF098158 | |
| ↑ | AURKA | aurora kinase A | NM_003600 | |
| ↑ | FBXO5 | F-box protein 5 | NM_012177 | |
| ↑ | MAD2L1 | MAD2 mitotic arrest deficient-like 1 (yeast) | NM_002358 | |
| ↑ | NUSAP1 | nucleolar and spindle associated protein 1 | NM_016359 | |
| ↑ | SMC4 | structural maintenance of chromosomes 4 | AL136877 | |
| ↑ | ZWINT | ZW10 interactor | NM_007057 | |
| ↑ | PRC1 | protein regulator of cytokinesis 1 | NM_003981 | |
| ↓ | GAS1 | growth arrest-specific 1 | NM_002048 | |
| ↓ | RABGAP1 | RAB GTPase activating protein 1 | AI922519 | |
| ↓ | CCNG1 | cyclin G1 | BC000196 | |
| ↓ | CCL2 | chemokine (C-C motif) ligand 2 | S69738 | |
| ↑ | PTTG1 | pituitary tumor-transforming 1 | NM_004219 | DNA metabolism/replication/recombination |
| ↑ | TK1 | thymidine kinase 1, solubile | NM_003258 | |
| ↑ | TOP2A | topoisomerase (DNA) II alpha 170 kDa | AL561834 | |
| ↑ | FEN1 | flap structure-specific endonuclease 1 | BC000323 | |
| ↑ | PRIM1 | primase, DNA, polypeptide 1 (49 kDa) | NM_000946 | |
| ↑ | CENPA | centromere protein A | NM_001809 | |
| ↑ | MSH6 | mutS homolog 6 (E. coli) | NM_000179 | |
| ↑ | KPNA2 | karyopherin alpha 2 (RAG cohort 1, importin alpha 1) | NM_002266 |
FC: fold change, arrows indicate the direction of gene modulation.
Genes are listed based on their functional category obtained from Gene Ontology annotations.
DISCUSSION
The formation of new bone is a complex cascade of events, involving cell proliferation, extracellular matrix (ECM) maturation, and matrix mineralization. Mammalian osteoblasts originate from hMSC that are competent to give rise to several cell types, including myocytes, chondrocytes, and adipocytes, along with nonmesodermal lineages. The tissue-specific commitment has been efficiently induced in hMSC, supporting their use for cell-based gene therapy applications aimed at tissue regeneration (27,29,31). The hMSC differentiation fate occurs through the reciprocal crosstalk of TGF-β, WNT, and BMP, signaling pathways, all involved in basic mechanisms regulating cell homeostasis (22,41). MAPK signaling also is required for osteoblast differentiation, as it drives the activation of molecules mediating matrix deposition (44). Therefore, hMSC undergo a specific differentiation as a result of a finely balanced regulation of this complex molecular network, through modulation of transcription, protein processing, and receptor binding, along with regulation of Smads function and extracellular antagonism of BMPs (22). In particular, TGF-β1 induces rapid nuclear translocation of β-catenin in hMSC in a Smad3-dependent manner. Functionally, this pathway is required for the stimulation of hMSC proliferation and the inhibition of osteogenic differentiation induced by TGF-β1, likely through the regulation of specific downstream target genes (13).
We demonstrated previously that exogenous human LMP3 induces the expression of bone-specific genes in hMSC, cooperating with the BMP-dependent osteogenic cascade, resulting in bone matrix mineralization in culture and efficient bone formation in vivo (16,28). In this study, the functional analysis of the microarray profile of hMSC during the early steps of osteogenic commitment induced by exogenous LMP3 suggested the activation of musculoskeletal and, more in general, connective developmental processes (see Figs. 5b and 6b, c). In particular, this seemed to occur via the upregulation of the TGF-β1 pathway (Fig. 5b, c), suggesting that LMP3 induces a switch of TGF-β1 signaling from cell proliferation to osteogenic differentiation of hMSC (10,13,22,23,40,41,43,44,46) (Fig. 5). These results are consistent with recent reports showing that both LMP1 and LMP3 interact and inhibit the activity of SMURF1, an E3 ligase that ubiquitinates many molecules in the TGF-β/BMP pathway and prevents their degradation by proteasomes (30). Therefore, these data suggest that the early transcriptional effects of LMP3 result in a shift from a proliferative state to a differentiated state in hMSC homeostasis. This illustrates the possible interplay of signaling cascades activated prior to the expression of markers associated to the osteogenic differentiation process, usually occurring after two or more days following the osteogenic induction in vitro (11,28). In fact, it was previously demonstrated that LMP is able to induce the expression of specific molecules associated to the osteogenic commitment of hMSC, including OSX, RUNX2, and certain BMPs (2,28,35,36). Taken together, these results suggest a role for LMP as an upstream regulator of the osteogenic cascade. The direct role of LMP3 in musculoskeletal connective tissue development was also suggested by the upregulation of genes involved in matrix deposition during bone and cartilage morphogenesis, including PCOLCE and PCOLCE2, which was previously found to be expressed during branch morphogenesis and in cartilaginous structures (34) (Fig. 6c). Conversely, genes involved in nonskeletal tissue differentiation and development, such as MYH9, KRT18, and COL15A1, were downregulated. Taken together, these observations suggest that LMP3 overexpression resulted in the early commitment of hMSC toward the skeletal tissue lineage differentiation.
In addition, IPA analysis also indicated the functional implication of cell proliferation in LMP3 transcriptional profiles (Fig. 6a). In fact, as TGF-β signaling plays several different roles in cell homeostasis, being also involved in apoptosis, the upregulation of TGF-β1 gene indicated that cell cycle was influenced by LMP3 expression in hMSC (13). This hypothesis was strengthened by the evidence of an important role of growth factors, such as BMP2, in cell cycle regulation during cell commitment, indicating the clear inverse relationship between cell cycling and differentiation (6,19,32).
In particular, the coordinate upregulation of CDKN3, a cyclin-dependent kinase inhibitor that prevents the activation of CDK2, CCNA2, and CCNB2, members of the B-type cyclins, could lead to the inhibition of the G2–M transition. Also, CCNB2 plays a leading role in the TGF-β-mediated cell cycle control, being responsible for the cycle arrest in the G1/S phase and involved in the epithelial–mesenchymal transition (17,38). In this respect, it was noteworthy the coordinate upregulation of genes inducing growth arrest such as GAS1 and G0S2. Although a role in osteogenic differentiation was not previously described for these molecules, their upregulation is consistent with involvement in cell cycle regulation and differentiation (14,20,47). Finally, the upregulation of PHLDA1 (see Table 1 and Fig. 6a), which is known to be induced by the insulin-like growth factor-1 through the p38/MAPK pathway and to mediate either pro- or antiapoptotic effects depending on the cell type, was consistent with LMP3-mediated growth inhibition (39). Interestingly, constitutive PHLDA1 expression was associated with a reduction in growth rate, a decrease in cloning efficiency, and an increased apoptosis sensitivity (24).
Interestingly, a significant transcriptional modulation of genes activating the cell cycle could be observed in Adψ5-transduced cells compared to control untransduced cells (Table 2). Nonetheless, no evident effects on cell viability and differentiation could be detected in hMSC upon empty adenoviral infection, suggesting that the observed transcriptional modulation could resemble the earliest phase of defective adenovirus type 5 infection, which is known to induce distinct biological events in the host cells (8,37).
In conclusion, the results of this research suggested that LMP3 could exert wider effects on cell homeostasis than was initially thought, as a result of the early modulation of a complex signaling network that inhibits proliferation to mediate the first steps in cell differentiation. The molecular events occurring during the full osteogenic commitment of hMSC, which are known to be induced by LMP3 at later time points (28), should be considered as downstream effects of the initial LMP3-mediated transcriptional modulation.
ACKNOWLEDGMENTS
This work was partially supported by the “Universitá Cattolica del Sacro Cuore” and the Musculo-Skeletal Tissue Bank of the Latium Region (Rome, IT).
REFERENCES
- 1. Ashburner M.; Ball C. A.; Blake J. A.; Botstein D.; Butler H.; Cherry J. M.; Davis A. P.; Dolinski K.; Dwight S. S.; Eppig J. T.; Harris M. A.; Hill D. P.; Issel-Tarver L.; Kasarskis A.; Lewis S.; Matese J. C.; Richardson J. E.; Ringwald M.; Rubin G. M.; Sherlock G. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25:25–29; 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Boden S. D.; Liu Y.; Hair G. A.; Helms J. A.; Hu D.; Racine M.; Nanes M. S.; Titus L. LMP-1, a LIM-domain protein, mediates BMP-6 effects on bone formation. Endocrinology 139:5125–5134; 1998. [DOI] [PubMed] [Google Scholar]
- 3. Boden S. D.; Titus L.; Hair G.; Liu Y.; Viggeswarapu M.; Nanes M. S.; Baranowski C. Lumbar spine fusion by local gene therapy with a cDNA encoding a novel osteoinductive protein (LMP-1). Spine 23:2486–2492; 1998. [DOI] [PubMed] [Google Scholar]
- 4. Brazma A.; Hingamp P.; Quackenbush J.; Sherlock G.; Spellman P.; Stoeckert C.; Aach J.; Ansorge W.; Ball C. A.; Causton H. C.; Gaasterland T.; Glenisson P.; Holstege F. C.; Kim I. F.; Markowitz V.; Matese J. C.; Parkinson H.; Robinson A.; Sarkans U.; Schulze-Kremer S.; Stewart J.; Taylor R.; Vilo J.; Vingron M. Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat. Genet. 29:365–371; 2001. [DOI] [PubMed] [Google Scholar]
- 5. Bünger M. H.; Langdahl B. L.; Andersen T.; Husted L.; Lind M.; Eriksen E. F.; B?nger C. E. Semiquantitative mRNA measurements of osteoinductive growth factors in human iliac-crest bone: Expression of LMP splice variants in human bone. Calcif Tissue Int. 73:446–454; 2003. [DOI] [PubMed] [Google Scholar]
- 6. Chalazonitis A.; Pham T. D.; Li Z.; Roman D.; Guha U.; Gomes W.; Kan L.; Kessler J. A.; Gershon M. D. Bone morphogenetic protein regulation of enteric neuronal phenotypic diversity: Relationship to timing of cell cycle exit. J. Comp. Neurol. 509(5):474–492; 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Friedenstein A. J.; Chailakhyan R. K.; Gerasimov U. V. Bone marrow osteogenic stem cells: In vitro cultivation and transplantation in diffusion chambers. Cell Tissue Kinetics 20:263–272; 1987. [DOI] [PubMed] [Google Scholar]
- 8. Granberg F.; Svensson C.; Pettersson U.; Zhao H. Adenovirus-induced alterations in host cell gene expression prior to the onset of viral gene expression. Virology 353(1):1–5; 2006. [DOI] [PubMed] [Google Scholar]
- 9. Gregory C. A.; Gunn W. G.; Reyes E.; Smolarz A. J.; Munoz J.; Spees J. L.; Prockop D. J. How Wnt signaling affects bone repair by mesenchymal stem cells from the bone marrow. Ann. NY Acad. Sci. 1049:97–106; 2005. [DOI] [PubMed] [Google Scholar]
- 10. Haag J.; Aigner T. Identification of calponin 3 as a novel Smad-binding modulator of BMP signaling expressed in cartilage. Exp. Cell Res. 313:3386–3394; 2007. [DOI] [PubMed] [Google Scholar]
- 11. Hamidouche Z.; Haÿ E.; Vaudin P.; Charbord P.; Schüle R.; Marie P. J.; Fromigué O. FHL2 mediates dexamethasone-induced mesenchymal cell differentiation into osteoblasts by activating Wnt/beta-catenin signaling-dependent Runx2 expression. FASEB J. 22:3813–3822; 2008. [DOI] [PubMed] [Google Scholar]
- 12. Irizarry R. A.; Hobbs B.; Collin F.; Beazer-Barclay Y. D.; Antonellis K. J.; Scherf U.; Speed T. P. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4:249–264; 2003. [DOI] [PubMed] [Google Scholar]
- 13. Jian H.; Shen X.; Liu I.; Semenov M.; He X.; Wang X. F. Smad3-dependent nuclear translocation of beta-catenin is required for TGF-beta1-induced proliferation of bone marrow-derived adult human mesenchymal stem cells. Genes Dev. 20(6):666–674; 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kitareewan S.; Blumen S.; Sekula D.; Bissonnette R. P.; Lamph W. W.; Cui Q.; Gallagher R.; Dmitrovsky E. G0S2 is an all-trans-retinoic acid target gene. Int. J. Oncol. 33(2):397–404; 2008. [PMC free article] [PubMed] [Google Scholar]
- 15. Lattanzi W.; Bernardini C.; Gangitano C.; Michetti F. Hypoxia-like transcriptional activation in TMT-induced degeneration: Microarray expression analysis on PC12 cells. J. Neurochem. 100(6):1688–1702; 2007. [DOI] [PubMed] [Google Scholar]
- 16. Lattanzi W.; Parrilla C.; Fetoni A.; Logroscino G.; Straface G.; Pecorini G.; Stigliano E.; Tampieri A.; Bedini R.; Pecci R.; Michetti F.; Gambotto A.; Robbins P. D.; Pola E. Ex vivo-transduced autologous skin fibroblasts expressing human Lim mineralization protein-3 efficiently form new bone in animal models. Gene Ther. 15(19):1330–1343; 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Liu J. H.; Wei S.; Burnette P. K.; Gamero A. M.; Hutton M.; Djeu J. Y. Functional association of TGF-beta receptor II with cyclin B. Oncogene. 18(1):269–275; 1999. [DOI] [PubMed] [Google Scholar]
- 18. Liu Y.; Hair G. A.; Boden S. D.; Viggeswarapu M.; Titus L. Overexpressed LIM mineralization proteins do not require LIM domains to induce bone. J. Bone Miner. Res. 17:406–414; 2002. [DOI] [PubMed] [Google Scholar]
- 19. Luppen C. A.; Leclerc N.; Noh T.; Barski A.; Khokhar A.; Boskey A. L.; Smith E.; Frenkel B. Brief bone morphogenetic protein 2 treatment of glucocorticoid-inhibited MC3T3-E1 osteoblasts rescues commitment-associated cell cycle and mineralization without alteration of Runx2. J. Biol. Chem. 278(45):44995–45003; 2003. [DOI] [PubMed] [Google Scholar]
- 20. Martinelli D. C.; Fan C. M. The role of Gas1 in embryonic development and its implications for human disease. Cell Cycle 6(21):2650–2655; 2007. [DOI] [PubMed] [Google Scholar]
- 21. Minamide A.; Boden S. D.; Viggeswarapu M.; Hair G. A.; Oliver C.; Titus L. Mechanism of bone formation with gene transfer of the cDNA encoding for the intracellular protein LMP-1. J. Bone Joint Surg. Am. 85:1030–1039; 2003. [DOI] [PubMed] [Google Scholar]
- 22. Miyazono K.; Kusanagi K.; Inoue H. Divergence and convergence of TGF-beta/BMP signaling. J. Cell Physiol. 187(3):265–276; 2001. [DOI] [PubMed] [Google Scholar]
- 23. Mukherjee A.; Rotwein P. Akt promotes BMP2-mediated osteoblast differentiation and bone development. J. Cell Sci. 122(Pt. 5):716–726; 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Neef R.; Kuske M. A.; Pröls E.; Johnson J. P. Identification of the human PHLDA1/TDAG51 gene: Down-regulation in metastatic melanoma contributes to apoptosis resistance and growth deregulation. Cancer Res. 62(20):5920–5929; 2002. [PubMed] [Google Scholar]
- 25. Parrilla C.; Lattanzi W.; Fetoni A. R.; Bussu F.; Pola E.; Paludetti G. Ex vivo gene therapy using autologous dermal fibroblasts expressing hLMP3 for rat mandibular bone regeneration. Head Neck 32:310–318; 2010. [DOI] [PubMed] [Google Scholar]
- 26. Pereira R. F.; Halford K. W.; O’Hara M. D.; Leeper D. B.; Sokolov B. P.; Pollard M. D.; Bagasra O.; Prockop D. J. Cultured adherent cells from marrow can serve as long-lasting precursor cells for bone, cartilage and lung in irradiated mice. Proc. Natl. Acad. Sci. USA 92:4857–4861; 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pittenger M. F.; Mackay A. M.; Beck S. C.; Jaiswal R. K.; Douglas R.; Mosca J. D.; Moorman M. A.; Simonetti D. W.; Craig S.; Marshak D. R. Multilineage potential of adult human mesenchymal stem cells. Science 284:143–147; 1999. [DOI] [PubMed] [Google Scholar]
- 28. Pola E.; Gao W.; Zhou Y.; Pola R.; Lattanzi W.; Sfeir C.; Gambotto A.; Robbins P. D. Efficient bone formation by gene transfer of human LIM mineralization protein-3. Gene Ther. 11(8):683–693; 2004. [DOI] [PubMed] [Google Scholar]
- 29. Prockop D. J. Marrow stromal cells as stem cells for nonhematopoietic tissues. Science 276:71–74; 1997. [DOI] [PubMed] [Google Scholar]
- 30. Sangadala S.; Boden S. D.; Viggeswarapu M.; Liu Y.; Titus L. Lim Mineralization protein-1 (LMP-1) potentiates bone morphogenetic protein responsiveness via a novel interaction with Smurf1 resulting in decreased ubiquitination of Smads. J. Biol. Chem. 281:17212–17219; 2006. [DOI] [PubMed] [Google Scholar]
- 31. Saulnier N.; Lattanzi W.; Puglisi M. A.; Pani G.; Barba M.; Piscaglia A. C.; Giachelia M.; Alfieri S.; Neri G.; Gasbarrini G.; Gasbarrini A. Mesenchymal stromal cells multipotency and plasticity: Induction toward the hepatic lineage. Eur. Rev. Med. Pharmacol. Sci. 13(Suppl. 1):71–78; 2009. [PubMed] [Google Scholar]
- 32. Sharov A. A.; Sharova T. Y.; Mardaryev A. N.; Tommasi di Vignano A.; Atoyan R.; Weiner L.; Yang S.; Brissette J. L.; Dotto G. P.; Botchkarev V. A. Bone morphogenetic protein signaling regulates the size of hair follicles and modulates the expression of cell cycle-associated genes. Proc. Natl. Acad. Sci. USA 103(48):18166–18171; 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Simon R.; Lam A. Technical reports. BRB Array Tools Users Guide. Biometric Research Branch, National Cancer Institute; http://linus.nci.nih.gov/brb/TechReport; 2006. [Google Scholar]
- 34. Steiglitz B. M.; Keene D. R.; Greenspan D. S. PCOLCE2 encodes a functional procollagen C-proteinase enhancer (PCPE2) that is a collagen-binding protein differing in distribution of expression and post-translational modification from the previously described PCPE1. J. Biol. Chem. 277:49820–49830; 2002. [DOI] [PubMed] [Google Scholar]
- 35. Strohbach C.; Kleinman S.; Linkhart T.; Amaar Y.; Chen S. T.; Mohan S.; Strong D. Potential involvement of the interaction between insulin-like growth factor binding protein (IGFBP)-6 and LIM mineralization protein (LMP)-1 in regulating osteoblast differentiation. J. Cell Biochem. 104:1890–1905; 2008. [DOI] [PubMed] [Google Scholar]
- 36. Strohbach C. A.; Rundle C. H.; Wergedal J. E.; Chen S. T.; Linkhart T. A.; Lau K. H.; Strong D. D. LMP-1 retroviral gene therapy influences osteoblast differentiation and fracture repair: A preliminary study. Calcif. Tissue Int. 83:202–211; 2008. [DOI] [PubMed] [Google Scholar]
- 37. Tamanini A.; Nicolis E.; Bonizzato A.; Bezzerri V.; Melotti P.; Assael B. M.; et al. Interaction of adenovirus type 5 fiber with the coxsackievirus and adenovirus receptor activates inflammatory response in human respiratory cells. J. Virol. 80(22):11241–11254; 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tavares A. L.; Mercado-Pimentel M. E.; Runyan R. B.; Kitten G. T. TGF beta-mediated RhoA expression is necessary for epithelial-mesenchymal transition in the embryonic chick heart. Dev. Dyn. 235(6):1589–1598; 2006. [DOI] [PubMed] [Google Scholar]
- 39. Toyoshima Y.; Karas M.; Yakar S.; Dupont J.; Lee H.; LeRoith D. TDAG51 mediates the effects of insulin-like growth factor I (IGF-I) on cell survival. J. Biol. Chem. 279(24):25898–25904; 2004. [DOI] [PubMed] [Google Scholar]
- 40. Tsuchida K.; Mathews L. S.; Vale W. W. Cloning and characterization of a transmembrane serine kinase that acts as an activin type I receptor. Proc. Natl. Acad. Sci. USA 90:11242–11246; 1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Varga A. C.; Wrana J. L. The disparate role of BMP in stem cell biology. Oncogene 24:5713–5721; 2005. [DOI] [PubMed] [Google Scholar]
- 42. Viggeswarapu M.; Boden S. D.; Liu Y.; Hair G. A.; Louis-Ugbo J.; Murakami H.; Kim H. S.; Mayr M. T.; Hutton W. C.; Titus L. Adenoviral delivery of LIM mineralization protein-1 induces new-bone formation in vitro and in vivo. J. Bone Joint Surg. Am. 83-A(3):364–376; 2001. [DOI] [PubMed] [Google Scholar]
- 43. Wilkinson L.; Kolle G.; Wen D.; Piper M.; Scott J. Little MCRIM1 regulates the rate of processing and delivery of bone morphogenetic proteins to the cell surface. J. Biol. Chem. 278(36):34181–34188; 2003. [DOI] [PubMed] [Google Scholar]
- 44. Xiao G.; Gopalakrishnan R.; Jiang D.; Reith E.; Benson M. D.; Franceschi R. T. Bone morphogenetic proteins, extracellular matrix, and mitogen-activated protein kinase signaling pathways are required for osteoblast-specific gene expression and differentiation in MC3T3-E1 cells. J. Bone Miner Res. 17(1):101–110; 2002. [DOI] [PubMed] [Google Scholar]
- 45. Yoon S. T.; Park J. S.; Kim K. S.; Li J.; Attallah-Wasif E. S.; Hutton W. C.; Boden S. D. ISSLS prize winner: LMP-1 upregulates intervertebral disc cell production of proteoglycans and BMPs in vitro and in vivo. Spine 29:2603–2611; 2004. [DOI] [PubMed] [Google Scholar]
- 46. Yu P. B.; Deng D. Y.; Lai C. S.; Hong C. C.; Cuny G. D.; Bouxsein M. L.; Hong D. W.; McManus P. M.; Katagiri T.; Sachidanandan C.; Kamiya N.; Fukuda T.; Mishina Y.; Peterson R. T.; Bloch K. D. BMP type I receptor inhibition reduces heterotopic [corrected] ossification. Nat. Med. 14(12):1363–1369; 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zandbergen F.; Mandard S.; Escher P.; Tan N. S.; Patsouris D.; Jatkoe T.; Rojas-Caro S.; Madore S.; Wahli W.; Tafuri S.; Müller M.; Kersten S. The G0/ G1 switch gene 2 is a novel PPAR target gene. Biochem. J. 392(Pt. 2):313–324; 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhang Q.; Wang X.; Chen Z.; Liu G.; Chen Z. Semi-quantitative RT-PCR analysis of LIM mineralization protein 1 and its associated molecules in cultured human dental pulp cells. Arch. Oral Biol. 52:720–726; 2007. [DOI] [PubMed] [Google Scholar]