Abstract
V-ets erythroblastosis virus E26 oncogene homolog2 (ETS2), located at chromosome 21 and overexpressed in Down’s syndrome (DS), has known cancer regulatory functions. Because leukemia is of common occurrence in DS subjects while solid tumors are rare, we have explored the role of ETS2 functional genetic polymorphisms in this differential oncological development. In silico methods were used for identifying deleterious SNPs, tagged SNPs, and linkage disequilibrium followed by genotyping of 14 SNPs in Indo-Caucasoid individuals (N = 668). Significantly different allelic frequencies for rs457705, rs1051420, and rs1051425 were observed in Indian controls (N = 149) compared to other ethnic groups. A heterozygous “T” insertion, between chromosomal contig positions 40195541 and 40195542, was observed in DS subjects and their parents. rs461155 showed significant allelic and genotypic association in breast and oral cancer patients. Significantly higher occurrence of G-C haplotype (rs461155-rs1051425) was also observed in these patients compared to DS and leukemic patients. This is the first report on this type of allelic discrimination pattern of ETS2 under different disease conditions. From the data obtained it may be proposed that allelic discrimination of deleterious SNPs in ETS2 may play a regulatory role in the differential development of malignancy in DS subjects.
Key words: rs1051420, rs1051425, rs461155, rs457705, Acute lymphoblastic leukemia (ALL), Breast cancer, Down’s syndrome, Oral cancer
INTRODUCTION
Malignancy is one of the predominant global health problems with varied paradoxical events in the initiation, invasion and promotion (2). One of the most prominent examples for this paradoxical state are subjects diagnosed with Down’ syndrome (DS); childhood onset of leukemia is a frequent problem while solid tumors, especially breast cancer, is of rare incidence in these individuals (2,13). Solid tumors, which have a lower risk in people with DS, are surrounded by stromal cells while leukemia and testicular cancers, showing higher frequencies in DS pro-bands, are either devoid of or have poorly developed stroma (13). The Tissue Organization Field Theory, formulated to rationalize this kind of situation, hypothesizes that malignancy develops as a result of miscommunication between different types of cells (2).
Down syndrome critical region (DSCR) is composed of a group of functionally important genes responsible for encoding transcription factors (TFs), phosphorylating and dephosphorylating enzymes, etc. An overdose of genes in the DSCR, due to triplication of this region, makes this site important while studying pathophysiology of DS (24). Among different genes located in the DSCR, V-ets erythroblastosis virus E26 oncogene homolog 2 (ETS2) at 21q22.3 is a TF responsible for expression of a number of cell cycle regulatory genes like BCL-XL, c-MYC, cyclin D1, and P53 (6,26).
The ETS group of genes, present throughout the body, control a number of functions including angiogenesis, cellular differentiation, cell cycle, migration, proliferation, and apoptosis. Expression of ETS2 is directly related to expression of P53 and BAX, and inversely related to expression of BCL2, thus increasing sensitivity to apoptosis (29). On the other hand, in breast cancer (BC), investigators have reported that ETS2 binds to the BRCA1 promoter and represses its expression (1). Further, ETS1 expression was found to be a good prognostic indicator for oral squamous cell carcinoma (OC) (22). Overexpression of ETS2 also showed association with DS associated neurocranial and cervical skeletal abnormalities (27).
Genetic translocations in ETS2 were found to be associated with DS-related leukemia (23) and in silico analyses of the dbSNP database (http://www.ncbi.nlm.nih.gov/projects/SNP) by our group revealed a number of SNPs in ETS2, which may disrupt function of the TF. Our preliminary investigation on ETS2 has also shown significant allelic and genotypic association of rs461155 with BC (7). Because functional genetic polymorphisms are useful measures of expression and function of protein, it was felt necessary to explore the role of functionally significant ETS2 genetic polymorphisms in DS and its associated malignancies. As has been already stated, leukemia is of common occurrence in DS children whereas solid tumors like BC and OC show rare incidence (2,13). Therefore, to determine whether ETS2 has any role in this differential malignant development of DS, genomic DNA samples from DS, acute lymphoblastic leukemia (ALL), BC, and OC individuals were analyzed to identify frequencies of functional polymorphisms in the gene.
MATERIALS AND METHODS
Phase I: In Silico Identification of Functional Variants
All the SNPs in the coding, regulatory, and intronic regions of ETS2 were retrieved from the NCBI database (http://www.ncbi.nlm.nih.gov/projects/SNP) and sorted on the bases of their function.
Analysis of Effects Caused by Base Substitution
Sorting Intolerant From Tolerant (SIFT, http://sift.jcvi.org), a sequence homology-based tool that shows tolerance index (TI, ranging from 0.00 to 1.00) for each amino acid substitution, was used for analyzing possible effect of nonsynonymous coding SNPs (16). TI of 0.00 to 0.05 point indicates an intolerant substitution while a score of 0.05 to 1.00 indicates tolerance towards the substitution.
The Polymorphism Phenotyping (PolyPhen) tool was also used (http://coot.embl.de/PolyPhen/), which helps in calculating position-specific independent count (PSIC) scores for every amino acid substitution. A PSIC score difference of 1.5 and above indicates damaging effect of an amino acid substitution, while a score below 1.5 implies that the variation is benign (25).
SNPs3D tool was used to obtain effect of allelic change in nonsynonymous SNPs; higher entropy value signifies more tolerant change. SNPs3D also provides Position Specific Scoring Matrix (PSSM) score, which is the exchangeability score generated from Psi-blast alignment. A higher PSSM score means the change is tolerable (http://www.snps3d.org/) (33).
Analysis of Function Played by Coding and Non-coding SNPs
To estimate the function of coding and noncoding SNPs, located at the TF binding sites, Pupasuite2 program (http://pupasuite.bioinfo.cipf.es) was used, which is based on Match program of the Transfac database. Putative exonic splicing enhancers (ESE), which are responsible for serine/arginine (SR)-rich protein-mediated splicing, were detected using scripts of Pupasuite. It gives a score for the alleles; a reduction in the score value is suggestive of decrease in SR protein-mediated splicing. DNA triplex that is stretched more than 10 polypurines or polypyrimidines in a gene sequence could also be detected by this program. Any SNP located in this triplex forming sequence may disrupt triplex formation and thus may interfere with normal genetic regulation (8). Input mode could be either list of gene, list of SNP IDs, or chromosomal region, and for the present investigation, we have enlisted gene IDs.
Identification of SNPs Altering TF Binding Sites
Functionally significant SNPs, located in the noncoding regulatory region between −5000 to +500 bp from the transcription initiation site, were identified by SNP@Promoter (http://variome.kobic.re.kr/SNPatPromoter) (15).
Estimation of Risk for a SNP
For calculating risk of a SNP, FastSNP tool was employed. This program uses TFSearch web service to follow the decision tree principle and SNPs are ranked based on the level of risk conferred (32). Score levels 0, 1, 2, 3, 4, or 5 signify no risk, very low, low, medium, high, and very high risk, respectively (http://fastsnp.ibms.sinica.edu.tw). Name of the gene was used to generate the input file.
Effect on Globular Domain Formation
GlobPlot was used to analyze the effect of a SNP in globular domain formation (20). For this analysis, one-letter amino acid code of the protein sequence was submitted as query. In the next step altered sequence of the protein, with amino acid substitution caused by a SNP at a specific position, was submitted as a fresh search and data obtained were compared with that observed for the wild type.
Effect of SNPs on MicroRNA Target Site
dbSMR tool (http://miracle.igib.res.in/dbSMR) was employed to search for SNPs that may alter the microRNA target site. The degree of change was calculated from the ratio of number of bases changing the conformation versus total number of bases involved in the binding (12). Gene symbol was used to predict the microRNA target sites.
Phase II: Analyses of Linkage Disequilibrium (LD) Block, Haplotype, and Tagged SNPs in Different Populations
Genotypic data of populations from Europe: Caucasians from Utah with ancestry from western and northern Europe (CEU), Centre d’Etude du Polymorphisme Human (CEPH); Africa: Yoruba from Ibadan, Nigeria (YRI); and Asia: Han Chinese from Beijing, China (HCB) and Japanese from Tokyo, Japan (JPT), were retrieved from the HapMap database (Release 24/phase II) and used for comparison with the data obtained for the eastern Indian population.
Haplotypes and LD patterns in each population were analyzed by Haploview 4.1. Input data source was the “SNP genotype data” of ETS2, downloaded from “HapMap Data Rel 24/phase II Nov 08, on NCBI B36 assembly, dbSNP b126” of HapMap Genome Browser (Phase I & II—full dataset). The Tagger in Haploview was used for tagSNP selection using pair-wise tagging with an r 2 cutoff value of 0.8.
Genotyping of Selected SNPs in the Indo-Caucasoid Population
A total five ethnically matched groups from the state of West Bengal in eastern India (23°N, 87°E) were recruited for analysis of SNPs. Healthy volunteers (N = 149), without any clinical history of intellectual disability or malignant disorder, were recruited as controls (IND-C). Nuclear families having a DS child (N = 132) were recruited from the outpatient department of Manovikas Kendra, Kolkata and trisomic status of probands was confirmed by karyotyping. OC patients (N = 54) were recruited by investigators of the Indian Institute of Chemical Biology, Kolkata, and genomic DNA isolated from peripheral blood was provided for the present study. ALL patients (N = 38) were recruited from the Kothari Clinic and Netaji Subhash Chandra Bose Cancer Research Institute, Kolkata. Genomic DNA from postoperative normal tissue, adjacent to malignant BC (N = 86), was collected by investigators from the Chittaranjan National Cancer Research Institute, Kolkata. All the samples were acquired from the respective individuals after obtaining informed written consent for participation. The Institutional Human Ethical Committee approved the study protocol.
Genotyping Procedure
Peripheral blood collected from normal volunteers, DS probands and their parents, as well as leukemia patients was used for extraction of genomic DNA as per the standard protocol (21). As mentioned above, genomic DNA was provided by respective investigators for BC and OC samples. Fourteen functionally important SNPs (rs457705, rs461155, rs34120017, rs35258008, rs1051420, rs1051425, rs11422952, rs34882229, rs35578874, rs13046062, rs3178021, rs3178022, rs3178023, and rs11540812) were genotyped using two set of primers. Primer sequences for rs457705, rs461155, rs34120017, and rs35258008 were: forward 5′-GTTGTCTTTGCCAGGGACTC-3′ and reverse 5′-CGGTGAATGTGGTACTGTGG-3′. For the rest 11 SNPs the primer sequences were: forward 5′-CAAGGGCCGACTAAGAGAAG-3′ and reverse 5′-GCATGCAAAGAAGTGGAAAA-3′. PCR amplicons were subjected to sequencing in ABI prism 3130 Genetic Analyzer using Big Dye sequencing kit v3.1 followed by analysis using Sequencing Analysis software v 5.2. Electropherograms obtained were further analyzed by Mutation Surveyor Demo V3.24 software to check for new mutation.
Statistical Analysis of Genotyped Data
Allelic and genotypic distributions of control, DS probands, their parents, ALL, BC, and OC patients were compared by simple r × c contingency table (http://www.physics.csbsju.edu/stats/contingency_NROW_NCOLUMN_form.html). LD values were measured by Haploview 4.1 using default settings. Allelic odds ratios of different diseased groups were calculated by Odds Ratio calculator (http://www.hutchon.net/ConfidORnulhypo.htm). Haplotype frequency distributions of polymorphic SNPs was inspected by Un-phased program (Version 2.403) (10). Piface program (18) was used to calculate power of all the chi-square tests.
RESULTS
Phase I
Analysis of all the SNPs in the dbSNP database (total number = 279) showed that ETS2 has three missense, four synonymous SNPs, and three frame shift changes in the coding region. A large number of intronic SNPs and noncoding SNPs in the 5′ and 3′ UTR were also observed.
Effects Caused by Base Substitution
By SIFT analysis, five nonsynonymous SNPs were detected. Among these, only rs1803557 revealed risk of damage; TI was found to be 1.00 for A (coding for glutamate) with a chance of being reduced to 0.01 by T substitution (encoding valine). Four other SNPs, rs34373350, rs61735785, rs66473060, and rs34472454, failed to show any risk (Table 1).
TABLE 1.
FUNCTIONAL SNPs IN ETS2 PREDICTED BY IN SILICO METHODS
| SNP ID (Alleles) | Type of SNP (Amino Acid Change) | Possible Functional Effect | Tool for Prediction | Specification of Functional Significance |
|---|---|---|---|---|
| rs1803557 (T/A) | Coding NS (E145V) | Damaging | SIFT | TI: 1.00 (E), 0.01 (V) |
| PolyPhen | PSIC score: 2.147 | |||
| SNPs3D | Entropy: 1.75 bits, PSSM score: −1 | |||
| FastSNP | High risk SNP (risk level: 3–4) | |||
| rs34373350 (A/G) | Coding NS (A64T) | Damaging | SIFT | TI: 1.00 (A), 0.55 (T) |
| PolyPhen | PSIC score: 0.189 | |||
| SNPs3D | Entropy: 2.30 bits, PSSM score: 0 | |||
| FastSNP | Medium risk SNP (risk level: 2–3) | |||
| GlobPlot | Disorder: 56–62 (A), 55–68 (T) | |||
| Globular domains: 3–188 (A), 69–188 (T) | ||||
| rs61735785 (A/C) | Coding NS (I217L) | Benign | PolyPhen | PSIC score: 0.183 |
| FastSNP | Medium risk SNP (risk level: 2–3) | |||
| rs66473060 (−/T) | Coding frame shift (P41S) | Damaging | PolyPhen | PSIC score: 1.756 |
| rs457705 (G/T) | Coding synonymous | Affects SR protein-mediated ESE activity | Pupasuite 2 | SR protein: SC35; Scores: 4.76 (T), 2.30 (G); Lose (−2.46) |
| FastSNP | Medium risk SNP (risk level 2–3) | |||
| rs461155 (G/A) | Coding synonymous | Affects SR protein-mediated ESE activity | Pupasuite 2 | SR protein; SRp40; Scores; 4.00 (A), 1.39 (G); Lose (−2.61) |
| FastSNP | Medium risk SNP (risk level: 2–3) | |||
| rs711(A/G) | 3′ UTR SNP | Affects SR protein-mediated ESE activity | Pupasuite 2 | SR protein: SC35, SF2; Scores: 2.73(A), 1.43 (G); Lose (−1.30) 1.99 (A), 1.26 (G); Lose (−0.73) |
| rs13046062 (T/G) | 3′ UTR SNP | Affects SR protein-mediated ESE activity | Pupasuite 2 | SR protein: SRp40; Scores: 2.77 (T), 1.21 (G); Lose (−1.56) |
| rs1051476 (C/G) | 3′ UTR SNP | Affects SR protein-mediated ESE activity | Pupasuite 2 | SR protein: SC35; Scores: 2.71 (C), 1.80 (G); Lose (−0.91) |
| rs3178021 (C/T) | 3′ UTR SNP | Affects SR protein-mediated ESE activity and disrupts TFO | Pupasuite 2 | SR protein: SRp40; Scores: 3.56 (C), 1.18 (T); Lose (−2.38) |
| rs3178022 (C/T) | 3′ UTR SNP | Affects SR protein-mediated ESE activity and disrupts TFO | Pupasuite 2 | SR protein: SRp40; Scores: 3.56 (C), 1.85 (T); Lose (−1.71) |
| rs3178023 (C/T) | 3′ UTR SNP | Affects SR protein-mediated ESE activity and disrupts TFO | Pupasuite 2 | SR protein: SRp40; Scores: 3.56 (C), 1.85 (T); Lose (−1.71) |
| rs11254 (C/T) | 3′ UTR SNP | ESE activity and microRNA target site changes | Pupasuite 2 dbSMR | SR protein: SC35; Scores: 2.54 (C), 1.97 (T); Lose (−0.57) |
| hsa-mir-136 microRNA target site gained (degree of change = 20–30%) | ||||
| rs11911369 (C/T) | 5′ UTR (−2216)* | Change at possible TF binding site | SNP@promoter and Pupasuite 2 | Myogenin/NF-1, Elk-1,c-Ets-1 p54, YY1† (C)‡ |
| rs34248663(−/C) | 5′ UTR (−2594)* | Change at possible TF binding site | SNP@promoter | PLZF (C)‡ |
| rs8129886 (C/G) | 5′ UTR (−2879)* | Change at possible TF binding site | SNP@promoter | Hand1:E47, PLZF† (C)‡ |
| rs8130465 (C/T) | 5′ UTR (−3204)* | Change at possible TF binding site | SNP@promoter | C/EBP† (T)‡ |
| rs8134441 (T/C) | 5′ UTR (−3955)* | Change at possible TF binding site | SNP@promoter | SMAD† (T)‡ |
| rs1209950 (C/T) | 5′ UTR (−4321)* | Change at possible TF binding site | Pupasuite 2 | CDX† (C)‡ |
| rs8129412 (C/A) | 5′ UTR (−3222)* | Change at possible TF binding site | Pupasuite 2 | HNF† (C):‡ |
ESE: exonic splicing enhancer; PSIC score: position specific independent counts; PSSM score: position specific scoring matrix; NS: nonsynonymous; SR protein: serine/arginine-rich protein; TFO: triplex forming oligonucleotide; TF: transcription factor; TI: tolerance index.
SNP position (−5000 to +500) relative to transcription start site.
TFs predicted to bind.
TF binding allele.
Analysis by PolyPhen revealed two SNPs, rs1803557 and rs66473060, as deleterious because their PSIC score difference exceeded the danger limit of 1.5 (Table 1).
SNPs3D analysis showed that rs1803557 has lower PSSM score (−1) and also a lower entropy value compared to others (Table 1). rs34373350, rs61735785, rs66473060, and rs34472454 failed to show any significant contribution by SNPs3D analysis.
Functional Analysis of Coding and Noncoding SNPs
Analysis of functional changes revealed alteration in TF binding sites by three SNPs in the ETS2 promoter; allelic changes induced at rs1209950 (−4321, C>T) and rs8129412 (−3222, C>A) were predicted to create binding sites for TF (CDX and HNF-1, respectively) while rs11911369 (−2215, C>T) may lead to loss of the Elk-1 binding site.
On the other hand, SNP@Promoter analysis revealed five SNPs (Table 1) that may alter binding sites for different TFs. The derived alleles of rs8130465 may generate site for C/EBP in ETS2. On the other hand, substitutions at rs11911369, rs34248663, rs8129886, and rs8134441 may abolish TF binding sites.
Identification of SNPs in the Promoter Region
Exonic splicing enhancer activity (ESE) was found to be altered by nine SNPs (Table 1). Sixteen SNPs (rs8129412, rs8129422, rs35548432, rs2283639, ENSSNP11887775, rs34005903, rs33953058, rs3833351, rs11288156, rs28565186, rs11702084, rs12483154, rs3178021, rs3178022, rs3178023, ENSSNP11887796), which may disrupt the triplex forming target sequence, were also identified by Pupasuite2.
Estimation of Risk Involved With a SNP
For the present analysis SNPs showing risk level below 2 by FastSNP were excluded. Five SNPs (rs34373350, rs11700777, rs457705, rs461155, rs17854245) were predicted to have chances of conferring low to medium risk (risk level 2–3). rs1803557 (risk level 3–4), which induces a missense alteration, showed a higher potency to modify splicing regulation.
Prediction of Effect on Globular Domain Formation
GlobPlot analysis revealed that while all the nonsynonymous SNPs may not be deleterious, rs34373350 may have some role in the formation of globular domain; order (56–62 for “A” residue; 55–68 for “T” residue) and potential globular domains (3–188 for “A” residue; 69–188 for “T” residue) were changed in comparison to the native protein.
Identification of SNPs Affecting MicroRNA Target Site
dbSMR analysis showed that a site of action for hsa-mir-136 at the 3′ region of both the transcripts of ETS2 (ENST00000360214 and ENST00000360938) was gained by rs11254 C/T substitution; the degree of change was 20–30%.
After analyses by all the SNP selection web servers, 20 sites were prioritized as functionally significant (Table 1).
Phase II: Analyses of Linkage Disequilibrium (LD) Block, Haplotype, and Tagged SNPs in ETS2 Among Different Populations
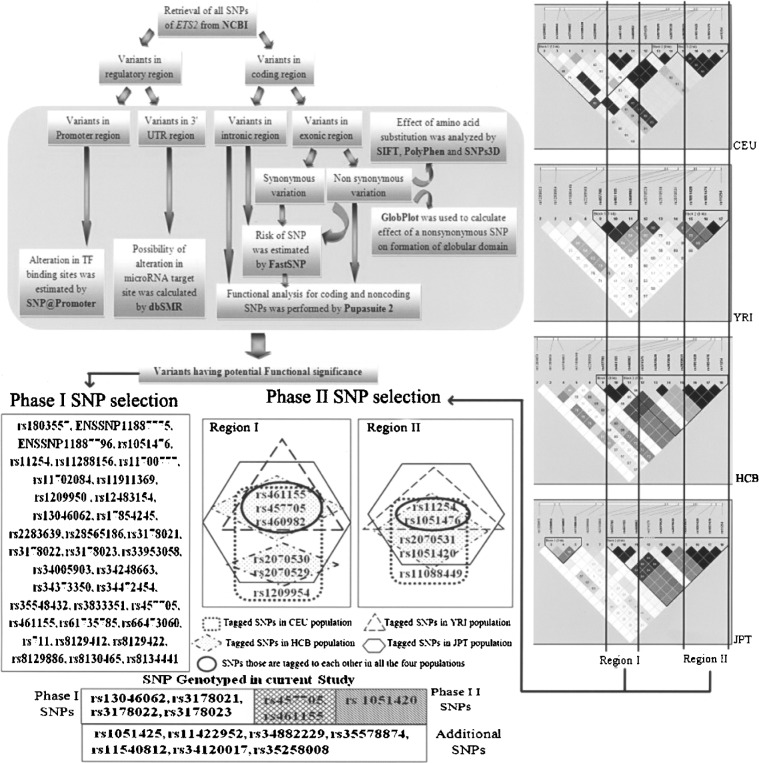
Hapmap Phase II genotype frequency dataset was used to explore LD block structure of ETS2 gene in CEU, YRI, HCB, and JPT. As different allelic frequencies among populations yield different LD structures, common block regions were inspected to target particular SNPs that may show similar allele frequency distribution in Indian population also. Tagger analysis identified two common regions (i.e., Region I and Region II), with rs461155, rs457705, rs460982, and rs11254, rs1051476 as common tagged SNPs (tSNPs) in all four populations (Fig. 1). SNPs like rs2070529, rs2070530, and rs1209954 from Region I and rs2070531, rs1051420, and rs11088449 from Region II were also identified as common tSNPs in CEU, HCB, and JPT, but not in YRI.
Figure 1.
Schematic representation of the selection procedure for Phase I and Phase II SNPs.
Fourteen SNPs distributed over Region I and Region II (Fig. 1) were genotyped in the eastern Indian population.
Comparative Analysis of Polymorphisms in IND and Other Different Populations
Out of 14 SNPs studied in the IND population, only rs461155 and rs1051425 were found to be polymorphic with a moderate LD between the two (D′ = 0.699; r 2 = 0.165). Ancestral alleles of rs1051425 were flipped to minor allele in the IND population. As was reported previously, the same type of flip was also observed for rs461155 (7). A significant difference between IND and CEPH population for rs1051425 alleles was also noticed (Table 2). Allelic and genotypic frequencies of rs457705 and rs1051420 also showed significant difference when compared with other populations; while the ancestral allele frequency of rs457705 was reduced in HCB and JPT compared to the Caucasian and African population, only the derived allele was detected in the IND population (Table 2).
TABLE 2.
COMPARISON OF ALLELIC AND GENOTYPIC FREQUENCIES IN DIFFERENT POPULATIONS
| SNP ID (A1/A2) | Ancestral Allele | Population | Allele | Chi-Square, p-Value | Genotype | Chi-Square, p-Value | |||
|---|---|---|---|---|---|---|---|---|---|
| A1 | A2 | A1A1 | A1A2 | A2A2 | |||||
| rs457705 (G/T) | G | Indian (N = 149) | 0.000 (0) | 1.000 (298) | — | 0.000 (0) | 0.000 (0) | 1.000 (149) | — |
| HCB | 0.389 | 0.611 | 48.4, <0.0001 | 0.200 | 0.378 | 0.422 | 81.7, <0.0001 | ||
| JPT | 0.364 | 0.636 | 43.9, <0.0001 | 0.091 | 0.545 | 0.364 | 94.1, <0.0001 | ||
| YRI | 0.525 | 0.475 | 72.1, <0.0001 | 0.250 | 0.550 | 0.200 | 133.0, <0.0001 | ||
| CEU | 0.833 | 0.167 | 142.0, <0.0001 | 0.667 | 0.333 | — | 200.0, <0.0001 | ||
| rs1051425 (T/C) | T | Indian (N = 149) | 0.171 (51) | 0.829 (247) | — | 0.758 (113) | 0.141 (21) | 0.101 (15) | — |
| CEPH | 0.480 | 0.520 | 21.9, <0.0001 | — | — | — | — | ||
| rs1051420 (A/C) | A | Indian (N = 149) | 1.000 (298) | 0.000 (0) | — | 1.000 (149) | 0.000 (0) | 0.000 (0) | — |
| HCB | 0.767 | 0.233 | 26.0, <0.0001 | 0.644 | 0.244 | 0.111 | 42.4, <0.0001 | ||
| JPT | 0.800 | 0.200 | 22.2, <0.0001 | 0.644 | 0.311 | 0.044 | 42.4, <0.0001 | ||
| YRI | 0.850 | 0.150 | 16.2, <0.0001 | 0.733 | 0.233 | 0.033 | 29.9, <0.0001 | ||
| CEU | 0.575 | 0.425 | 53.2, <0.0001 | 0.350 | 0.450 | 0.200 | 96.3, <0.0001 | ||
Distribution of ETS2 SNPs Among Different Disease Groups
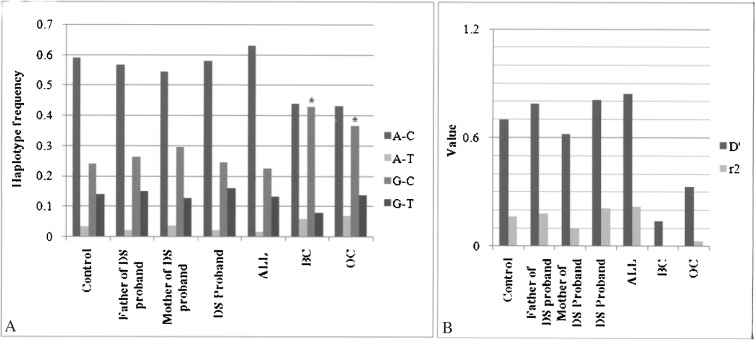
A significantly higher allelic and genotypic frequency for “G” allele of rs461155, with a strong study power, was observed in the genomic DNA samples of patients with solid tumor (BC and OC) compared to controls (Table 3) as well as ALL individuals and families with DS probands. The G-C haplotype was also present in significantly higher frequency in BC (χ2 = 15.23, p = 9.50E-05, power = 70.34%) and OC (χ2 = 4.83, p = 0.02798, power = 56.15%), while A-C was found to be the prevalent haplotype in other groups including control (Fig. 2A).
TABLE 3.
ALLELIC AND GENOTYPIC DISTRIBUTION OF rs461155, rs1051425, AND INSERTION OF “T” AMONG DIFFERENT STUDY GROUPS
| SNP ID | Allele/Genotype Frequency | IND-C (N = 149) | Father of DS (N = 91) | Mother of DS (N = 118) | DS (N = 132) | ALL (N = 38) | BC (N = 86) | OC (N = 54) |
|---|---|---|---|---|---|---|---|---|
| rs461155 (A/G) | A | 0.621 (185) | 0.588 (107) | 0.581 (137) | 0.629 (249) | 0.645 (49) | 0.494 (85) | 0.500 (54) |
| G | 0.379(113) | 0.412(75) | 0.419 (99) | 0.371 (147) | 0.355 (27) | 0.506 (87) | 0.500 (54) | |
| Chi-square, p-value, Power (%) | — | 0.188, 0.664, 6.19 | 0.333, 0.564, 6.63 | 0.213E-01, 0.884, 5.34 | 0.194, 0.659, 7.97 | 7.15, 0.007, 53.15 | 4.78, 0.029, 55.72 | |
| OR (CI) | — | 1.1477 (0.787–1.6738) | 1.183 (0.8347–1.6766) | 1.0982 (0.7822–1.542) | 0.9031 (0.5371–1.5187) | 1.6757 (1.1464–2.4494) | 1.6449 (1.0522–2.5715) | |
| AA | 0.369 (55) | 0.286 (26) | 0.271 (32) | AAA 0.258 (34) | 0.395 (15) | 0.105 (9) | 0.037 (2) | |
| AG | 0.503 (75) | 0.604 (55) | 0.619 (73) | AAG 0.432 (57)*
AGG 0.250 (33)* |
0.500 (19) | 0.779 (67) | 0.926 (50) | |
| GG | 0.128(19) | 0.110(10) | 0.110 (13) | GGG 0.060 (8) | 0.105 (4) | 0.116 (10) | 0.037 (2) | |
| Chi-square, p-value, Power (%) | — | 2.37, 0.305, 16.01 | 3.68, 0.159, 18.39 | 9.81, 0.007, 38.92 | 0.176, 0.916, 6.81 | 20.9, <0.0001, 88.73 | 30.2, <0.0001, 99.85 | |
| rs1051425 (C/T) | C | 0.829(247) | 0.830(151) | 0.839 (198) | 0.848 (336) | 0.855 (65) | 0.866 (149) | 0.796 (86) |
| T | 0.171 (51) | 0.170 (31) | 0.161 (38) | 0.152 (60) | 0.145 (11) | 0.134 (23) | 0.204 (22) | |
| Chi-square, p-value, Power (%) | — | 0.525E-03, 0.982, 5.16 | 0.818, 0.366, 9.06 | 1.19, 0.276, 10.3 | 1.64, 0.201, 31.2 | 1.15, 0.283, 12.94 | 0.570, 0.450, 11.23 | |
| OR (CI) | — | 0.9943 (0.6095–1.6221) | 0.9298 (0.5882–1.4697) | 1.0489 (0.6781–1.6225) | 0.8266 (0.4203–1.6257) | 0.7476 (0.4389–1.2735) | 1.2464 (0.7031–2.2096) | |
| CC | 0.758(113) | 0.747(68) | 0.738 (87) | CCC 0.712 (94) | 0.816 (31) | 0.814 (70) | 0.722 (39) | |
| CT | 0.141 (21) | 0.165 (15) | 0.203 (24) | CCT 0.189 (25)†
CTT 0.030 (4)† |
0.079 (3) | 0.105 (9) | 0.148 (8) | |
| TT | 0.101 (15) | 0.088 (8) | 0.059 (7) | TTT 0.068 (9) | 0.105 (4) | 0.081 (7) | 0.130 (7) | |
| Chi-square, p-value, Power (%) | — | 0.320, 0.852, 6.35 | 2.93, 0.23, 15.45 | 3.51, 0.173, 16.26 | 1.04, 0.594, 16.61 | 0.995, 0.608, 9.62 | 0.390, 0.823, 7.82 | |
| Novel heterozygous insertion of of “T” | Without Ins “T” | 0.993 (148) | 0.989 (90) | 0.966 (114) | 0.970 (128) | 1.000 (38) | 1.000 (86) | 1.000 (54) |
| Ins “T” | 0.007 (1) | 0.011 (1) | 0.033 (4) | 0.030 (4) | 0.000 (0) | 0.000 (0) | 0.000 (0) | |
| Chi-square, p-value, Power (%) | — | 0.125, 0.724, 5.79 | 2.65, 0.104, 18.52 | 2.23, 0.135, 15.1 | — | — | — | |
| OR (CI) | — | 1.6744(0.0957–29.3118) | 4.3664(0.7375–25.8518) | 3.8384(0.6545–22.5118) | — | — | — |
OR: odds ratio, CI: confidence interval.
Combined: 0.682 (90).
Combined: 0.220 (29).
Figure 2.
Haplotype frequency (A) and linkage disequilibrium (B) of rs461155 and rs1051425 in eastern Indian control and disease populations (*p-values for the G-C haplotypes are 9.50E-05 and 0.02798 in BC and OC, respectively).
Comparison of global D′ and r 2 value for rs461155 and rs1051425 among different groups revealed that BC and OC have significantly lower LD values for these two SNPs (D′ = 0.139 and 0.3), although father of DS probands, DS probands, and ALL cases showed a high LD value (D′ is near to 0.8) with control and DS mother showing moderate LD (D′ = 0.3) (Fig. 2B).
Novel Insertion Detected in the ETS2 3′ UTR
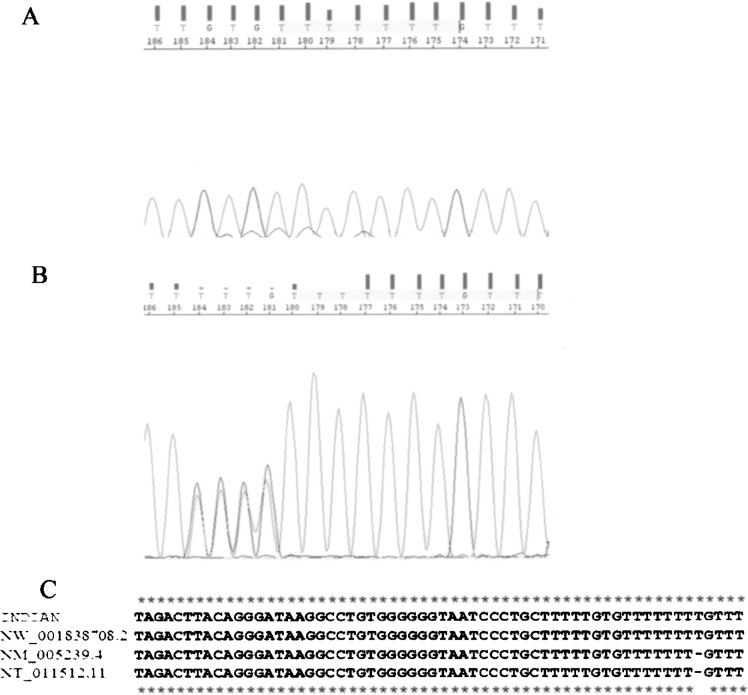
Mutation Surveyor analyses showed a rare variation (Fig. 3A) between chromosomal contig position 40195541 and 40195542 (heterozygous insertion of a “thymidine” residue; presented in Fig. 3B) in only 10 out of 668 eastern Indian individuals studied. One of them was a 26-year-old nulliparous healthy female volunteer. Among the other nine individuals harboring “insertion,” one was father and four were mothers of DS proband and four were DS probands. The insertion was never detected in homozygous condition. Overall frequency of the variation was 0.0158 in the eastern Indian subjects. Although chi-square and p-values calculated for the differences between control and DS probands or their mothers were not significant, odds ratio was very high (Table 3).
Figure 3.
Representative electropherogram for (A) wild-type ETS2 sequence; (B) heterozygous insertion of “T”; (C) multiple alignment of the 3′ UTR (40195494 to 40195554 chromosomal contig position) Indian: sequence with “T” insertion, NW_001838708.2: sequence from a Caucasian male, NM_005239.4: mRNA reference sequence, NT_011512.11: Homo sapiens chromosome 21 genomic contig (GRCh37 primary assembly).
BLAST search (http://blast.ncbi.nlm.nih.gov/Blast.cgi) followed by multiple sequence alignment (Multalin version 5.4.1; http://www-archbac.u-psud.fr/genomics/multalin.html) (9) revealed that the “T” is absent in ETS2 mRNA reference sequence (NM_005239.4) and Homo sapiens chromosome 21 genomic contig (GRCh37 reference primary assembly, NT_011512.11). However, it was observed in the ETS2 sequence of a Caucasian male chromosome 21 genomic contig (HuRef; NW_001838708.2) (Fig. 3C).
DISCUSSION
Differential expression of TFs in different tissues has been well documented in the literature (31). However, it is still unknown whether aberrant expression of TFs can lead to uncontrolled growth in certain tissues generating different types of malignancies. In the present investigation, we have attempted to explore the role of ETS2 SNPs under different disease conditions and, interestingly, significantly altered frequencies of two SNPs were noticed in samples obtained from OC and BC patients compared to control, DS probands, parents of DS probands, and leukemic groups.
Analysis of base substitution using various in silico tools (i.e., SIFT, Polyphen, SNPs3D) indicated that rs1803557 may contribute to an intolerant substitution (TI 0.01 for the T allele) as well as splicing regulation. Similar substitution of glutamate by valine, under homozygous condition, is well known for causing a conformational change in the hemoglobin beta subunit molecule leading to sickle cell anemia (11). Among the other four nonsynonymous substitutions, only rs66473060 points to a chance of being damaging by causing change in TF binding. During our phase I SNP selection, more than one in silico analysis also revealed that rs11911369, rs34373350, rs 457705, rs461155, rs11254, rs3178021, rs3178022, and rs3178023 were associated with deleterious effects and could be considered to confer higher risk. However, to date, these SNPs have never been explored in association with disorders and it would be worth looking into these under disease conditions.
The present study has also revealed the presence of several SNPs containing sites for SR protein-mediated ESE activity (Table 1). Among them, rs13046062, rs3178021, rs3178022, and rs3178023 were nonpolymorphic in the IND population and therefore may not participate in differential splicing. Other SNPs, which may hamper SR protein-mediated functioning, deserve further attention because altered expression of SRp40 was observed in breast cancer and metastasized lymph node tissues compared to adjacent normal tissue and it was hypothesized that SR protein-mediated alternative splicing of pre-mRNA could be associated with malignant development (14).
In silico analysis also predicted rs711 and rs457705 as damaging SNPs. In the eastern Indian population, rs457705 showed 100% frequency for the derived “T” allele, which may not have a major role in the disease process. In the Korean population also, rs457705 failed to show any contribution while increased risk for acute myeloid leukemia was found to be associated with two SNPs, rs711 and rs530 (17). In the present study, rs711 was not explored and further investigation on these sites would be required to understand whether these SNPs have any role in disease development.
We have also attempted to identify the status of ETS2 SNPs in genomic DNA samples from patients with DS and leukemia (ALL) as well as solid tumor (BC and OC). Significantly higher occurrence of the “G” allele of rs461155 was noticed in the solid tumor groups (BC and OC) compared to control, DS subjects, their parents, and ALL patients (p < 0.0001). It is well known that BC has a higher familial occurrence and, as was observed in the present study, DS individuals may get some protection from BC due to lower occurrence as well was transmission of the “G” allele from their parents. Further, the A-C haplotype of rs461155 and rs1051425 had significantly lower frequency in BC and OC (χ2 = 8.048, 6.654; p = 0.004554, 0.009891, respectively) with a concomitantly higher G-C compared to other groups including control (Fig. 2A). As was reported previously by our group, rs461155 “G” allele reduces SRp40-mediated splicing of ETS2 pre-mRNA and may thus reduce ETS2 activity by generating splice variants (7). rs1051425 was found have a potency to alter transcriptional regulation. Therefore, it may be inferred that the G-C haplotype formed between rs461155 and rs1051425 may confer risk of BC and OC by altering ETS2 function. Observation made in the present investigation also supports a risk of oral squamous cancer conferred by ETS2 as was reported previously (22). It would be interesting to find out whether the tagged SNPs of rs461155 (rs460982, rs2070529, and rs2070530) also bear risk of BC.
Analysis of LD between rs461155 and rs1051425 showed a different pattern in the solid tumor groups compared to the others (Fig. 2B); while moderate LD was observed in all other groups, BC and OC showed almost absence of LD between the two sites. The two sites are present in separate LD blocks (HapMap data explored by Haploview 4.1) at a distance of 3.846 kb. Whether there is any recombination hotspot between the two sites is not known yet. Further, due to limitation in the number of samples analyzed for the solid tumor groups, it would be premature to make any inference on the LD structure.
Because trisomic TDT analysis require information regarding specific stage of occurrence of meiotic nondisjunction for each and every case of DS (30), transmission pattern of alleles from parents to offspring was not performed. The present study was confined to investigating biasness in occurrence of alleles among the parent populations and DS probands. Data obtained indicate lack of any specific association of ETS2 SNPs investigated with DS. Comparison of allelic frequencies revealed no statistically significant difference. However, genotype analysis revealed statistically higher occurrence of heterozygous genotypes, which could be due to clubbing of two groups (AAG and AGG) together.
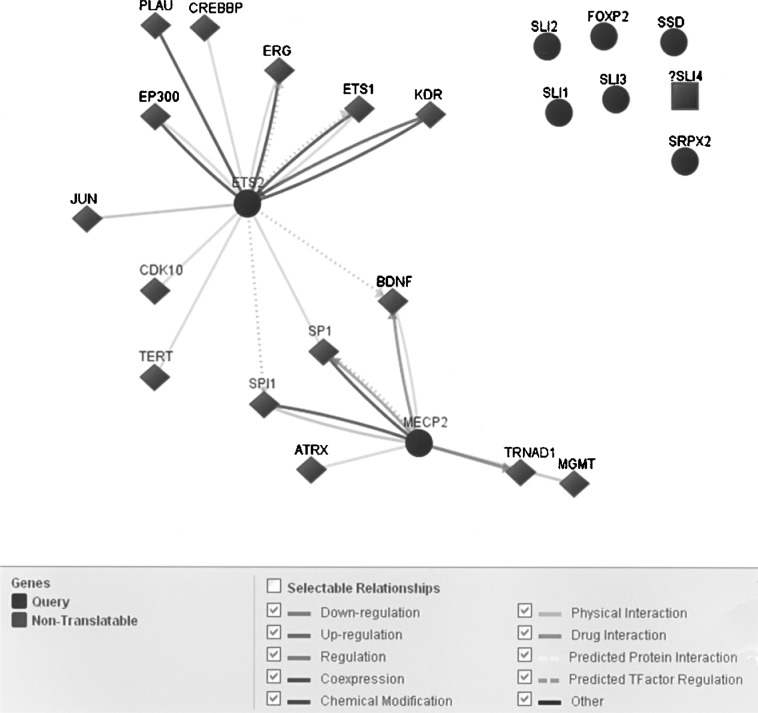
A heterozygous insertion of “T” allele was identified in eastern Indian nuclear family members with DS probands except for one control female individual. As the frequency of this insertion was rare, effective sample size became low and odds ratio was high in DS and their mothers with a broad range of confidence interval (Table 3). Analyses of phenotypes revealed that out of the four DS probands containing the insertion, three exhibited significant speech articulation problem. Further in silico analysis using RNA Analyzer tool (http://wb2x01.biozentrum.uni-wuerzburg.de/) (3) showed that the ETS2 RNA region bearing the “T” insertion faces a change in energy of stem-loop formation (−41.31) compared to the wild-type RNA (−39.91). Physical interaction and regulation of genes like MECP2 by ETS2 via SPI1, SP1, BDNF, etc., which are involved in proper development of speech, were also observed by in silico analysis (Gene Network Central Pro™) (Fig. 4). Articulation is a common disability of DS children and it could be hypothesized from the present preliminary investigation that the change in structural conformation of ETS2 induced by the insertion may be related to the observed speech problem besides other causes. A cleavage stimulating factor (CstF), element 2a binding motif was also predicted to be created due to the insertion. Earlier investigations have shown that the CstF element 2a acts during 3′ end processing of pre-mRNA molecule (4). It may be inferred that in presence of the insertion, the 3′ end processing schedule of ETS2 mRNA may get hampered. This pioneering report on the ETS2 3′ UTR sequence variation, which may have an important role in ETS2 expression and therefore can have a regulatory role in ETS2 regulated disease pathogenesis, could be helpful in understanding the role played by this TF in DS pathophysiology.
Figure 4.
In silico analysis of the interactive pathway formed between ETS2 and speech-related genes by Gene Network Central Pro™.
Overexpression of ETS2 was reported in oral, breast, esophageal as well as prostate cancer tissues (5,19,27,28). On the other hand, DS individuals with gene overdosage as well as overexpression of ETS2 are protected from development of these solid tumors. Although from the present investigation it is not clear whether the stromal theory (13) or miscommunication between different types of cells (2) lead to the development of malignancy, it can be hypothesized that differential expression of ETS2, caused by the presence of different allelic variants, may at least partly be responsible for the process. An in-depth analysis of these variants may help us to understand the regulatory procedure better.
This is the first report on the identification of functional SNPs in ETS2 in the eastern Indian population. rs461155 showed allelic and genotypic association with BC and OC. Higher occurrence of the G-C haplotype of rs461155 and rs1051425 was also found in BC and OC. A heterozygous insertion of “T” was found for the first time in DS family members of eastern Indian origin. From the result obtained it may be predicted that ETS2 expression may get hampered in the presence of risk haplotypes in BC and OC but not in DS or ALL. Differential expression of ETS2 in different malignant groups and DS may be regulated by marked difference in the frequency of risk alleles. To resolve the role of ETS2 in the paradoxical events of DS-related malignancy, further extensive investigation on deleterious SNPs is needed in a larger cohort of DS and its related malignant conditions.
ACKNOWLEDGMENT
The authors are thankful to all the individuals for their participation in the study.
REFERENCES
- 1. Baker K. M.; Wei G.; Schaffner A. E.; Ostrowski M. C. Ets-2 and Components of mammalian SWI/SNF form a repressor complex that negatively regulates the BRCA1 promoter. J. Biol. Chem. 278:17876–17884; 2003. [DOI] [PubMed] [Google Scholar]
- 2. Baker S. G.; Kramer B. S. Paradoxes in carcinogenesis: New opportunities for research directions. BMC Cancer 7:151; 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bengert P.; Dandekar T. A software tool-box for analysis of regulatory RNA elements. Nucl. Acids Res. 31:3441–3445; 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Beyer K.; Dandekar T.; Keller W. RNA ligands selected by cleavage stimulation factor contain distinct sequence motifs that function as downstream elements in 3′-end processing of pre-mRNA. J. Biol. Chem. 272:26769–26779; 1997. [DOI] [PubMed] [Google Scholar]
- 5. Buggy Y.; Maguire T. M.; McDermott E.; Hill A. D.; O’Higgins N.; Duffy M. J. Ets2 transcription factor in normal and neoplastic human breast tissue. Eur. J. Cancer 42:485–491; 2006. [DOI] [PubMed] [Google Scholar]
- 6. Carbone G. M.; Napoli S. Triplex DNA-mediated downregulation of Ets2 expression results in growth inhibition and apoptosis in human prostate cancer cells. Nucl. Acids Res. 32:4358–4367; 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chatterjee A.; Mukherjee N.; Panda C. K.; Mukhopadhyay K. Could allelic discrimination in ETS2 predispose individuals to breast cancer? Open Breast Cancer J. 2:1–3; 2010. [Google Scholar]
- 8. Conde L.; Vaquerizas J. M.; Dopazo H.; Arbiza L.; Reumers J.; Rousseau F.; et al. PupaSuite: Finding functional single nucleotide polymorphisms for large-scale genotyping purposes. Nucl. Acids Res. 34: W621–625; 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Corpet F. Multiple sequence alignment with hierarchical clustering. Nucl. Acids Res. 16(22):10881–10890; 1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dudbridge F. Pedigree disequilibrium tests for multi-locus haplotypes. Genet. Epidemiol. 25(2):115–121; 2003. [DOI] [PubMed] [Google Scholar]
- 11. Finch J. T.; Perutz M. F.; Bertles J. F.; Doblert J. Structure of sickled erythrocytes and of sickle-cell hemoglobin fibers. Proc. Natl. Acad. Sci. USA 70:718–722; 1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hariharan M.; Scaria V.; Brahmachari S. K. dbSMR: A novel resource of genome-wide SNPs affecting microRNA mediated regulation. BMC Bioinformatics 10:108; 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hasle H.; Clemmensen I. H.; Mikkelsen M. Risks of leukemia and solid tumors in individuals with Down’s syndrome. Lancet 355:165–169; 2000. [DOI] [PubMed] [Google Scholar]
- 14. Huang C. S.; Shen C. Y.; Wang H. W.; Wu P. E.; Cheng C. W. Increased expression of SRp40 affecting CD44 splicing is associated with the clinical outcome of lymph node metastasis in human breast cancer. Clin. Chim. Acta 384:69–74; 2007. [DOI] [PubMed] [Google Scholar]
- 15. Kim B. C.; Kim W. Y.; Park D.; Chung W. H.; Shin K.; Bhak J. SNP@Promoter: A database of human SNPs single nucleotide polymorphisms within the putative promoter regions. BMC Bioinformatics 9(Suppl. 1):S2; 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kumar P.; Henikoff S.; Ng P. C. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nature Protocols 4:1073–1082; 2009. [DOI] [PubMed] [Google Scholar]
- 17. Lee I. K.; Choi J. H.; Kim Y. K.; Kim H. N.; Park K. S.; Lee J. J.; et al. Two single nucleotide polymorphisms of the ETS2 transcriptional factor gene predispose individuals to high-risk acute myelogenous leukemia AML. Blood ASH Annual Meeting Abstracts 106, Abstract 2729; 2005.
- 18. Lenth R. V. Statistical power calculations. J. Anim. Sci. 85:E24–29; 2007. [DOI] [PubMed] [Google Scholar]
- 19. Li X.; Lu J. Y.; Zhao L. Q.; Wang X. Q.; Liu G. L.; Liu Z.; et al. Overexpression of ETS2 in human esophageal squamous cell carcinoma. World J. Gastroenterol. 9:205–208; 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Linding R.; Russell R. B.; Neduva V.; Gibson T. J. GlobPlot: Exploring protein sequences for globularity and disorder. Nucl. Acids Res. 31:3701–3708; 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Miller S. A.; Dykes D. D.; Polesky H. F. A simple salting out procedure for extracting DNA from human nucleated cells. Nucl. Acid Res. 16:1215; 1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pande P.; Chakravarti N.; Soni S.; Mathur M.; Shukla N. K.; Ralhan R. ETS-1 expression in human oral squamous cell carcinoma: Potential prognostic implications. Cancer Detect. Prev. 24(Suppl. 1); 2000. [Google Scholar]
- 23. Papas T. S.; Watson D. K.; Sacchi N.; et al. ETS family of genes in leukemia and Down syndrome. Am. J. Med. Genet. 7:251–261; 1990. [DOI] [PubMed] [Google Scholar]
- 24. Rachidi M.; Lopes C. Mental retardation in Down syndrome: From gene dosage imbalance to molecular and cellular mechanisms. Neurosci. Res. 59:349–369; 2007. [DOI] [PubMed] [Google Scholar]
- 25. Ramensky V.; Pork P.; Sunyaev S. Human non-synonymous SNPs: Server and survey. Nucl. Acids Res. 30:3894–3900; 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sementchenko V. I.; Watson D. K. Ets target genes: Past, present and future. Oncogene 19:6533–6548; 2000. [DOI] [PubMed] [Google Scholar]
- 27. Sumarsono S. H.; Wilson T. J.; Tymms M. J.; et al. Down’s syndrome-like skeletal abnormalities in Ets2 transgenic mice. Nature 379:534–538; 1996. [DOI] [PubMed] [Google Scholar]
- 28. Turner D. P.; Moussa O.; Sauane M.; Fisher P. B.; Watson D. K. Prostate-derived ETS factor is a mediator of metastatic potential through the inhibition of migration and invasion in breast cancer. Cancer Res. 67:1618–1625; 2007. [DOI] [PubMed] [Google Scholar]
- 29. Wolvetang E. J.; Wilson T. J.; Sanij E.; Busciglio J.; et al. ETS2 overexpression in transgenic models and in Down syndrome predisposes to apoptosis via the p53 pathway. Hum. Mol. Genet. 12:247–255; 2003. [DOI] [PubMed] [Google Scholar]
- 30. Xu Z.; Kerstann K. F.; Sherman S. L.; Chakravarti A.; Feingold E. A trisomic transmission disequilibrium test. Genet. Epidemiol. 26:125–131; 2004. [DOI] [PubMed] [Google Scholar]
- 31. Yanai I.; Benjamin H.; Shmoish M.; Chalifa-Caspi V.; Shklar M.; Ophir R.; Bar-Even A.; Horn-Saban S.; Safran M.; Domany E.; Lancet D.; Shmueli O. Genome-wide midrange transcription profiles reveal expression level relationships in human tissue specification. Bioinformatics 21:650–659; 2005. [DOI] [PubMed] [Google Scholar]
- 32. Yuan H. Y.; Chiou J. J.; Tseng W. H.; Liu C. H.; Liu C. K.; Lin Y. J.; et al. FASTSNP: An always uptodate and extendable service for SNP function analysis and prioritization. Nucl. Acids Res. 34:W635–641; 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yue P.; Melamud E.; Moult J. SNPs3D: Candidate gene and SNP selection for association studies. BMC Bioinformatics 7:166; 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]