Abstract
The objective of this study was to perform global gene expression profiling of patients affected by narcolepsy with cataplexy (NRLCP). This enabled identifying new potential biomarkers and relevant molecules possibly involved in the disease pathogenesis. In this study 10 NRLCP patients and 10 healthy controls were compared. Total RNA isolated from blood specimens was analyzed using microarray technology followed by statistical data analysis to detect genome-wide differential gene expression between patients and controls. Functional analysis of the gene list was performed in order to interpret the biological significance of the data. One hundred and seventy-three genes showed significant (p < 0.01) differential expression between the two tested conditions. The biological interpretation allowed categorizing differentially expressed genes involved in neurite outgrowth/extension and brain development, which could be possibly regarded as peripheral markers of the disease. Moreover, the NRLCP-related gene expression profiles indicated a dysregulation of metabolic and immune-related mechanisms. In conclusion, the gene expression profile associated to NRLCP suggested that molecular markers of neurological impairment, dysmetabolic and immune-related mechanisms, can be detected in blood of NRLCP patients.
Key words: Microarray, Narcolepsy, Cataplexy, Gene expression
INTRODUCTION
Narcolepsy with cataplexy (NRLCP) is a rare sleep disorder characterized by excessive daytime sleepiness; cataplexy (sudden loss of muscle tone triggered by emotions); other abnormal rapid eye movement (REM) sleep phenomena, such as sleep paralysis and hypnagogic hallucinations, and disturbed nocturnal sleep, along with additional clinical symptoms (2). Patients often share a dysmetabolic phenotype: many studies showed an increased body weight or body mass index (BMI). An increased prevalence of type 2 diabetes in this population have been also described in recent years (10,13,20,40,44). Human NRLCP is regarded as a multifactorial disorder, with multiple genetic and environmental factors involved in the etiopahtogenesis (22). A genetic susceptibility factor associated with the disorder has been found in the human leukocyte antigen (HLA) class II region, mapped on 6p21.3: the HLA-DRB1*1501-DQB1* 0602 haplotype [HLA-DRB1 (MIM 142857) and HLA-DQB1 (MIM 604305)] (14,30). Although NRLCP does not seem to be a classical autoimmune disorder, this evidence allowed to hypothesize that some immunological dysregulations may occur (31,47,48). These alterations are likely to be reflected in the gene expression changes occurring in white blood cells.
Additional human NRLCP candidate regions and genes have been investigated in association studies, indicating at least three susceptibility loci (NRCLP1, 17q21; NRCLP2, 4p13-q21; NRCLP3, 21q11.2), and some possible candidate genes, including the narcolepsy candidate region gene 1A (NLC1A), the myxovirus resistance 2 gene (MX2), the carnitine O-palmitoyltransferase 1 (CPT1B), and the cholin kinase beta (CHKB) (11,22,32). Moreover, growing evidences are supporting the role of the hypocretin receptor 2 (HCRTR2, orexin) as a NRLCP candidate gene. Mutation in HCRTR2 was found to be responsible for the disorder in an autosomal recessive canine model of NRLCP (25). In humans, hypocretin concentration in cerebrospinal fluid was reduced in sporadic forms and the number of hypothalamic hypocretin neurons was decreased in postmortem NRLCP brains, suggesting an imbalance in hypothalamic neurotransmission (35,39,50).
Despite this intense research effort, clear explanations of the disease mechanisms along with the availability of specific molecular biomarkers are still lacking.
Many essential genes and signaling cascades involved in the pathogenesis of neurological disorders are expressed in blood cells, even in the absence of a clinically relevant hematological involvement (42). Based on such evidences, we hypothesized that blood gene expression profiles could help to elucidate the molecular pathways underlying NRLCP etiopathogenesis and provide some useful hints towards the definition of new potential biomarkers. Therefore, we attempted to identify differentially expressed genes across the whole genome of the white blood cells of NRLCP using microarray technology.
MATERIALS AND METHODS
Patients
A case-control study was performed comparing 10 narcoleptic patients (both sexes, mean age 50, median age 50) with 10 age- and sex-matched healthy controls. The entire study protocol was approved by the Ethical Committee of the IRCCS–Oasi Institute for Research on Mental Retardation and Brain Aging (IRCCS), Troina, Italy. All patients were of Caucasian origin, in a postpubertal state, and drug naive. The NRLCP diagnosis was made according to the International Classification of Sleep Disorders, as previously described (40). All subjects were enrolled by the Sleep Disorders Center, Department of Neurological Sciences, University of Bologna (Italy), upon providing a written informed consent according to local ethics committee. Briefly, NRLCP patients had excessive daytime sleepiness for at least 3 months, as assessed with an Epworth Sleepiness Scale pathologic score (i.e., ≥ 11). Clear-cut cataplexy, defined as sudden bilateral loss of muscle tone triggered by sudden emotions, was clinically diagnosed by a neurologist with a structured interview focused on detailed descriptions of the attacks, their length, frequency, trigger factors, and parts of the body involved. The presence of possible hypnagogic hallucinations and sleep paralysis was ascertained by history taking. Confirmation of instrumental diagnosis included continuous 48-h polysomnography performed in a free-running condition, followed by a diagnostic multiple sleep latency test (MSLT) (i.e., at least two sleep-onset REM periods and a mean sleep latency within 8 min). All NC patients were diagnosed as cataleptic; based on the BMI four patients were obese, five were overweight, and one normal weighted. One NC patient was diabetic. HLA haplotyping was performed on all individuals enrolled in the study. All narcoleptic patients and one control subject were HLA-DQB1*0602 positive (Table 1). The healthy controls did not experience excessive daytime sleepiness or exhibit any signs of immunological abnormalities at the time of blood collection. Moreover blood count was performed on all subjects, revealing no significant differences in total leukocyte counts.
TABLE 1.
PATIENTS AND CONTROLS HAPLOTYPES
| Cod. | Age | Sex | Type | DRB1 | DQB1 | HR |
|---|---|---|---|---|---|---|
| n_1 | 40 | M | NRLCP | 11 15 | 03 06 | 0602 |
| n_2 | 38 | M | NRLCP | 11 15 | 03 06 | 0602 |
| n_3 | 73 | M | NRLCP | 11 15 | 06 | 0602/0604 |
| n_4 | 60 | M | NRLCP | 15 | 05 06 | 0602 |
| n_5 | 12 | F | NRLCP | n.d. | 02 06 | 0602 |
| n_6 | 53 | M | NRLCP | 11 15 | 03 06 | 0602 |
| n_7 | 60 | M | NRLCP | 07 15 | 03 06 | 0602 |
| n_8 | 30 | M | NRLCP | 04 15 | 03 06 | 0602 |
| n_9 | 31 | F | NRLCP | 15 16 | 05 06 | 0602 |
| n_10 | 39 | M | NRLCP | 04 15 | 03 06 | 0602 |
| c_1 | 50 | M | CTRL | 03 11 | 03 | n.d. |
| c_2 | 58 | F | CTRL | 01 07 | 02 05 | n.d. |
| c_3 | 42 | M | CTRL | 13 15 | 06 | 0609 |
| c_4 | 60 | M | CTRL | 11 | 03 | n.d. |
| c_5 | 40 | M | CTRL | 04 11 | 03 | n.d. |
| c_6 | 39 | F | CTRL | 07 11 | 02 03 | n.d. |
| c_7 | 54 | M | CTRL | 03 08 | 02 | n.d. |
| c_8 | 55 | M | CTRL | 01 15 | 06 | 0601/0603 |
| c_9 | 29 | M | CTRL | 01 16 | 05 | n.d. |
| c_10 | 52 | M | CTRL | 15 | 05 06 | 0602 |
NRLCP, narcoleptic; CTRL, healthy control; n.d.: not detected.
Gene Expression Profiling
A blood specimen of 2.5 ml was collected from each subject using PAXgene Blood RNA tubes (Becton Dickinson, Franklin Lakes, NJ, USA) and total RNA was isolated using PAXgene Blood RNA Kit (Qiagen, Valencia, CA, USA), according to the manufacturer’s protocols. Total RNA was quantified using spectrophotometry (Beckman Coulter, Brea, CA, USA). Quality of total RNA was checked using 2100 Bioanalyzer (Agilent, Palo Alto, CA, USA).
Gene expression profiling was then performed using the Genechip microarray technology (Affymetrix, Santa Clara, CA, USA), as described elsewhere (4,23). Briefly, the syntheses of cDNA and biotinylated cRNA was performed according to the protocols provided by the manufacturer. Biotinylated fragmented cRNA probes were hybridized to the Gene-Chip Human Genome U133A 2.0 Array (Affymetrix), allowing the simultaneous analysis of over 18,000 transcripts. Hybridization was performed at 45°C for 16 h in a hybridization oven (Affymetrix). The Genechips were then automatically washed and stained with streptavidin-phycoerythrin conjugate using the Genechip Fluidics Station (Affymetrix). Fluorescence intensities were scanned with the Gene-Chip Scanner 3000-7G (Affymetrix). Hybridizations was carried out independently for each condition, according to the “minimum information about a microarray experiment” (MIAME) guidelines (www.mged.org/Workgroups/MIAME/miame.html) (9).
Data Analysis
Preprocessing
Gene expression data were analyzed using Partek Genomics Suite software (version 6.4 Copyright ©2009 Partek Inc., St Louis, MO, USA). For this purpose CEL files were imported using the default Partek normalization parameters. Probe-level data were preprocessed, including background correction, normalization, and summarization (54), using robust multiarray average (RMA) analysis; subsequent data normalization was performed across all arrays using quantile normalization (7,21). The normalized probe intensity values were then compiled, or summarized, within each probe set, using the median polish technique, to generate a single measure of expression (21). These expression measures were then log transformed, base 2.
Differential Expression Analysis
Differential expression levels were computed using algorithms implemented in the Partek Genomic Suite software, which allowed to perform a one-way analysis of variance (ANOVA) with a significance level of p< 0.01. This enabled obtaining a list of the top significantly modulated genes between patients and controls. The resulting list was annotated according to functional roles or biological processes according to the Gene Ontology Consortium directions (3).
The gene list was further analyzed by Ingenuity Pathway Analysis (IPA; Ingenuity Systems, Inc., Redwood City, CA, USA), aimed at the biological interpretation of the microarray results. This generated functional networks and canonical pathways that connect the differentially expressed genes, using the IPA Knowledge base, in which the interactions are supported by peer-reviewed publications and which contains over 1.4 million interactions between genes, proteins, and drugs. Scores were assigned, allowing ranking of the networks, using a Fisher’s right-tailed exact test. The functional analysis of the gene list was accomplished using the Core Analysis workflow, enabling to associate biological functions and diseases to the experimental results by assigning a p-value, indicating the statistical significance of the association. Moreover, IPA network functions allowed to observe the networking of all genes included in the gene list resulting from the micro-array data analysis.
Gene Expression Validation
The gene expression results obtained by means of microarray analysis were validated using quantitative real-time PCR to amplify selected transcripts. For this purpose total RNA was isolated and reverse-transcribed from all patients and controls, as above mentioned. Real-time PCR was carried out using SybrGreen as already described previously (24).
The following genes were selected for validation, as either being indicative of the main functional groups of modulated transcripts or exerting the highest levels of modulation. The glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene was amplified by real-time PCR from the same RNA samples and used as a housekeeping control gene. The sequences of the oligonucleotide primers, designed using Primer 3 software (http://primer3.sourceforge.net/), were as follows: ISG15_F, 5′-gacaaatgcgacgaacctct-3′ and ISG15_R, 5′-gcccttgttattcctcacca-3′; TTC3_F, 5′-ccctgactgtgaaggtgtca-3′ and TTC3_R, 5′-caccaccactgct gaagatg-3; TNC_F, 5′-aatctcccagtgacaacatcg-3′ and TNC_R, 5′-tcccagagccacctaagaga-3′; PAFAH1B1_F, 5′-caaatcactattcgtttttggtg-3′ and PAFAH1B1_R, 5′-atgcacattgcacatcccta-3′; WNK1_F, 5′-cttgtgtttctgctgcttgg-3′ and WNK1_R, 5′-tgaaaatggactgcgtgaaa-3′; TNFAIP3_F, 5′-gggaaaatgccacagtgttc-3′ and TNFAIP3_R, 5′-agcctgactcaaagcaagga-3′; SMARCA2_F, 5′-gtcatcaatggggctgaact-3′ and SMARCA2_R, 5′-agcagctcaaacttccctga-3′; MED1_F, 5′-atgcatttgcatgaaggttg-3′ and MED1_R, 5′-ccatgactcaaacggacaact-3′; COX 6A1-F, 5′-gtgtacctgaagtcgcacca-3′ and COX6Al_R, 5′-atgaactcgggtctctcgtg-3′; GAPDH_F, 5′-tggaaggactcatgaccaca-3′ and GAPDH_R, 5′-gtcttctgggtggcagt gat-3.
The analysis was performed on a Step One realtime thermal cycler (Applied Biosystem, Foster City, CA). The PCR conditions were as follows: an initial incubation of 50°C for 2 min and 95°C for 10 min followed by 40 cycles of 94°C for 15 s and 60°C for 1 min. Standard curves were generated for all the assays to verify PCR efficiency. The threshold cycle (CT) which correlates inversely with the levels of target mRNA was measured as the cycle number at which the reporter fluorescence emission exceeded a preset threshold level. The amplified transcripts were quantified using the comparative CT method as previously described (26), with the formula for relative quantity (RQ) = 2−ΔΔCT, where ΔΔCT = [ΔCT gene of interest (treated sample) − ΔCT 5S rRNA (treated sample)]−]ΔCT gene of interest (control sample) − ΔCT 5S rRNA (control sample)]. ΔCT represents the mean CT value of each sample. An unpaired t-test among ΔCT values with a cut-off of p < 0.05 was used to assess the statistical significance of qPCR results.
RESULTS
Molecular Profiling and Functional Interpretation
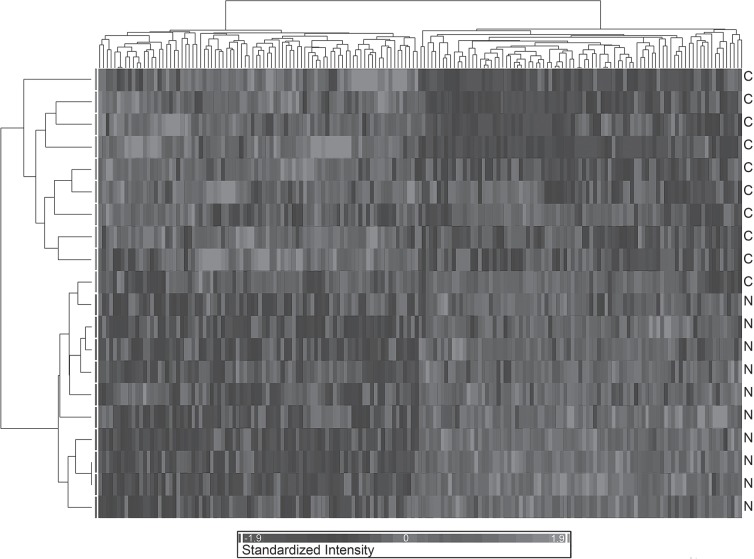
The comparative gene expression profiling of NRLCP versus controls was performed by means of microarray analysis using RNA isolated from blood lymphomonocites. Raw data (.cel files) have been submitted to the Gene Expression Omnibus (GEO) international web-based database (http://www.ncbi.nlm.nih.gov/geo; GEO accession number GSE21592). Data resulting from microarray analysis were normalized and summarized as described in Materials and Methods, then analyzed using ANOVA with a cutoff value of p < 0.01. This allowed to obtain a list of 173 genes being differentially expressed across the dataset (data not shown). These genes represented the molecular signature of the disease resulting from the analysis. Based on them, it was possible to efficiently segregate the two classes of individuals (NRCLP vs, CTRL) (Fig. 1). The top modulated genes, scored based on their expression value (i.e., fold change) are reported in Table 2.
Figure 1.
The hierarchical dendrogram represents the “condition tree” where the subjects are in row and the genes in column (gray scale of expression on the bottom side). The individual expression signal of the statistically significantly modulated transcripts across the categories (i.e., C: CTRL, N: NRLCP) was clustered: the transcripts with most similar expression blueprints across the different subjects were placed adjacent to each other. The clustering algorithm allowed the identification of two main trends in gene expression, discriminating the control from the narcoleptic individuals.
TABLE 2.
TOP MODULATED GENES
| Fold Change | Molecules |
|---|---|
| Upregulated | |
| TTC3 | 1.562 |
| PAFAH1B1 | 1.462 |
| APP | 1.462 |
| CD44 | 1.450 |
| WNK1 | 1.440 |
| PALM2-AKAP2 | 1.427 |
| ROD1 | 1.422 |
| BPTF | 1.416 |
| ZNF652 | 1.389 |
| LARP4B | 1.386 |
| Downregulated | |
| ISG15 | −1.783 |
| COX5B | −1.326 |
| UBL5 | −1.322 |
| COX6A1 | −1.310 |
| FAM128B | −1.268 |
| DDT | −1.240 |
| OASL | −1.230 |
| DRAP1 | −1.219 |
| GCHFR | −1.215 |
| GPX4 | −1.211 |
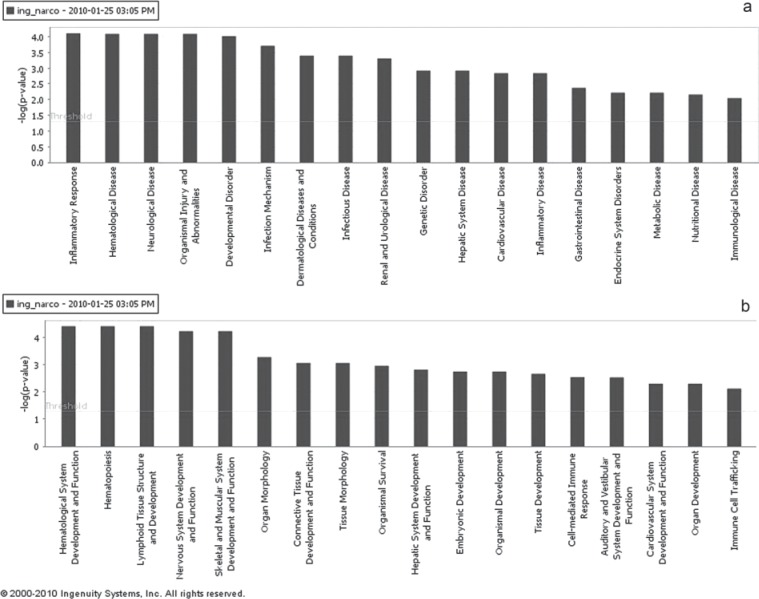
The biological interpretation of microarray results by means of IPA software, allowed to categorize significantly modulated genes into functional classes, based on their biological function. This provided an overview of the cellular and disease phenotypes most significantly associated to the resulting genes, enabling to hypothesize how they impact the phenotype. This allowed identifying the top biological functions (including Diseases and disorders and Physiological System Development and Function subcategories) that were most significant to the data set (p < 0.05), based on IPA Knowledge Base (www.ingenuity.com). In particular, among the Diseases and Disorders category genes mainly involved in inflammatory response, hematological diseases, neurological diseases, organism injury and abnormalities, and developmental disorders were annotated (Fig. 2a). Also, among the Physiological System Development and Function were categorized molecules mainly implicated in the development and function of the hematological system and hematopoiesis, lymphoid tissue, nervous system, and skeletal-muscular system (Fig. 2b). The overall number of genes involved in the above mentioned biological functions with corresponding statically significance are listed in Table 3.
Figure 2.
Top biological functions resulting from IPA analysis. (a) Diseases and Disorders categories that are involved in the gene list resulting from microrray analysis are displayed along the x-axis, the y-axis displays the significance (-log p-value). The threshold line is set as default at p = 0.05; (b) Physiological System Development and Function are displayed along the x-axis, the y-axis displays the significance (-log p-value).The threshold line is set as default at p = 0.05.
TABLE 3.
TOP BIOLOGICAL FUNCTIONS
| Name | p-Value | No. Genes |
|---|---|---|
| Diseases and disorders | ||
| Inflammatory response | 7.80E-05–1.13E-02 | 13 |
| Hematological disease | 8.13E-05–1.39E-02 | 6 |
| Neurological disease | 8.13E-05–1.13E-02 | 10 |
| Organismal injury and abnormalities | 8.13E-05–9.70E-03 | 14 |
| Developmental disorder | 9.65E-05–9.05E-03 | 17 |
| Physiological system development and function | ||
| Hematological system development and function | 3.93E-05–1.67E-02 | 25 |
| Hematopoiesis | 3.93E-05–1.67E-02 | 19 |
| Lymphoid tissue structure and development | 3.93E-05–1.26E-02 | 4 |
| Nervous system development and function | 5.92E-05–1.53E-02 | 21 |
| Skeletal and muscular system development and function | 5.92E-05–1.39E-02 | 17 |
On the whole, the biological interpretation allowed to categorize differentially expressed genes into three main functional groups: brain development, immunomodulation, and metabolism (Table 4). In particular, among neurological-related upregualted genes, the following genes are noteworthy. PAFAH1B1 (encoding the noncatalytic alpha subunit of the intracellular Ib isoform of platelet-activating factor acteylhydrolase) is involved in the abnormal neuronal migration phenotypes. Increased PAFAH1B1 dosage causes mild brain structural abnormalities, moderate to severe developmental delay, and failure to thrive. In fact, transgenic mice overexpressing PAFAH1B1 presented with smaller brains and neuronal migration abnormalities (6). TTC3 gene maps on the Down syndrome critical region and encodes a protein of unknown fucntion. TTC3 mRNA is progressively enriched in post-mitotic regions of the central nervous system (CNS) (27,41), thus suggesting its possible involvement in neuronal differentiation. Accordingly, it has been found that TTC3 overexpression in PC12 cells strongly inhibits the neurite extension induced by NGF, whereas TTC3 RNAi consistently increases neurite length (5). SPOCK2 modulates cell adhesion, neurite growth, and signal transduction during the development of the nervous system. An inhibition of neurite growth was observed when SPOCK2 was added to primary cerebellar neurons (43). WNK1 codes for a serine-threonine kinase expressed in brain; mutations in this gene were found to cause hereditary sensory and autonomic neuropathy type II (45). A recent study demonstrated that WNK-1 is indirectly involved in the inhibition of neurite extension (55). In addition, the amyloid precursor protein (APP) gene resulted upregulated in the dataset; APP is present in the amyloid plaques in the brain of Alzheimer disease and Down syndrome patients. Also, the downregulation of selected neurodevelopment-related genes was observed in the dataset: TNC, PLP1, OCL (Table 4). In particular, the tenascin-C (TNC) gene, coding an extracellular matrix (ECM) glycoprotein, is abundantly expressed in neural and nonneural tissues in remodeling processes like embryogenesis, wound healing, and tumorigenesis (15). In the CNS, TNC is produced and released by glial cells and is involved in both glial proliferation/differentiation and neuronal migration (36). The effects of reduced dosage of this gene on neurodevelopmental processes have been analyzed in Tnc-deficient mice, where structural and functional brain cortex aberrations, along with abnormal behavior and neurotransmission, were observed (16). The proteolipid protein 1 gene (PLP1) encodes a predominant myelin transmembrane protein present in the central nervous system, playing a role in the formation of myelin sheaths, as well as in oligoden-drocyte development and axonal survival (1). The tight junction protein occludin (OCL) is involved in the immunomodulation of CNS vascular permeability, as it decreases during CD8 T-cell-mediated BBB damage, occurring in several neurological disorders (46).
TABLE 4.
FUNCTIONAL GROUPS
| Gene | FC | p-Value |
|---|---|---|
| Brain development | ||
| TTC3 | 1.56 | 0.006 |
| PAFAH1B1 | 1.24 | 0.009 |
| SPOCK2 | 1.19 | 0.009 |
| SSTR2 | −1.15 | 0.007 |
| APP | 1.46 | 0.008 |
| TNC | −1.12 | 0.0001 |
| WNK1 | 1.44 | 0.002 |
| PLP1 | −1.15 | 0.007 |
| MARK4 | 1.22 | 0.001 |
| NENF | −1.11 | 0.003 |
| OCLN | −1.21 | 0.002 |
| Immunomodulation | ||
| HLADQB1* | 4.90 | 0.028 |
| IFNA1* | −1.06 | 0.016 |
| IFNA2* | −1.11 | 0.045 |
| IFNA5 | −1.08 | 0.006 |
| IFNA14 | −1.09 | 0.007 |
| ISG15 | −1.78 | 0.003 |
| UBE2D1 | −1.09 | 0.010 |
| UBL5 | −1.32 | 0.010 |
| Metabolism | ||
| CHKB | −1.13 | 0.004 |
| FABP1 | −1.16 | 0.008 |
| MNAT1 | −1.12 | 0.007 |
| MED1 | 1.37 | 0.008 |
| SMARCA2 | 1.26 | 0.006 |
| HMGCS1 | 1.11 | 0.005 |
| INSR | 1.18 | 0.005 |
| TWIST1 | 1.07 | 0.009 |
| TNFAIP3 | 1.23 | 0.010 |
p < 0.05.
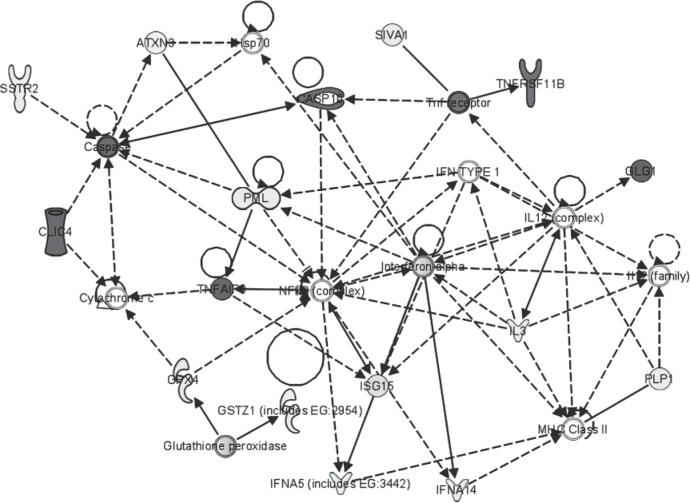
Furthermore, among the top-modulated molecules, two genes coding for cyclo-oxygenases (COX5B and COX6A1) were both downregulated (Table 2). In order to focus on functionally relevant genes and gene interactions within the gene list, data have been categorized using the networks function implemented in IPA Core Analysis, enabling the study of interactions and regulatory events. In particular, the key molecular events seemed to involve immunoregulatory molecules, including interferon alpha cluster genes (IFNA5 and IFNA14), IFN-inducible gene ISG15, and TNFAIP3 (Fig. 3). These molecules are involved in the immunoregulatory functions of T lymphocites (52). The actual role of all these genes in the disease mechanism needs to be further investigated by means of functional validation.
Figure 3.
Network of interacting genes resulting from IPA analysis. Genes significantly associated to the main biological functions resulted from the functional analysis were associated in the graphical representation of their reciprocal interactions; interactions are represented as nodes and the biological relationship between two nodes is represented as an edge (line). All edges are supported by at least one reference from the literature, from a textbook, or from canonical information stored in the Ingenuity Pathways Knowledge Base (www.ingenuity.com). The gray scale intensity of the node indicates the degree of upregulation or downregulation. Nodes are displayed using various shapes that represent the functional class of the gene product.
Interestingly, analyzing an extended gene list, which included less significant extent of differential expression (p < 0.05, data not shown), additional molecules could be included in the above-specified functional categories. In particular, other IFNA cluster genes (namely IFNA 1 and 2) were also downregulated, while the HLADQB1 gene, coding for the class II MHC molecule, appeared to be strongly upregulated (FC = 4.90).
Gene Expression Validation
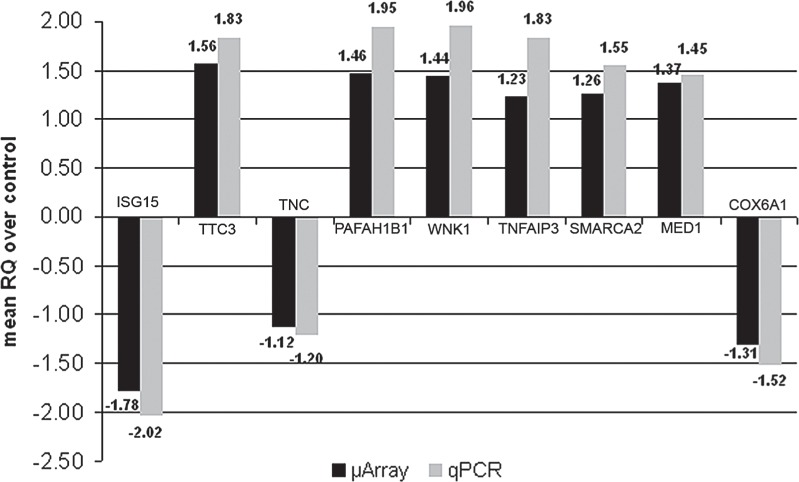
Gene expression results obtained by microarray analysis were also validated by real-time PCR performed on all patients and controls in triplicate experiments. The assay confirmed the modulated expression of the target genes tested, showing consistent alignment with the trends shown in the microarray analysis (Fig. 4).
Figure 4.
Real-time PCR validation. Results of real-time PCR on selected genes. Relative quantity values (RQ) obtained in qPCR are compared to fold changes obtained with microarray analysis. Results for all genes are statistically significant (p < 0.05).
DISCUSSION
This study attempted to identify a specific blood expression profile associated to narcolepsy, aimed at the identification of potential biomarkers and at delineating possible hints towards a better explanation of the disease pathogenesis. The biological interpretation of the gene list allowed focusing on selected functionally related genes, which were categorized into at least three distinct functional groups: brain development, immunomodulation, and metabolism (Table 4). Among these transcripts, the downregulation of TNC along with the upregulation of TTC3, SPOCK2, and WNK1 altogether pointed toward the inhibition of neurite outgrowth extension, essential for the correct brain development (5,6,36,43,55). Undoubtedly, the actual role of these genes in the disease mechanism needs further functional investigations.
The involvement of the PAFAH1B1 gene in the pathogenesis of neurological disorders has been already described. This gene maps within a region on chromosome 17p13.3 deleted in an isolated lissencephaly sequence, known as Miller-Dieker Syndrome (MDS; MIM#247200). Complete trisomy of 17p was associated to a MDS-like phenotype, suggesting that increased PAFAH1B1 gene dosage could be involved in the development of human brain diseases. In fact, individuals with gene duplication showed cognitive, neurobehavioral and subtle brain abnormalities, while triplication of PAFAH1B1 resulted in a more severe phenotype with mental retardation and muscle hypotonia. Furthermore, the cellular phenotype of trangenic mice overexpressing PAFAH1B1 indicated the impairment of cell polarity and neuronal migration (6).
It has been reported that OCLN levels decrease during blood–brain barrier damage through an immune-mediated mechanism (46); thus, the down-regulation of OCLN, could possibly suggest an impairment of CNS vascular permeability. It may be speculated that this could be a significant immune-related event that independently contributes to brain damage in narcolepsy. In fact, recent findings failed to demonstrate any evidence of hypothalamic-specific autoimmunity in narcoleptic patients (29).
The COX gene downregulation observed in the dataset could in part reflect possible metabolic alterations occurring also in the nervous system of affected individuals. In fact, it has been proposed that cellular COX activity changes in response to metabolic demands both in the intact brain and dissociated neurons in culture. Therefore, it seems that modulation of COX activity in the nervous system is directly related to energy demand through neural activity (37).
APP was among the top 10 upregulated genes detected in the molecular signature described in this study. A previous study documented the correlation between sleep alterations and APP overexpression in neurons; this could possibly affect networks involved directly in memory processing and cognition during the different sleep stages (12).
Recently, Honda and colleagues attempted to categorize the narcolepsy-associated gene expression signature, by means of microarray analysis of human postmortem posterior hypothalamic regions (19). Their findings were limited to the description of downregulated genes, with special regard to the role of the insulin-like growth factor binding protein 3 (IGFBP3). This molecule, produced by hepatocytes, was downregulated in both human and mouse NRLCP models and colocalizes with hypocretin in the mouse neurons (19). The same authors also found an increase in CSF levels of IGFBP3, suggesting the possible translocation of the blood–brain barrier. Nonetheless, our results demonstrated that IGFBP3 expression is not significantly dysregulated in the blood of affected individuals. Interestingly, IGFBP3 was found to inhibit the protective role of insulin growth factor 1 (IGF1) against APP in an Alzhemier disease model (33). Therefore, we could speculate that the upregulation of the APP gene in patients’ blood reflects the APP-mediated toxicity acting during the pathogenesis of selected damage of the posterior hypothalamus in NRLCP patients, where the IGFBP-3/IGF1 balance is altered.
Taken together, these data may suggest that the blood gene expression signature of NRLCP patients could, to some extent, reveal the alteration of brain homeostasis and function. Although the expression of genes involved in CNS function and development is not expected to be reflected in circulating cells, a proof-of-principle study of blood gene expression profiling in neurological disease was originally performed in a rat model (49). Additional studies showed further evidence that peripheral blood gene expression can be used as a fingerprint of selected neurological diseases, including Huntington’s disease, Alzheimer’s disease, and autism (8,28,34). More recently a large-scale blood gene expression study revealed ALS-specific profiles, providing new insights into the molecular mechanisms of the disease (42).
The functional analysis of the gene list also indicated that selected differentially expressed genes, including TWIST1, MED1, SMARCA2, and MNAT1 (Table 4), are involved in the maintenance of energy homeostasis, metabolic control, and adipogenesis (17,18,38,51). These data are consistent with previous studies reporting a definite pattern of dysmetabolic alterations associated to NRLCP (40). NRLCP patients, including those enrolled in this study, are typically obese. Moreover, the NRLCP candidate gene hypocretin 1/orexin-A, is implicated in the regulation of feeding behavior and energy balance (HCR) (53). Taken together, these data could imply that the altered expression of genes involved in distinct metabolic pathways could represent the blood signature of the dysmetabolic syndrome associated to NRLCP.
Additional functionally significant genes shared functions implicated in the modulation of the immune response (Fig. 3). In particular, the downregulation of interacting genes, such as the IFNA-1, -2, -5, and -14 and the top downregulated ISG15, could imply some kind of dysregulation of immunomodulatory networks. The biological significance and possible clinical consequences of this finding need further investigation.
Although NRLCP does not seem to be a classical autoimmune disorder, the involvement of some immunological dysregulations in NRLCP pathogenesis has been previously postulated (31,48).
On the whole these data seemed to indicate a neurological immune molecular profile in blood of NRLCP patients, which provides original insights toward the definition of a panel of potential diagnostic biomarkers. The correct functional interpretation of these molecular profiles in the light of understanding the disease pathogenesis or their prognostic significance deserves additional investigation.
ACKNOWLEDGMENTS
This study was supported by grants to R.F. by the National Center for Rare Diseases of the Italian Institute of Health (ISS).
Footnotes
Neither any conflict of interest nor off-label/investigational use of data applies to this work.
REFERENCES
- 1. Albertson D. N.; Pruetz B.; Schmidt C. J.; Kuhn D. M.; Kapatos G.; Bannon M. J. Gene expression profile of the nucleus accumbens of human cocaine abusers: Evidence for dysregulation of myelin. J. Neurochem. 88:1211–1219; 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. American Academy of Sleep Medicine. International classification of sleep disorders, 2nd ed: Diagnostic and coding manual. Westchester, IL: American Academy of Sleep Medicine; 2005. [Google Scholar]
- 3. Ashburner M.; Ball C. A.; Blake J. A.; et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25:25–29; 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bernardini C.; Lattanzi W.; Businaro R.; et al. Transcritpional effects of S100B on neuroblastoma cells: Perturbation of cholesterol homeostasis and interference on the cell cycle. Gene Expr. 14:345–359; 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Berto G.; Camera P.; Fusco C.; et al. The Down syndrome critical region protein TTC3 inhibits neuronal differentiation via RhoA and citron kinase. J. Cell Sci. 120:1859–1867; 2007. [DOI] [PubMed] [Google Scholar]
- 6. Bi W.; Sapir T.; Shchelochkov O. A.; et al. Increased LIS1 expression affects human and mouse brain development. Nat. Genet. 41:168–77; 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bolstad B. M.; Irizarry R. A.; Astrand M.; Speed T. P. A comparison of normalization methods for high density oligonucleotidearray data based on variance and bias. Bioinformatics 19:185–193; 2003. [DOI] [PubMed] [Google Scholar]
- 8. Borovecki F.; Lovrecic L.; Zhou J.; et al. Genome-wide expression profiling of human blood reveals biomarkers for Huntington’s disease. Proc. Natl. Acad. Sci. USA 102:11023–11028, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Brazma A.; Hingamp P.; Quackenbush J.; et al. Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat. Genet. 29:365–371; 2001. [DOI] [PubMed] [Google Scholar]
- 10. Cave H. Narcolepsy. Arch. Neurol. Psychiatr. 13:50–101; 1931. [Google Scholar]
- 11. Caylak E. The genetics of sleep disorders in humans: Narcolepsy, restless legs syndrome, and obstructive sleep apnea syndrome. Am. J. Med. Genet. A 149A:2612–2626; 2009. [DOI] [PubMed] [Google Scholar]
- 12. Colas D.; Valletta J. S.; Takimoto-Kimura R.; Nishino S.; Fujiki N.; Mobley W. C.; Mignot E. Sleep and EEG features in genetic models of Down syndrome. Neurobiol. Dis. 30:1–7; 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Daniels L. Narcolepsy. Medicine 13:1–122; 1934. [Google Scholar]
- 14. Dauvilliers Y.; Arnulf I.; Mignot E. Narcolepsy with cataplexy. Lancet 369:499–511; 2007. [DOI] [PubMed] [Google Scholar]
- 15. Faissner A.; Clement A.; Lochter A.; Streit A.; Mandl C.; Schachner M. Isolation of a neural chondroitin sulfate proteoglycan with neurite outgrowth promoting properties. J. Cell Biol. 126:783–799; 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fukamauchi F.; Aihara O.; Kusakabe M. Internalization and down-regulation of muscarinic acetylcholine receptors in cerebellar granule cells of tenascin-gene deficient mice. Neurochem. Int. 36:153–158; 2000. [DOI] [PubMed] [Google Scholar]
- 17. Ge K.; Guermah M.; Yuan C. X.; Ito M.; Wallberg A. E.; Spiegelman B. M.; Roeder R. G. Transcription coactivator TRAP220 is required for PPAR gamma 2-stimulated adipogenesis. Nature 417:563–567; 2002. [DOI] [PubMed] [Google Scholar]
- 18. Helenius K.; Yang Y.; Alasaari J.; Mäkelä T. P. Mat1 inhibits peroxisome proliferator-activated receptor gamma-mediated adipocyte differentiation. Mol. Cell. Biol. 29:315–323; 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Honda M.; Eriksson K. S.; Zhang S.; Tanaka S.; Lin L.; Salehi A.; Hesla P. E.; Maehlen J.; Gaus S. E.; Yanagisawa M.; Sakurai T.; Taheri S.; Tsuchiya K.; Honda Y.; Mignot E. IGFBP3 colocalizes with and regulates hypocretin (orexin). PLoS One 4(1):e4254; 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Honda Y.; Doi Y.; Ninomiya R.; Ninomiya C. Increased frequency of noninsulin-dependent diabetes mellitus among narcoleptic patients. Sleep. 9:254–259; 1986. [DOI] [PubMed] [Google Scholar]
- 21. Irizarry R. A.; Hobbs B.; Collin F.; Beazer-Barclay Y. D.; Antonellis J.; Scherf U.; Speed T. P. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4:249–264; 2003. [DOI] [PubMed] [Google Scholar]
- 22. Kawashima M.; Tamiya G.; Oka A.; et al. Genome-wide association analysis of human narcolepsy and a new resistance gene. Am. J. Hum. Genet. 79:252–263; 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lattanzi W.; Bernardini C.; Gangitano C.; Michetti F. Hypoxia-like transcriptional activation in TMT-induced degeneration: Microarray expression analysis on PC12 cells. J. Neurochem. 100:1688–1702; 2007. [DOI] [PubMed] [Google Scholar]
- 24. Lattanzi W.; Parrilla C.; Fetoni A.; et al. Ex vivo-transduced autologous skin fibroblasts expressing human Lim mineralization protein-3 efficiently form new bone in animal models. Gene Ther. 15:1330–1343; 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lin L.; Faraco J.; Li R.; Kadotani H.; Rogers W.; Lin X.; Qiu X.; de Jong P. J.; Nishino S.; Mignot E. The sleep disorder canine narcolepsy is caused by a mutation in the hypocretin (orexin) receptor 2 gene. Cell 98(3):365–376; 1999. [DOI] [PubMed] [Google Scholar]
- 26. Livak K. J.; Schmittgen T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25:402–408; 2001. [DOI] [PubMed] [Google Scholar]
- 27. Lopes C.; Rachidi M.; Gassanova S.; Sinet P. M.; Delabar J. M. Developmentally regulated expression of mtprd, the murine ortholog of tprd, a gene from the Down syndrome chromosomal region 1. Mech. Dev. 84:189–193; 1999. [DOI] [PubMed] [Google Scholar]
- 28. Maes O. C.; Xu S.; Yu B.; Chertkow H. M.; Wang E.; Schipper H. M. Transcriptional profiling of Alzheimer blood mononuclear cells by microarray. Neurobiol. Aging 28:1795–1809; 2007. [DOI] [PubMed] [Google Scholar]
- 29. Martínez-Rodríguez J. E.; Sabater L.; Graus F.; Iranzo A.; Santamaria J. Evaluation of hypothalamic-specific autoimmunity in patients with narcolepsy. Sleep. 30(1):27–28; 2007. [DOI] [PubMed] [Google Scholar]
- 30. Matsuki K.; Grumet F. C.; Lin X.; Guilleminault C.; Dement W. C.; Mignot E. HLADQB1-0602, rather than HLA DRw15(DR2), is the disease susceptibility gene in Black narcoleptics. Lancet 339:1052; 1992. [DOI] [PubMed] [Google Scholar]
- 31. Mignot E.; Tafti M.; Dement W. C.; Grumet F. C. Narcolepsy and immunity. Adv. Neuroimmunol. 5:23–37; 1995. [DOI] [PubMed] [Google Scholar]
- 32. Miyagawa T.; Kawashima M.; Nishida N.; et al. Variant between CPT1B and CHKB associated with susceptibility to narcolepsy. Nat. Genet. 40:1324–1328; 2008. [DOI] [PubMed] [Google Scholar]
- 33. Niikura T.; Hashimoto Y.; Okamoto T.; Abe Y.; Yasukawa T.; Kawasumi M.; Hiraki T.; Kita Y.; Terashita K.; Kouyama K.; Nishimoto I. Insulin-like growth factor I (IGF-I) protects cells from apoptosis by Alzheimer’s V642I mutant amyloid precursor protein through IGF-I receptor in an IGF-binding protein-sensitive manner. J. Neurosci. 21(6):1902–1910; 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nishimura Y.; Martin C. L.; Vazquez-Lopez A.; et al. Genome-wide expression profiling of lymphoblastoid cell lines distinguishes different forms of autism and reveals shared pathways. Hum. Mol. Genet. 16:1682–1698; 2007. [DOI] [PubMed] [Google Scholar]
- 35. Nishino S.; Ripley B.; Overeem S.; Lammers G. J.; Mignot E. Hypocretin (orexin) deficiency in human narcolepsy. Lancet. 355:39–40; 2000. [DOI] [PubMed] [Google Scholar]
- 36. Nishio T.; Kawaguchi S.; Iseda T.; Kawasaki T.; Hase T. Secretion of tenascin-C by cultured astrocytes: Regulation of cell proliferation and process elongation. Brain Res. 990:129–140; 2003. [DOI] [PubMed] [Google Scholar]
- 37. Ongwijitwat S.; Wong-Riley M. T. Functional analysis of the rat cytochrome c oxidase subunit 6A1 promoter in primary neurons. Gene. 337:163–71; 2004. [DOI] [PubMed] [Google Scholar]
- 38. Pan D.; Fujimoto M.; Lopes A.; Wang Y. X. Twist-1 is a PPARdelta-inducible, negative-feedback regulator of PGC-1alpha in brown fat metabolism. Cell 137:73–86; 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Peyron C.; Faraco J.; Rogers W.; et al. A mutation in a case of early onset narcolepsy and a generalized absence of hypocretin peptides in human narcoleptic brains. Nat. Med. 6:991–997; 2000. [DOI] [PubMed] [Google Scholar]
- 40. Poli F.; Plazzi G.; Di Dalmazi G.; et al. Body mass index-independent metabolic alterations in narcolepsy with cataplexy. Sleep 32:1491–1497; 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rachidi M.; Lopes C.; Gassanova S.; et al. Regional and cellular specificity of the expression of TPRD, the tetratricopeptide Down syndrome gene, during human embryonic development. Mech. Dev. 93:189–193; 2000. [DOI] [PubMed] [Google Scholar]
- 42. Saris C. G.; Horvath S.; van Vught P. W.; et al. Weighted gene co-expression network analysis of the peripheral blood from Amyotrophic Lateral Sclerosis patients. BMC Genomics 10:405; 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Schnepp A.; Komp Lindgren P.; Hülsmann H.; Kröger S.; Paulsson M.; Hartmann U. Mouse testican-2. Expression, glycosylation, and effects on neurite outgrowth. J. Biol. Chem. 280:11274–11280; 2005. [DOI] [PubMed] [Google Scholar]
- 44. Schuld A.; Hebebrand J.; Geller F.; Pollmächer T. Increased body-mass index in patients with narcolepsy. Lancet 355:1274–1275; 2000. [DOI] [PubMed] [Google Scholar]
- 45. Shekarabi M.; Girard N.; Rivière J. B.; et al. Mutations in the nervous system—specific HSN2 exon of WNK1 cause hereditary sensory neuropathy type II. J. Clin. Invest. 118(7):2496–2505; 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Suidan G. L.; Dickerson J. W.; Chen Y.; et al. CD8 T cell-initiated vascular endothelial growth factor expression promotes central nervous system vascular permeability under neuroinflammatory conditions. J. Immunol. 184:1031–1040; 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tafti M.; Nishino S.; Aldrich M. S.; Liao W.; Dement W. C.; Mignot E. Major histocompatibility class II molecules in the CNS: increased microglial expression at the onset of narcolepsy in canine model. J. Neurosci. 16:4588–4595; 1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tanaka S.; Honda Y.; Honda M. Identification of differentially expressed genes in blood cells of narcolepsy patients. Sleep 30:974–979; 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tang Y.; Lu A.; Aronow B. J.; Sharp F. R. Blood genomic responses differ after stroke, seizures, hypoglycemia, and hypoxia: Blood genomic fingerprints of disease. Ann. Neurol. 50:699–707; 2001. [DOI] [PubMed] [Google Scholar]
- 50. Thannickal T. C.; Moore R. Y.; Nienhuis R.; et al. Reduced number of hypocretin neurons in human narcolepsy. Neuron 27:469–474; 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. van den Berg S. W.; Dollé M. E.; Imholz S.; van der A D. L.; van’t Slot R.; Wijmenga C.; Verschuren W. M.; Strien C.; Siezen C. L.; Hoebee B.; Feskens E. J.; Boer J. M. Genetic variations in regulatory pathways of fatty acid and glucose metabolism are associated with obesity phenotypes: A population-based cohort study. Int. J. Obes. (Lond.) 33:1143–1152; 2009. [DOI] [PubMed] [Google Scholar]
- 52. Vereecke L.; Beyaert R.; van Loo G. The ubiquitin-editing enzyme A20 (TNFAIP3) is a central regulator of immunopathology. Trends Immunol. 30:383–391; 2009. [DOI] [PubMed] [Google Scholar]
- 53. Willie J. T.; Chemelli R. M.; Sinton C. M.; Yanagisawa M. To eat or to sleep?. Orexin in the regulation of feeding and wakefulness Annu. Rev. Neurosci. 24:429–458; 2001. [DOI] [PubMed] [Google Scholar]
- 54. Wu Z.; Irizarry R. A. A statistical framework for the analysis of microarray probelevel data. Johns Hopkins University, Department of Biostatistics Working Papers, Working Paper 73, 2005. Retrieved from http://www.bepress.com/jhubiostat/paper73
- 55. Zhang Z.; Xu X.; Zhang Y.; Zhou J.; Yu Z.; He C. LINGO-1 interacts with WNK1 to regulate nogo-induced inhibition of neurite extension. J. Biol. Chem. 284:15717–15728; 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]