Abstract
Background
Bovine tuberculosis (bTB) caused by Mycobacterium bovis is the most serious endemic disease affecting livestock in the UK. The European badger (Meles meles) is the most important wildlife reservoir of bTB transmission to cattle, making eradication particularly difficult. In this respect, oral vaccination with the attenuated M. bovis vaccine Bacillus Calmette-Guerin (BCG) has been suggested as a wide-scale intervention to reduce bTB infection in badgers. However, experimental studies show variable protection. Among the possibilities for this variation is that the resident gut bacteria may influence the success of oral vaccination in badgers; either through competitive exclusion and/or inhibition, or via effects on the host immune system. In order to explore this possibility, we have tested whether typical gut commensals such as Lactic Acid Bacteria (LAB) have the capacity to impact on the viability and survival rate of BCG and to modulate the immune response to BCG using an in vitro model.
Results
Twelve LAB isolated from badger faeces displayed inhibitory activity to BCG that was species-dependent. Weissella had a bacteriostatic effect, whereas isolates of enterococci, lactobacilli and pediococci had a more bactericidal activity. Furthermore, BCG-induced activation of the pro-inflammatory transcription factor NF-κB in human THP-1 macrophages was modulated by LAB in a strain-dependent manner. Most pediococci enhanced NF-κB activation but one strain had the opposite effect. Interestingly, isolates of enterococci, lactobacilli and weissella had different effects as immunomodulators of BCG-induced macrophage responses as some had no significant influence on NF-κB activation, but others increased it significantly.
Conclusions
Our in vitro results show that LAB isolated from badgers exhibit significant inhibitory activity against BCG and influence the immune activation mediated by BCG in a human macrophage assay. These findings suggest that gut commensal bacteria could play a role in influencing the outcome of oral BCG vaccination. Inactivated cells of LAB, or LAB that are bacteriostatic but have a synergistic immunostimulatory effect with BCG, could be potential adjuvants to be used for oral vaccination in badgers. Further work is needed to take into account the complex nature of the gut microbiome, specific immunity of the badger and the in vivo context.
Electronic supplementary material
The online version of this article (10.1186/s12866-018-1210-z) contains supplementary material, which is available to authorized users.
Keywords: Lactic acid bacteria, Bovine tuberculosis, BCG, Oral vaccination, Badgers
Background
Bovine tuberculosis (bTB) caused by Mycobacterium bovis is the most serious endemic disease affecting livestock in England and Wales [1]. Scientific evidence suggests that the European badger (Meles meles) is one of the most significant wildlife reservoirs of M. bovis in parts of the UK and Ireland [2]. The cases of bTB infection across England and Wales appear to occur more frequently in regions with higher densities of badgers [3], which have led both governments to implement comprehensive control strategies, including tighter cattle movement controls, biosecurity measures, culling and vaccination of badgers [4].
Biosecurity measures intended to keep badgers and cattle apart can be hard to implement at pasture [5]. In this respect, vaccination with the attenuated M. bovis vaccine Bacillus Calmette-Guerin (BCG) could be a viable alternative to control M. bovis infection in badgers [6]. However, the use of injectable BCG is limited and expensive due to the need to trap animals in order to vaccinate them [7, 8]. Oral BCG vaccination could bring the prospect of a wider-scale implementation and has been reported to reduce not only bTB infection in badgers but also their susceptibility to infection [9, 10]. However, protection with oral BCG is variable, at least in experimental models [11]. Although the reasons for this variability in protection have not been accounted for conclusively, one possibility could be the influence of gut resident bacteria on the efficacy of orally administered BCG due to competitive exclusion and/or inhibition in the gut; or after uptake by the oral mucosa; or through direct and indirect effects on immune cells [12] .
The composition of the mammalian intestinal microflora varies considerably over time and between individuals [13], and badgers are likely to be no different. Therefore, we sought to focus on Lactic Acid Bacteria (LAB) for the purposes of our initial investigations. LAB are gut commensal bacteria that provide a beneficial effect on intestinal epithelial cells in numerous ways: they metabolize indigestible compounds; provide the host with essential nutrients; contribute to the development of the intestinal architecture; and prevent against colonization of opportunistic pathogens [14]. It is very well documented that LAB produce antimicrobial compounds such as organic acids, hydrogen peroxide, ethanol and bacteriocins [15], which can be active against a broad range of bacteria including mycobacteria [16, 17]. Furthermore, LAB are key to maintaining the intestinal microbial balance in mammals, as well as to generate beneficial host immunomodulation [13]. The gut immune system recognizes LAB via the ligation of microbe-associated molecular patterns (MAMPs) to pattern recognition receptors (PRRs) such as Toll-like receptors (TLRs) that are expressed on immune cells and epithelial cells [18]. Upon MAMP recognition, PRRs activate an intracellular signalling cascade converging on the pro-inflammatory transcription factor NF-κB, which is crucial in maintaining immune homeostasis and regulating immunological and anti-microbial responses in barrier tissues such as the intestine [19]. Therefore, LAB play a role in innate immune responses and modulate responses to oral vaccination [20].
The aim of this study was to isolate LAB naturally excreted by badgers in order to identify those isolates that display inhibitory activity to BCG and assess their ability to modulate BCG-mediated pro-inflammatory responses in an established surrogate macrophage model. LAB isolated from badger faeces were initially screened for inhibitory activity against non-pathogenic Mycobacterium smegmatis, with a total of 40 LAB showing this activity. From these isolates, 12 were selected to be most representative and co-cultured with BCG to evaluate their antagonistic influence on the viability and survival rate of BCG. To study their potential as immunodulators, the human macrophage line THP-1 was exposed to the 12 LAB strains in isolation and in the presence of BCG, and the inflammatory responses recorded in the form of NF-κB activation and TNF-α production. Our results demonstrate that numerous LAB isolated from badgers have the capacity to reduce the viability and survival of BCG in vitro and to modulate the immune response of human THP-1 macrophages exposed to BCG. These fundamental findings suggest that gut commensal bacteria could play a role in influencing the outcome of oral BCG vaccination in badgers. In this respect, in vivo work will be needed to further explore this possibility.
Results
Isolation of LAB from badgers with antimycobacterial activity
A total of 40 LAB isolated from badger faeces showed antimicrobial activity against Mycobacterium smegmatis and were identified as enterococci, lactobacilli, pediococci and weissella (Figure 1). At this stage M. smegmatis was used as a fast-growing mycobacteria species to facilitate rapid antimycobacterial screening [21]. As indicated in Table 1, these 40 isolates with antimicrobial activity against M. smegmatis were randomly distributed between sexes and the weight of the badgers and/or the absolute or relative total counts observed on selective agar plates for LAB (De Man, Rogosa, Sharpe, MRS) and total viable counts (Plate Count Agar, PCA).
Fig. 1.

Agar-well Diffusion Test showing the antimicrobial activity of cell-free supernatants collected from cultures of 40 LAB isolated from badgers against M. smegmatis. These 40 isolates were identified by 16S rRNA sequencing and named as: (1) Pediococcus acidilactici A5; (2) P. lolii A6; (3) Enterococcus faecalis A7; (4) Weissella cibaria A23; (5) W. paramesenteroides A37; (6) P. pentosaceus B4; (7) P. acidilactici B41; (8) P. lolii B53; (9) E. faecalis C34; (10) Lactobacillus reuterii D4; (11) P. acidilactici E12; (12) P. lolii E23; (13) P. acidilactici E24; (14) P. acidilactici E48; (15) P. lolii F7; (16) P. acidilactici F20; (17) P. acidilactici F24; (18) P. acidilactici F44; (19) E. faecalis F48; (20) P. lolii G23; (21) P. pentosaceus G24; (22) P. acidilactici G41; (23) E. faecalis G42; (24) P. lolii G54; (25) P. acidilactici H33; (26) E. faecalis H34; (27) P. acidilactici I32; (28) E. faecium I47; (29) L. plantarum J2; (30) P. acidilactici J26; (31) P. acidilactici L4; (32) P. acidilactici M16; (33) P. acidilactici M17; (34) W. paramesenteroides N43; (35) E. faecium O44; (36) L. plantarum P5; (37) P. acidilactici Q16; (38) L. plantarum R20; (39) L. plantarum S48; (40) L. plantarum T17. All supernatants showed very acidic pH ranging between 3.6–-4.0 for lactobacilli and pediococci, and 4.2–-4.5 for weissella and enterococci. MRS broth (MRS) and supernatants from cultures of L. lactis NZ9700 (700) and L. lactis NZ9800 (800) were used as controls. Both L. lactis strains reduce pH in MRS but no lower than pH 4.5
Table 1.
Total counts of microaerophilic bacteria and bacterial isolates displaying antimycobacterial activity from faecal samples of the 26 badgers selected for this study
| Badgers: | Total Counts: | Isolates with antimycobacterial activityf | ||||
|---|---|---|---|---|---|---|
| ICa | Sexb | Weight (Kg) | MRSc | PCAd | MRS:PCAe | |
| A | f | 8.3 | 5.61 | 5.56 | 1.14 | 5, 6, 7, 8, 23, 37 |
| B | m | 10.1 | 5.98 | 5.93 | 1.12 | 4, 41, 53 |
| C | f | 8.2 | 6.36 | 6.38 | 0.96 | 34 |
| D | m | 10.2 | 7.54 | 7.61 | 0.85 | 4 |
| E | f | 7.5 | 5.72 | 5.71 | 1.02 | 12, 23, 24, 48 |
| F | m | 12.1 | 7.32 | 6.34 | 9.54 | 7, 20, 24, 44, 48 |
| G | f | 9.1 | 7.46 | 6.45 | 10.35 | 23, 24, 41, 42 |
| H | f | 8.8 | 6.70 | 6.40 | 2.00 | 33, 34 |
| I | f | 11.5 | 6.08 | 6.26 | 0.67 | 32, 47 |
| J | f | 10.7 | 5.38 | 5.40 | 0.96 | 2, 26 |
| K | f | 10.1 | 7.92 | 7.76 | 1.43 | – |
| L | f | 8.8 | 8.86 | 8.87 | 0.97 | 4 |
| M | m | 12.3 | 8.45 | 8.43 | 1.04 | 16, 17 |
| N | m | 8.4 | 8.95 | 8.08 | 0.75 | 43 |
| O | f | 8.0 | 8.76 | 8.87 | 0.78 | 44 |
| P | m | 10.8 | 8.38 | 8.53 | 0.71 | 5 |
| Q | f | 8.4 | 8.72 | 8.87 | 0.72 | 16 |
| R | f | 8.8 | 8.13 | 8.15 | 0.96 | 20 |
| S | m | 9.9 | 7.66 | 7.65 | 1.02 | 48 |
| T | f | 10 | 7.41 | 7.32 | 1.24 | 17 |
| U | m | 10.5 | 8.23 | 8.40 | 0.68 | – |
| V | m | 9 | 8.48 | 8.51 | 0.94 | – |
| W | f | 10 | 7.46 | 6.86 | 4.03 | – |
| X | m | 10 | 7.96 | 6.94 | 10.46 | – |
| Y | m | 12.4 | 7.08 | 6.56 | 3.33 | – |
| Z | m | 9.9 | 7.94 | 7.49 | 2.8 | – |
| AA | m | 9.1 | 7.93 | 6.92 | 10.24 | – |
| AB | f | 8.8 | 7.72 | 6.04 | 4.82 | – |
aIdentification code
bFemale (f) or Male (m)
cTotal counts on MRS agar plates expressed as log10
dTotal counts on PCA plates expressed as log10
eTotal Counts ratio between MRS and PCA plates
fIsolates from MRS that display antimicrobial activity against M. smegmatis on a stab-on-agar test. These isolates were named with the numbers indicated in the column and preceded with a letter from their corresponding animal IC
Viability of BCG in combination with LAB
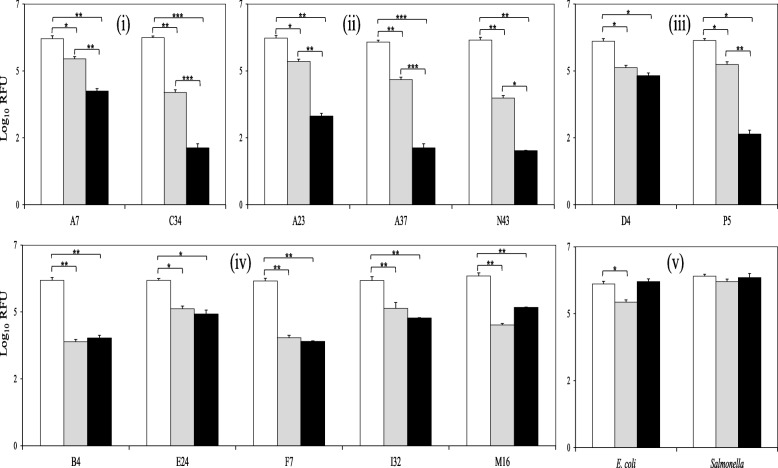
Twelve out the 40 LAB isolates with antimycobacterial activity were selected to test their influence on the fitness of BCG (referred as BCG viability in this study). These twelve LAB isolates were: 1, Enterococcus faecalis A7; 2, Weissella cibaria A23; 3, W. paramesenteroides A37; 4, Pediococcus pentosaceus B4; 5, E. faecalis C34; 6, Lactobacillus reuterii D4; 7, P. acidilactici E24; 8, P. lolii F7; 9, P. acidilactici I32; 10, P. acidilactici M17; 11, W. paramesenteroides N43; and 12, L. plantarum P5; which represented four different genera – Enterococcus, Weissella, Pediococcus and Lactobacillus - with two strains of the same genus and/or species from at least two different animals, as well as two or more different strains within the most predominant genus and/or species. The 12 selected isolates were then cultured with a BCG strain carrying a Green Fluorescence Protein (GFP) expression vector in order to monitor the fluorescence emission for 48 h (h), as a marker of cell viability. E. coli and Salmonella were also co-cultured with BCG to provide results with other typical fast-growing enteric bacteria. As illustrated in Figure 2., all LAB isolates reduced BCG viability significantly after 24 and 48 h, especially isolates of enterococci and weissella. No significant differences were observed in the E. coli co-cultures after 48 h, nor with Salmonella at either time point (24 or 48 h), demonstrating that the activity was specific to LAB and not due to trivial factors, such as competition for nutrients with other faster-growing bacteria.
Fig. 2.
BCG viability when combined with badger isolates of enterococci (i), weissella (ii), lactobacilli (iii) and pediococci (iv) after 0 (white bars), 24 (grey bars) and 48 (black bars) h of incubation. Co-cultures of BCG with gut-associated, Gram-negative bacteria (v) were also monitored for comparative purposes. BCG viability is expressed as log10 Relative Fluorescence Units (RFU). Data are mean ± SD with statistical analysis (Student’s t-test, *p < 0.05, **p < 0.01, ***p < 0.005) and are representative of 3 biological replicates each
Survival of BCG in combination with LAB
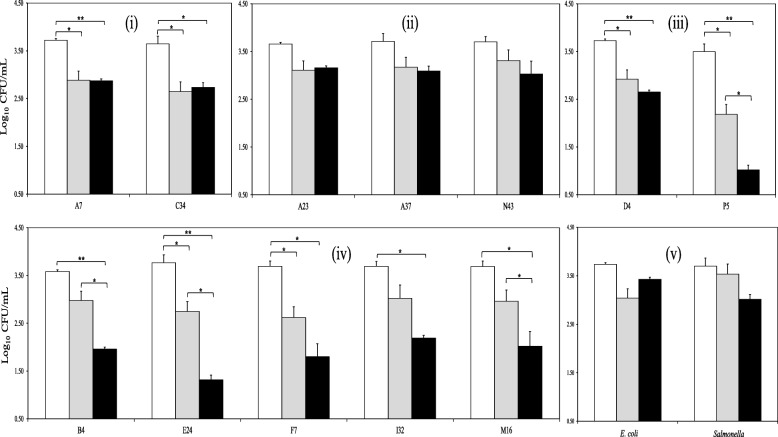
The survival of BCG when co-cultured with LAB, E. coli or Salmonella, was determined by total bacterial counts. With the exception of the weissella isolates, all the LAB combinations resulted in a significant reduction in BCG survival (Figure 3); being particularly evident in the case of the lactobacilli isolates and most of the pediococci. Neither the weissella, E. coli nor Salmonella had a significant effect on the BCG counts. Taken together with the results for GFP expression, we have shown that isolates of enterococci, lactobacilli and pediococci reduce significantly the viability and survival of BCG, whereas co-cultures with weissella or enterobacteria do not. We next sought potential explanations for these results; whether altered pH or metabolites from LAB could account for the results we observed.
Fig. 3.
BCG survival rate when combined with badger isolates of enterococci (i), weissella (ii), lactobacilli (iii) and pediococci (iv) after 0 (white bars), 24 (grey bars) and 48 (black bars) h of incubation. Co-cultures of BCG with gut-associated, Gram-negative bacteria (v) were also monitored for comparative purposes. BCG survival is expressed as log10 CFU/mL. Data are mean ± SD with statistical analysis (Student’s t-test, *p < 0.05, **p < 0.01, ***p < 0.005) and are representative of three biological replicates each
Effect of pH on BCG with or without LAB
The pH of the co-cultures described above was monitored over 48 h. A significant decrease in pH was recorded in all co-cultures after 24 and 48 h but no significant differences were observed between cultures, including LAB, E. coli and Salmonella (Additional file 1: Figure S1). The only exception was isolate P5 after 24 h for reasons that remain unknown. Furthermore, both the viability (GFP expression) and survival rate (bacterial counts) of BCG as a monoculture showed no significant changes at pH 7 or pH 5 after 24 and 48 h (Additional file 1: Figure S2). With the exception of isolate P5, all these results confirm that low pH may be insufficient on its own to cause a detrimental effect on BCG.
Effect of metabolites from LAB on the viability and survival rate of BCG
In order to test whether the antimicrobial effects observed against BCG in the presence of LAB was due to an accumulation of toxic metabolites from LAB, we monitored the viability and survival rate of BCG in cell-free supernatants obtained from mono-cultures of LAB after 48 h of incubation. Supernatants taken from 48 h cultures of E. coli and Salmonella were also used as controls. No significant reductions were observed with any of the culture supernatants, either in terms of viability or as survival rate (Additional file 1: Figure S3 and S4, respectively). This demonstrated that the negative effect of LAB on BCG was only observed when the bacteria were present together, rather than through the indirect accumulation of antimicrobial metabolites derived from the LAB, such as organic acids (e.g. lactic acid).
NF-κB response in macrophages exposed to LAB
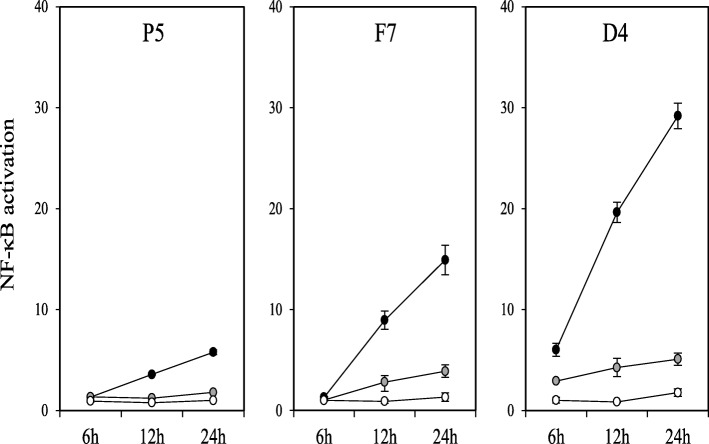
To study the effect of LAB on NF-κB activation we employed THP-1 macrophages carrying a NF-κB luciferase reporter as a model system. Cells were differentiated with phorbol 12-myristate 13-acetate (PMA) for 48 h and subsequently exposed to the 12 LAB isolates. We assessed NF-κB responses at 100 bacteria per macrophage, a dose that had been previously observed to trigger activation by LAB (data not shown). We found that exposure to inactivated weissella and enterococci isolates did not trigger NF-κB luciferase activation. However, a significant increase in the reporter activity was recorded upon exposure to pediococci and lactobacilli isolates (see white bars in Figure 4). Interestingly, activation was strain-specific. Isolates P. pentosaceus B4, P. acidilactici I32 and L. plantarum P5 showed a low NF-κB activation that is directly comparable to that observed with the human probiotic L. plantarum WCFS-1, with no significant differences between them. By contrast, isolates P. acidilactici E24, E. faecalis F7 and P. acidilactici M16 displayed a significantly higher pro-inflammatory effect by comparison with WCFS-1 (p < 0.05), with a moderate NF-κB luciferase activation as observed for LPS at 0.2 μg/mL. Interestingly, isolate L. reuterii D4 induced a NF-κB response that was as high as that observed with other bacteria associated with a high pro-inflammatory profile such as E. coli and a BCG Pasteur strain, with no significant differences between them. We then assessed activation of the NF-κB-driven luciferase at different time points (6, 12 and 24 h post-exposure) and doses (100, 10 and 1 bacteria per macrophage) with three representative LAB (low, medium, and high NF-κB induction) (Figure 5). In all cases, LAB activated NF-κB in a dose-dependent manner.
Fig. 4.
NF-κB activation (white bars) and TNF-α production (black bars) in PMA-differentiated THP-1 macrophages exposed to inactivated cells from LAB isolates of weissella, pediococci, enterococci and lactobacilli at a ratio of 1 macrophage per 100 bacteria for 24 h. NF-κB activation is presented as a fold increase over a non-stimulated condition on the primary Y axis (left hand), while TNF-α production is presented in ng/mL on the secondary Y axis (right hand). Lactobacillus plantarum WCFS-1, E. coli K12, BCG and LPS at 0.2 μg/mL were included as controls
Fig. 5.
NF-κB activation in PMA-differentiated THP-1 macrophages exposed to inactivated cells of Lactobacillus plantarum P5, Pediococcus lolii F7 and Lactobacillus reuterii D4 at a bacteria:macrophage ratio of 100:1 (black dots); 10:1 (grey dots); or 1:1 (white dots) for 24 h. NF-κB activation is presented as a fold increase over a non-stimulated condition and the selected LAB strains represent a low (P5), moderate (F7), and high (B4) NF-κB response in THP-1 macrophages. Data represent at least two biological replicates
Production of TNF-α in macrophages exposed to LAB
To confirm that the NF-κB activation observed by the luciferase reporter led to a production of an inflammatory cytokine, the levels of tumour necrosis factor alpha (TNF-α) were measured in the medium of macrophages exposed to LAB. TNF-α is a critical NF-κB-dependent cytokine commonly found as a host inflammatory marker in response to bacteria [22]. PMA-differentiated THP-1 cells were exposed to 100 bacteria per macrophage for 24 h and supernatants were tested in an anti-TNF-α ELISA. TNF-α production correlated positively with NF-κB activation (Additional file 1: Figure S5). Enterococci and weissella were unable to induce any TNF-α production, whereas pediococci and lactobacilli mediated variable amounts of TNF-α production (black bars in Figure 4). These results confirmed not only that the pro-inflammatory role of our LAB isolates is dependent on the species but also that isolates of pediococci and lactobacilli induce variable pro-inflammatory levels in a strain-specific manner.
Influence of LAB on BCG-induced NF-κB activation
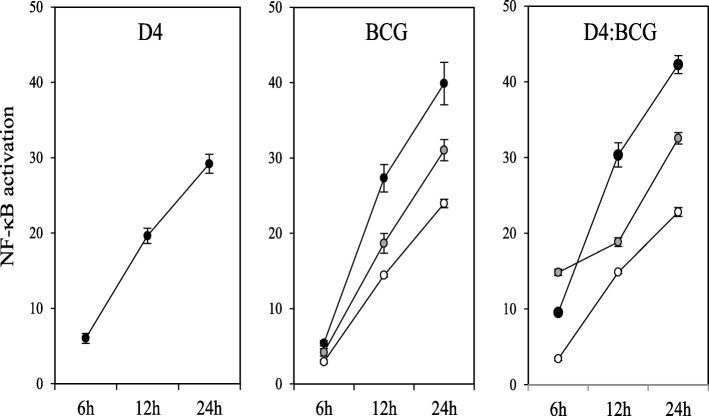
In order to test whether LAB isolates had a significant effect on the response of macrophages to BCG, we monitored NF-κB changes in macrophages exposed to a BCG Pasteur strain in combination with each of our LAB isolates. BCG is a potent activator of NF-κB and exposure to PMA-differentiated THP-1 cells for as little as 6 h is sufficient to detect significant changes in NF-κB reporter activity, either alone or in combination with LAB, especially at a ratio of 10 LAB per 1 BCG (Figure 6). Thus, we studied the effect of LAB on BCG-mediated NF-κB activation at a ratio of 100:10:1 (LAB:BCG:macrophage) after 6 h of co-incubation. In order to facilitate data interpretation, the results obtained from the macrophages exposed to the LAB-BCG combinations were divided into two groups based on the intensity of the pro-inflammatory profile of the LAB isolates on their own (Figure 7). Most LAB isolates with no, or very little, effect on NF-κB activation had a negligible impact on BCG-induced reporter activity (Figure 7a). However, isolates E. faecalis C34 and W. paramesenteroides N43, which had no effect on the NF-κB response in isolation for even 24 h (Figure 4), significantly increased NF-κB activation when combined with BCG. On the other hand, nearly all the isolates that clearly activated the NF-κB pathway on their own significantly enhanced the pro-inflammatory response of macrophages when combined with BCG (Fig. 7b). The exceptions were isolates P. acidilactici E24 and L. plantarum P5. Both P5 and E24 increased NF-κB activation when incubated in isolation with macrophages for 24 h (Fig. 4), but this was not observed after 6 h of co-incubation with BCG. The BCG-induced response remained unaffected by P5, but was surprisingly reduced by E24 and this was statistically significant.
Fig. 6.
NF-κB activation in PMA-differentiated THP-1 macrophages exposed to inactivated cells of Lactobacillus reuterii D4, BCG and combinations of D4 with BCG (D4:BCG) for 24 h. When the bacteria were tested on their own, macrophages were exposed to bacteria at a bacteria:macrophage ratio of 100:1 (black dots); 10:1 (grey dots); and 1:1 (white dots); for the D4:BCG combinations, the macrophages were challenged with bacteria at a LAB:BCG:macrophage ratio of 100:100:1 (back dots); 100:10:1 (grey dots); and 100:1:1 (white dots). NF-κB activation is presented as a fold increase over a non-stimulated condition and data represent at least two biological replicates
Fig. 7.
NF-κB activation in PMA-differentiated THP-1 macrophages exposed to inactivated cells from badger LAB isolates with no (or very low) pro-inflammatory (a) or pro-inflammatory (b) profile, in the absence (white bars) or presence (black bars) of BCG at a ratio of 100:10:1 (LAB:BCG:macrophage) for 6 h. A medium-only condition (RPMI) was included as a control. Comparative statistical analysis was carried out between BCG and all the LAB-BCG combinations using the Student t-test (*p < 0.05, **p < 0.01, ***p < 0.005). Data are mean ± SD representing three biological replicates each
Discussion
This is the first publication reporting the isolation and identification of LAB from faeces of badgers. Previous works have reported only a single LAB isolate from the intestine of a badger [23] or a LAB population detected in faeces of other mustelids [24]. In our study we show that different LAB species isolated from different badgers can exert in vitro inhibitory activity against M. smegmatis and M. bovis BCG. The effect of the LAB isolates seems to be species-dependent. The weissella isolates only reduced BCG viability as measured by GFP expression, but had no significant effect on the survival rate of BCG as measured by bacterial counts, suggesting a more bacteriostatic, rather than bacteriocidal, effect. In contrast, isolates of enterococci, lactobacilli and pediococci displayed a bactericidal effect since they were capable of reducing significantly both the viability and survival rate of BCG.
We suspected that the activity that our LAB isolates displayed against M. smegmatis were due to the combination of acidic pH and metabolites such as lactic acid [25]. However, low pH and accumulation of organic acids, either alone or in combination, were insufficient to induce a decrease in the viability and survival of M. bovis BCG when grown as a monoculture or in acidic cell-free supernatants from LAB. Our finding is in agreement with a previous study that reported that M. bovis tolerates low pH and organic acids; being able to survive and remain virulent in very acidic environments such as cheese [26]. Interestingly, the 12 LAB isolates selected in our study caused a significant reduction in the viability and the survival of BCG when cultured together in vitro. These results are consistent with two previous studies that reported a reduction in M. bovis counts in co-cultures with LAB in milk [27, 28]. We also observed that this significant BCG reduction only took place when BCG and LAB were present in cultures together, suggesting the direct inhibition of BCG by LAB, rather than an indirect effect due to acidic conditions or the accumulation of toxic metabolites derived from LAB. Whether this inhibition is due to antimicrobial metabolites that are only produced in the presence of BCG is a question that remains to be elucidated. However, other studies have reported that the efficacy of LAB metabolites against bacterial pathogens is more evident when competing simultaneously in the same environment [29]. The relevance of our findings depends on the extent to which opportunities exist in vivo for resident LAB and orally administered BCG vaccine to come into intimate contact with one another. This is currently unknown and would need to be tested using more complex model systems, such as in vitro organ culture models. Nonetheless, LAB may still contribute to a gastric environment that is not conducive to BCG survival.
The reasons why our LAB isolates differ in their activity against BCG may lie in the differing metabolic profiles of the isolates. Weissella are obligate heterofermenters, able to produce not only lactic acid as the major end product from energy metabolism but also other metabolites such as acetic acid and ethanol [30]. The remainder of the isolates produce lactic acid as the main end product since they are obligate or facultative heterofermenters [31]. The amount of lactic acid produced by the isolates in vitro may change depending on the oxygen availability, but this organic acid seems to have a key role as the main antimicrobial metabolite. Besides, strains of pediococci, enterococci and lactobacilli have been reported to produce antimicrobial peptides such as pediocins [32], enterocins [33] and plantaricins [34], as well as reuterin [35], which could act synergistically with lactic acid against BCG.
Oral administration of probiotics has been suggested as a convenient and safe way to improve the efficacy of conventional vaccines within communities [36]. Surface and/or secreted proteins and metabolites of LAB act as inflammatory mediators, interacting with cells of the mammalian immune system to stimulate and modify immunity [37–39], typified by activation of the NF-κB pathway in macrophages. Whilst studies are relatively limited in the context of oral vaccination, it would appear that efficient activation of the NF-κB pathway following oral immunisation results in strong systemic immunity [40]. Much of the research on the role of probiotics in the immune response has been conducted with lactobacilli, but other studies using other LAB that are in close contact with the mucus layer and epithelial crypts of the small intestine such as enterococci and pediococci, have also demonstrated an immune modulation that enhanced vaccine efficacy [41–43]. Therefore, we wanted to determine the impact that different badger LAB isolates had on the inflammatory response, on their own, and in the context of BCG vaccination.
In this study we have observed that inactivated cells of pediococci and lactobacilli induce NF-κB activation and TNF-α production in the human THP-1 macrophage cell line. On the contrary, inactivated cells of weissella and enterococci had no effect either on the NF-κB pathway or on TNF-α production. As previously reported by Jensen et al. [38], our Lactobacillus reuterin isolate induced the highest pro-inflammatory response, with TNF-α production, whereas L. plantarum was a much lower inducer, as observed with the human probiotic L. plantarum WCFS-1. When compared to E. coli and BCG, the pediococci isolates induced a moderate pro-inflammatory response in THP-1 macrophages. This response was observed to be strain-dependent as some of the isolates were much higher inducers than others. More detailed studies are needed to fully understand the pro-inflammatory effect of pediococci but a recent study has reported that exopolysaccharides from a Pediococcus pentosaceus strain induce the production of pro-inflammatory cytokines via NF-κB [44]. Furthermore, our experiments with inactivated cells of enterococci did not show any NF-κB activation although heat-killed enterococci can activate the NF-κB pathway in murine macrophages [39]. This difference could be explained by the fact that our bacteria and macrophages derive from strains and species that are different to the previous work.
Combinations of BCG with inactivated cells from almost all our lactobacilli and pediococci resulted in enhanced NF-κB activation, or at least, in a non-detrimental effect on this pro-inflammatory pathway. On the basis of other work [45], it is possible this is effect is mediated through TLR-2. The only exception was P. acidilactici E24, which induced NF-κB activation in isolation, but reduced the extent of activation triggered by BCG. Takata et al. reported that P. acidilactici inoculation in mice suppressed the production of inflammatory cytokines, whilst increasing the number of interleukin (IL)-10-producing T cells [46]. It is well-known that IL-10 and other anti-inflammatory cytokines have the capacity to modulate NF-κB activation [47]. Further investigation is required to determine whether the effects observed for P. acidilactici in mice also occur in badgers.
Although inactivated cells of weissella and enterococci were unable to cause any pro-inflammatory activation in THP-1 macrophages, co-cultures of certain strains with BCG resulted in a synergistic enhancement of NF-κB responses. Previous studies have reported that heat-killed weissella and E. faecalis proteins block the secretion of pro-inflammatory cytokines and attenuate NF-κB activation [48, 49], but also that lipoteichoic acids of weissella and enterococci significantly increase the secretion of pro-inflammatory cytokines and contribute to host inflammatory responses [48, 50]. Therefore, there could be a significant variability between weissella and enterococci strains, with some of them able to modulate the outcome of BCG-induced inflammatory responses; possibly via TLR2, as both LAB and BCG activate macrophages in a TLR-2-dependent manner [37, 45].
Despite the fact that we did not use monocytes from badgers, our model has proved to be informative; generating preliminary data that show the potential for badger-derived LAB to cause immunomodulation, either alone or in combination with BCG. This now needs to be tested using macrophages isolated from badgers. In addition, although the mechanisms that our isolates utilize as immunomodulators remain to be elucidated, our data also raise the interesting possibility of their potential exploitation as adjuvants to BCG if used as inactivated cells.
Conclusions
We started from the premise that the variation and activity of commensal gut bacteria could play an important, and hitherto underappreciated role in the outcome of oral BCG vaccination in badgers. We found that LAB isolated from badger faeces could influence the viability of BCG vaccine in vitro and modify the pro-inflammatory response using an established macrophage-like model. These preliminary results, although generated from reductionist in vitro assays, suggest our premise is worthy of further investigation. More extensive evaluation of the badger gut microbiome and its impact on the efficacy of oral vaccination in vivo appears warranted.
Methods
Badgers and sampling
Faecal samples were collected from a total of 26 badgers kept in captivity at the Animal Plant and Health Agency for other scientific projects authorised under a UK Home Office Project Licence and Ethical approval by the Agency. These badgers were initially taken from a bTB-free wild population and they all tested negative to M. bovis infection following a quarantine procedure that consisted of bTB testing on three sequential occasions. The TB testing was carried out at approximately one month intervals using the badger interferon (IFN)-γ ELISPOT, the badger specific IFN-γ release assay (IGRA) and a standard M. bovis culture with clinical samples. Three negative tests were required before an animal was considered free of bTB and admissible to the study. All the badgers were on a diet that mainly consisted of commercial dog food with no probiotics, a very high source of carbohydrates from wholegrain maize and wheat (58%), a high source of protein from fish (20%), a low level of fat (7%) -most of it as polyunsaturated fatty acids; and enriched with fibre (4%) and essential vitamins and minerals. The badgers were also provided with commercial dry dog food (no LAB in its composition), peanuts, apples and sweet chestnuts, when available, as well as any food from foraging activities such as insects, worms and mice. Fresh water was available ad libitum. Faecal samples were taken from badgers using rectal swabs following intramuscular administration of ketamine hydrochloride (10 mg/kg, Vetalar, Boehringer Ingelheim), medetomidine hydrochloride (0.1 mg/kg, Domitor, Pfizer), and butorphanol (0.1 mg/kg, Torbugesic R, Zoetis UK Ltd) as described previously [11]. Badgers were then weighed, examined for any signs of disease or injury and their corresponding sex noted to ascertain whether these factors could affect the different levels of LAB found in the collected faecal samples.
Isolation of LAB from faecal samples
The rectal swabs were immersed in maximum recovery diluent (Oxoid) to prepare serial dilutions that were spread onto MRS agar plates (Oxoid) for the isolation of LAB. The serial dilutions were also spread onto PCA plates (Oxoid) to estimate the percentage of LAB when compared to total microaerophilic viable bacteria. Plates were incubated at 37 °C for 48–60 h under microaerophilic conditions. A total of 50 different colonies were then selected from MRS-agar plates containing approximately 20–100 colonies from each of the 26 collected faecal samples (a total of 1300 colonies), and cultivated in Thermo Scientific Nunc MicroWell™ 96-well plates with MRS broth at 37 °C for 48–60 h under microaerophilic conditions.
Antimycobacterial screening of LAB
The supernatants of the MRS cultures present in the 96-well plates were tested against Mycobacterium smegmatis mc2 155 [51] using the spot-on-agar test. Briefly, the 96-well plates were centrifuged for 10 min at 5000 rpm to deposit 5 μL of the culture supernatants on TSA plates which had been swabbed with M. smegmatis. The MRS cultures whose supernatants showed antimicrobial activity against M. smegmatis were selected to confirm their antimycobacterial activity using the Agar-well Diffusion Test (ADT) [52]. The nisin-producer Lactococcus lactis NZ9700 and its corresponding knock-out strain NZ9800 were used as positive and negative controls, respectively [53].
Bacterial strains and growth conditions
All LAB, including the badger isolates and the human probiotic L. plantarum WCFS-1 [54], were grown in MRS broth/agar (Oxoid) at 37 °C without any aeration for 24 h. The L. lactis strains NZ9700 and NZ9800 [55] were also grown static in MRS for 24 h but at 30 °C. The strains E. coli ATCC25922, E. coli K12 [56] and S. enterica ATCC14028 were propagated in Trypticase Soy Broth/Agar (TSB/TSA, Oxoid) at 37 °C for 24 h. M. smegmatis mc2 155 was also cultured in TSB at 37 °C but supplemented with 0.05% Tween 80 (Sigma Aldrich) and for 48 h. The two M. bovis strains BCG ΔleuD pASOriMXF [57] and BCG Pasteur [58] were cultured in Middlebrook 7H9 (Difco) supplemented with 10% Oleic acid albumin dextrose-catalase (OADC) enrichment (BD BBL™), 0.1% Tween 80 (Sigma Aldrich) and 0.2% glycerol (Sigma Aldrich) or Middlebrook 7H11/7H10 (Difco) medium supplemented with 5% OADC and glycerol (2 mL/L) at 37 °C. BCG ΔleuD pASOriMXF was previously generated by transforming BCG Pasteur ΔleuD with the mycobacterial episomal vector pASOriMXF that complements the leucine mutation to correct auxotrophy and enable stable expression of GFP under the control of the constitutively expressed promoter pL5X [59]. All enterobacteria and mycobacteria were grown with aeration in an orbital shaker at 225 rpm and all bacterial strains were maintained as − 80 °C frozen stocks in their appropriate media with the addition of 15% glycerol.
Identification and selection of LAB with antimycobacterial activity
Colonies from MRS cultures whose anti-mycobacterial activity was confirmed by ADT were identified based on their morphology (small, circular, white/creamy colonies), gram staining (positive) and oxidase/catalase tests (negative). Further identification was carried out by 16S rRNA gene sequencing (LGC Genomics) before a final selection of the 12 most representative LAB species comprising different bacterial species isolated from different animals but also different strains representing the most predominant bacterial species found.
Co-cultures of BCG and LAB
Cell pellets of the selected LAB and the leucine auxotrophic strain BCG ΔleuD pASOriMXF were obtained from cultures at their exponential phase of growth. The auxotrophy correction allowed us to perform co-cultures of LAB and BCG without using antibiotics while maintaining the GFP plasmid to monitor BCG viability over time. In order to prepare the co-cultures, 12 BCG pellets were resuspended with each LAB pellet using Mueller-Hinton (MH) broth supplemented with 10% Oleic acid-Albumin-Dextrose-Catalase (OADC) enrichment, 0.1% Tween 80 and 0.2% glycerol, to generate 12 co-cultures at an initial concentration of 5 × 103 CFU/mL for BCG and 5 × 105 CFU/mL for the LAB. Our own preliminary studies showed that MH broth supplemented with OADC, tween and glycerol is the optimal to support the growth of LAB without altering the viability and survival rate of BCG for 48 h when grown as mono-cultures. The co-cultures were then grown in an orbital shaker at 37 °C for 48 h and samples were collected at 0, 24 and 48 h post-incubation to determine BCG viability and survival rate. Co-cultures of BCG with the enterobacteria E. coli (ATCC 25922) and S. enterica serovar Typhimurium (ATCC 14028) at an initial concentration of 5 × 105 CFU/mL were also included in the study for comparative purposes.
Viability and survival rate experiments
In order to monitor BCG viability in the co-cultures, aliquots of 100 μl were transferred into Thermo Scientific Nunc MicroWell™ 96-well plates to measure fluorescence emission at 485/535 nm in a DTX 880 Multimode Detector microplate reader (Beckman Coulter). Simultaneously, BCG survival rate was determined by total bacterial counts on BCG-selective agar plates (Middlebrook 7H10 supplemented with 5% OADC and 0.2% glycerol). Bacterial counts were also carried out for LAB and the enterobacteria on their corresponding selective agar plates: MRS and Violet Red Bile Glucose (Oxoid), respectively. BCG viability was expressed as log10 RFU (Relative Fluorescence Units), where measured fluorescence units (FU) were multiplied by the log10 increase in the CFU/mL of the competitor and normalized with the highest increase in CFU/mL observed for any of the competitors. These relative units let us compare GFP expression between the co-cultures at each time point over 48 h since the bacterial counts varied amongst the LAB and enterobacteria (Additional file 1: Figure S6). The BCG survival rate was presented as CFU/mL.
pH in co-cultures
The pH of all co-cultures was measured using a Hanna pH meter at time points 0, 24 and 48 h, starting from an initial pH of 7 for the media, to determine the effect of pH on the viability and survival of BCG while co-cultured with LAB. The viability and survival rate of BCG was also monitored at pH 7 and pH 5 as explained above but as a monoculture to evaluate the influence of pH on the growth of BCG.
Metabolite accumulation experiments
In order to determine the effect of LAB metabolites on BCG growth, cell pellets of BCG ΔleuD pASOriMXF were resuspended in cell-free supernatants collected from 48 h cultures of LAB, E. coli ATCC25922 and S. enterica ATCC14028 to monitor viability, and survival rate as explained above. The BCG survival rate was indicated as CFU/mL and the viability as log10 FU (Fluorescence Units).
Culture and differentiation of THP-1 cells
The THP-1 Lucia™ NF-κB monocyte cell line (Invivogen) was used to monitor the NF-κB signal transduction pathway in macrophages when exposed to inactivated cells obtained from the badger LAB isolates. These macrophages secrete Lucia luciferase under the control of the NF-κB promoter. The cells were grown in Roswell Park Memorial Institute (RPMI) 1640 (Life Technologies) supplemented with 15% foetal bovine serum (FCS, Seralab) and 1% Penicillin/Streptomycin (Pen/Strep, Life Technologies) at 37 °C in an atmosphere of 5% CO2. PMA (Santa Cruz Biotechnology) was dissolved in DMSO at 10 mg/mL. 5 × 104 cells per well were seeded in with RPMI supplemented with 20 ng/mL PMA in Thermo Scientific Nunc MicroWell™ 96-well plates. After 48 h, medium was replaced with RPMI containing 2% FCS and 1% Pen/Strep supplemented with inactivated cells of LAB.
Luciferase measurements from THP-1 cells exposed to LAB
PMA-differentiated THP-1 cells were exposed to heat-inactivated (70 °C, 2 h) LAB pellets that were resuspended in RPMI containing 2% FCS and 1% Pen/Strep. At the indicated times, supernatants were transferred to white-bottom 96-well plates and luciferase activity was measured in the presence of 2 μg/mL of coelenterazine (NanoLigh Technology) using a Clariostar plate reader (BMG Biotech). The BCG Pasteur strain, E. coli K12 and L. plantarum WCFS1, all as inactivated cells, were used as controls, as well as LPS at 0.2 μg/μl (Sigma Aldrich). The BCG Pasteur strain was also used to test whether LAB isolates had a significant effect on the response of macrophages to BCG. NF-κB activation was calculated as a fold increase over the measurements recorded for unchallenged macrophages.
ELISA for TNF-α detection
TNF-α production was detected and quantified in the supernatants of PMA-differentiated THP-1 cells using the eBioscience Human TNF-α ELISA Ready-SET-Go kit as indicated by the manufacturer’s instructions.
Statistical analysis
Statistical analysis was performed using GraphPad Prism version 7.00 for Windows (GraphPad Software, La Jolla California USA, www.graphpad.com). Data are mean ± SD, representing three biological replicates, unless indicated. Differences between time-points for the same samples were analysed by the Student t-test, while differences between samples at different time points were analyzed by one way ANOVA followed by Fisher’s Least Significant Difference (LSD) Test.
Additional file
Figure S1. pH of BCG co-cultures with LAB isolates or E. coli and Salmonella after 0 (white bars), 24 (grey bars) and 48 (black bars) h of incubation. Different letters signify statistical differences between mean values using ANOVA followed by LSD (p < 0.05).
Figure S2. Viability (A) and survival rate (B) of BCG when grown as a monoculture at pH 5 or pH 7 after 0 (white bars), 24 (grey bars) and 48 (black bars) h of incubation. Viability and survival rate are expressed as log10 Fluorescence Units (FU) and log10 CFU/mL, respectively.
Figure S3. BCG viability when grown in cell-free supernatants from 48 h-cultures of enterococci (i), weissella (ii), lactobacilli (iii), pediococci (iv) and gut-associated Gram-negative bacteria (v) after 0 (white bars), 24 (grey bars) and 48 (black bars) h of incubation. BCG viability is expressed as log10 Fluorescence Units (FU).
Figure S4. Survival rate of BCG when grown in cell-free supernatants from 48 h-cultures of enterococci (i), weissella (ii), lactobacilli (iii), pediococci (iv) and gut-associated Gram-negative bacteria (v) after 0 (white bars), 24 (grey bars) and 48 (black bars) h of incubation. BCG survival rate is expressed as log10 CFU/mL.Figure S5. Significant positive correlation between mean values of NF-κB activation and TNFα production in PMA-differentiated THP-1 macrophages exposed to inactivated bacterial strains and LPS, at a ratio of 1:100 (macrophage::bacteria) and 0.2 μg/μl, respectively (p < 0.0001).
Figure S6. Growth rate of enterococci (i), weissella (ii), lactobacilli (iii), pediococci (iv) and gut-associated Gram-negative bacteria (v) after 0 (white bars), 24 (grey bars) and 48 (black bars) h of incubation when co-cultured with BCG. Growth rate is expressed as log10 CFU/mL. (DOCX 217 kb)
Acknowledgments
We would like to express our gratitude to Prof Kleerebezem (Wageningen University, the Netherlands) and Prof Kuipers (Groningen University, the Netherlands) for providing us with the strains L. plantarum WCFS-1, L. lactis NZ9700 and L. lactis NZ9800. We would like to also thank Pablo Zubiaur-Precioso for his help with THP-1 experiments.
Funding
This project was funded by the Morris Animal Foundation (D15ZO-020), with no role in the design of the study and collection, analysis, interpretation of data or in writing the manuscript.
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- BCG
Bacillus Calmette-Guerin
- bTB
Bovine tuberculosis
- LAB
Lactic acid bacteria
Authors’ contributions
AS planned, performed experiments and edited the manuscript. CMM designed experiments, aided in data analysis, and aided in preparing and editing the manuscript. SL and DD conducted the sampling and edited the manuscript. MC designed experiments and aided in preparing and editing the manuscript. JGM designed and performed experiments, aided in data analysis, and prepared and edited the manuscript. All authors read and approved the final manuscript.
Ethics approval
All animal procedures were covered by a licence issued by the UK Home Office under the Animal [Scientific Procedures] Act1986, and approved by the Animal Welfare and Ethical Review Board at the Animal and Plant Health Agency (APHA).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s12866-018-1210-z) contains supplementary material, which is available to authorized users.
Contributor Information
Anna Stedman, Email: anna.stedman@pirbright.ac.uk.
Carlos Maluquer de Motes, Email: c.maluquerdemotes@surrey.ac.uk.
Sandrine Lesellier, Email: Sandrine.Lesellier@apha.gsi.gov.uk.
Deanna Dalley, Email: Deanna.Dalley@apha.gsi.gov.uk.
Mark Chambers, Email: m.chambers@surrey.ac.uk.
Jorge Gutierrez-Merino, Email: j.gutierrez@surrey.ac.uk.
References
- 1.Harris KA, Brunton L, Brouwer A, Garcia MP, Gibbens JC, Smith NH, Upton PA. Bovine TB infection status in cattle in great Britain in 2015. Vet Rec. 2017;180(7):170–175. doi: 10.1136/vr.j759. [DOI] [PubMed] [Google Scholar]
- 2.Hardstaff JL, Marion G, Hutchings MR, White PC. Evaluating the tuberculosis hazard posed to cattle from wildlife across Europe. Res Vet Sci. 2014;97(Suppl):S86–S93. doi: 10.1016/j.rvsc.2013.12.002. [DOI] [PubMed] [Google Scholar]
- 3.Wilson GJ, Carter SP, Delahay RJ. Advances and prospects for management of TB transmission between badgers and cattle. Vet Microbiol. 2011;151(1–2):43–50. doi: 10.1016/j.vetmic.2011.02.024. [DOI] [PubMed] [Google Scholar]
- 4.Chambers MA, Carter SP, Wilson GJ, Jones G, Brown E, Hewinson RG, Vordermeier M. Vaccination against tuberculosis in badgers and cattle: an overview of the challenges, developments and current research priorities in great Britain. Vet Rec. 2014;175(4):90–96. doi: 10.1136/vr.102581. [DOI] [PubMed] [Google Scholar]
- 5.Ward AI, Judge J, Delahay RJ. Farm husbandry and badger behaviour: opportunities to manage badger to cattle transmission of Mycobacterium bovis? Prev Vet Med. 2010;93(1):2–10. doi: 10.1016/j.prevetmed.2009.09.014. [DOI] [PubMed] [Google Scholar]
- 6.O'Connor CM, Haydon DT, Kao RR. An ecological and comparative perspective on the control of bovine tuberculosis in great Britain and the Republic of Ireland. Prev Vet Med. 2012;104(3–4):185–197. doi: 10.1016/j.prevetmed.2011.11.010. [DOI] [PubMed] [Google Scholar]
- 7.Robinson PA, Corner LA, Courcier EA, McNair J, Artois M, Menzies FD, Abernethy DA. BCG vaccination against tuberculosis in European badgers (Meles meles): a review. Comp Immunol Microbiol Infect Dis. 2012;35(4):277–287. doi: 10.1016/j.cimid.2012.01.009. [DOI] [PubMed] [Google Scholar]
- 8.Chambers MA, Rogers F, Delahay RJ, Lesellier S, Ashford R, Dalley D, Gowtage S, Dave D, Palmer S, Brewer J, et al. Bacillus Calmette-Guerin vaccination reduces the severity and progression of tuberculosis in badgers. Proc Biol Sci. 2011;278(1713):1913–1920. doi: 10.1098/rspb.2010.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Corner LA, Costello E, O'Meara D, Lesellier S, Aldwell FE, Singh M, Hewinson RG, Chambers MA, Gormley E. Oral vaccination of badgers (Meles meles) with BCG and protective immunity against endobronchial challenge with Mycobacterium bovis. Vaccine. 2010;28(38):6265–6272. doi: 10.1016/j.vaccine.2010.06.120. [DOI] [PubMed] [Google Scholar]
- 10.Aznar I, Frankena K, Morea SJ, O’Keeffe J, McGratha G, de Jong MCM. Quantification of Mycobacterium bovis transmission in a badger vaccine field trial. Prev Vet Med. 2018;149:29–37. doi: 10.1016/j.prevetmed.2017.10.010. [DOI] [PubMed] [Google Scholar]
- 11.Chambers MA, Aldwell F, Williams GA, Palmer S, Gowtage S, Ashford R, Dalley DJ, Dave D, Weyer U, Salguero FJ, et al. The effect of oral vaccination with Mycobacterium bovis BCG on the development of tuberculosis in captive European badgers (Meles meles) Front Cell Infect Microbiol. 2017;7:6. doi: 10.3389/fcimb.2017.00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sindhu KN, Cunliffe N, Peak M, Turner M, Darby A, Grassly N, Gordon M, Dube Q, Babji S, Praharaj I, et al. Impact of maternal antibodies and infant gut microbiota on the immunogenicity of rotavirus vaccines in African, Indian and European infants: protocol for a prospective cohort study. BMJ Open. 2017;7(3):e016577. doi: 10.1136/bmjopen-2017-016577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chin J, Turner B, Barchia I, Mullbacher A. Immune response to orally consumed antigens and probiotic bacteria. Immunol Cell Biol. 2000;78(1):55–66. doi: 10.1046/j.1440-1711.2000.00883.x. [DOI] [PubMed] [Google Scholar]
- 14.Martin R, Miquel S, Ulmer J, Kechaou N, Langella P, Bermudez-Humaran LG. Role of commensal and probiotic bacteria in human health: a focus on inflammatory bowel disease. Microb Cell Factories. 2013;12:71. doi: 10.1186/1475-2859-12-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pessione E. Lactic acid bacteria contribution to gut microbiota complexity: lights and shadows. Front Cell Infect Microbiol. 2012;2:86. doi: 10.3389/fcimb.2012.00086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carroll J, Draper LA, O'Connor PM, Coffey A, Hill C, Ross RP, Cotter PD, O’Mahony J. Comparison of the activities of the lantibiotics nisin and lacticin 3147 against clinically significant mycobacteria. Int J Antimicrob Agents. 2010;36(2):132–136. doi: 10.1016/j.ijantimicag.2010.03.029. [DOI] [PubMed] [Google Scholar]
- 17.Donaghy J. Lantibiotics as prospective antimycobacterial agents. Bioeng Bugs. 2010;1(6):437–439. doi: 10.4161/bbug.1.6.13855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ren C, Zhang Q, de Haan BJ, Zhang H, Faas MM, de Vos P. Identification of TLR2/TLR6 signalling lactic acid bacteria for supporting immune regulation. Sci Rep. 2016;6:34561. doi: 10.1038/srep34561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pasparakis M. Role of NF-kappaB in epithelial biology. Immunol Rev. 2012;246(1):346–358. doi: 10.1111/j.1600-065X.2012.01109.x. [DOI] [PubMed] [Google Scholar]
- 20.Yan F, Polk DB. Probiotics and immune health. Curr Opin Gastroenterol. 2011;27(6):496–501. doi: 10.1097/MOG.0b013e32834baa4d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sharma S, Gelman E, Narayan C, Bhattacharjee D, Achar V, Humnabadkar V, Balasubramanian V, Ramachandran V, Dhar N, Dinesh N. Simple and rapid method to determine antimycobacterial potency of compounds by using autoluminescent Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2014;58(10):5801–5808. doi: 10.1128/AAC.03205-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Newton K, Dixit VM. Signaling in innate immunity and inflammation. Cold Spring Harb Perspect Biol. 2012;4(3):a006049. [DOI] [PMC free article] [PubMed]
- 23.Collins MD, Hutson RA, Foster G, Falsen E, Weiss N. Isobaculum melis gen. Nov., sp. nov., a Carnobacterium-like organism isolated from the intestine of a badger. Int J Syst Evol Microbiol. 2002;52(Pt 1):207–210. doi: 10.1099/00207713-52-1-207. [DOI] [PubMed] [Google Scholar]
- 24.Vulfson L, Pedersen K, Chriel M, Holmen Andersen T, Dietz HH. Assessment of the aerobic faecal microflora in mink (Mustela vison Schreiber) with emphasis on Escherichia coli and Staphylococcus intermedius. Vet Microbiol. 2003;93(3):235–245. doi: 10.1016/S0378-1135(03)00037-3. [DOI] [PubMed] [Google Scholar]
- 25.Ross RP, Morgan S, Hill C. Preservation and fermentation: past, present and future. Int J Food Microbiol. 2002;79(1–2):3–16. doi: 10.1016/S0168-1605(02)00174-5. [DOI] [PubMed] [Google Scholar]
- 26.de la Rua-Domenech R. Human Mycobacterium bovis infection in the United Kingdom: incidence, risks, control measures and review of the zoonotic aspects of bovine tuberculosis. Tuberculosis (Edinb) 2006;86(2):77–109. doi: 10.1016/j.tube.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 27.Mariam SH. Interaction between lactic acid bacteria and Mycobacterium bovis in Ethiopian fermented milk: insight into the fate of M. bovis. Appl Environ Microbiol. 2009;75(6):1790–1792. doi: 10.1128/AEM.01943-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Macuamule CL, Wiid IJ, van Helden PD, Tanner M, Witthuhn RC. Effect of milk fermentation by kefir grains and selected single strains of lactic acid bacteria on the survival of Mycobacterium bovis BCG. Int J Food Microbiol. 2016;217:170–176. doi: 10.1016/j.ijfoodmicro.2015.10.024. [DOI] [PubMed] [Google Scholar]
- 29.Arques JL, Rodriguez E, Langa S, Landete JM, Medina M. Antimicrobial activity of lactic acid bacteria in dairy products and gut: effect on pathogens. Biomed Res Int. 2015;2015:584183. doi: 10.1155/2015/584183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fusco V, Quero GM, Cho GS, Kabisch J, Meske D, Neve H, Bockelmann W, Franz CM. The genus Weissella: taxonomy, ecology and biotechnological potential. Front Microbiol. 2015;6:155. doi: 10.3389/fmicb.2015.00155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carr FJ, Chill D, Maida N. The lactic acid bacteria: a literature survey. Crit Rev Microbiol. 2002;28(4):281–370. doi: 10.1080/1040-840291046759. [DOI] [PubMed] [Google Scholar]
- 32.Porto MC, Kuniyoshi TM, Azevedo PO, Vitolo M, Oliveira RP. Pediococcus spp.: an important genus of lactic acid bacteria and pediocin producers. Biotechnol Adv. 2017;35(3):361–374. doi: 10.1016/j.biotechadv.2017.03.004. [DOI] [PubMed] [Google Scholar]
- 33.Khan H, Flint S, Yu PL. Enterocins in food preservation. Int J Food Microbiol. 2010;141(1–2):1–10. doi: 10.1016/j.ijfoodmicro.2010.03.005. [DOI] [PubMed] [Google Scholar]
- 34.Diep DB, Straume D, Kjos M, Torres C, Nes IF. An overview of the mosaic bacteriocin pln loci from Lactobacillus plantarum. Peptides. 2009;30(8):1562–1574. doi: 10.1016/j.peptides.2009.05.014. [DOI] [PubMed] [Google Scholar]
- 35.Talarico TL, Casas IA, Chung TC, Dobrogosz WJ. Production and isolation of reuterin, a growth inhibitor produced by Lactobacillus reuteri. Antimicrob Agents Chemother. 1988;32(12):1854–1858. doi: 10.1128/AAC.32.12.1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ouwehand A, Isolauri E, Salminen S. The role of the intestinal microflora for the development of the immune system in early childhood. Eur J Nutr. 2002;41(Suppl 1):I32–I37. doi: 10.1007/s00394-002-1105-4. [DOI] [PubMed] [Google Scholar]
- 37.Bermudez-Brito M, Plaza-Diaz J, Munoz-Quezada S, Gomez-Llorente C, Gil A. Probiotic mechanisms of action. Ann Nutr Metab. 2012;61(2):160–174. doi: 10.1159/000342079. [DOI] [PubMed] [Google Scholar]
- 38.Jensen H, Dromtorp SM, Axelsson L, Grimmer S. Immunomodulation of monocytes by probiotic and selected lactic acid bacteria. Probiotics Antimicrob Proteins. 2015;7(1):14–23. doi: 10.1007/s12602-014-9174-2. [DOI] [PubMed] [Google Scholar]
- 39.Itoh T, Miyake Y, Onda A, Kubo J, Ando M, Tsukamasa Y, Takahata M. Immunomodulatory effects of heat-killed Enterococcus faecalis TH10 on murine macrophage cells. Microbiologyopen. 2012;1(4):373–380. doi: 10.1002/mbo3.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eom JS, Seok Kim J, Im Jang J, Kim BH, Young Yoo S, Hyeon Choi J, Bang IS, Lee IS, Keun Park Y. Enhancement of host immune responses by oral vaccination to Salmonella enterica serovar typhimurium harboring both FliC and FljB flagella. PLoS One. 2013;8(9):e74850. doi: 10.1371/journal.pone.0074850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Talebi A, Amirzadeh B, Mokhtari B, Gahri H. Effects of a multi-strain probiotic (PrimaLac) on performance and antibody responses to Newcastle disease virus and infectious bursal disease virus vaccination in broiler chickens. Avian Pathol. 2008;37(5):509–512. doi: 10.1080/03079450802356995. [DOI] [PubMed] [Google Scholar]
- 42.Gareau MG, Sherman PM, Walker WA. Probiotics and the gut microbiota in intestinal health and disease. Nat Rev Gastroenterol Hepatol. 2010;7(9):503–514. doi: 10.1038/nrgastro.2010.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sekirov I, Russell SL, Antunes LC, Finlay BB. Gut microbiota in health and disease. Physiol Rev. 2010;90(3):859–904. doi: 10.1152/physrev.00045.2009. [DOI] [PubMed] [Google Scholar]
- 44.Shin JS, Jung JY, Lee SG, Shin KS, Rhee YK, Lee MK, Hong HD, Lee KT. Exopolysaccharide fraction from Pediococcus pentosaceus KFT18 induces immunostimulatory activity in macrophages and immunosuppressed mice. J Appl Microbiol. 2016;120(5):1390–1402. doi: 10.1111/jam.13099. [DOI] [PubMed] [Google Scholar]
- 45.Kumar P, Tyagi R, Das G, Bhaskar S. Mycobacterium indicus pranii and Mycobacterium bovis BCG lead to differential macrophage activation in toll-like receptor-dependent manner. Immunology. 2014;143(2):258–268. doi: 10.1111/imm.12306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Takata K, Kinoshita M, Okuno T, Moriya M, Kohda T, Honorat JA, Sugimoto T, Kumanogoh A, Kayama H, Takeda K, et al. The lactic acid bacterium Pediococcus acidilactici suppresses autoimmune encephalomyelitis by inducing IL-10-producing regulatory T cells. PLoS One. 2011;6(11):e27644. doi: 10.1371/journal.pone.0027644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Driessler F, Venstrom K, Sabat R, Asadullah K, Schottelius AJ. Molecular mechanisms of interleukin-10-mediated inhibition of NF-kappaB activity: a role for p50. Clin Exp Immunol. 2004;135(1):64–73. doi: 10.1111/j.1365-2249.2004.02342.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hong YF, Lee YD, Park JY, Kim S, Lee YW, Jeon B, Jagdish D, Kim H, Chung DK. Lipoteichoic acid isolated from Weissella cibaria increases cytokine production in human monocyte-like THP-1 cells and mouse Splenocytes. J Microbiol Biotechnol. 2016;26(7):1198–1205. doi: 10.4014/jmb.1601.01047. [DOI] [PubMed] [Google Scholar]
- 49.Zou J, Baghdayan AS, Payne SJ, Shankar N. A TIR domain protein from E. faecalis attenuates MyD88-mediated signaling and NF-kappaB activation. PLoS One. 2014;9(11):e112010. doi: 10.1371/journal.pone.0112010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Baik JE, Ryu YH, Han JY, Im J, Kum KY, Yun CH, Lee K, Han SH. Lipoteichoic acid partially contributes to the inflammatory responses to Enterococcus faecalis. J Endod. 2008;34(8):975–982. doi: 10.1016/j.joen.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 51.Snapper SB, Melton RE, Mustafa S, Kieser T, Jacobs WR., Jr Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol Microbiol. 1990;4(11):1911–1919. doi: 10.1111/j.1365-2958.1990.tb02040.x. [DOI] [PubMed] [Google Scholar]
- 52.Gutierrez J, Larsen R, Cintas LM, Kok J, Hernandez PE. High-level heterologous production and functional expression of the sec-dependent enterocin P from Enterococcus faecium P13 in Lactococcus lactis. Appl Microbiol Biotechnol. 2006;72(1):41–51. doi: 10.1007/s00253-005-0233-1. [DOI] [PubMed] [Google Scholar]
- 53.Montville TJ, Chung HJ, Chikindas ML, Chen Y. Nisin a depletes intracellular ATP and acts in bactericidal manner against Mycobacterium smegmatis. Lett Appl Microbiol. 1999;28(3):189–193. doi: 10.1046/j.1365-2672.1999.00511.x. [DOI] [PubMed] [Google Scholar]
- 54.Kleerebezem M, Boekhorst J, van Kranenburg R, Molenaar D, Kuipers OP, Leer R, Tarchini R, Peters SA, Sandbrink HM, Fiers MW, et al. Complete genome sequence of Lactobacillus plantarum WCFS1. Proc Natl Acad Sci U S A. 2003;100(4):1990–1995. doi: 10.1073/pnas.0337704100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kuipers OP, Beerthuyzen MM, Siezen RJ, De Vos WM. Characterization of the nisin gene cluster nisABTCIPR of Lactococcus lactis. Requirement of expression of the nisA and nisI genes for development of immunity. Eur J Biochem. 1993;216(1):281–291. doi: 10.1111/j.1432-1033.1993.tb18143.x. [DOI] [PubMed] [Google Scholar]
- 56.Blattner FR, Plunkett G, 3rd, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF, et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277(5331):1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 57.Stedman A. BCG as a vaccine vehicle to deliver porcine immunity to African swine fever. Doctoral Thesis: University of Surrey; 2017.
- 58.Brosch R, Gordon SV, Buchrieser C, Pym AS, Garnier T, Cole ST. Comparative genomics uncovers large tandem chromosomal duplications in Mycobacterium bovis BCG Pasteur. Yeast. 2000;17(2):111–123. doi: 10.1002/1097-0061(20000630)17:2<111::AID-YEA17>3.0.CO;2-G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Borsuk S, Mendum TA, Fagundes MQ, Michelon M, Cunha CW, McFadden J, Dellagostin OA. Auxotrophic complementation as a selectable marker for stable expression of foreign antigens in Mycobacterium bovis BCG. Tuberculosis (Edinb) 2007;87(6):474–480. doi: 10.1016/j.tube.2007.07.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. pH of BCG co-cultures with LAB isolates or E. coli and Salmonella after 0 (white bars), 24 (grey bars) and 48 (black bars) h of incubation. Different letters signify statistical differences between mean values using ANOVA followed by LSD (p < 0.05).
Figure S2. Viability (A) and survival rate (B) of BCG when grown as a monoculture at pH 5 or pH 7 after 0 (white bars), 24 (grey bars) and 48 (black bars) h of incubation. Viability and survival rate are expressed as log10 Fluorescence Units (FU) and log10 CFU/mL, respectively.
Figure S3. BCG viability when grown in cell-free supernatants from 48 h-cultures of enterococci (i), weissella (ii), lactobacilli (iii), pediococci (iv) and gut-associated Gram-negative bacteria (v) after 0 (white bars), 24 (grey bars) and 48 (black bars) h of incubation. BCG viability is expressed as log10 Fluorescence Units (FU).
Figure S4. Survival rate of BCG when grown in cell-free supernatants from 48 h-cultures of enterococci (i), weissella (ii), lactobacilli (iii), pediococci (iv) and gut-associated Gram-negative bacteria (v) after 0 (white bars), 24 (grey bars) and 48 (black bars) h of incubation. BCG survival rate is expressed as log10 CFU/mL.Figure S5. Significant positive correlation between mean values of NF-κB activation and TNFα production in PMA-differentiated THP-1 macrophages exposed to inactivated bacterial strains and LPS, at a ratio of 1:100 (macrophage::bacteria) and 0.2 μg/μl, respectively (p < 0.0001).
Figure S6. Growth rate of enterococci (i), weissella (ii), lactobacilli (iii), pediococci (iv) and gut-associated Gram-negative bacteria (v) after 0 (white bars), 24 (grey bars) and 48 (black bars) h of incubation when co-cultured with BCG. Growth rate is expressed as log10 CFU/mL. (DOCX 217 kb)
Data Availability Statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.