Abstract
Nkx3.1 is a well-conserved homeobox gene that is involved in development, differentiation and maintenance of prostate epithelial cells. Nkx3.1 expression is induced by androgen in prostate epithelia and, as such, our interest is to understand the mechanism(s) for this androgen-dependent expression in normal epithelial cells. In this report, we show that the region of DNA sequence 2.7 kilobases in front of the mouse Nkx3.1 gene drives enhanced transcription in prostate epithelia cells; however, this segment was not capable of androgen-directed regulation. Among the multiple, potential androgen response elements (AREs) identified by scanning sequences near and within the gene, two sequences within the intron of the murine Nkx3.1 gene were demonstrated to confer androgen-dependent transcription in reporter gene transfection experiments. Each of the elements, termed ARE A and ARE B, contained a 6-base pair core sequence, TGTTCT, that has been described as an androgen receptor half-site binding sequence, separated by 498 base pairs of DNA. Both of the intronic half-sites bind activated androgen receptor from a variety of sources, albeit with different apparent affinities. This region of the Nkx3.1 gene demonstrates a high degree of conservation among diverse species and mutagenesis experiments demonstrated that both elements are required for androgen stimulation. Taken together, our study shows that androgen-dependent transcription of the mouse Nkx3.1 gene is conferred through a noncanonical element within the intron of the gene.
Key words: Nkx3.1, Androgen receptor, Steroid regulation, Prostate, Cancer, Epithelia, Androgen response element (ARE)
INTRODUCTION
Nkx3.1 belongs to the Nk2 family (25,56) of homeodomain proteins, which contain a conserved tyrosine at position 54 of the homeodomain. The Nk2 proteins contain mutiple conserved domains, including the TN and NK2-SD, which are important for promoting interactions with other proteins and the regulation of target genes (7,32,55,58,70). Nkx3.1 expression is first observed in multiple tissues during development (1,5,26,51,54,57,60), while in the adult Nkx3.1 expression is predominant in the male urogenital system, including testis, seminal vesicle, and prostate (5,6,20,26,48,59). Loss of Nkx3.1 expression is associated with tissue dedifferentiation and initiation of prostate cancer, leading to the suggestion that it may be a tumor suppressor-like gene (1,2,5,34,42). Additionally, in the mouse, targeted disruption of Nkx3.1 leads to tissue dedifferentiation, prostate hyperplasia, and dysplasia (2,5,34,42,51). Thus, NKX3.1 is a critical regulator of prostate epithelial cell function; however, there is currently limited information on how the gene is regulated.
Maintenance of Nkx3.1 expression in prostate epithelia is highly dependent upon androgen signaling. Nkx3.1 expression has been shown to be significantly downregulated in prostate epithelial cells, which have been removed from androgen signaling both in vivo (6,26,48) and in responsive prostate cancer cell lines when androgen is removed from the culture media (6,18,26,48). Further, removal of androgen receptor expression in mature mouse prostate epithelia leads to a reduction in expression of several genes, including Nkx3.1, and these studies demonstrated that receptor-deficient cells were less differentiated and exhibited a hyperproliferative phenotype compared to wild-type littermates (45,66). Although several studies have identified basal promoter elements and DNA segments responsible for directing developmental prostate Nkx3.1 expression (10,27–29), to date it is not clear how androgen signaling directs/maintains prostate-specific expression of the gene. Androgen signaling occurs through a cytoplasmic receptor that becomes activated upon binding steroid. The activated receptor translocates into the nucleus where it binds specific DNA sequences, called the androgen response element (ARE). The androgen receptor belongs to a class of nuclear receptors termed the class I steroid receptors, which includes glucocorticoid, progesterone, and mineralocorticoid receptors that bind to DNA containing the core sequence 5′-TGTTCT-3′ for regulatory activity (12,22,49,64,65). While the class I steroid receptors all recognize similar DNA elements, it has been shown that androgen receptor possesses the capability to recognize sequences that vary from the consensus (12,22), indicating that gene-specific regulation by this receptor may be due to subtle sequence variations of the class I consensus.
We have examined the mechanism(s) by which androgen signaling affects Nkx3.1 gene expression because of the apparent relationship between the loss of androgen responsiveness and reduction of Nkx3.1 expression in metastatic prostate cancer. In the studies presented here, we show that there are multiple potential AREs near and within the gene. However, only DNA segments containing two of these core sequences found within the intron of the gene are capable of supporting functional androgen-dependent transcriptional activation. These core sequences are separated by 491 base pairs (bp) of DNA, bind activated androgen receptor, and both are required for androgen responsiveness. Thus, we have identified a nonconsensus ARE within the intron of the murine Nkx3.1 gene, further supporting the concept that androgen-dependent regulation is likely due to gene-specific location of DNA elements.
MATERIALS AND METHODS
Bioinformatics
rVISTA 2.0 (http://rvista.dcode.org/) was employed to produce local sequence alignments and to predict the regulatory potential of noncoding sequences (39,40).
Recombinant DNAs
Six different segments of the Nkx3.1 5′ promoter were amplified by PCR using specific primers with a bacterial artificial chromosome (BAC) containing the murine Nkx3.1 gene (RP23-116H24) as template. These PCR products were purified by column purification (Stratagene, La Jolla, CA, USA), and then cloned into the pGL3-Basic, promoterless luciferase vector. Several other regions of the Nkx3.1 gene, including intervening sequence (intron), exon 2 including the 3′ UTR (exon2_3′UTR), and sequences 3′ to the gene (Down5708bp), were obtained by PCR and then cloned into the Nkx3.1-up2716 bp promoter plasmid behind the luciferase gene and into the pGL3-promoter vector, 5′ to the SV40 promoter. Mutagenesis of the luciferase constructs was accomplished by PCR as previously described (9,18,70), using Nkx3.1-2716bp-intron as a template.
Cell Cultures, DNA Transfection, and Luciferase Assays
All cell lines in this study were obtained from the American Type Culture Collection (ATCC, Manassas, VA). Human prostate carcinoma cell lines LNCaP (ATCC; CRL-1740) and PC-3 (ATCC; CRL-1435), and monkey kidney COS-7 cells were maintained as previously described (18). The mouse prostate adenocarcinoma cell line TRAMP-C1 (ATCC; CRL-2730) was cultured in DMEM containing 0.5 mg/ml insulin, 10 nM dehydroisoandrosterone, 5% FBS, 5% Nu-serum IV, and 1% penicillin/streptomycin.
Transcriptional activation was measured by DNA transfection analysis essentially as described by Filmore et al. (18). Cells were plated onto a 24-well dish at a density of ∼50,000 cells/well and transfection was carried out with the Lipofectamine reagent (Invitrogen Life Technologies, Carlsbad, CA) using 0.25 μg of the luciferase construct per well. For experiments examining androgen sensitivity, cells were maintained in phenol red-free media containing charcoal-stripped FBS for 24 h prior to transfection. Androgen response was measured by adding 10 nM of the synthetic androgen R1881 (PerkinElmer, Waltham, MA) in ethanol, with control cells receiving a like amount of ethanol vehicle. Forty-eight hours posttransfection, the cells were harvested in 100 μl luciferase lysis buffer and 20 μl of the lysate assayed using a Turner Model 20 luminometer (Promega, Sunnyvale, CA). Protein concentration of the lysate was determined by the Bradford method (Bio-Rad, Hercules, CA) and used to normalize the luciferase activities, which were plotted as the mean activity ± SEM. To examine androgen stimulation, the normalized luciferase activities obtained from R1881-treated samples were compared with activities measured in lysate derived from cells receiving the same plasmid reporter constructs treated with ethanol vehicle.
Electrophoresis Mobility Shift Assay
TRAMP-C1 and LNCaP cells were used as a source of androgen receptor. Cells were plated onto 100-mm dishes and, 24 h postplating, the cells were treated with 10 nM R1881 for 48 h at 5% CO2 and 37°C. Protein lysates were prepared in PBS containing proteinase inhibitor cocktail by sonication and following removal of insoluble material, protein concentration of the lysate was determined by Bradford assay (Bio-Rad, Hercules, CA). Positive controls containing activated androgen receptor from the MCF-7 cell line were obtained from Santa Cruz Biological and used essentially as specified by the manufacturer (Santa Cruz Biotechnology, Santa Cruz, CA, USA). Double-stranded oligonucleotides containing potential AREs surrounding Nkx3.1 were synthesized as probes. The sequences are: ARE A forward 5′-aaattatggatgttcttttaagtctt-3′, ARE B forward 5′-tctctccttctgttcttctccagatg-3′, ARE C forward 5′-tgtgtaaggaagaacaggacaaaagg-3′, and ARE consensus from the human PSA gene (positive control) forward 5′-cttgcagaacagcaagtgctagctg-3′ (69). Oligonucleotides were end-labeled with [γ32P]ATP as described previously (8,9). Binding reactions were performed in a 25-μl reaction volume at room temperature. Each reaction mix contained 3 μg of double-stranded poly(dI-dC) and 30 μg of protein lysate. Assembled mixtures were incubated for 10 min in the absence of probe, and the reactions were then incubated for an additional 30 min at room temperature following addition of probe sequences (12,000 cpm). In supershift experiments, an androgen antibody (SC 7305X; Santa Cruz Biotechnology) was added to the reaction mix just prior to adding the oligonucleotide probe and in competition experiments unlabeled competitor DNA was added during preincubation as described (8,9). The samples were loaded onto a 5% polyacrylamide gel, and electrophoresis of samples carried out for 90 min at 180 V. The gel was dried onto filter paper and binding complexes were visualized by autoradiography on a Fuji FLA-5100 phosphoimager.
Statistics
Statistical analyses for luciferase assay were carried out using the Statistix for Windows software (Analytical Software, Tallahassee, FL) as previously described (9,18). The analyses performed utilized the Student t-test and AOV method of comparison of the means for statistical significance at p < 0.05.
RESULTS
Nkx3.1 Promoter Activity
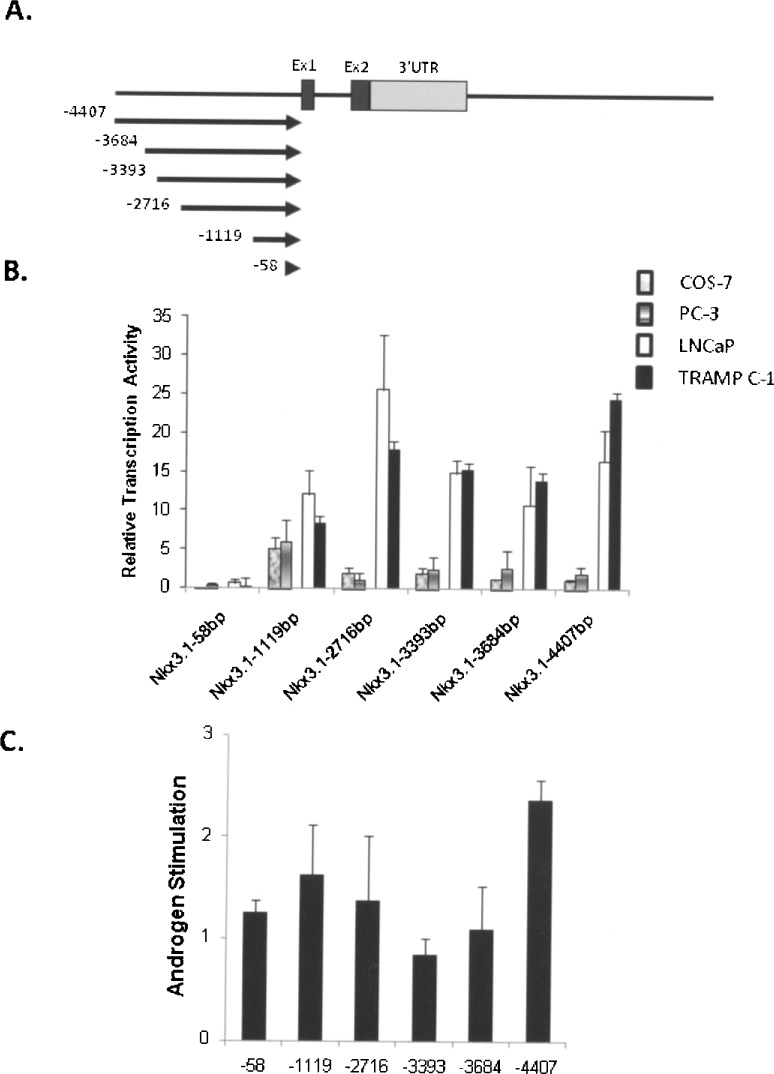
We initiated an assessment of the murine Nkx3.1 gene by analysis of sequences 5′ to this two-exon gene (Fig. 1A). To determine the basal transcription activity of Nkx3.1 upstream promoter, constructs containing serially deleted sequences 5′ to the gene were transiently transfected into prostate carcinoma cell lines, and non-prostate cells as controls. Previous studies demonstrated that the androgen-dependent LNCaP cells express Nkx3.1 in a dose-dependent fashion, while no expression is observed in the androgen-independent PC3 cells (18). As shown in Figure 1B, PC3 cells maintained Nkx3.1 promoter activity around fivefold relative to the vector-only with constructs containing ∼1100 bp or more of sequence. In androgen-responsive LNCaP cells, the 1119-bp construct stimulated transcription by 12.5-fold, which was increased to 25-fold with additional sequence to −2716 bp. This increased transcriptional response was maintained throughout the rest of promoter constructs to ∼4.4 kb of 5′ sequence (Fig. 1B). Overall, promoter activity was higher in LNCaP cells than observed in PC3 cells. These results suggest that Nkx3.1 prostate enhanced expression is governed by sequences within the upstream 2716 bp of the coding region. The mouse prostate carcinoma cell line TRAMP-C1 also expresses androgen receptor (19). The mouse cells demonstrated a similar result as the human LNCaP cells (Fig. 1B), indicating that the initial ∼2.7 Kb adjacent to the gene demonstrates enhanced transcriptional activity in androgen-responsive prostate cells. Only marginal activity was observed in non-prostate cells (COS-7) with the Nkx3.1 1119-bp construct, indicating a preference for prostate cells in the transcriptional activation. Taken together, these data indicate that prostate-selective expression of Nkx3.1 is directed by DNA sequence between −2716 and −1119 bp upstream (5′) of Nkx3.1 coding sequence.
Figure 1.
Analysis of mouse Nkx3.1 promoter. The transcriptional capacity of the 5′ promoter region of the mouse Nkx3.1 gene was assayed in androgen-responsive prostate cell lines, LNCaP and TRAMP C-1; nonresponsive prostate cells, PC-3; and non-prostate cells, COS-7, by transfection as described in Materials and Methods. (A) A diagram of the two-exon Nkx3.1 gene is shown with transcriptional initiation site in exon 1. The relative positions of the six promoter deletion fragments cloned into the pGL3-basic luciferase vector is denoted by the arrows, each containing a common 3′ end within the gene (+35) and the different 5′ end positions shown relative to the transcriptional start site. (B) The Nkx3.1 promoter deletion constructs were placed into LNCaP (open bars), TRAMP C-1 (filled bars), PC-3 (stripped bars), and COS-7(gray bars) cells and the resultant luciferase activity measured in cellular lysates measured 48 h after transfection as described in Materials and Methods. Results are shown as the relative transcriptional activity by each promoter fragment compared to the activity generated by the empty vector (pGL3-basic) in each cell line. Results were derived from a minimum of three separate experiments performed in triplicate and the measured activity is shown ± SEM shown by the error bars. (C) Androgen stimulation of the 5′ promoter deletion fragments was measured in the androgen-responsive LNCaP cell line. The results are shown as the fold stimulation of each construct in cells treated with the synthetic androgen R1881 compared to the activity derived from the same construct in cells treated with vehicle (ethanol) alone. Results were derived from a minimum of three experiments performed in triplicate for each condition with the SEM denoted by the error bars.
There are several potential AREs within this prostate-selective region that may participate in androgen-dependent activation of Nkx3.1 gene expression. To locate AREs within this segment of the Nkx3.1 locus, we transfected the same promoter constructs in LNCaP cells with or without synthetic androgen, R1881. Reporters containing solely DNA 5′ to the Nkx3.1 gene were induced by one- to twofold in androgen receptor-positive cells (Fig. 1C), suggesting that there are no functional AREs in the upstream (5′) region that greatly facilitate androgen-dependent Nkx3.1 gene regulation.
Functional AREs Are Located Within the Nkx3.1 Gene
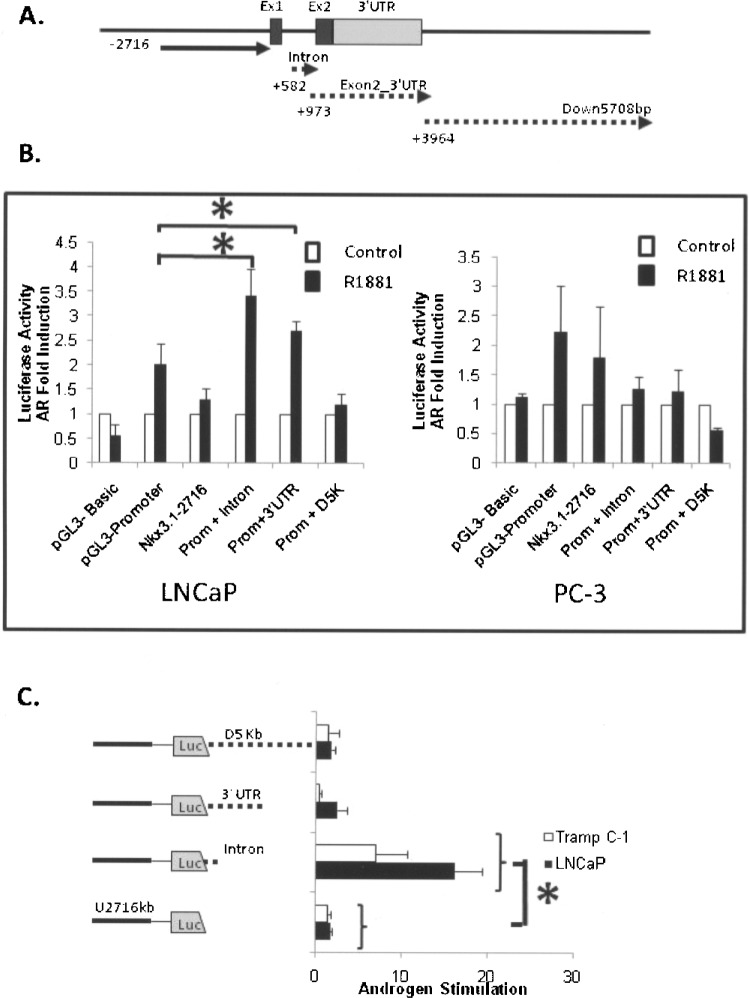
We reasoned that androgen-regulated Nkx3.1 transcription might involve DNA elements within or 3′ to the gene because DNA 4.4 kb in front of the gene was not capable of significant androgen-directed activation. To identify potential AREs in DNA within the Nkx3.1 gene and 3′ surrounding DNA, we cloned three separate segments from the Nkx3.1 locus into luciferase vectors and tested for their capability to drive androgen-dependent transcription. As shown in Figure 2A, these DNA segments were from within (intron and Exon2_3′UTR fragments) and just 3′ (Down5708bp fragment, Fig 2A) to the Nkx3.1 gene. These DNA regions each contain multiple potential ARE core sequences as determined by computer analyses. In order to evaluate if any of the DNA sequences within the gene are capable of functional androgen stimulation, they were cloned into the multi-cloning site adjacent to the SV40 minimal promoter of pGL3-promoter vector (Fig. 2B) and 3′ to the luciferase gene in the Nkx3.1 promoter vector −2716-luc (Fig. 2C). The latter vectors were constructed so that the DNAs were within an approximate context found within the native Nkx3.1 genomic locus; that is, 3′ to the prostate selective promoter and the structural Nkx3.1 segment (Fig. 2A). Androgen-dependent stimulation (2.5–4-fold) was observed in constructs containing the intron and the Exon2_3′UTR segments in responsive LNCaP cells not found in the androgen-deficient PC-3 prostate cells (Fig. 2B). As indicated in Figure 2C, the activity of Nkx3.1 −2716 bp + intron construct was stimulated by 10 nM R1881 approximately 20-fold in the LNCaP and sevenfold in TRAMP C-1 androgen receptor-positive cells, while other regions had less than threefold induction by androgen. Taken together, these data indicate that the intron and 3′ UTR regions contain functional ARE(s) and the transcriptional ability of these elements is influenced by the sequence location and/or context.
Figure 2.
Androgen transcriptional responses elicited by DNA fragments within and 3′ to the mouse Nkx3.1 gene. (A) A diagram of the gene with the relative position of the cloned DNA fragments is shown. The dotted lines denote the fragments cloned into the pGL3-promoter in front of the minimal SV-40 promoter and into the Nkx3.1-2716 vector behind the luciferase coding segment. (B) The three gene fragments shown in (A) (intron, Exon2_3′UTR, and Down 5708bp) cloned into the pGL3-promoter vector were assayed for androgen stimulation in LNCaP cells as described in Materials and Methods. Following transfection the cells were treated with 10 nM synthetic androgen R1881 (filled bars) or vehicle (open bars) and lystaes from the cells analyzed for luciferase activity. The activity derived from androgen-treated cells was compared with the activity derived from vehicle-treated cells and the resultant data plotted as an androgen (AR) fold induction. As controls, plasmids with no promoter (pGL3-basic), the minimal SV40 promoter (pGL3-promoter), and the 5′ Nkx3.1 promoter segment (Nkx3.1-2716) were transfected into LNCaP cells and the entire complement of vectors were transfected into the non-androgen-responsive PC-3 cell line. The asterisks denote statistical difference in androgen induction between the test construct and the parental pGL3-promoter plasmid (p < 0.05). (C) The three DNA fragments shown in (A) were cloned into the Nkx3.1-2716 vector after the luciferase coding sequence and the resulting clones evaluated for androgen stimulation in LNCaP cells (black bars) and TRAMP-C1 cell (open bars) as described above. Androgen stimulation was determined as the activity derived from transfected cells treated with R1881 compared to cells containing the same promoter/DNA luciferase vector that were treated with vehicle alone. The asterisk denotes statistical differences between the test plasmid and the parental control plasmid Nkx3.1-2716 (p < 0.05).
AR Binds to ARE(s) Located in Intron of Nkx3.1
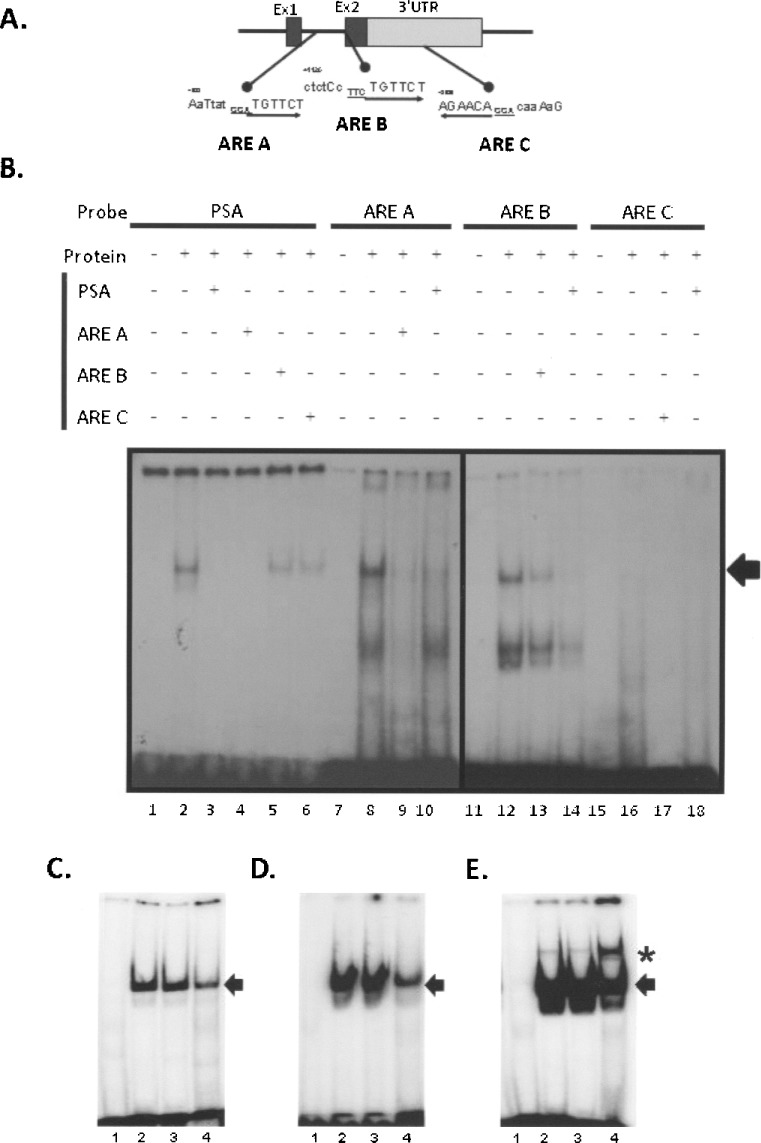
Having determined that functional activation of Nkx3.1 by androgen was dependent on intronic DNA segments, we next assessed if sequences within this region was bound by AR using electrophoretic mobility shift assay (EMSA). Within the intron, there are two potential androgen receptor binding sites (ARE half-sites) predicted by rVISTA (illustrated in Fig. 3A) and we termed them ARE A and ARE B. Additionally, there is a potential ARE identified by bioinformatics within the 3′ UTR segment of the gene (ARE C), conserved in sequence and location to the human Nkx 3.1 gene (61,62). All the potential AREs contained the core steroid receptor binding sequence (6 bp: TGTTCT) and we tested their capacity to bind androgen receptor by fashioning oligonucleotide probes that contained 10 bases before and after the core binding sequence for a total probe length of 26 bases. Whole cell lysate from TRAMP-C1 cells treated with the synthetic androgen R1881 was used in initial experiments (Fig. 3B). An oligonucleotide probe containing the AR binding sequence for the PSA gene served as a positive control for these experiments. As shown in Figure 3B, a single complex was observed with the PSA ARE, while multiple species were observed with Nkx 3.1 ARE A, and ARE B, but no binding was observed with ARE C probe (Fig. 3B, lanes 2, 8, 12, and 16). We next examined sequence-specific binding using competition experiments. Cold PSA probe effectively inhibited complex formation with the labeled PSA sequence (lane 3) and like migrating complexes formed with ARE A (lane 10) and ARE B (lane 14) probes. Additionally, unlabeled ARE A (50-fold excess) was able to effectively compete the positive control PSA probe binding (lane 4); however, ARE B reduced but did not totally fully inhibit PSA complex formation (lane 5). These data show that Nkx3.1 ARE A and B are capable of binding activated androgen receptor, albeit with different apparent affinities.
Figure 3.
Evaluation of androgen receptor binding to potential AREs in the Nkx3.1 gene intron and 3′ UTR. (A) The location of potential AREs within the Nkx3.1 gene relative to the intron and second exon containing the 3′ UTR are shown in this diagram. The sequence of each potential ARE is indicated with the capital letters showing sequence identity with recognized AREs and the arrow denotes the sequence polarity (5′ to 3′). Oligonucleotides for each of the potential Nkx3.1 AREs, ARE A, ARE B, and ARE C, were constructed to test their ability for androgen receptor binding. (B) Androgen receptor binding was evaluated using protein lysate derived from Tramp C-1 cells activated with R1881 as detailed in Materials and Methods. Oligonucleotides representing each potential Nkx3.1 ARE along with a positive control ARE from the human PSA gene were radiolabeled and incubated with no protein lysate (lanes 1, 7, 11, and 15), with protein lysate alone (lanes 2, 8, 12, and 16), with lysate and a 500-fold excess of unlabeled self-probe (lanes 3, 9, 13, and 17), or with 500-fold excess of unlabeled PSA probe (lanes 10, 14, and 18). To further assess specific androgen receptor binding, the labeled PSA reaction was incubated with 500-fold excess unlabeled ARE A (lane 4), ARE B (lane 5), and ARE C (lane 6). The arrow at the right of the autoradiogram denotes the specific complexes formed with androgen receptor. (C) The probes were assayed with lysate from androgen receptor-positive control MCF-7 cells. Shown is an autoradiogram derived from incubating the lysate with ARE C (lane 1), ARE B (lane 2), ARE A (lane 3), and PSA ARE (positive control, lane 4) with the arrow denoting the position of the androgen binding complex. (D) Lysate derived from R1881-stimulated LNCaP androgen-responsive cells was used to probe for androgen receptor binding. The resultant reactions shown are ARE C (lane 1), ARE B (lane 2), ARE A (lane 3), and PSA ARE (positive control, lane 4) and the arrow shows the position of ARE binding complex. (E) A supershift experiment using an anti-androgen receptor antibody is shown. The oligonucleotide probes were incubated with LNCaP cell lysates as in (D) and in each reaction there was added 1 μl of androgen receptor antibody as described in Materials and Methods. The arrow shows the position of the androgen receptor binding complex and the asterisk shows the position of the supershifted band in response to added antibody.
We further assessed androgen receptor binding by examining the binding of the murine Nkx 3.1 probes along with the PSA ARE-positive control using lysates from a commercial source and derived from LNCaP cells. The MCF7 androgen receptor-positive control lysate (Fig. 3C) and cell extract from LNCaP human prostate cancer cells (Fig. 3D) showed significant binding to the ARE A and B probes (lanes 3 and 2 respectively, Fig. 3C and D) consistent with the positive control PSA ARE (lane 4, Fig. 3C and D). Additionally, we preformed a supershift assay using LNCaP lysates and an androgen receptor antibody that has been previously shown to identify androgen receptor complexes (37,41). As illustrated in Figure 3E, inclusion of this antibody into the reaction caused a supershift of the PSA probe (lane 4) and to a lesser extent the Nkx3.1 intronic ARE core sequences (lanes 2 and 3). The difference in apparent antibody binding and supershift may be due to the difference in the binding sites, with ARE A and ARE B being nonexact palindrome androgen half-sites and the PSA control being a consensus ARE binding for dimerized receptor. Taken together, these competition and supershift experiments demonstrate that ARE core sequences conserved within the intron of the murine Nkx3.1 gene are capable of androgen receptor complex binding.
Two ARE Half-Sites Are Responsible for Androgen Transcriptional Enhancement
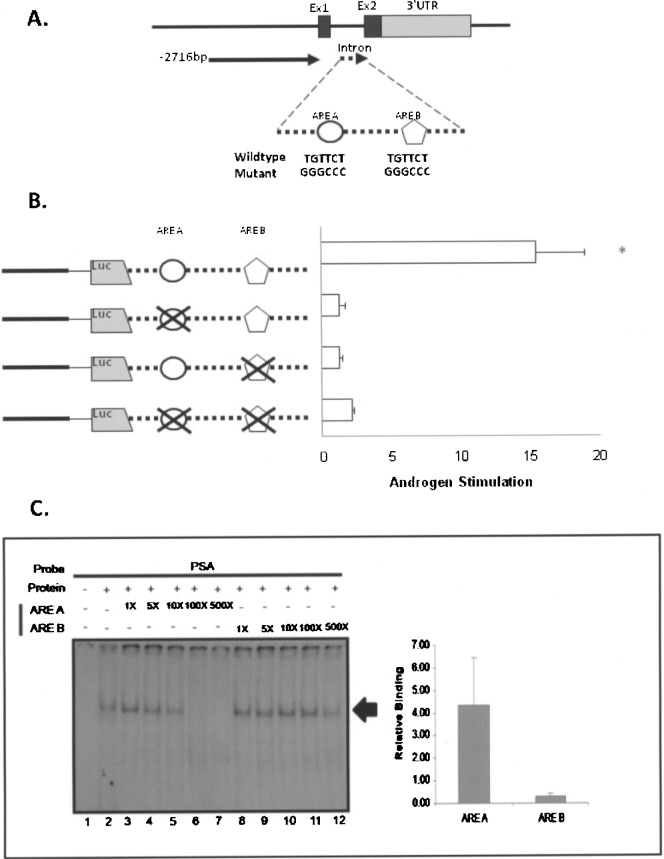
To directly examine the contribution of the intronic AREs to androgen activation of Nkx3.1 transcription, plasmid Nkx3.1 −2716 promoter + intron (Fig. 2) was utilized to create individual mutations of ARE A, ARE B, and a vector in which both elements were mutated. These DNAs transfected into androgen-responsive prostate cell lines and evaluated for androgen activation by adding the synthetic steroid R1881. Individual mutation of ARE A or ARE B consistently abolished all androgen responsiveness, indicating that both ARE A and ARE B are required for androgen-dependent stimulation of the Nkx3.1 intron (Fig. 4B). This was also observed for the promoter/intron doubly mutated DNA. Thus, with mutagenesis, we have established that both the AREs within the Nkx3.1 gene intron are required for androgen responsiveness in prostate epithelial cells.
Figure 4.
Relative affinity of androgen receptor binding to Nkx3.1 ARE A and ARE B. (A) A diagram of Nkx3.1 intron with two potential AREs (ARE A and ARE B) is shown. The WT and mutated sequences for each ARE (ARE A and ARE B) are shown. Mutations were constructed in the Nkx3.1-2716 + intron vector shown in Figure 2. (B) Androgen Stimulation of Nkx3.1-2716 + intron, ARE A mutant, ARE B mutant, and double mutant was tested in LNCaP cells. The results shown are calculated as the fold induction of each construct in cells treated with R1881 relative to cells treated with vehicle (ethanol) alone. Data are generated from three experiments performed in quadruplet for each condition and were plotted as the mean ± SEM and the asterisk indicates significantly different from other mutant constructs (p > 0.05). (C) Radiolabeled PSA ARE was incubated with lysate derived from the Tramp C-1 cell line and tested for relative binding affinity by competition experiments. The experiment included the PSA ARE probe incubated with no protein lysate (lane 1), or with protein lysate alone (lane 2), and with protein lysate containing 1-fold, 5-fold, 10-fold, 100-fold, and 500-fold molar excess of unlabeled ARE A (lanes 3–7) or ARE B (lanes 8–12). The resultant bands were visualized on a Fuji 5100 phosphoimager and the amount of radioactivity associated with each band was quantified by scanning densitometry. For graphical analysis, the band intensity of each lane was quantified, the values plotted versus the molar excess of added unlabeled probe, and the slope of the resultant line was calculated for ARE A and ARE B, respectively, and plotted as relative binding. The values obtained for three separate experiments were averaged and plotted ±SEM.
Although these experiments indicated that the functional AR regulation of Nkx3.1 expression is due to AR binding, the apparent affinity of AR to the elements appeared different (Fig. 3). With excess cold competitor, ARE A effectively abolished the observed binding complex of PSA ARE, while ARE B did not. To examine the relative affinity of AR to ARE A and ARE B, the PSA probe was examined by competition with increasing amounts (1–500×) of ARE A or ARE B (Fig. 4C). With ARE A, the PSA ARE complex intensity decreased as the amount of unlabeled ARE A increased. The binding complex totally disappears with the addition of 100× and 500× of ARE A. However, with ARE B, although the band intensity is decreased gradually with increased unlabeled ARE B, it never completely competes away binding (Fig. 4C). The intensity of the binding complex was semiquantified by scanning densitometry and image analysis, which indicates that AR binds to ARE A with a higher affinity compared to ARE B (Fig. 4C). Therefore, our experiments demonstrate that AR is binding to both ARE A and ARE B, and AR has a higher apparent affinity for ARE A compared to ARE B.
DISCUSSION
Nkx3.1 is a homeobox gene that is a homolog of Drosophila bagpipe and has been implicated in prostate organogenesis and carcinogenesis. Because androgen is known to play a significant role in these processes, the observation that Nkx3.1 is AR responsive demonstrates that it is likely a critical component of prostate epithelial cell homeostasis. In this study, we characterized the androgen regulation of Nkx3.1 gene in prostate epithelial cells. DNA sequences within the 5′ segment of the murine Nkx3.1 gene (4.3 kb) was screened for its promoter activity in AR-positive LNCaP and Tramp C-1 cell lines and AR-negative PC3 cells. We identified a 2.7-kb region 5′ to the gene that demonstrates enhanced transcription in prostate cells; however, this region was not associated with androgen-driven gene transcription. Subsequent analysis of DNA within and surrounding the Nkx3.1 gene showed that the intron region exhibited androgen-dependent transcriptional activation in transfection analyses. The androgen activation was dependent upon two specific DNA elements that contain the well-conserved half-site core binding sequence, TGTTCT separated by 491 bp of DNA. Both are bound by activated AR, and both sites are required for functional gene activation by AR. Thus, androgen-enhanced expression of Nkx3.1, a critical gene product for prostate development and maturation, is manifested through specific DNA elements within the intron of this two-exon gene.
Our first question was to ascertain if the sequences 5′ to the Nkx3.1 gene contained functional ARE(s) to direct prostate-specific and androgen-responsive transcription. There are many sequences within the DNA surrounding the gene as well as within the Nkx3.1 gene that conform to consensus ARE, specifically the half-site core binding sequence. Although several of the potential AREs are found within sequences 5′ to the gene, a position conventionally considered being critical for appropriate gene transcription, our studies demonstrate that 5′ DNA up to ∼4.5 kb is not capable of conferring AR-dependent transcriptional activity to the Nkx3.1 gene. Yoon and Wong (67) have shown that DNA 5′ to the human Nkx3.1 gene contains androgen receptor binding sites using chromatin immunoprecipitation (CHiP) analyses; however, this binding was not evaluated for function with regard to Nkx3.1 gene transcription. Similar results were observed using a computational analysis of the Nkx3.1 promoter for GATA factor and androgen receptor binding sites, albeit predicting a different AR binding site than that of Yoon and Wong (67) and again no functional measures were determined (43). Our experiments examined equivalent segments of DNA from the murine Nkx3.1 gene and found no functional androgen-dependent transcriptional activity resided within this gene segment. This is consistent with functional analyses of the human Nkx3.1 promoter, which found no functional AREs within DNA 5′ to the gene (61,62). Thus, these 5′ binding sites may not be needed for functional activity, or more likely are not sufficient for androgen-dependent transcriptional activation.
Our studies showing that an appropriate context was necessary to elicit enhanced AR transcriptional responses in our reporter gene constructs (Fig. 2) might indicate that the 5′ promoter sequences work in conjunction with other DNA regions for AR-dependent transcriptional activation. Moreover, in transgenic animal studies a potential enhancer for prostate cell regulation has been identified in DNA sequences ∼5 kb 3′ to the end of the gene (10). This previously identified prostate-specific segment might also participate with our prostate-selective 5′ segment to provide prostate-specific basal transcription activation. The DNA sequence 5′ to the gene did elicit a transcriptional response in prostate cell lines, indicating that this segment housed a prostate cell-selective transcriptional capacity. In particular, our studies demonstrated that the initial 2.7 kb of DNA adjacent to the gene exhibited an enhanced transcriptional response in LNCaP and Tramp C-1, both of which are androgen receptor-positive prostate carcinoma cell lines (Fig. 2). This basal activation was found to be significantly different in magnitude than that observed in androgen receptor-negative PC-3 cells. The PC-3 cell line represents a terminal stage of prostate cancer in which the cells are aggressive in metastasis and are no longer androgen responsive. Thus, our results indicate that in the process of prostate cancer progression there is a fundamental change in the basal transcriptional capacity of prostate genes likely due to changes in transcriptional activator complexes within these cells. Similar changes in basal activators have been found in other systems commensurate with the epithelial–mesenchymal transition (EMT) demonstrated in cancer progression (35). Taken together, our studies support the concept that the appropriate regulation of Nkx3.1 in prostate development and maturation requires input from multiple DNA segments surrounding and within the structural gene.
Functional ARE(s) were not found within the prostate-specific 2.7-kb region of the murine Nkx3.1 promoter; instead, we mapped them to the intronic DNA. Our results are consistent with recent studies of the human Nkx3.1 gene (61,62), which found that while there were no retinoic acid response elements within or near the gene, DNA comprising the intron and second exon was able to elicit an androgen response. Here we have localized this response to sequences within the intron of the murine gene. While different in exact locations within the Nkx3.1 gene, the mouse and human genes are both activated by internal ARE binding sequences that are half-site consensus (TGTTCT) separated by significant sequence (61). Interestingly, the one sequence segment demonstrating the highest degree of conservation in location and structure, ARE 3 in humans (61) and ARE C in mouse, is capable of influencing androgen transcriptional responses without androgen receptor binding (Figs. 2 and 3). Although there are many examples of androgen regulation via receptor binding to a canonical ARE palindromic sequence, both within the 5′ promoter (13,14,33,36,47,69) and in other locations with the gene locus (53,65), it has been suggested that androgen response DNA elements can significantly vary from this consensus. This includes sequence differences within the ARE element palindrome and the presence of singular core binding elements, termed the ARE half-sites (3,11,12,65). Further, it has been suggested that the context and placement of half-site ARE within or near a gene can contribute specificity to androgen selectivity of transcription. For example, the murine androgen receptor gene contains two functional half-site AREs (ARE-1, TGTCCT; ARE-2, AGTACTCC) within its coding sequences that are separated by 182 bp and these AREs synergistically regulate the AR mRNA expression in a spatiotemporal manner (15). As suggested by in vitro mutagenesis experiments (21,22,64,65), the separation of half-site AREs can contribute to androgen selectivity. Thus, our demonstration of intronic half-sites separated by ∼500 bp that govern Nkx3.1 androgen-specific transcription indicates that this gene is regulated by a mechanism different from the consensus steroid receptor activation in which a receptor dimmer binds to a palindrome sequence. We would suggest that the observed intense androgen-driven Nkx3.1 transcriptional response is derived from the unique character of the intronic half-site ARE core elements.
Our experiments demonstrated a relatively weak affinity for androgen receptor binding by the individual half-site core sequences (ARE A and ARE B) and an apparent difference in binding affinity between these two core binding elements (Figs. 3 and 4). Thus, it is the inherent qualities of the androgen receptor binding to the intronic Nkx3.1 ARE half-sites that likely define the ability of androgen to selectively activate transcription of this gene in the maturing prostate epithelia. This appears to be the case with the murine pem and the human secretory compartment (sc) gene, both of which contain weak binding half-site core sequences that are critical for androgen selectivity and responsiveness (3,24,63). In a spacio- and temporal-specific manner, different affinities of AR could be serving an important role through cooperative interactions with adjacent or nearby transcription factors (38,46). In the case of Nkx3.1, both ARE A and ARE B contain ARE half-site sequence, TGTTCT, that are separated by 491 bp. The sequences adjacent to the ARE, TGTTCT, were not found to contain any known AR dimer binding sequences by homology. Based on previous studies, sequences surrounding functional AREs could have effect on the AR affinity and gene regulation (16,22,68). Further, it has been suggested that AR monomers or dimmers may bind depending availability of other active transcription factors that influence the gene expression (4,17,23,30,31,44,46,50,52). The association of coactivators that augment or alter the activity of nuclear receptors in response to steroid ligand seems to be a common regulatory paradigm, including AR, ER, and other ligand-dependent nuclear receptors (38,46). Thus, in both mice and humans (61,62) the Nkx3.1 gene is androgen responsive through internal AREs, and what coactivators, if any, participate in this process remains to be elucidated.
In conclusion, our data demonstrate that two functional AREs located in the intron region are critical for androgen-dependent transcriptional activation of the Nkx3.1 gene. There were no recognizable sites for dimerized ARs, which indicates a mechanism whereby sequences separated by ∼500 bp come together for receptor dimerization and could suggest that there are other transcription factors that are involved in the androgen regulation of Nkx3.1 gene through direct or indirect interaction with AR. An identification and characterization of Nkx3.1 upstream regulators that work with the intronic androgen response element will help elucidate the molecular mechanism controlling appropriate Nkx3.1 gene expression and eventually reveal the pathogenic defects that result in loss of Nkx3.1 expression during prostate carcinogenesis.
ACKNOWLEDGMENTS
This work was conducted in partial fulfillment of the Ph.D. requirements for Chinatsu Kojioma. The authors wish to acknowledge the members of the Zimmer laboratory for their excellent technical help and many suggestions during the course of this work.
REFERENCES
- 1. Abate-Shen C.; Shen M. M.; Gelmann E. Integrating differentiation and cancer: The Nkx3.1 homeobox gene in prostate organogenesis and carcinogenesis. Differentiation 76:717–727; 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Abdulkadir S. A.; Magee J. A.; Peters T. J.; Kaleem Z.; Naughton C. K.; Humphrey P. A.; Milbrandt J. Conditional loss of Nkx3.1 in adult mice induces prostatic intraepithelial neoplasia. Mol. Cell. Biol. 22:1495–1503; 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Barbulescu K.; Geserick C.; Schuttke I.; Schleuning W. D.; Haendler B. New androgen response elements in the murine pem promoter mediate selective transactivation. Mol. Endocrinol. 15:1803–1816; 2001. [DOI] [PubMed] [Google Scholar]
- 4. Bennett N. C.; Gardiner R. A.; Hooper J. D.; Johnson D. W.; Gobe G. C. Molecular cell biology of androgen receptor signalling. Int. J. Biochem. Cell Biol. 42:813–827; 2010. [DOI] [PubMed] [Google Scholar]
- 5. Bhatia-Gaur R.; Donjacour A. A.; Sciavolino P. J.; Kim M.; Desai N.; Young P.; Norton C. R.; Gridley T.; Cardiff R. D.; Cunha G. R.; Abate-Shen C.; Shen M. M. Roles for Nkx3.1 in prostate development and cancer. Genes Dev. 13:966–977; 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bieberich C. J.; Fujita K.; He W. W.; Jay G. Prostate-specific and androgen-dependent expression of a novel homeobox gene. J. Biol. Chem. 271:31779–31782; 1996. [DOI] [PubMed] [Google Scholar]
- 7. Bowen C.; Stuart A.; Ju J. H.; Tuan J.; Blonder J.; Conrads T. P.; Veenstra T. D.; Gelmann E. P. NKX3.1 homeodomain protein binds to topoisomerase I and enhances its activity. Cancer Res. 67:455–464; 2007. [DOI] [PubMed] [Google Scholar]
- 8. Browning C. L.; Culberson D. E.; Aragon I. V.; Fillmore R. A.; Croissant J. D.; Schwartz R. J.; Zimmer W. E. The developmentally regulated expression of serum response factor plays a key role in the control of smooth muscle-specific genes. Dev. Biol. 194:18–37; 1998. [DOI] [PubMed] [Google Scholar]
- 9. Carson J. A.; Fillmore R. A.; Schwartz R. J.; Zimmer W. E. The smooth muscle gamma-actin gene promoter is a molecular target for the mouse bagpipe homologue, mNkx3-1, and serum response factor. J. Biol. Chem. 275:39061–39072; 2000. [DOI] [PubMed] [Google Scholar]
- 10. Chen H.; Mutton L. N.; Prins G. S.; Bieberich C. J. Distinct regulatory elements mediate the dynamic expression pattern of Nkx3.1. Dev. Dyn. 234:961–973; 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Claessens F.; Verrijdt G.; Schoenmakers E.; Haelens A.; Peeters B.; Verhoeven G.; Rombauts W. Selective DNA binding by the androgen receptor as a mechanism for hormone-specific gene regulation. J. Steroid Biochem. Mol. Biol. 76:23–30; 2001. [DOI] [PubMed] [Google Scholar]
- 12. Claessens F.; Verrijdt G.; Haelens A.; Callewaert L.; Moehren U.; d’Alesio A.; Tanner T.; Schauwaers K.; Denayer S.; Van Tilborgh N. Molecular biology of the androgen responses. Andrologia 37:209–210; 2005. [DOI] [PubMed] [Google Scholar]
- 13. Cleutjens K. B.; van der Korput H. A.; van Eekelen C. C.; van Rooij H. C.; Faber P. W.; Trapman J. An androgen response element in a far upstream enhancer region is essential for high, androgen-regulated activity of the prostate-specific antigen promoter. Mol. Endocrinol. 11:148–161; 1997. [DOI] [PubMed] [Google Scholar]
- 14. Crozat A.; Palvimo J. J.; Julkunen M.; Janne O. A. Comparison of androgen regulation of ornithine decarboxylase and S-adenosylmethionine decarboxylase gene expression in rodent kidney and accessory sex organs. Endocrinology 130:1131–1144; 1992. [DOI] [PubMed] [Google Scholar]
- 15. Dai J. L.; Burnstein K. L. Two androgen response elements in the androgen receptor coding region are required for cell-specific up-regulation of receptor messenger RNA. Mol. Endocrinol. 10:1582–1594; 1996. [DOI] [PubMed] [Google Scholar]
- 16. Darne C. H.; Morel L.; Claessens F.; Manin M.; Fabre S.; Veyssiere G.; Rombauts W.; Jean C. L. Ubiquitous transcription factors NF1 and Sp1 are involved in the androgen activation of the mouse vas deferens protein promoter. Mol. Cell. Endocrinol. 132:13–23; 1997. [DOI] [PubMed] [Google Scholar]
- 17. Denayer S.; Helsen C.; Thorrez L.; Haelens A.; Claessens F. The rules of DNA recognition by the androgen receptor. Mol. Endocrinol. 24:898–913; 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Filmore R. A.; Dean D. A.; Zimmer W. E. The smooth muscle gamma-actin gene is androgen responsive in prostate epithelia. Gene Expr. 10:201–211; 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Foster B. A.; Gingrich J. R.; Kwon E. D.; Madias C.; Greenberg N. M. Characterization of prostatic epithelial cell lines derived from transgenic adenocarcinoma of the mouse prostate (TRAMP) model. Cancer Res. 57:3325–3330; 1997. [PubMed] [Google Scholar]
- 20. Gelmann E. P.; Bowen C.; Bubendorf L. Expression of NKX3.1 in normal and malignant tissues. Prostate 55:111–117; 2003. [DOI] [PubMed] [Google Scholar]
- 21. Geserick C.; Meyer H. A.; Barbulescu K.; Haendler B. Differential modulation of androgen receptor action by deoxyribonucleic acid response elements. Mol. Endocrinol. 17:1738–1750; 2003. [DOI] [PubMed] [Google Scholar]
- 22. Geserick C.; Meyer H. A.; Haendler B. The role of DNA response elements as allosteric modulators of steroid receptor function. Mol. Cell. Endocrinol. 236:1–7; 2005. [DOI] [PubMed] [Google Scholar]
- 23. Glass C. K. Differential recognition of target genes by nuclear receptor monomers, dimers, and heterodimers. Endocr. Rev. 15:391–407; 1994. [DOI] [PubMed] [Google Scholar]
- 24. Haelens A.; Verrijdt G.; Schoenmakers E.; Alen P.; Peeters B.; Rombauts W.; Claessens F. The first exon of the human sc gene contains an androgen responsive unit and an interferon regulatory factor element. Mol. Cell. Endocrinol. 153:91–102; 1999. [DOI] [PubMed] [Google Scholar]
- 25. Harvey R. P. NK-2 homeobox genes and heart development. Dev. Biol. 178:203–216; 1996. [DOI] [PubMed] [Google Scholar]
- 26. He W. W.; Sciavolino P. J.; Wing J.; Augustus M.; Hudson P.; Meissner P. S.; Curtis R. T.; Shell B. K.; Bostwick D. G.; Tindall D. J.; Gelmann E. P.; Abate-Shen C.; Carter K. C. A novel human prostate-specific, androgen-regulated homeobox gene (NKX3.1) that maps to 8p21, a region frequently deleted in prostate cancer. Genomics 43:69–77; 1997. [DOI] [PubMed] [Google Scholar]
- 27. Jiang A. L.; Zhang J. Y.; Young C.; Hu X. Y.; Wang Y. M.; Liu Z. F.; Hao M. L. Molecular cloning and characterization of human homeobox gene Nkx3.1 promoter. Acta Biochim. Biophys. Sin. (Shanghai) 36:64–67; 2004. [DOI] [PubMed] [Google Scholar]
- 28. Jiang A. L.; Zhang P. J.; Hu X. Y.; Chen W. W.; Kong F.; Liu Z. F.; Yuan H. Q.; Zhang J. Y. Identification of a positive Cis-element upstream of human NKX3.1 gene. Acta Biochim. Biophys. Sin. (Shanghai) 37:773–778; 2005. [DOI] [PubMed] [Google Scholar]
- 29. Jiang A. L.; Zhang P. J.; Chen W. W.; Liu W. W.; Yu C. X.; Hu X. Y.; Zhang X. Q.; Zhang J. Y. Effects of 9-cis retinoic acid on human homeobox gene NKX3.1 expression in prostate cancer cell line LNCaP. Asian J. Androl. 8:435–441; 2006. [DOI] [PubMed] [Google Scholar]
- 30. Jiang M.; Ma Y.; Chen C.; Fu X.; Yang S.; Li X.; Yu G.; Mao Y.; Xie Y.; Li Y. Androgen-responsive gene database: Integrated knowledge on androgen-responsive genes. Mol. Endocrinol. 23:1927–1933; 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jin F.; Fondell J. D. A novel androgen receptor-binding element modulates Cdc6 transcription in prostate cancer cells during cell-cycle progression. Nucleic Acids Res. 37:4826–4838; 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ju J. H.; Maeng J. S.; Lee D. Y.; Piszczek G.; Gelmann E. P.; Gruschus J. M. Interactions of the acidic domain and SRF interacting motifs with the NKX3.1 homeodomain. Biochemistry 48:10601–10607; 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kasper S.; Rennie P. S.; Bruchovsky N.; Lin L.; Cheng H.; Snoek R.; Dahlman-Wright K.; Gustafsson J. A.; Shiu R. P.; Sheppard P. C.; Matusik R. J. Selective activation of the probasin androgen-responsive region by steroid hormones. J. Mol. Endocrinol. 22:313–325; 1999. [DOI] [PubMed] [Google Scholar]
- 34. Kim M. J.; Bhatia-Gaur R.; Banach-Petrosky W. A.; Desai N.; Wang Y.; Hayward S. W.; Cunha G. R.; Cardiff R. D.; Shen M. M.; Abate-Shen C. Nkx3.1 mutant mice recapitulate early stages of prostate carcinogenesis. Cancer Res. 62:2999–3004; 2002. [PubMed] [Google Scholar]
- 35. Laffin B.; Wellberg E.; Kwak H. I.; Burghardt R. C.; Metz R. P.; Gustafson T.; Schedin P.; Porter W. W. Loss of singleminded-2s in the mouse mammary gland induces an epithelial-mesenchymal transition associated with up-regulation of slug and matrix metalloprotease 2. Mol. Cell. Biol. 28:1936–1946; 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lareyre J. J.; Reid K.; Nelson C.; Kasper S.; Rennie P. S.; Orgebin-Crist M. C.; Matusik R. J. Characterization of an androgen-specific response region within the 5′ flanking region of the murine epididymal retinoic acid binding protein gene. Biol. Reprod. 63:1881–1892; 2000. [DOI] [PubMed] [Google Scholar]
- 37. Li B. Y.; Liao X. B.; Fujito A.; Thrasher J. B.; Shen F. Y.; Xu P. Y. Dual androgen-response elements mediate androgen regulation of MMP-2 expression in prostate cancer cells. Asian J. Androl. 9:41–50; 2007. [DOI] [PubMed] [Google Scholar]
- 38. Lonard D. M.; O’Malley B. W. Nuclear receptor coregulators: Judges, juries, and executioners of cellular regulation. Mol. Cell 27:691–700; 2007. [DOI] [PubMed] [Google Scholar]
- 39. Loots G. G.; Ovcharenko I.; Pachter L.; Dubchak I.; Rubin E. M. rVista for comparative sequence-based discovery of functional transcription factor binding sites. Genome Res. 12:832–839; 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Loots G. G.; Ovcharenko I. rVISTA 2.0: Evolutionary analysis of transcription factor binding sites. Nucleic Acids Res. 32:W217–221; 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Maffey A. H.; Ishibashi T.; He C.; Wang X.; White A. R.; Hendy S. C.; Nelson C. C.; Rennie P. S.; Ausio J. Probasin promoter assembles into a strongly positioned nucleosome that permits androgen receptor binding. Mol. Cell. Endocrinol. 268:10–19; 2007. [DOI] [PubMed] [Google Scholar]
- 42. Magee J. A.; Abdulkadir S. A.; Milbrandt J. Haploin-sufficiency at the Nkx3.1 locus. A paradigm for stochastic, dosage-sensitive gene regulation during tumor initiation. Cancer Cell 3:273–283; 2003. [DOI] [PubMed] [Google Scholar]
- 43. Masuda K.; Werner T.; Maheshwari S.; Frisch M.; Oh S.; Petrovics G.; May K.; Srikantan V.; Srivastava S.; Dobi A. Androgen receptor binding sites identified by a GREF_GATA model. J. Mol. Biol. 353:763–771; 2005. [DOI] [PubMed] [Google Scholar]
- 44. Moore T. W.; Mayne C. G.; Katzenellenbogen J. A. Minireview: Not picking pockets: Nuclear receptor alternate-site modulators (NRAMs). Mol. Endocrinol. 24:683–695; 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Niu Y.; Altuwaijri S.; Lai K. P.; Wu C. T.; Ricke W. A.; Messing E. M.; Yao J.; Yeh S.; Chang C. Androgen receptor is a tumor suppressor and proliferator in prostate cancer. Proc. Natl. Acad. Sci. USA 105:12182–12187; 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. O’Malley B. W.; McKenna N. J. Coactivators and co-repressors: What’s in a name? Mol. Endocrinol. 22:2213–2214; 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Patrikainen L.; Shan J.; Porvari K.; Vihko P. Identification of the deoxyribonucleic acid-binding site of a regulatory protein involved in prostate-specific and androgen receptor-dependent gene expression. Endocrinology 140:2063–2070; 1999. [DOI] [PubMed] [Google Scholar]
- 48. Prescott J. L.; Blok L.; Tindall D. J. Isolation and androgen regulation of the human homeobox cDNA, NKX3.1. Prostate 35:71–80; 1998. [DOI] [PubMed] [Google Scholar]
- 49. Roche P. J.; Hoare S. A.; Parker M. G. A consensus DNA-binding site for the androgen receptor. Mol. Endocrinol. 6:2229–2235; 1992. [DOI] [PubMed] [Google Scholar]
- 50. Sadar M. D. Androgen-independent induction of prostate-specific antigen gene expression via cross-talk between the androgen receptor and protein kinase A signal transduction pathways. J. Biol. Chem. 274:7777–7783; 1999. [DOI] [PubMed] [Google Scholar]
- 51. Schneider A.; Brand T.; Zweigerdt R.; Arnold H. Targeted disruption of the Nkx3.1 gene in mice results in morphogenetic defects of minor salivary glands: Parallels to glandular duct morphogenesis in prostate. Mech. Dev. 95:163–174; 2000. [DOI] [PubMed] [Google Scholar]
- 52. Schule R.; Muller M.; Kaltschmidt C.; Renkawitz R. Many transcription factors interact synergistically with steroid receptors. Science 242:1418–1420; 1988. [DOI] [PubMed] [Google Scholar]
- 53. Schuur E. R.; Henderson G. A.; Kmetec L. A.; Miller J. D.; Lamparski H. G.; Henderson D. R. Prostate-specific antigen expression is regulated by an upstream enhancer. J. Biol. Chem. 271:7043–7051; 1996. [DOI] [PubMed] [Google Scholar]
- 54. Sciavolino P. J.; Abrams E. W.; Yang L.; Austenberg L. P.; Shen M. M.; Abate-Shen C. Tissue-specific expression of murine Nkx3.1 in the male urogenital system. Dev. Dyn. 209:127–138; 1997. [DOI] [PubMed] [Google Scholar]
- 55. Simmons S. O.; Horowitz J. M. Nkx3.1 binds and negatively regulates the transcriptional activity of Sp-family members in prostate-derived cells. Biochem. J. 393:397–409; 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Stanfel M. N.; Moses K. A.; Schwartz R. J.; Zimmer W. E. Regulation of organ development by the NKX-homeodomain factors: An NKX code. Cell. Mol. Biol. (Noisy-le-grand) Suppl. 51:OL785–799; 2005. [PubMed] [Google Scholar]
- 57. Stanfel M. N.; Moses K. A.; Carson J. A.; Zimmer D. B.; DeMayo F.; Schwartz R. J.; Zimmer W. E. Expression of an Nkx3.1-CRE gene using ROSA26 reporter mice. Genesis 44:550–555; 2006. [DOI] [PubMed] [Google Scholar]
- 58. Sun Q.; Taurin S.; Sethakorn N.; Long X.; Imamura M.; Wang D. Z.; Zimmer W. E.; Dulin N. O.; Miano J. M. Myocardin-dependent activation of the CArG box-rich smooth muscle gamma-actin gene: Preferential utilization of a single CArG element through functional association with the NKX3.1 homeodomain protein. J. Biol. Chem. 284:32582–32590; 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Tanaka M.; Lyons G. E.; Izumo S. Expression of the Nkx3.1 homobox gene during pre and postnatal development. Mech. Dev. 85:179–182; 1999. [DOI] [PubMed] [Google Scholar]
- 60. Tanaka M.; Komuro I.; Inagaki H.; Jenkins N. A.; Copeland N. G.; Izumo S. Nkx3.1, a murine homolog of Drosophila bagpipe, regulates epithelial ductal branching and proliferation of the prostate and palatine glands. Dev. Dyn. 219:248–260; 2000. [DOI] [PubMed] [Google Scholar]
- 61. Thomas M. A.; Preece D. M.; Bentel J. M. Androgen regulation of the prostatic tumour suppressor NKX3.1 is mediated by its 3′ untranslated region. Biochem. J. 425:575–583; 2010. [DOI] [PubMed] [Google Scholar]
- 62. Thomas M. A.; Hodgson M. C.; Loermans S. D.; Hooper J.; Endersby R.; Bentel J. M. Transcriptional regulation of the homeobox gene NKX3.1 by all-trans retinoic acid in prostate cancer cells. J. Cell Biochem. 99(3):1409–1419; 2006. [DOI] [PubMed] [Google Scholar]
- 63. Verrijdt G.; Schoenmakers E.; Alen P.; Haelens A.; Peeters B.; Rombauts W.; Claessens F. Androgen specificity of a response unit upstream of the human secretory component gene is mediated by differential receptor binding to an essential androgen response element. Mol. Endocrinol. 13:1558–1570; 1999. [DOI] [PubMed] [Google Scholar]
- 64. Verrijdt G.; Haelens A.; Claessens F. Selective DNA recognition by the androgen receptor as a mechanism for hormone-specific regulation of gene expression. Mol. Genet. Metab. 78:175–185; 2003. [DOI] [PubMed] [Google Scholar]
- 65. Verrijdt G.; Tanner T.; Moehren U.; Callewaert L.; Haelens A.; Claessens F. The androgen receptor DNA-binding domain determines androgen selectivity of transcriptional response. Biochem. Soc. Trans. 34:1089–1094; 2006. [DOI] [PubMed] [Google Scholar]
- 66. Wu C. T.; Altuwaijri S.; Ricke W. A.; Huang S. P.; Yeh S.; Zhang C.; Niu Y.; Tsai M. Y.; Chang C. Increased prostate cell proliferation and loss of cell differentiation in mice lacking prostate epithelial androgen receptor. Proc. Natl. Acad. Sci. USA 104:12679–12684; 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Yoon H. G.; Wong J. The corepressors silencing mediator of retinoid and thyroid hormone receptor and nuclear receptor corepressor are involved in agonist- and antagonist-regulated transcription by androgen receptor. Mol. Endocrinol. 20:1048–1060; 2006. [DOI] [PubMed] [Google Scholar]
- 68. Zhang J.; Zhang S.; Murtha P. E.; Zhu W.; Hou S. S.; Young C. Y. Identification of two novel cis-elements in the promoter of the prostate-specific antigen gene that are required to enhance androgen receptor-mediated transactivation. Nucleic Acids Res. 25:3143–3150; 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Zhang S.; Murtha P. E.; Young C. Y. Defining a functional androgen responsive element in the 5′ far upstream flanking region of the prostate-specific antigen gene. Biochem. Biophys. Res. Commun. 231:784–788; 1997. [DOI] [PubMed] [Google Scholar]
- 70. Zhang Y.; Fillmore R. A.; Zimmer W. E. Structural and functional analysis of domains mediating interaction between the bagpipe homologue, Nkx3.1 and serum response factor. Exp. Biol. Med. (Maywood) 233:297–309; 2008. [DOI] [PubMed] [Google Scholar]