Abstract
MicroRNAs (miRNAs) play a major role in cancer development and also act as a key factor in many other diseases. In this investigation, we propose three methods for handling miRNA expressions. The first two methods determine whether a miRNA is indicating normal or cancer condition, and the third one determines how many miRNAs are supporting the cancer sample/patient. While Method 1 acts as a two-class classifier and is based on normalized average expression value, Method 2 also does the same and is based on the normalized average intraclass distance. Method 3 checks whether a miRNA belongs to the cancer class or not, provides the percentage of supporting miRNAs for a cancer patient, and is based on weighted normalized average intraclass distance. The values of the weights are determined using exhaustive search by maximizing the accuracy in training samples. The proposed methods are tested on the differentially regulated miRNAs in three types of cancers (breast, colon, and melanoma cancer). The performances of Method 1 and Method 2 are evaluated by F score, Matthews Correlation Coefficient (MCC), and plotting “1 − specificity versus sensitivity” in Receiver Operating Characteristic (ROC) space and are found to be superior to the kNN and SVM classifiers for breast, colon, and melanoma cancer data sets. It is also observed that both the sensitivity and the specificity of Method 1 and Method 2 are higher than 0.5. For the same data sets, Method 3 achieved an average accuracy of more than 98% in detecting the miRNAs, supporting the cancer condition.
Key words: Bioinformatics, MicroRNA expressions, Cancer, Pattern recognition
INTRODUCTION
MicroRNAs (miRNAs) are a special type of noncoding RNAs (13,17), which are found in every living organism having eucaryotic cell and directly work with messenger RNAs (mRNA) (2,6,9,19). Noncoding RNAs are those RNAs that are not directly associated with protein coding functions. miRNAs indirectly take part in various biological and pathological process by inhibiting the translation process (the process of generation of protein from mRNA) of mRNA. In this inhibiting process, mature miRNAs create a bond with RNA Induced Silencing Complex (RISC) and produce miRISC (20). This miRISC binds its 5′ untranslated region (5′ UTR) to the 3′ untranslated region (3′ UTR) of the targeted mRNA by Watson-Crick base pairing mechanism (20) and the translation process does not proceed further (3,8,15).
Deregulation of miRNA expression is one of the major causes of the development of cancers in an animal body (10). It causes the cells of a tissue to fail to exit from cell cycle in proper time (18) and the cells go for uncontrolled divisions and suppress the activities of the other cells. As a consequence, an animal body becomes very sick and unable to survive without any treatment. miRNAs are also considered as major biomarkers of various other human diseases, like viral infection, metabolic disorders, etc. Now, given a sample (patient) with expression values of miRNAs, the task can be the classification of each miRNA as either normal or cancer. In this regard, we propose two methods (Method 1 and Method 2) by viewing the problem as two-class classification problem, where the task is to check how many miRNAs are indicating the normal and the cancer conditions of a given patient. On the other hand, the problem can be also be viewed as identifying miRNAs supporting the cancerous condition of a given cancer patient. To handle this problem we propose Method 3, where the aim is to determine how many miRNAs are supporting the condition of a cancer patient.
The rest of this article is organized as follows. In Section 2, some existing approaches are described. The details about the miRNA generation, miRNA expression, data sets, and the proposed investigation are described in Section 3. In Section 4 experimental results are reported. Finally, Section 5 concludes this investigation.
EXISTING APPROACHES
The first investigation in the field of miRNA and its role on cancers was investigated in 2002 (4). The application of computational techniques comes in the scenario for their inexpensiveness and time-saving benefit (10).
In Lu et al. (10), 334 mammalian tissue samples, including both the normal and the cancer samples, were collected and 217 different miRNAs were extracted from the collected tissues. Hierarchical clustering was then performed (using Pearson correlation and average linkage) on the generated miRNA expressions. The method separates the expressions according to the location of their origins. It is also shown that miRNA expressions are more informative than the mRNA expressions, even in the case of very little sign of cancer.
Investigations were also conducted (3) to extract the normal and the cancer miRNA expressions. It is shown that the normal and the cancer miRNA expressions are making different clusters using average linkage hierarchial clustering with Pearson correlation as similarity measure. It is also observed that miRNAs express themselves differently for different breast cancer subtypes.
From previous investigations (3,10,22), it is observed that miRNA expression values are determined using biochemical methods and then clustering techniques are applied to differentiate the origin of their tissue locations. Later, emphasis is given on the separation of the normal and the cancer miRNA expressions (3,10,22) and the separation of the subclasses for a particular type of cancer (3). In Leidinger et al. (12), miRNAs responsible for the melanoma cancer were identified and 16 miRNAs were pointed out, which show significant deregulation in the cancer patients. Reviews on some other existing methodologies are also available (5,16,21).
Most of the existing investigations mainly focused on determining the nature (i.e., expression values) of the miRNAs in different stages (normal and cancer) and classifying samples by using miRNAs as features. In this article, we deal with the problem of predicting the condition (normal or cancer) of miRNAs (i.e., considering miRNAs as patterns) and also finding the supporting miRNAs for a given cancer patient.
MATERIALS AND METHODS
The natural biochemical process, by which generation of miRNA from miRNA genes takes place, involves four steps (5,8,11,20–22). These are as follows:
Generation of primary miRNA transcripts: At the first step, primary miRNA transcripts (pri-miRNA), of length ∼1000 nt, are generated from the miRNA genes in the nucleolus.
Generation of precursor miRNA: pri-miRNA is then cleaved by RNase endonuclease-III enzyme Drosha and its partner DGCR8/Pasha in the nucleus and generates precursor miRNA (pre-miRNA) of length ∼60–100 nt.
Transportation of pre-miRNA into cytoplasm: The generated pre-miRNA is transported from nucleus to cytoplasm through the pores of the nuclear membrane by the proteins RanGTP and exprotin-5.
Generation of mature miRNA: Pre-miRNAs are further cleaved by Dicer enzyme and generate ∼22 nt mature miRNA duplex, containing a guide stand and a passenger stand. From this duplex, passenger stand degrades and the guide stand generates simplex mature miRNA.
The mature miRNAs can be classified into intergenic and intragenic miRNAs, based on the location of miRNA-coding genes. While in intergenic miRNA, miRNA-coding genes are located in between protein-coding genes, in intragenic miRNA, miRNA-coding genes are located within their host protein-coding genes.
miRNA Expression Generation
There are three major processes, by which miRNA expressions can be obtained (7). They are as follows:
miRNA expression profiling by cloning and sequencing: This process is accomplished by isolation of mature miRNA, adaptor ligation, reverse transcription, and polymerase chain reaction (PCR) amplification.
Microarray analysis: The steps to prepare microarray for miRNA expressions involve oligonucleotide probe design, preparation of labeled material from RNA samples (with amplification or without amplification), and microarray preparation.
Microbead expression analysis: One of the successful methodologies in this type of technology is xMAP, where 100 different miRNAs can be analyzed in one reaction. Each miRNA is treated as a microbead and each microbead has its own identity as a color (fluorescent dye) code. The amount of a particular miRNA can be scanned as the intensity value of the color, and this intensity value is stored as the expression of that miRNA.
Data Sets
In this investigation we used three different types of cancer data sets: breast (3), colon (1), and melanoma (12). While the breast cancer data set consists of 98 (5 normal + 93 cancer) samples and 309 miRNA expressions, the colon cancer data set consists 66 (8 normal + 58 cancer) samples and 287 miRNA expression values, and the melanoma cancer data consists of 57 (22 normal + 35 cancer) samples and 866 miRNA expressions. In Blenkiron et al. (3), out of 309 miRNAs, 38 miRNAs were pointed out as differentially expressed in the normal and the cancerous breast samples. Similarly, in Arndt et al. (1) and Leidinger et al. (12), 37 out of 287 and 51 out of 866 miRNAs were identified as differentially expressed between the normal and the cancer samples in colon and melanoma, respectively. Hence in our investigation those differentially expressed miRNAs are only considered for further study. Table 1 summarizes the details of the data sets.
TABLE 1.
SUMMARY OF THE USED DATA SETS
| Cancer Type | Total No. of Human miRNAs | No. of Cancer-Related miRNAs | No. of Normal Samples | No. of Cancer Samples |
|---|---|---|---|---|
| Breast cancer | 309 | 38 | 5 | 93 |
| Colon cancer | 287 | 37 | 8 | 58 |
| Melanoma cancer | 866 | 51 | 22 | 35 |
Proposed Approaches
As stated earlier, the main issues tackled in this investigation are: i) to check how many miRNAs are indicating the normal and the cancer conditions of a given patient, and ii) to check how many miRNAs are supporting a cancer patient’s condition.
In this regard we proposed three methods, among which Method 1 and Method 2 deal with the first issue and Method 3 deals with the second issue. For all of these methods, we used leave-one-out cross-validation procedure, where at a particular instance one sample is kept for testing purpose and all other samples are used for training. The process is then repeated for all the samples one by one. The performance of a method is then judged by the average result over all the samples. Now we discuss the proposed methods in detail.
Method 1
In this method, given a miRNA with the normal and the cancer samples, we calculated the normal representative by taking the ratio of mean and standard deviation of the normal expressions of that miRNA. The cancer representative is also calculated in a similar manner by using the cancerous expressions of the same miRNA. Now for the test sample first we consider one of its expressions corresponding to the given miRNA and two values are generated from that expression by normalizing it with standard deviation of the normal and the cancer expressions, respectively. For these two values, city block distances are then calculated from the normal and the cancer representatives, respectively, and the decision for the miRNA expression, chosen from the test sample, is taken according to the closeness of those values to the representative of each class (normal or cancer). To find how many miRNAs of a test sample are normal and how many are cancerous, we repeat the process for all the miRNAs.
Let N, M, and L be the total number of normal samples, cancer samples, and miRNAs, respectively, in a data set and x k i corresponds to the ith (i = 1, 2, . . . N) normal expression value of the kth (k = 1, 2, . . . L) miRNA, and y k j represents the jth (j = 1, 2, . . . M) cancer expression value of the kth miRNA. According to the leave-one-out cross-validation method, at a particular instance one sample from the whole set is selected for testing and other samples are used for training. So, there will be N − 1 and M numbers of training samples in the normal and the cancer training sets, respectively, if we chose the test sample (say T n for normal) from the set of the normal samples. There will be N and M − 1 numbers of training samples in the normal and the cancer training set, respectively, if we chose the miRNA expressions of the test sample (say T c for cancer) from the set of the cancer samples.
The steps for Method 1 are given below:
Step 1. In the training phase, calculate the representative of the kth miRNA in the normal and the cancer classes, if test sample is chosen from the set of the normal samples, as
| (1) |
| (2) |
respectively. If the test sample is chosen from the cancer samples, then calculate the representative of the kth miRNA in the normal and the cancer classes as
| (3) |
| (4) |
respectively, where, σk n and σk c represent standard deviations of the normal and the cancer expression values, respectively, of the kth miRNA.
Step 2. In the testing phase, the goal is to find whether an unknown miRNA expression for the test sample is normal or cancer. In this regard perform the following task:
a) Normalize the kth miRNA expression of the test sample with σk n and σk c and represent them as
| (5) |
| (6) |
respectively, where, u k is the expression value of the kth miRNA of the test sample.
b) Select the kth miRNA of the test sample as the normal one if it satisfies the condition:
| (7) |
and select the kth miRNA as cancerous if it satisfies the condition
| (8) |
Step 3. Repeat steps 1 to 2 for all k (i.e., for all the miRNAs in the test sample), where, k = 1, 2, . . . L.
Step 4. Repeat steps 1 to 3, for all the samples considering as test sample one by one.
Step 5. Evaluate the performance of the this method in terms of F score, Mathews Correlation Coefficient (MCC), and by plotting “1 − specificity versus sensitivity” in receiver operating characteristic (ROC) space.
The F score is defined as:
| (9) |
where the sensitivity (Sn) is defined as
| (10) |
and the specificity (Sc) is defined as
| (11) |
The MCC is defined as
| (12) |
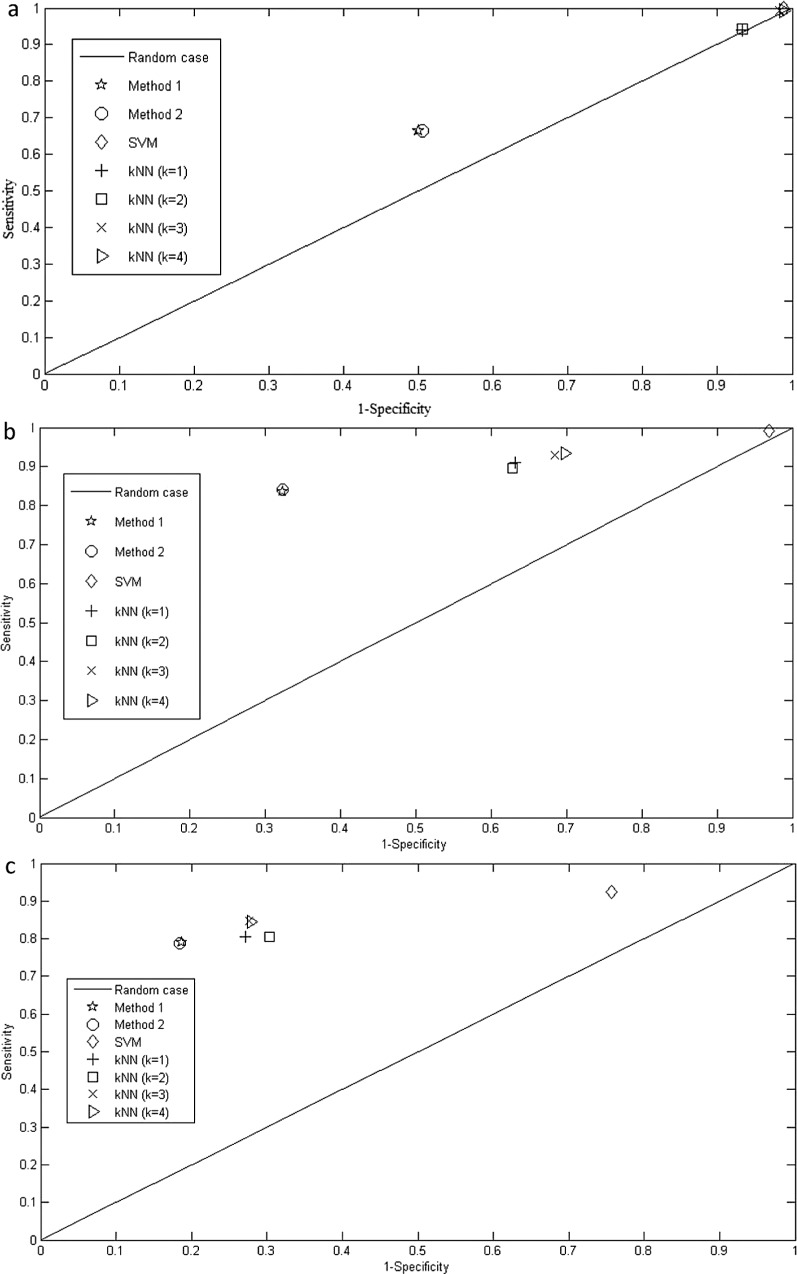
Here, the true positive refers to the number of correctly detected cancer miRNA expressions and false negative refers to the number of undetected cancer miRNA expressions. True negative implies the number of correctly detected normal miRNA expressions and false positive implies the wrongly detected cancer miRNA expressions (i.e., detected as cancer expressions, but actually they are normal expressions). The value of MCC lies between −1 to +1, where MCC value less than zero implies prediction capability worse than random prediction and greater than zero indicates the prediction capability better than random prediction. In the ROC space, any point on the straight line, passing through the coordinates (0,0) and (1,1) (see Fig. 2a–c), indicates that the prediction performance is the same as that of random prediction. On the other hand, coordinate (0,1) implies a perfect prediction and the coordinate (1,0) indicates a totally wrong prediction.
Figure 2.
(a) Comparison of the proposed two class classifiers with SVM and kNN in ROC space for (a) breast cancer data, (b) colon cancer data, and (c) melanoma cancer data.
Method 2
In this method, the average intraclass distance for the normal class and the cancer class are calculated by using the corresponding class mean and all expression values for that miRNA in that class. Then the average intraclass distances, for normal and cancer class, of that miRNA are normalized with the standard deviation of the normal and the cancer expressions of that miRNA, respectively. For the same miRNA, the city block distance between the unknown expression (test sample) and the class mean of the normal training samples is calculated and normalized by the standard deviation of the normal training samples of that miRNA. Similarly, for the same miRNA the process is repeated for the unknown expression and the class mean of the cancer training samples. Decision (normal or cancer) for the unknown miRNA in the test sample is then taken according to the closeness of the normalized city block distances with the normalized intraclass distances. The whole process is then repeated for all the miRNAs of a given test sample.
Let, N, M, and L be the total number of the normal samples, cancer samples, and miRNAs, respectively, in a data set. The ith (i = 1, 2, . . . N) normal expression value of the kth (k = 1,2, . . . L) miRNA is represented as x k i, and the jth (j = 1, 2, . . . M) cancerous expression of the kth miRNA is represented as y k j. As we are using leave-one-out cross-validation, the numbers of training samples in the normal and the cancer samples are N − 1 and M, respectively, when the test sample (say T n) is selected from the normal samples. Similarly, there are N and M − 1 numbers of training samples, respectively, if the test sample (say T c) is selected from the cancer samples.
The steps for Method 2 are as follows:
Step 1. If the test sample is chosen from the normal samples, calculate the normalized average intraclass distance of the kth miRNA in the normal class as
| (13) |
where μk n and σk n represent the mean and the standard deviation of the normal expression values of the kth miRNA, respectively. The normalized average intraclass distance of the kth miRNA in the cancer class is calculated as,
| (14) |
where the mean and the standard deviation of the expression values of the kth miRNA in the cancer class are represented as μk c and σk c, respectively.
If the test sample is chosen from the cancer class then calculate the normalized average intraclass distance of the kth miRNA in the normal and the cancer classes as
| (15) |
and
| (16) |
respectively, where μk n, σk n, μk c, and σk c represents the same variables mentioned previously.
Step 2. Calculate the city block distance between the unknown expression (in the test sample) and the class mean of the kth miRNA in the normal training samples and normalize the city block distance by the standard deviation of that miRNA in the same samples as
| (17) |
Similarly, calculate the city block distance between the unknown expression and the cancer class mean of the kth miRNA and normalize it by the standard deviation of the kth miRNA in cancer training samples as
| (18) |
where u k represents the expression value of the kth miRNA in the test sample.
Step 3. Consider the kth miRNA as normal if
| (19) |
and cancerous if
| (20) |
Step 4. Repeat steps 1 to 3 for all the values of k (k = 1, 2, . . . L) of a given sample.
Step 5. According to the leave-one-out cross-validation procedure, select all samples from the whole set one by one as test sample and repeat steps 1 to 4.
Step 6. Evaluate the performance of the method by F score, MCC value, and by plotting “1 − specificity versus sensitivity” in ROC space in a similar way to Method 1.
Method 3
As mentioned in section 1, here we discuss Method 3, which can be used to identify the miRNAs, supporting cancerous condition of a given cancer patient. In this regard, in the training phase we only used known cancer samples, and in testing phase we checked whether a miRNA expression is supporting cancerous condition or not. Finally, we determined how many miRNAs are supporting cancerous condition of a sample. Here, we introduced a weight factor that is determined through exhaustive search by maximizing the predicting accuracy, using the training samples. The steps for determining the condition of a miRNA by this method are given below.
Let M and L be the total number of the cancer samples and miRNAs, respectively, in a data set. Hence, the number of the training samples and the test sample will be M − 1 and 1, respectively, according to leave-one-out cross-validation method. The expression value of the jth (j = 1, 2, . . . M) cancer sample of the kth (k = 1, 2, . . . L) miRNA is represented as y k j.
The steps for Method 3 are as follows:
Step 1. In a way similar to step 2 in Method 2, in the training process, calculate the normalized average intraclass distance of kth miRNA in the cancer class as
| (21) |
where μk c and σk c represent the mean and the standard deviation of the expression values, respectively, in the cancer training samples of the kth miRNA and T c represents the test sample.
Step 2. Determine the weighted normalized average intraclass distance (t ′k c), for the kth miRNA in the cancer class, as
| (22) |
where w k c is the weight factor for the kth miRNA.
Step 3. Assign the initial value of w k c as 1 and increment the value of w k c in steps of 0.1, until the accuracy is maximized in detecting all the training samples (j = 1, . . . M − 1) for the kth miRNA.
Step 4. Repeat steps 1 to 3 for all the values of k (k = 1, . . . L) and store the values of t ′k c (k = 1, . . . L).
Step 5. In the testing phase, calculate the normalized city block distance between the expression value of the kth miRNA in the test sample (say, ith sample) and the mean expression value of the kth miRNA in the training samples, as
| (23) |
where u k c represents the expression value of the kth miRNA in test sample and μk c and σk c are the mean and the standard deviation of the expression values, respectively, in the training samples of the kth miRNA.
Step 6. A miRNA expression will be considered as cancerous expression if it satisfies the condition
| (24) |
Step 7. Repeat steps 5 to 6 for all values of k (i.e., for all miRNAs), where k varies from 1 to L.
Step 8. Calculate the percentage of supporting miRNA for the ith sample as
| (25) |
where C i is the number of correctly detected supporting miRNAs for cancer.
Step 9. Select all the samples from the whole set one by one as test sample and repeat steps 1 to 8 and calculate the average (say p c) of the obtained results as
| (26) |
where M is the number of samples.
EXPERIMENTAL RESULTS
The proposed methods are tested on subsets of miRNAs, which are identified as differentially expressed in breast, colon, and melanoma cancer, are only considered. First, the performances of Method 1 and Method 2 are compared with the performance of the fold change of miRNAs in normal and cancer cells, k-nearest neighbor (kNN) classifier, and SVM classifier, and then the performance of Method 3 is compared with only fold change based method as Method 3 does not handle the problem as a two-class classification problem, like kNN and SVM classifiers.
Fold change (14) (say F) of a miRNA is defined as the ratio between its normalized mean expression values of the cancer class (say t c) and its normalized mean expression value of the normal class (say t n). These fold change values indicate whether the fold change is positive or negative for a particular miRNA. For an unknown miRNA, we generated two values (say u n and u c) by normalizing its expression with the standard deviation of the normal and the cancer class of that miRNA, respectively. Now for a miRNA with positive fold change (i.e., F > 1), it is considered as cancerous if the ratio of u c and t n is greater than or equal to F (i.e., ) and it is considered as normal for the opposite condition (i.e., ). For a miRNA with negative fold change (i.e., 0 < F < 1), we calculated the ratio of u c and t n, and if it is less than or equal to F (i.e., ) it is considered as cancerous. A miRNA (with negative fold change value) is selected as normal if it satyisfies the opposite condition (i.e., ). Finally, leave-one-out cross-validation procedure is used for evaluating the performance of the method.
The comparison between Method 1 and fold change-based method on three different data sets, in terms of F score is presented in Figure 1a. Similarly, Figure 1b represents the comparison between Method 2 and fold change-based method on three different data sets in terms of the same measure. It is observed that the F score values for breast, colon, and melanoma cancer data sets are 0.5703, 0.7487, and 0.8324, respectively, for Method 1, and for Method 2 F scores are 0.5669, 0.7506, and 0.8324 for breast, colon, and melanoma cancer data sets, respectively. On the other hand, values of F score for breast, colon, and melanoma cancer data sets are 0.5038, 0.6090, and 0.5319, respectively, in fold change based method. Hence, it can be said that Method 1 and Method 2 perform better than fold change-based method in terms of F score. It is also seen, for different data sets, while the sensitivity varies from 0.6637 to 0.8372 and 0.6643 to 0.8420 for Method 1 and Method 2, respectively, the specificity varies from 0.5100 to 0.8128 and 0.5044 to 0.8155, respectively.
Figure 1.
Comparison of F score, with different types of data sets, between Method 1 (a) and Method 2 (b) and fold change-based method.
The comparisons of Method 1 and Method 2 with SVM and kNN (where k = 1, 2, 3, 4), in terms of F score, is reported in Table 2. It is obtained from the table that F score varies from 0.5768 to 0.8324 and from 0.5734 to 0.8342 for Method 1 and Method 2, respectively. On the other hand, F score for SVM varies from 0.0220 to 0.3853 for the different data sets and F score for kNN varies from 0.0328 to 0.7808 for the different values of k and different data sets. A similar type of comparison, in terms of MCC value, is reported in Table 3. From this table it is observed that MCC value for Method 1 varies from 0.1660 to 0.6029 for different types of data sets. The range of MCC values lies between 0.1611 and 0.6029 for Method 2, using different data sets. It is observed that using SVM classifier, MCC value from 0.0718 to 0.2299 is achieved for different data sets and using kNN classifier, MCC value from 0.0156 to 0.5758 is achieved for different values of k with different data sets. It is seen from the Tables 2 and 3 that Method 1 and Method 2 work better, in terms of F score and MCC value, than SVM and kNN in terms of average F score and MCC. Even for individual data set, Method 1 and Method 2 perform better than SVM and kNN in terms of the mentioned performance measuring values (i.e., F score and MCC).
TABLE 2.
COMPARISON OF F SCORES FOR DIFFERENT ALGORITHMS
| Data Set | Method 1 | Method 2 | SVM | k − NN (k = 1) | k − NN (k = 2) | k − NN (k = 3) | k − NN (k = 4) |
|---|---|---|---|---|---|---|---|
| Breast cancer | 0.5768 | 0.5734 | 0.0220 | 0.1245 | 0.1245 | 0.0328 | 0.0328 |
| Colon cancer | 0.7487 | 0.7506 | 0.0606 | 0.5242 | 0.5254 | 0.4718 | 0.4566 |
| Melanoma cancer | 0.8324 | 0.8342 | 0.3853 | 0.7644 | 0.7476 | 0.7808 | 0.7783 |
| Average | 0.7193 | 0.7194 | 0.1560 | 0.4710 | 0.4710 | 0.4285 | 0.4226 |
TABLE 3.
COMPARISON OF MCC VALUES FOR DIFFERENT ALGORITHMS
| Data Set | Method 1 | Method 2 | SVM | k − NN (k = 1) | k − NN (k = 2) | k − NN (k = 3) | k − NN (k = 4) |
|---|---|---|---|---|---|---|---|
| Breast cancer | 0.1660 | 0.1611 | 0.0718 | 0.0156 | 0.0168 | 0.0492 | 0.0492 |
| Colon cancer | 0.5210 | 0.5262 | 0.0809 | 0.3315 | 0.3157 | 0.3125 | 0.3060 |
| Melanoma cancer | 0.6029 | 0.6029 | 0.2299 | 0.5342 | 0.5062 | 0.5758 | 0.5708 |
| Average | 0.4300 | 0.4301 | 0.1275 | 0.2938 | 0.2796 | 0.3125 | 0.3087 |
We also tested the performance of Method 1, Method 2, SVM, and kNN in receiver operating characteristic (ROC) space. As mentioned earlier we plotted “1 − specificity versus sensitivity” in this space. Here, any point on the straight line, passing through the coordinates (0,0) and (1,1), indicates that the prediction performance is the same as that of random prediction. On the other hand, coordinate (0,1) implies a perfect prediction and the coordinate (1,0) indicates a totally wrong prediction. The results in ROC space for the above mentioned methods are shown in Figure 2a–c for breast cancer, colon cancer, and melanoma cancer, respectively. It can be observed from the figure that Method 1 and Method 2 perform better than the SVM and kNN (for k = 1, 2, 3, 4) in terms of specificity as the x-axis represents “1 − specificity.” Hence, the higher value in the x-axis represents low specificity.
As mentioned earlier, given a cancer patient, Method 3 provides the number of miRNAs, supporting the cancerous condition of a sample. Note that in a part of the process Method 3 also identifies those miRNAs. Hence, the performances of Method 3 is compared with the performance of the fold change-based technique in terms of the percentage of supporting miRNAs in the cancer sample. In the fold change-based method, a miRNA, with positive fold change value is selected as supporting miRNA for cancer sample if the ratio of u c and t n is greater than or equal to F (i.e., ), where u c is the unknown expression normalized with the standard deviation of the cancer class and t n is the average normal expression normalized with the standard deviation of the normal class. A miRNA, with negative fold change value is considered as supporting miRNA if the ratio of u c and t n is less than or equal to F (i.e., ).
Table 4 shows the comparative performance of Method 3 and fold change-based technique. From the table, it is observed that while the percentage of supporting miRNAs varies from 98.77% to 99.50% for Method 3 for different data sets, the percentage of supporting miRNAs lies between 48.40% and 60.17% for fold change-based method. Note that the main advantage of Method 3 lies in finding the similarity of unknown expression with the cancer class, which does not require any normal expression information, like the fold change-based technique.
TABLE 4.
COMPARISON OF METHOD 3 WITH FOLD CHANGE-BASED METHOD: PERCENTAGE OF SUPPORTING miRNAs
| Data Set | Method 3 | Fold Change-Based Method |
|---|---|---|
| Breast cancer | 99.50% | 49.67% |
| Colon cancer | 99.19% | 60.17% |
| Melanoma cancer | 98.77% | 48.40% |
CONCLUSIONS
In this article, we proposed two approaches (Method 1 and Method 2) for identifying whether a miRNA is indicating normal or cancer condition, and one approach (Method 3) to check how many miRNAs are supporting the condition of a cancer sample. While the first method is based on normalized average expression value, the second method deals with the normalized average intraclass distance and the third method is based on weighted normalized average intraclass distance. Experiments are performed on breast, colon, and melanoma cancer data and it is observed that sensitivity, specificity, and F score for Methods 1 and 2 are above 0.66, 0.50, and 0.56, respectively. Hence, these methods can also be used for condition prediction of an unknown patient. MCC values for Method 1 and Method 2 are found to be positive for all the data sets. It is also observed that the first two methods perform better than kNN and SVM classifiers in terms of average F score, average MCC, and plots in ROC space for all types of data sets. Note that for results of SVM depends on the proper kernel selection and also on the selection of the parameters of the kernel, if applicable, and the results of kNN also depends on the selection of value of k. In the contrary, Method 1 and Method 2 do not have any parameter to set manually and they also consider the expression variation among the samples. Experimental results, on the same three data sets, show that the supporting miRNAs predicted by Method 3 is above 98% for all the data sets. Although the SVM and kNN classifiers are not compared with the Method 3, as this method does not handle the issue as a two-class problem, experimental results on multiple date sets revealed the potential value of our approach.
ACKNOWLEDGMENTS
The authors are thankful to the Department of Science and Technology, Government of India, for establishing the Center for Soft Computing Research at Indian Statistical Institute, Kolkata. The work was done when S. K. Pal was a J. C. Bose fellow of the Government of India.
REFERENCES
- 1. Arndt G. M.; Dossey L.; Cullen L. M.; Lai A.; Druker R.; Eisbacher M.; Zhang C.; Tran N.; Fan H.; Retzlaff K.; Bittner A.; Raponi M. Characterization of global microRNA expression reveals oncogenic potential of miR-145 in metastatic colorectal cancer. BMC Cancer 9:374; 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bartel D. P. MicroRNAs: Genomics, biogenesis, mechanism and function. Cell 116:281–297; 2004. [DOI] [PubMed] [Google Scholar]
- 3. Blenkiron C.; Goldstein L. D.; Thorne N. P.; Spiteri I.; Chin S.; Dunning M. J.; Barbosa-Morais N. L.; Teschendorff A. E.; Green A. R.; Ellis I. O.; Tavar S.; Caldas C.; Miska E. A. MicroRNA expression profiling of human breast cancer identifies new markers of tumor subtype. Genome Biol. 8:R214–R214.16; 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Calin G. A.; Dumitru C. D.; Shimizu M.; Bichi R.; Zupo S.; Noch E.; Aldler H.; Rattan S.; Keating M.; Rai K.; Rassenti L.; Kipps T.; Negrini M.; Bullrich F.; Croce C. M. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. USA 99:15524–15529; 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Calin G. A.; Croce C. M. MicroRNA signatures in human cancers. Nature 6:857–866; 2006. [DOI] [PubMed] [Google Scholar]
- 6. Chen F.; Chen Y. P. Exploring cross-species-related miRNAs based on sequence and secondary structure. IEEE Trans. Biomed. Eng. 57:1547–1553; 2010. [DOI] [PubMed] [Google Scholar]
- 7. Einat P. Methodologies for high-throughput expression profiling of microRNAs. In: MicroRNA protocols. New Jersey: Springer Verlag; 2006:139–157. [DOI] [PubMed] [Google Scholar]
- 8. Iorio M. V.; Ferracin M.; Liu C.; Veronese A.; Spizzo R.; Sabbioni S.; Magri E.; Pedriali M.; Fabbri M.; Campiglio M.; Mnard S.; Palazzo J. P.; Rosenberg A.; Musiani P.; Volinia S.; Nenci I.; Calin G. A.; Querzoli P.; Negrini M.; Croce C. M. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 65:7065–7070; 2005. [DOI] [PubMed] [Google Scholar]
- 9. Khvorova A.; Reynolds A.; Jayasena S. D. Functional siRNA and miRNAs exibit stand bias. Cell 115:209–216; 2003. [DOI] [PubMed] [Google Scholar]
- 10. Lu J.; Getz G.; Miska E. A.; Alvarez-Saavedra E.; Lamb J.; Peck D.; Sweet-Cordero A.; Ebert1 B. L.; Mak R. H.; Ferrando A. A.; Downing J. R.; Jacks T.; Horvitz H. R.; Golub T. R. MicroRNA expression profiles classify human cancers. Nature 435:834–838; 2005. [DOI] [PubMed] [Google Scholar]
- 11. Lund E.; Gttinger S.; Calado A.; Dahlberg J. E.; Kutay U. Nuclear export of microRNA precursors. Science 303:95–98; 2003. [DOI] [PubMed] [Google Scholar]
- 12. Leidinger P.; Keller A.; Borries A.; Reichrath J.; Rass K.; Jager S. U.; Lenhof H. P.; Meese E. High-throughput miRNA profiling of human melanoma blood samples. BMC Cancer 10:262; 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Madden S. F.; Carpenter S. B.; Jeffery I. B.; Bjrkbacka H.; Fitzgerald K. A.; O’Neill L. A.; Higgins D. G. Detecting microRNA activity from gene expression data. BMC Bioinform. 11:257–271; 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Navon R.; Wang H.; Steinfeld I.; Tsalenko A.; Ben-Dor A.; Yakhini Z. Novel Rank-Based Statistical methods reveal microRNAs with differential expression in multiple cancer types. Plos One 4:1–10; 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Olsen P. H.; Ambros V. The lin-4 regulatory RNA controls developmental timing in Caenorhabditis elegans by blocking LIN-14 protein synthesis after the initiation of translation. Dev. Biol. 216:671–680; 1999. [DOI] [PubMed] [Google Scholar]
- 16. Oulas A.; Reczko M.; Poirazi P. MicroRNAs and cancer—the search begins!. IEEE Trans. Inform. Technol. Biomed. 13:67–77; 2009. [DOI] [PubMed] [Google Scholar]
- 17. Rajewsky N.; Socci N. D. Computational identification of microRNA targets. Dev. Biol. 267:529–535; 2004. [DOI] [PubMed] [Google Scholar]
- 18. Reinhart B. J.; Slack F. J.; Basson M.; Pasquinelli A. E.; Bettinger J. C.; Rougvie A. E.; Horvitz H. R.; Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans . Nature 403:901–906; 2000. [DOI] [PubMed] [Google Scholar]
- 19. Selbach M.; Schwanhusser B.; Thierfelder N.; Fang Z.; Khanin R.; Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature 455:58–63; 2008. [DOI] [PubMed] [Google Scholar]
- 20. Wang Z.; Yang B. Detection, profiling, and quantification of miRNA expression. In: MicroRNA expression detection methods, 1st ed. Heidelberg: Springer Verlag; 2010:3–53. [Google Scholar]
- 21. Wu W.; Sun M.; Zou G.; Chen J. MicroRNA and cancer: Current status and prospective. Int. J. Cancer 120:953–960; 2006. [DOI] [PubMed] [Google Scholar]
- 22. Yang H.; Kong W.; He L.; Zhao J.; O’Donnell J. D.; Wang J.; Wenham R. M.; Coppola D.; Kruk P. A.; Nicosia S. V.; Cheng J. Q. MicroRNA expression profiling in Hhman ovarian cancer: miR-214 induces cell survival and cisplatin resistance by targeting PTEN. Cancer Res. 68:425–433; 2008. [DOI] [PubMed] [Google Scholar]