Abstract
Ubiquitin conjugating enzyme UbcM2 (Ubiquitin-conjugating enzymes from Mice, the number reveals the identification order) has been implicated in many critical processes, such like growth-inhibiting, mediating cell proliferation and regulation of some transcription factor, but the expression profile during mouse embryo development remains unclear. Hereby, during mid-later embryonic stage, the expression patterns of UbcM2 were examined using in situ hybridization and quantitative real-time PCR (qRT-PCR). The signals were significantly intense in central nervous system and skeletal system, weak in tongue, heart, lung, liver, and kidney. In the central nervous system, UbcM2 was principally expressed in thalamus, external germinal layer of cerebellum (EGL), mitral cell layer of olfactory bulb, hippocampus, marginal zone and ventricular zone of cerebral cortex, and spinal cord. In the skeletal system, UbcM2 was primarily expressed in proliferating cartilage. Furthermore, qRT-PCR analysis displayed that the expression of UbcM2 was ubiquitous at E15.5, most prominent in brain, weaker in lung liver and kidney, accompanied by the lowest level in tongue and heart. During brain development, the expression level of UbcM2 first ascended and then decreased from E12.5 to E18.5, the peak of which sustained starting at E14.5 until E16.5. Together, these results suggest that UbcM2 may play potential roles in the development of mouse diverse tissues and organs, particularly in the development of brain and skeleton.
Key words: UbcM2, In situ hybridization, Mouse embryo development
INTRODUCTION
The ubiquitin (Ub) system has a greater-than-average involvement in regulating numerous crucial cellular processes, such as protein quality control (26) and transcription (11,22) and so on. Most proteins are targeted for proteasome-mediated degradation by covalent attachment of multiple ubiquitin molecules (24). Ubiquitination of a protein requires an enzyme cascade composed of a Ub-activating enzyme (E1), a Ub-conjugating enzyme (E2), and a Ub protein ligase (E3) (10). Ub is first activated in an ATP-dependent manner by the E1, the cysteine active site of which forms a thiol ester intermediate with the C-terminal glycine residue of ubiquitin. After that, Ub is transferred to the active site of the E2 cysteine residue. An E3 ligase facilitates the transfer of ubiquitin from the E2 to the substrate (6,24).
UbcM2, one of class II E2s that have amino-terminal but lack carboxyl-terminal extensions, is located in mouse chromosome 2. UbcM2 was first identified as the closest homologs of yeast UBC4/UBC5, sharing an exceptionally highly conserved domain called UBC with its counterparts UBE2E3 from humans. The UBC domain is all E2s’ characteristic that encompasses ∼150 residues to form a globular domain, containing an active site of catalytic cysteine (13,23). This domain is necessary to mediate the interaction with Nedd4-2, consequently involved in the regulation of ENaC cell surface expression (3). So far, the field of UbcM2’s characteristics and functions has been widely digged up. UbcM2 localizes to the nucleus and can shuttle in and out of the nucleus in the Ub-charging state by importin-11 (20,21). It is identified as a growth-inhibitory gene that enzymatic activity of UbcM2 may lead to inhibition of cell cycle progression, affecting the activity of some critical proteins that are involved in cell cycle (8,16). Moreover, UbcM2’s counterpart UBE2E3 is required for retinal pigment epithelial cell proliferation (18), and it is necessary to protect photoreceptors from acute photo-oxidative toxicity (14). UbcM2 is overexpressed in cancer compared with normal prostate tissue, becoming a specific marker of prostate cancer (1). Although Western blotting demonstrates that UbcM2 is most prominently expressed in the retina and ubiquitously expressed in most tissues at lower levels in 7-month-old mice (14), the expression patterns of UbcM2 during mouse mid-late embryonic stage have not been reported thus far.
In this study, we examined the expression of UbcM2 in diverse tissues and developmental stages by in situ hybridization and qRT-PCR. The result showed that UbcM2 was ubiquitously expressed during mouse embryonic stage, with significantly heavy signals in the central nervous system (CNS) and skeletal system. In addition, the data of qRT-PCR displayed that the expression of UbcM2 was ubiquitous at E15.5, most abundant in the brain. During the mid-late embryonic stage, the expression of UbcM2 in the brain formed a crest between E14.5 and E16.5. These data suggested that UbcM2 may be involved in the development of diverse mouse tissues or organs, particularly brain and skeleton.
MATERIALS AND METHODS
Animal
Mice (C57BL/6 J) were time-mated and pregnant females were sacrificed to collect the embryos and tissues at distinct embryonic stages. The age of the embryos was determined by the presence of a vaginal plug representing embryonic day 0.5 (E0.5). The day of birth was designated as postnatal day (P). All procedures were carried out following the “Rules for experiments’ animals” promulgated by Chinese Government (Beijing, China).
In Situ Hybridization (ISH)
The whole embryos (E10.5, E12.5, E14.5, and E16.5) were fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS) at 4°C overnight. Embryos were dehydrated, paraffin embedded, and subsequently sectioned at 9 μm. For preparing UbcM2 probe, a mouse cDNA fragment (sense: 5′-aagatgtccagtgacaggcaaag-3′, antisense: 5′-tctgctcctt cacactgcatac-3′) was subcloned into pBluescript II KS(+) (Stratagene, CA, USA) T-vector by using DNA Ligation Kit (TARAKA, Dalian, China) following the manufacturer’s instructions. Antisense and sense RNA probes specific to UbcM2 gene were synthesized by in vitro transcription in the presence of digoxygenin-UTP (Roche, Mannheim, Germany). ISH (section and whole-mount) was performed as previously described (2,27).
RNA Preparation and Quantitative Real-Time-PCR (qRT-PCR)
Total RNAs were extracted from diverse tissues of E15.5 and brain tissues from E12.5 to E18.5 by Trizol reagent (Invitrogen, Eugene, OR, USA). The cDNAs were synthesized using a SuperScript™III RNase H− Reverse Transcriptase kit (Invitrogen). Subsequently, the cDNAs were used for UbcM2 expression analysis by qRT-PCR. Sequences of UbcM2 primers used in qRT-PCR were: 5′-caggttctg tatatgaaggtg-3′ (sense), 5′-gtcaaatactgagtggctatg-3′ (antisense). qRT-PCR analysis was performed by the ABI Prism 7500 Real-Time PCR System, and reactions were prepared using the SYBR Green PCR master mix kit (Applied Biosystems, CA, USA) according to manufacturer’s instructions. The expression level of UbcM2 gene was normalized by that of the glyceraldehyde-3-phosphate dehydrogenase (Gapdh) gene. All samples were analyzed in triplicate at least.
RESULTS
UbcM2 Gene Structure and Amino Acid Sequence Analysis of UbcM2 With Its Orthologues
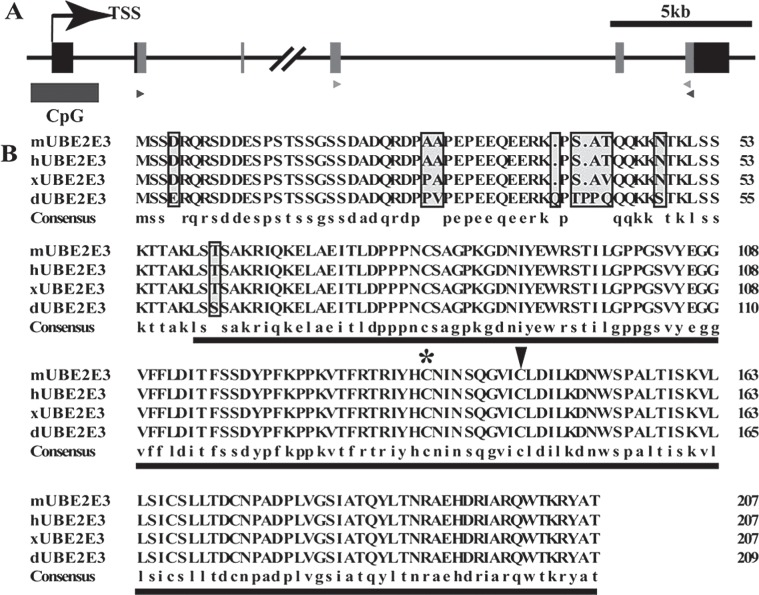
The Mus musculus UbcM2 gene (Gene ID: 22193) is located in mouse chromosome 2 2D region, includes 6 exons, and encodes a 207-amino acid protein. A predicted CpG island is situated from −507 bp upstream to +969 bp downstream harboring 140 CpG sites in a stretch of the UbcM2 promoter region, the exon 1 and intron 1 (Fig. 1A).
Figure 1.
UbcM2 was highly conserved in vertebrates. (A) Schematic representation of the UbcM2 gene structure (Mus musculus, chromosome 2 2D). Gray boxes represent the coding region of UbcM2. Dark rectangle demonstrates a predicted CpG island harboring a stretch of 140 CpG sites (http://genome.ucsc.edu). The black and gray triangles label the primers for specific sense and antisense probe preparation and qRT-PCR, respectively. (B) Amino acid sequence alignment of the primary sequences of mouse UbcM2 (mUBE2E3) and its orthologues in human (hUBE2E3), Xenopus laevis (xUBE2E3), D. rerio (dUBE2E3). The highly conserved UBC core domain is underlined. The conserved catalytic cysteine (145) is indicated by an arrowhead. The noncatalytic cysteine (136), which is identified as a putative redox sensor, is indicated by an asterisk.
The UBC core domain is located at amino acids 59 to 207. It contains a conserved catalytic cysteine (145) that is required for enzyme-ubiquitin thiol ester formation. The noncatalytic cysteine (136) was identified as a putative redox sensor (19). The alignment of mUBE2E3 sequences from other species indicated that this domain is highly conserved, only a serine (S) representing a threonine (T) in Danio rerio (dUBE2E3) (Fig. 1B). Furthermore, it was more notable that the amino acid sequences of mouse (mUBE2E3) and human (hUBE2E3) exhibited 100% identity, demonstrating the conservative property of UbcM2 enzyme in the phylogenetic evolution. Comparison of the conservation among these orthologues was generated by multiple amino acid sequence alignment using DNAMAN (Fig. 1B).
Expression of UbcM2 During Mouse Embryonic Development
To analyze the expression of UbcM2 in mouse embryos during mid-later gestation period, whole-mount and section in situ hybridizations were carried out, respectively. Specific RNA probe using the primers, shown as blue triangles in Figure 1A, was prepared.
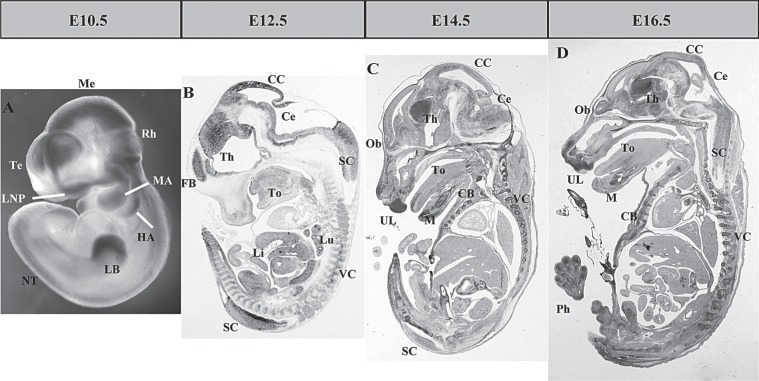
By E10.5, the strong expression signals were detected in the cortical anlage and mesenchyma of the telencephalon (Te), mesencephalon (Me), and limb buds (LB) (Fig. 2A). The signals can be observed at the rhombencephalon (Rh) and neural tube (NT) from dorsal of the identical embryo as well. In addition, UbcM2 was also detected in the mandibular arch (MA), hyoid arch (HA), and lateral nasal process (LNP), consistent with the signals viewed at the upper lip (UL) and tongue (To) of subsequent section in situ hybridization. As a negative control, no signals were detected using the sense probe (data not shown).
Figure 2.
UbcM2 overall expression pattern during mid-later gestation period. (A) The expression of UbcM2 in E10.5 embryo by whole-mount ISH. (B–D) The expression of UbcM2 in E12.5, E14.5, and E16.5 embryo by section ISH. FB, forebrain; Th, thalamus; CC, cerebral cortex; Ce, cerebellum; SC, spinal cord; Th, thalamus; To, tongue; Li, liver; Lu, lung; VC, vertebral column; Ob, olfactory bulb; UL, upper lip; M, Meckel’s cartilage; CB, costal bone; Ph, phalange.
From E12.5 to E16.5, the expression signals of UbcM2 mainly distributed in the CNS and skeletal system. By E12.5, the signals were primarily detected in brain (Fig. 3A, a) liver (Li, Fig. 3A, e) lung (Lu, Fig. 3A, d), and spinal cord (SC), weaker in the tongue (To, Fig. 3A, b), kidney (Fig. 3A, f), and vertebral column (VC), while no signals were detected in heart (Fig. 3A, c). UbcM2 in the brain was mainly located at the fore brain (FB), thalamus (Th), cerebral cortex (CC), and cerebellum (Ce) (Fig. 2B), most of which was in accordance with those derived from the signal location of E10.5.
Figure 3.
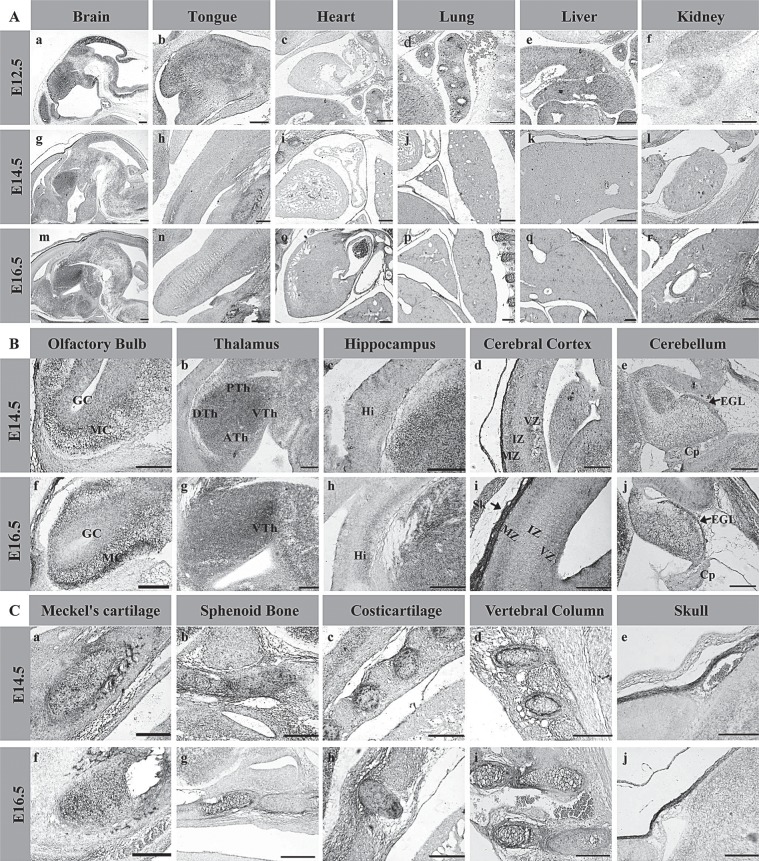
The expression of UbcM2 in diverse tissues of different development stages. (A) The expression pattern of UbcM2 in main tissues of E12.5, E14.5, and E16.5. (B) The expression pattern of UbcM2 in the brain of E14.5 and E16.5. GC, granule cell layer; MC, mitral cell layer (a, f); DTh, the dorsal area of thalamus; ATh, the anterior area of thalamus; VTh, the ventral area of thalamus; PTh, the posterior area of thalamus (b, g); Hi, immature hippocampus (c, h); MZ, marginal zone; VZ, ventricular zone; IZ, intermedial zone (d, i); EGL, external germinal layer; Cp, choroid plexus (e, j). (C) The expression pattern of UbcM2 in the skeletal system of E14.5 and E16.5. Scale bars: 200 μm.
By E14.5 and E16.5, the expression of UbcM2 became more intensive by in situ hybridization (Fig. 2C, D), principally in the CNS and skeletal system, with low expression level of tongue (Fig. 3A, h, n), heart (Fig. 3A, i, o), liver (Fig. 3A, j, p), lung (Fig. 3A, k, q), and kidney (Fig. 3A, l, r). In the developing CNS, the strong signal was detected in the regions of cerebral cortex (CC), thalamus (Th), cerebellum (Ce), olfactory bulb (OB), hippocampus (Hi), and spinal cord (SC). In cerebral cortex, the signals of UbcM2 were detected mainly in the marginal zone (MZ) and ventricular zone (VZ), accompanied by low expression in intermedial zone (IZ) (Fig. 3B, d, i). The strongest expression signals were observed in thalamus (Fig. 3B, b, g). By 14.5, the signals appeared in the whole regions of thalamus, including the dorsal (DTh), anterior (ATh), ventral (VTh), and posterior (PTh) areas, which was different from the appearance of individual signals in ventral thalamus by E16.5. In cerebellum, only the external granular layer (EGL) exhibited UbcM2’s expression signal (Fig. 3B, e, j). In olfactory bulb (Fig. 3B, a, f), UbcM2 mainly distributed in the mitral cell layer (MC), with weaker signals detected in the granule cell layer (GC). UbcM2 also showed a high expression level in immature hippocampus (Fig. 3B, c, h) and in the central nucleus of the dorsal horn, gelatinous substance, motoneurons of spinal cord (SC). In addition to the expression in CNS, the evident signals also presented in Meckel’s cartilage (Fig. 3C, a, f), sphenoid bone (Fig. 3C, b, g), costicartilage (Fig. 3C, c, h), cartilages of vertebral column (Fig. 3C, d, i), and skull (Fig. 3C, e, j), primarily visible in proliferating chondrocytes, rather than in the hypertrophic chondrocytes.
Expression Analysis of UbcM2 in Different Tissues
To further investigate UbcM2 expression pattern, we performed qRT-PCR to detect its expression among diverse tissues. At E15.5, UbcM2 was most abundant in brain, moderate in lung, liver, and kidney, accompanied by the lowest level in tongue and heart. This result was in accordance with the trends we discovered by ISH in mouse embryos. The prominent signals of UbcM2 in developmental brain showed high specificity, prompting us to examine its expression in brain in more detail. We then analyzed the expression of UbcM2 by qRT-PCR during mouse brain development. The result showed that the UbcM2 dynamic expression ascended from E12.5, reached a plateau at E14.5, maintained the status until E16.5, and then decreased during the mid-later embryonic stage (Fig. 4B).
Figure 4.
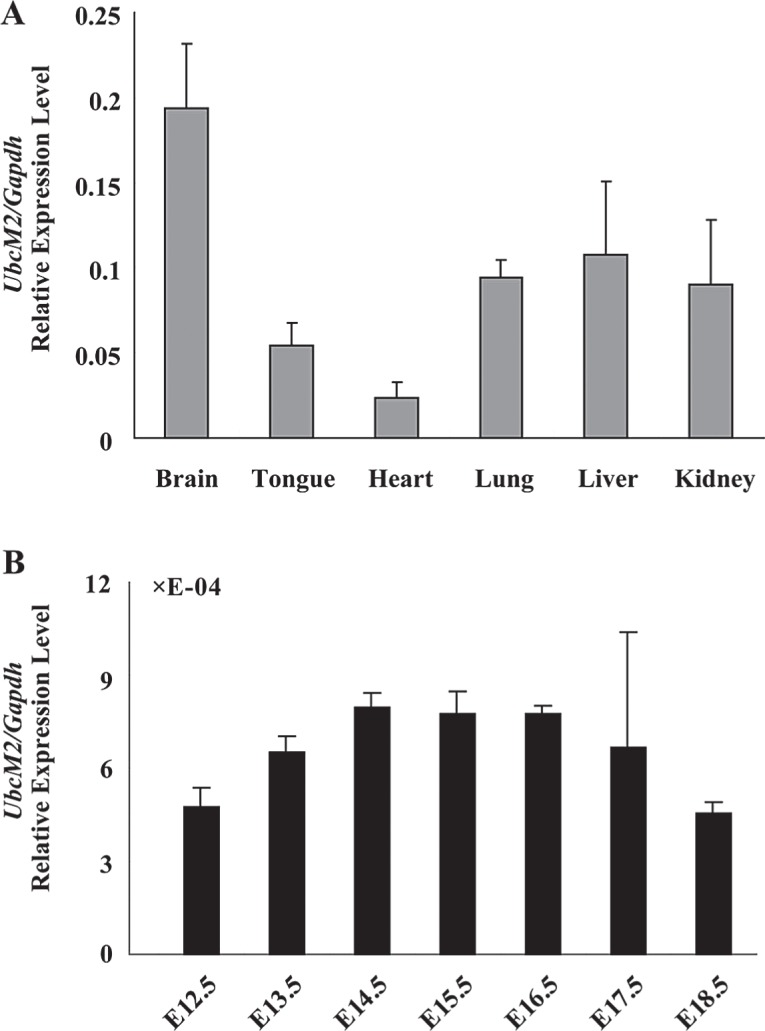
The qRT-PCR analysis of UbcM2 in E15.5 and in developing brain. (A) The expression of UbcM2 in different tissues of E15.5. (B) The expression profiles of UbcM2 in developing brain from E12.5 to E18.5. Data were expressed relative to the corresponding values for Gapdh. Error bars represented the standard deviation in three independent experiments.
DISCUSSION
Despite the discovery of numerous events UbcM2 can be involved in (3,10,20,21), to date the expression profile of this enzyme still needs to be identified. In this report, we performed in situ hybridization to analyze the expression patterns of UbcM2 during mouse embryogenesis. It exhibited intense signals in developing CNS and skeletal system, compared to those signal-nonsignificant tissues or organs. This result is in accordance with the Western blot data of adult mouse tissues, which show a strong expression in retina and brain, but weak in heart, lung, and liver (14).
The CNS of vertebrates (such as mouse) consists of the brain and spinal cord. The first sign of the nervous system is the appearance of the neural plate. The inner portion of the neural plate is destined to turn into the CNS, which is organized around a rostral–caudal axis of bilateral symmetry. With the development of the neural plate, it gets surrounded by neural folds, which finally become the neural tube. In our study, the embryo of E10.5 revealed a strong expression in the CNS, including telencephalon, mesencephalon, rhombencephalon, and neural tube, which indicated that UbcM2 may involve in the morphogenesis or development of the CNS in midgestation embryos. As the CNS develops, telencephalon, diencephalon, mesencephalon, and rhombencephalon differentiate into the hippocampus and neocortex, the thalamus, the tectum, the cerebellum, and the medulla oblongata, respectively, in accordance with the signals detected in those parts of brain by ISH from E14.5 to adulthood. At the E14.5 stage, organogenesis is largely complete, applicable to study organ architecture, with cell differentiation still occurring. In addition, early corticogenesis is taking place at this stage, whereas middle telencephalic development is occurring at E16.5.
These results indicated that UbcM2 could act a role in the developing brain as well as the spinal cord. Because of the character E2 enzyme UbcM2 played, the expression patterns may relate to the distribution of E3 ligases and relevant substrates. Nedd4-1, as a HECT E3 ubiquitin ligase, is abundant in mammalian neurons (12). It is a positive regulator of dendrite development; moreover, HECT-type ubiquitin-protein ligases (Nedd4 and Nedd4-2) can have an interaction with UBE2E3, a homolog of UbcM2 (5). It interprets the expression of UbcM2 in brain to some extent. It also seemed possible that these expression patterns in brain appeared due to the relevance between UbcM2 and Mdm2, a negative regulator of p53 activity (7,9). Mdm2 id involved in the DDD–E2 complex, which consists of UbcM2 as a critical member (17). Mdm2 is expressed in the early CNS (4); in addition, loss of Mdm2 lead to the neonatal lethality (28). UbcM2 null mice as disruption of both alleles is embryonic lethal (14), implicating the important role UbcM2 played during mouse embryogenesis. We also used qRT-PCR to examine UbcM2’s expression in brain, the tendency may result from the differentiation and proliferation of neurocytes. UbcM2 is required for RPE cell proliferation, and UbcM2 is downregulated during RPE maturation (18), which is applicative in the brain neurocytes. These inferences still need to be investigated in our further study.
In the skeletal system, the signals of UbcM2 are located in the proliferating chondrocytes, which may infer that UbcM2 is involved in skeletogenesis. Nedd4 is expressed abundantly in proliferating chondrocytes, but is undetectable in hypertrophic chondrocytes (25), which is in accord with our results. Moreover, knocking down an E2 enzyme Ubc9 in zebrafish will lead to defects in cranial cartilage, and it is required during vertebrate organogenesis (15), which makes our data evident as well.
In conclusion, the present research reveals that the UbcM2 is ubiquitously expressed during mouse embryonic stage and it is developmentally regulated in the CNS and skeletal system. Our data also indicate a spatial and temporal expression pattern of UbcM2 within various structures of the developing brain. This may be helpful for dissection of the regulation mechanism of the proteasome proteolytic system in mouse brain in the future.
ACKNOWLEDGMENTS
This work was supported by grants from the Fundamental Research Funds for the Central Universities (Grant No. HIT.NSRIF.2010027) and the National Natural Science Foundation of China (No. 30971645 and 31171383).
REFERENCES
- 1. Bull J. H.; Ellison G.; Patel A.; Muir G.; Walker M.; Underwood M.; Khan F.; Paskins L. Identification of potential diagnostic markers of prostate cancer and prostatic intraepithelial neoplasia using cDNA microarray. Br J. Cancer 84:1512–1519; 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Correia K. M.; Conlon R. A. Whole-mount in situ hybridization to mouse embryos. Methods 23:335–338; 2001. [DOI] [PubMed] [Google Scholar]
- 3. Debonneville C.; Staub O. Participation of the ubiquitin-conjugating enzyme UBE2E3 in Nedd4-2-dependent regulation of the epithelial Na+ channel. Mol. Cell Biol. 24:2397–2409; 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. de Oca Luna R. M.; Tabor A. D.; Eberspaecher H.; Hulboy D. L.; Worth L. L.; Colman M. S.; Finlay C. A.; Lozano G. The organization and expression of the mdm2 gene. Genomics 33:352–357; 1996. [DOI] [PubMed] [Google Scholar]
- 5. Fotia A. B.; Cook D. I.; Kumar S. The ubiquitin-protein ligases Nedd4 and Nedd4-2 show similar ubiquitin-conjugating enzyme specificities. Int. J. Biochem. Cell Biol. 38:472–479; 2006. [DOI] [PubMed] [Google Scholar]
- 6. Glickman M. H.; Ciechanover A. The ubiquitin-proteasom proteolytic pathway: Destruction for the sake of construction. Physiol. Rev. 82:373–428; 2002. [DOI] [PubMed] [Google Scholar]
- 7. Haupt Y.; Maya R.; Kazaz A.; Oren M. Mdm2 promotes the rapid degradation of p53. Nature 387:296–299; 1997. [DOI] [PubMed] [Google Scholar]
- 8. Hochstrasser M. Ubiquitin, proteasomes, and the regulation of intracellular protein degradation. Curr. Opin. Cell Biol. 7:215–223; 1995. [DOI] [PubMed] [Google Scholar]
- 9. Honda R.; Tanaka H.; Yasuda H. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 420:25–27; 1997. [DOI] [PubMed] [Google Scholar]
- 10. Jentsch S. The ubiquitin-conjugation system. Annu. Rev. Genet. 26:179–207; 1992. [DOI] [PubMed] [Google Scholar]
- 11. Kaiser P.; Flick K.; Wittenberg C.; Reed S. I. Regulation of transcription by ubiuitination without proteolysis: Cdc34/SCF(Met30)-mediated inactivation of the transcription factor Met4. Cell 102:303–314; 2000. [DOI] [PubMed] [Google Scholar]
- 12. Kawabe H.; Neeb A.; Dimova K.; Young S. M. Jr.; Takeda M.; Katsurabayashi S.; Mitkovski M.; Malakhova O. A.; Zhang D. E.; Umikawa M.; Regulation of Rap2A by the ubiquitin ligase Nedd4-1 controls neurite development. Neuron 65:358–372; 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Matuschewski K.; Hauser H. P.; Treier M.; Jentsch S. Identification of a novel family of ubiquitin-conjugating enzymes with distinct amino-terminal extensions. J. Biol. Chem. 271:2789–2794; 1996. [DOI] [PubMed] [Google Scholar]
- 14. Mirza S.; Plafker K. S.; Aston C.; Plafker S. M. Expression and distribution of the class III ubiquitin-conjugating enzymes in the retina. Mol. Vis. 16:2425–2437; 2010. [PMC free article] [PubMed] [Google Scholar]
- 15. Nowak M.; Hammerschmidt M. Ubc9 regulates mitosis and cell survival during zebrafish development. Mol. Biol. Cell. 17:5324–5336; 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pestov D. G.; Grzeszkiewicz T. M.; Lau L. F. Isolation of growth suppressors from a cDNA expression library. Oncogene. 17:3187–3197; 1998. [DOI] [PubMed] [Google Scholar]
- 17. Pick E.; Lau O. S.; Tsuge T.; Menon S.; Tong Y.; Dohmae N.; Plafker S. M.; Deng X. W.; Wei N. Mammalian DET1 regulates Cul4A activity and forms stable complexes with E2 ubiquitin-conjugating enzymes. Mol. Cell. Biol. 27:4708–4719; 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Plafker K. S.; Farjo K. M.; Wiechmann A. F.; Plafker S. M. The human ubiquitin conjugating enzyme, UBE2E3, is required for proliferation of retinal pigment epithelial cells. Invest. Ophthalmol. Vis. Sci. 49:5611–5618; 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Plafker K. S.; Nguyen L.; Barneche M.; Mirza S.; Crawford D.; Plafker S. M. The ubiquitin-conjugating enzyme UbcM2 can regulate the stability and activity of the antioxidant transcription factor Nrf2. J. Biol. Chem. 285:23064–23074; 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Plafker S. M.; Macara I. G. Importin-11, a nuclear import receptor for the ubiquitin-conjugating enzyme, UbcM2. EMBO J. 19:5502–5513; 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Plafker S. M.; Plafker K. S.; Weissman A. M.; Macara I. G. Ubiquitin charging of human class III ubiquitin-conjugating enzymes triggers their nuclear import. J. Cell Biol. 167:649–659; 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Salghetti S. E.; Caudy A. A.; Chenoweth J. G.; Tansey W. P. Regulation of transcriptional activation domain function by ubiquitin. Science. 293:1651–1653; 2000. [DOI] [PubMed] [Google Scholar]
- 23. VanDemark A. P.; Hill C. P. Structural basis of ubiquitylation. Curr. Opin. Struct. Biol. 12:822–830; 2002. [DOI] [PubMed] [Google Scholar]
- 24. Weissman A. M. Themes and variations on ubiquitylation. Nat. Rev. Mol. Cell Biol. 2:169–178; 2001. [DOI] [PubMed] [Google Scholar]
- 25. Weston A. D.; Underhill T. M. Analysis of Nedd4 expression during skeletal development in the mouse limb. Mech. Dev. 94:247–250; 2000. [DOI] [PubMed] [Google Scholar]
- 26. Wickner S.; Maurizi M. R.; Gottesman S. Posttranslational quality control: Folding, refolding, and degrading proteins. Science 286:1888; 1999. [DOI] [PubMed] [Google Scholar]
- 27. Wilkinson D. G. Whole mount in situ hybridization of vertebrate embryos. In situ hybridization: A practical approach. 109:75–83; 1992. [Google Scholar]
- 28. Xiong S.; Van Pelt C. S.; Elizondo-Fraire A. C.; Liu G.; Lozano G. Synergistic roles of Mdm2 and Mdm4 for p53 inhibition in central nervous system development. Proc. Natl. Acad. Sci. USA 103:3226–3231; 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]