Abstract
Functional MRI (fMRI) is modeled as a convolution of the hemodynamic response function (HRF) and an unmeasured latent neural signal. However, HRF itself is variable across brain regions and subjects. This variability is induced by both neural and non-neural factors. Aberrations in underlying neurochemical mechanisms, which control HRF shape, have been reported in autism spectrum disorders (ASD). Therefore, we hypothesized that this will lead to voxel-specific, yet systematic differences in HRF shape between ASD and healthy controls. As a corollary, we also hypothesized that such alterations will lead to differences in estimated functional connectivity in fMRI space compared to latent neural space. To test these hypotheses, we performed blind deconvolution of resting-state fMRI time series acquired from large number of ASD and control subjects obtained from the Autism Brain Imaging Data Exchange (ABIDE) database (N = 1102). Many brain regions previously implicated in autism showed systematic differences in HRF shape in ASD. Specifically, we found that precuneus had aberrations in all HRF parameters. Consequently, we obtained precuneus-seed-based functional connectivity differences between ASD and controls using fMRI as well as using latent neural signals. We found that non-deconvolved fMRI data failed to detect group differences in connectivity between precuneus and certain brain regions that were instead observed in deconvolved data. Our results are relevant for the understanding of hemodynamic and neurochemical aberrations in ASD, as well as have methodological implications for resting-state functional connectivity studies in Autism, and more generally in disorders that are accompanied by neurochemical alterations that may impact HRF shape.
Keywords: Resting-state fMRI, Deconvolution, Autism, Hemodynamic response function (HRF), HRF variability, Seed-based functional connectivity
Highlights
-
•
We demonstrate that HRF variability negatively impacts fMRI functional connectivity.
-
•
HRF variability confounds functional connectivity differences between groups.
-
•
We provide evidence for this in Autism Spectrum Disorders using the ABIDE database.
-
•
Most studies do not account for this HRF variability: implications in neuroimaging.
1. Introduction
Resting-state functional magnetic resonance imaging (fMRI) is widely used to examine brain networks by investigating temporal correlations of the blood oxygen level dependent (BOLD) signals in different brain regions (Biswal et al., 1995; Cordes et al., 2001). Specifically in the case of Autism Spectrum Disorder (ASD), resting-state fMRI (rs-fMRI) based functional connectivity (FC) studies could be used to identify potential biomarkers (Nielsen et al., 2014; Cheng et al., 2015; Abraham et al., 2017). For example, under-connectivity of the superior temporal sulcus that predicts emotion recognition deficits in ASD have been reported (Alaerts et al., 2013). Also deficits in the somatosensory, default mode, and visual regions have been highlighted in characterizing ASD (Chen et al., 2015).
Notwithstanding the strides made in understanding the neurobiology underlying ASD using rs-fMRI FC, one drawback of the method is that the BOLD signal only provides an indirect measurement of neural activity (Ogawa and Lee, 1990), i.e. the observed BOLD signal is a convolution of latent neural activity with the hemodynamic response function (HRF). HRF is the transfer function that broadly represents neurovascular coupling. This could raise many issues while using the BOLD signal as a tool for examining and inferring neural activity. Specifically problematic is the large variability of the HRF across subjects, as well as across brain regions in the same subject (Handwerker et al., 2004; Handwerker et al., 2012; Aguirre et al., 1998). Further, the HRF seems to vary across pathological populations (Reynell and Harris, 2013; Duarte et al., 2015) and can be influenced by the composition of the genome (Shan et al., 2016). With specific reference to ASD, prior studies have shown alterations in neurotransmitters which control neurovascular coupling (Reynell and Harris, 2013), and this could potentially cause changes in the HRF. Therefore, if the HRF varies due to any factors other than underlying neural activity, it could lead to false inferences of FC, because synchronicity between observed BOLD data in any two given brain regions may or may not exist in latent neural data depending on the differences in HRF shape between the two given regions (Fig. 1) (Rangaprakash et al., 2017a, Rangaprakash et al., 2018a). Our recent research also suggests that HRF variability confounds FC estimates by 15% on average (Rangaprakash et al., 2018b, Rangaprakash et al., 2018c). Additionally, we have recently reported alterations in HRF parameters as well as in associated seed-based FC in post-traumatic stress disorder (Rangaprakash et al., 2017b, Rangaprakash et al., 2017c), and hence we believe that a similar investigation in ASD may be timely. Therefore, our objective in this work is to investigate the effects of HRF variability on resting-state fMRI FC estimates in the ASD population. In order to do so, we utilized blind deconvolution of rs-fMRI data (Wu et al., 2013) from ASD and control populations and characterized FC differences before and after deconvolution.
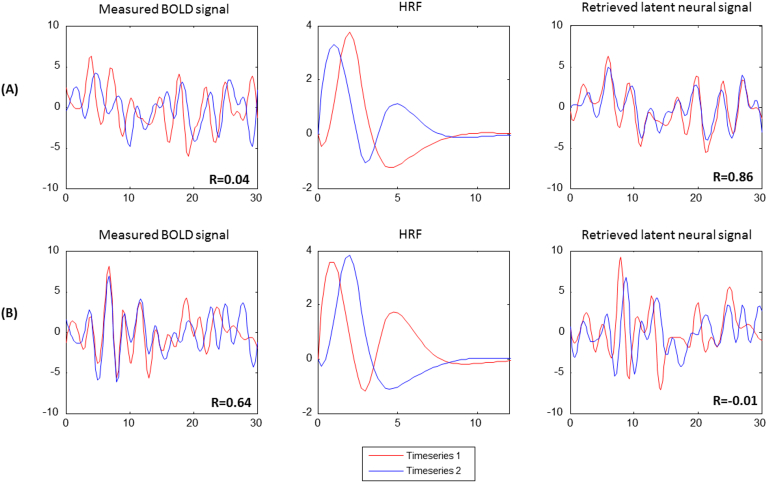
Fig. 1.
Illustration of the impact of HRF variability on functional connectivity analysis. The measured BOLD signal, retrieved latent neural signal and the voxel-level HRF are shown: (A) Two fMRI data timeseries that have low correlation while the correlation between underlying neural signals is high. (B) Two fMRI data timeseries that are highly correlated while the correlation between underlying neural signals is low. This apparent dissociation between BOLD and latent neural space is induced by the spatial variability of the HRF shape (especially, its latency), as illustrated. The figure was generated using two example fMRI time series and the corresponding derived HRF, obtained from the experimental fMRI data used in this study. We performed a brute-force search of connectivity and HRF differences between raw and deconvolved data to find this illustrative example.
In fMRI task paradigms, the neural activity as well as the BOLD response is entrained to the external sensory input or the motor output. Hence, it is relatively straightforward to deconvolve the HRF and recover latent neural activity, as the timing of neural events is known (Havlicek et al., 2011; Grant et al., 2014; Deshpande et al., 2013; Grant et al., 2015; Karahanoǧlu et al., 2013). However, this is not true in case of resting state wherein the neural events must be estimated from the data (Rangaprakash et al., 2017c; Wu and Marinazzo, 2014) or inferred from independent measurements of electrical activity (David et al., 2008) before deconvolution is performed. In this study, we employed the rs-fMRI deconvolution method proposed by Wu et al. (2013), which is based on assuming resting-state data to be generated by neural events at random times and then performing Wiener deconvolution.
In ASD, abnormalities of the neurotransmitters which control neurovascular coupling are well established (Reynell and Harris, 2013; Fatemi et al., 2009). Thus, we hypothesized that the HRF, which depends on cerebrovascular reactivity and neurovascular coupling, may be altered between ASD and healthy controls, thereby altering inferred group differences in resting-state FC. We examined voxel-specific HRFs obtained by deconvolving each voxel time series and characterizing group differences of HRF shape in terms of three parameters: time-to-peak (TTP), response height (RH), and full-width at half-max (FMHW) (see Fig. 2). We determined brain regions with significantly altered HRF between the ASD and control groups. Further, we examined possible impacts of the altered HRF on the rs-fMRI FC differences between groups.
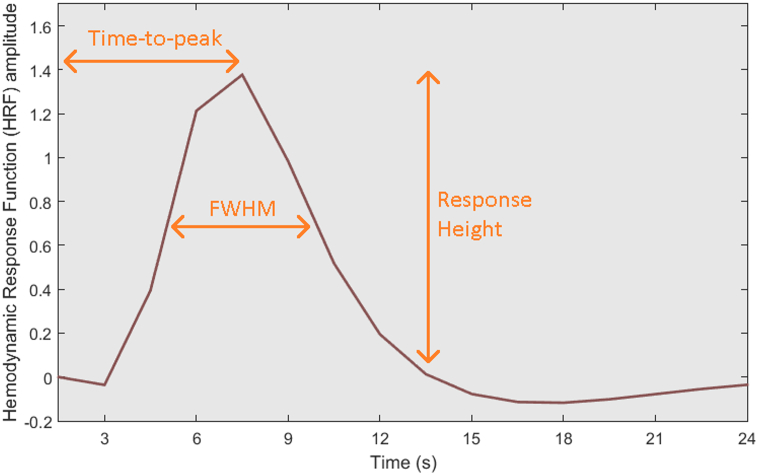
Fig. 2.
Illustration of response height (RH), time-to-peak (TTP), and full-width at half-max (FMHW) from a HRF derived from our experimental data (an arbitrary autism subject), from an arbitrary voxel in the left angular gyrus.
2. Materials and methods
2.1. Resting-state fMRI data
The Autism Brain Imaging Data Exchange (ABIDE) (Di Martino et al., 2014) consists of rs-fMRI data from 1102 subjects contributed by 17 different institutions, including 531 individuals with ASD and 571 age- and sex-matched typical controls. Of these subjects, 739 were males, and 363 were females (Table 1). The data from each subject consisted of resting functional MRI acquisitions and a volumetric magnetization-prepared rapid acquisition with gradient echo (MPRAGE) image. Local Institutional Review Boards (IRBs) approved the study protocol at each institution, the subjects provided informed consent, and the data was fully anonymized in accordance with Health Insurance Portability and Accountability Act (HIPAA) guidelines. Details of acquisition, informed consent, and site-specific protocols are available at http://fcon_1000.projects.nitrc.org/indi/abide/.
Table 1.
Gender distribution of data from autism spectrum disorder (ASD) and healthy control groups in the ABIDE database, acquired at 17 different institutions.
| No. | Institutions | Male | Female | ASD | Control | Total |
|---|---|---|---|---|---|---|
| 1 | California Institute of Technology | 30 | 8 | 19 | 19 | 38 |
| 2 | Kennedy Krieger Institute | 42 | 13 | 22 | 33 | 55 |
| 3 | University of Leuven | 56 | 8 | 29 | 35 | 64 |
| 4 | Ludwig Maximilians University Munich | 50 | 7 | 24 | 33 | 57 |
| 5 | Oregon Health and Science University | 28 | 0 | 13 | 15 | 28 |
| 6 | University of Pittsburgh School of Medicine | 49 | 8 | 30 | 27 | 57 |
| 7 | Social Brain Lab UMC Groningen NIN | 30 | 0 | 15 | 15 | 30 |
| 8 | San Diego State University | 14 | 24 | 14 | 22 | 36 |
| 9 | Stanford University | 20 | 20 | 20 | 20 | 40 |
| 10 | Trinity College Dublin | 49 | 0 | 24 | 25 | 49 |
| 11 | University of California, Los Angeles | 55 | 44 | 62 | 47 | 99 |
| 12 | University of Michigan | 117 | 28 | 68 | 77 | 145 |
| 13 | NYU Langone Medical Center | 79 | 105 | 79 | 105 | 184 |
| 14 | Olin, Institute of Living at Hartford Hospital | 20 | 16 | 20 | 16 | 36 |
| 15 | University of Utah School of Medicine | 58 | 43 | 58 | 43 | 101 |
| 16 | Yale Child Study Center | 28 | 28 | 28 | 28 | 56 |
| 17 | Carnegie Mellon University | 14 | 13 | 14 | 13 | 27 |
2.2. Pre-processing
Pre-processing of rs-fMRI data was performed in Data Processing Assistant for Resting-State fMRI (DPARSF) (Chao-Gan and Yu-Feng, 2010) toolbox, which is based on Statistical Parametric Mapping (SPM8) (Penny and Harrison, 2006) and Resting-State fMRI Data Analysis Toolkit (Song et al., 2011). For each individual participant's data set, the first 4 image volumes were discarded. Remaining volumes underwent the following pre-processing steps. Slice time correction was performed by shifting the signal measured in each slice relative to the acquisition of the slice at the mid-point of each TR. Realignment of all the images by using six rigid body motion parameters was followed by spatial normalization to the Montreal Neurological Institute (MNI) template using 4th degree B-spline interpolation. Then we regressed out head motion effects with a 24-parameter (6 head motion parameters, 6 head motion parameters one time point before, and the 12 corresponding squared items) model (Friston et al., 1996). Signals from the white matter and cerebrospinal fluid were regressed out to reduce respiratory and cardiac effects.
At this point, the processing pipeline was split into two. In the first pipeline, the data was deconvolved using the method proposed by Wu et al. (2013), and the resulting latent neural variables were temporally bandpass filtered in the (0.01–0.1 Hz) range. We will refer to this as the deconvolved (DC) dataset. In the second pipeline, the data was not deconvolved (hitherto referred to as non-deconvolved or NDC dataset), but was subjected to temporal bandpass filtering in the (0.01–0.1 Hz) range.
2.3. Blind deconvolution and HRF estimation
In order to characterize HRF variability in the data (Handwerker et al., 2012), we employed a blind deconvolution technique developed for rs-fMRI by Wu et al. (2013). This method has been validated using both simulations and experimental data (Wu et al., 2013; Rangaprakash et al., 2018b). It is based on the idea that the resting-state BOLD signal [f(t)] could be considered as the convolution of the voxel-specific HRF [h(t)] and spontaneous neural events [n(t)] occurring at random times (Tagliazucchi et al., 2012).
| (1) |
where e(t) is noise. Accordingly, spontaneous neural events (defined next in Eq. (2)) were determined from BOLD fluctuations of relatively large amplitude. “Relatively large” was quantified as one standard deviation away from the mean, as prescribed in (Wu et al., 2013; Tagliazucchi et al., 2011). This was done after pre-processing procedures (elucidated earlier), which reduced or eliminated potential sources of noise. This ensured that spikes contributed by noise sources were not mistaken for neural events. Specifically, a temporal mask with frame-wise displacement (FD) <0.3 mm was added to avoid pseudo point process events induced by motion artifacts. Using denoised and pre-processed data, the spontaneous neural events were defined as pseudo neural events modeled as a train of Dirac delta functions (Tagliazucchi et al., 2012, Tagliazucchi et al., 2011).
| (2) |
Then, a general linear model (GLM) was fitted by adjusting the delay between pseudo neural events and the BOLD peaks as a free parameter. Estimation of the HRF was then performed by fitting the canonical double-gamma HRF model along with its two derivatives to these delay-adjusted neural events. Once the estimated HRF was available along with the BOLD time series, the latent neural time series was derived using a standard Wiener filter (Glover, 1999):
| (3) |
where FT−1 is the inverse Fourier transform operator, and W(w) is the Fourier transform of the Wiener filter defined as follows
| (4) |
The estimated HRFs were characterized by three parameters: response height (RH), time-to-peak (TTP), and full-width at half-wax (FWHM). The HRF parameters were further Z-scored. Statistical two sample t-tests were performed to investigate between-group differences in HRF parameters. Also, a two-way ANOVA analysis on HRF parameters with data acquisition sites and groups as two factors was performed in order to investigate inter-site variability and its interaction with group differences. Significant main effect of site or interaction between group and site factors were not found. The deconvolution code in MATLAB is publicly available at http://users.ugent.be/~dmarinaz/HRF_deconvolution.html.
2.4. Seed region selection
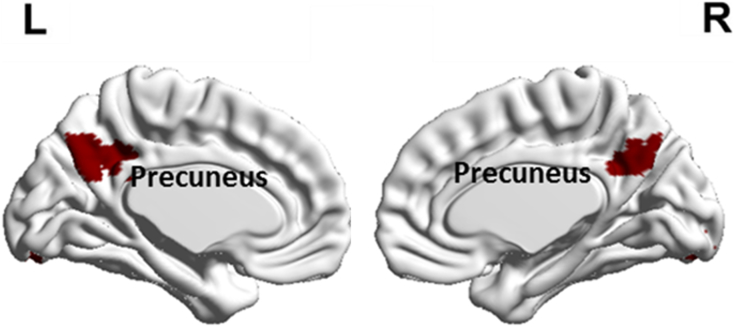
Our motive was to find brain regions that had alterations in all the three HRF parameters (TTP, FWHM and RH) in ASD compared to control subjects and use these regions as seeds to perform FC mapping with DC and NDC data. Two sample two-tailed t-tests were conducted using ASD and control samples for the three parameters separately to obtain maps indicating voxels with statistically significant difference (FDR corrected p-value < 0.05, cluster size > 50 voxels chosen based on AlphaSim correction) between the groups. FDR corrections were implemented by the Benjamini–Hochberg procedure. These maps were overlapped (intersection) to obtain brain regions that had alterations in all the three HRF parameters. We found that this corresponded to the precuneus where in all the three HRF parameters were greater in controls compared to ASD. Therefore, precuneus was selected as the seed region of interest (seed ROI) for calculating FC with the remaining brain regions (Fig. 4). Table S1 in supplementary material lists the details of the chosen seed region, including the Montreal Neurological Institute (MNI) coordinates of the cluster and cluster size.
Fig. 4.
The cluster within the precuneus which showed alterations in all three HRF parameters with RH, TTP and FWHM being higher in Controls compared to ASD.
2.5. Seed-based functional connectivity
For each participant, seed-based connectivity maps were obtained by evaluating Pearson's correlation coefficient between the mean time series from the precuneus seed ROI and the rest of the pre-processed voxel time series in the brain. A Fisher's z-transform was applied to improve the normality of these correlation coefficients (Press et al., 1992; Lowe et al., 1998). The converted z-score maps are hereafter referred to as “the correlation maps”. This pipeline was implemented separately for the two datasets: (i) NDC: data pre-processed without deconvolution, and (ii) DC: data pre-processed with deconvolution.
It is critical to note that connectivity differences obtained in the latent neural space are not obtained from neural event data. The fixed neural model (unit spiking before threshold crossing) is indeed used to estimate the neural event time series and the HRF. The utility of this model is in estimating the proper HRF lags for deconvolution. However, once the voxel-specific HRF is obtained, the latent neural time series is estimated using Wiener deconvolution. According to the model by Karahanoğlu et al. (Karahanoǧlu et al., 2013) as well as that used by Dynamic Causal Modeling (Friston et al., 2003), the former signal is the “innovation signal” and is constrained by fixed amplitudes (i.e. either ON or OFF). The latter latent neural time series used in connectivity analysis is the “activity-inducing signal” which is a continuous time series with amplitude variations as shown in the right-most column of Fig. 1.
2.6. Group-level analyses
The z-score maps from individual subjects were entered into a random effect one-sample t-test to determine the brain regions showing significant connectivity to the precuneus within each group. They were also entered into a random effect two-sample t-test to identify the regions showing significant differences in connectivity to the precuneus between control and ASD groups (Holmes and Friston, 1998). These procedures were also implemented separately for both DC and NDC datasets.
2.7. The effect of deconvolution
To investigate the effect of deconvolution on between-group differences in FC, a two-way repeated-measures ANOVA was performed within each voxel connected with the precuneus seed. We considered the groups (Control and ASD) as one factor and with/without applying deconvolution as the other factor. The voxels showing a significant interaction between the two factors (FDR corrected, p < 0.05) were identified. Statistical tests were performed using SPSS (version 20, IBM Inc., USA).
3. Results
Whole brain voxel-specific maps of HRF parameters for each individual subject in both Autism and healthy control groups have been shared publicly elsewhere (Yan et al., 2018).
3.1. Inter-group HRF differences
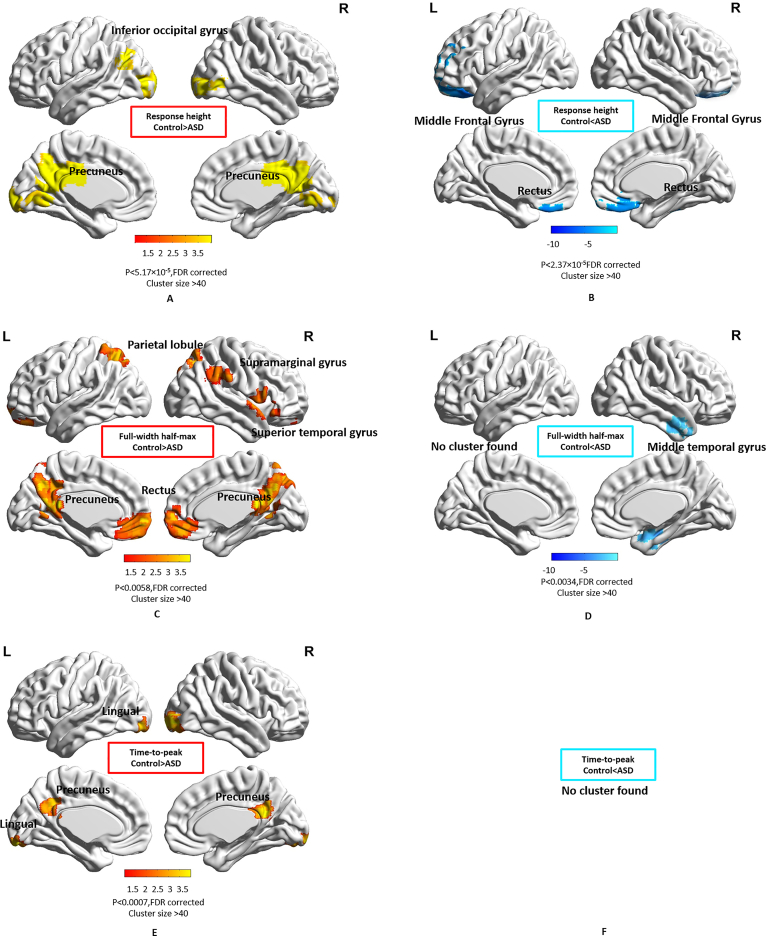
We found that the bilateral inferior occipital gyrus and precuneus had significantly higher RH (Fig. 3A) in the control group while the middle frontal gyrus and bilateral rectus had significantly higher RH (Fig. 3B) in the ASD group. The bilateral parietal lobule, bilateral rectus, supramarginal gyrus, superior temporal gyrus, and precuneus exhibited significantly higher FWHM (Fig. 3C) in the control group while the middle temporal gyrus exhibited significantly higher FWHM (Fig. 3D) in the ASD group. The left lingual gyrus and precuneus showed significantly higher TTP (Fig. 3E) in the control group. Detailed information such as cluster sizes, cluster centroids etc. are represented in Table S1 in supplementary material. We found that only one cluster within the precuneus showed alterations in all three HRF parameters (Fig. 4) (note: RH, TTP and FWHM were higher in Control group in this region compared to ASD). Also, a two-way ANOVA analysis on HRF parameters with data acquisition sites and groups as two factors did not show significant main effect of site or interaction between group and site factors.
Fig. 3.
Spatial maps showing regions with significantly different HRF parameters in ASD compared to the control group. Color bars represent the T-value. (A) Response height, Control > ASD, (B) Response height, ASD > Control, (C) FWHM, Control > ASD, (D) FWHM, ASD > Control, (E) Time-to-peak, Control > ASD (F) Time-to-peak, ASD > Control.
3.2. Precuneus-based functional connectivity within groups
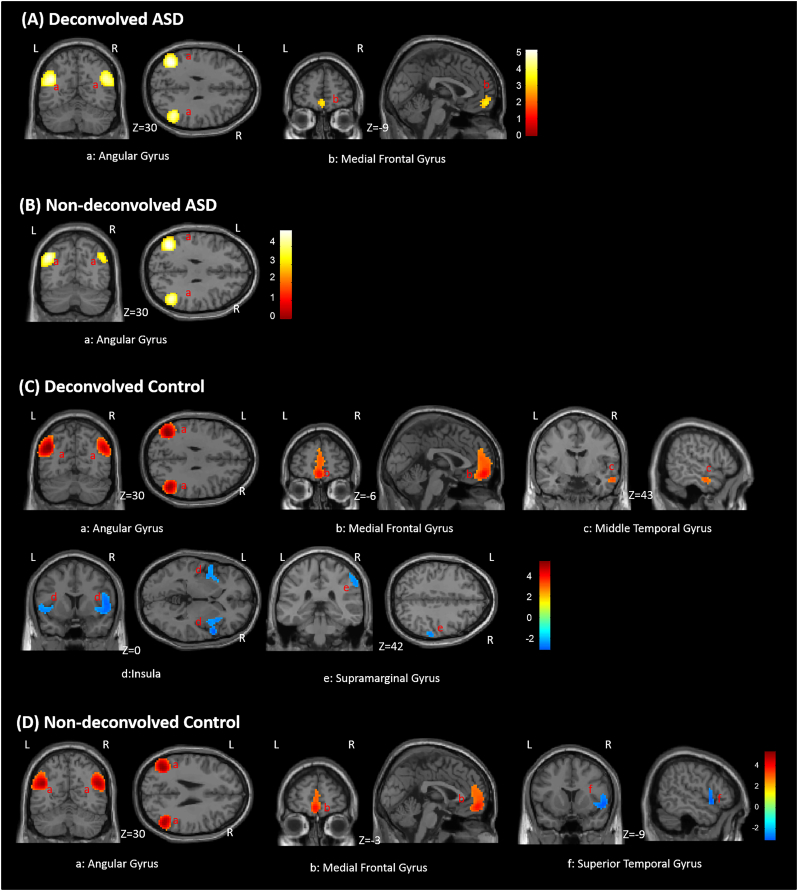
In the NDC dataset, positive FC between the precuneus seed and bilateral angular gyrus were observed in both control and ASD groups (Fig. 5B and D). In the control group, the positive FC between precuneus and the medial frontal lobe as well as negative FC between precuneus and right superior temporal gyrus (also partly containing the Insula) were detected, while these were not detected in the ASD group.
Fig. 5.
Within group functional connectivity maps with precuneus seed. (A) Deconvolved ASD group. (B) Non-deconvolved ASD group. (C) Deconvolved Control group. (D) Non-deconvolved Control group. Red indicates area of the significant positive functional connectivity while blue indicates significant negative functional connectivity. Color bars represent Z-values. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
In the DC dataset, more clusters significantly connected with the precuneus were detected in both groups (Fig. 5A and C). In the ASD group, in addition to precuneus – bilateral angular gyrus connectivity that was observed with NDC data, additional significant positive FC between precuneus and the medial frontal gyrus was detected. Likewise, in the control group, functional connectivities identified with NDC data were also identified using DC data. Additionally, positive FC between the precuneus and right middle temporal gyrus as well as negative FC with right supramarginal gyrus and bilateral insula were also detected. It is noteworthy that among regions identified to be functionally connected to precuneus in both DC and NDC datasets, those obtained from the DC dataset had a larger spatial extent (Table S2 in supplementary material).
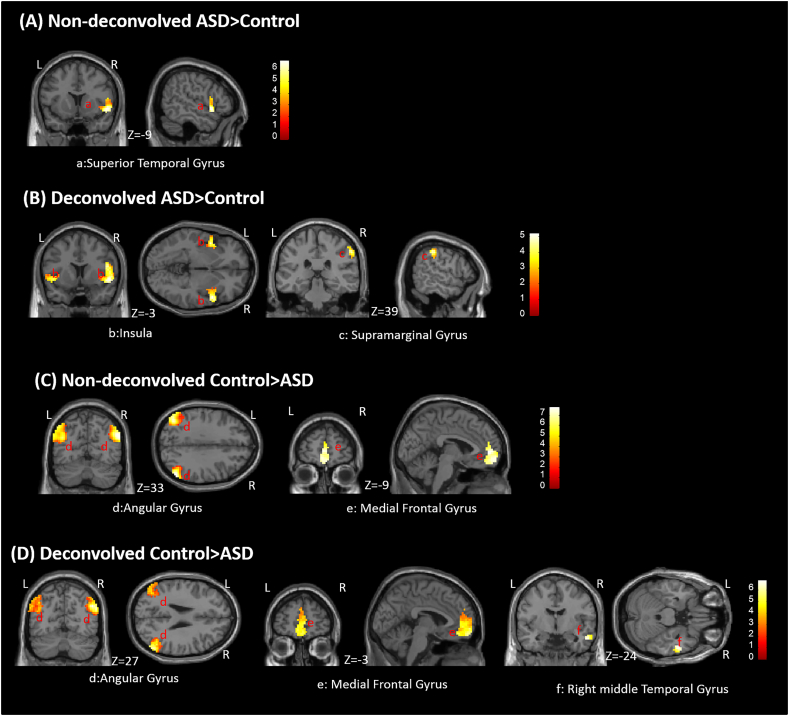
3.3. Precuneus-based connectivity differences between groups
We quantified the differences in seed-based FC between ASD and control groups, using both DC and NDC datasets separately. In the NDC dataset, the superior temporal gyrus showed higher negative connectivity with precuneus in the control group, while bilateral angular gyrus and medial frontal gyrus showed higher positive connectivity in the control group compared to ASD (Fig. 6, Table S3 in supplementary material). In the DC dataset, the superior temporal gyrus, insula and right supramarginal gyrus showed higher negative connectivity with precuneus in the control group while bilateral angular gyrus, medial frontal gyrus and right middle temporal gyrus showed higher positive connectivity in the control group compared to ASD (Fig. 6, Table S3 in supplementary material).
Fig. 6.
Between-group (ASD vs. control) differences in seed-based functional connectivity estimated from a seed in precuneus. Results are shown from both DC and NDC datasets. (A) Non-deconvolved ASD > Control, (B) Deconvolved ASD > Control, (C) Non-deconvolved Control > ASD, (C) Deconvolved Control > ASD. Color bars represent T-values. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.4. The effect of deconvolution
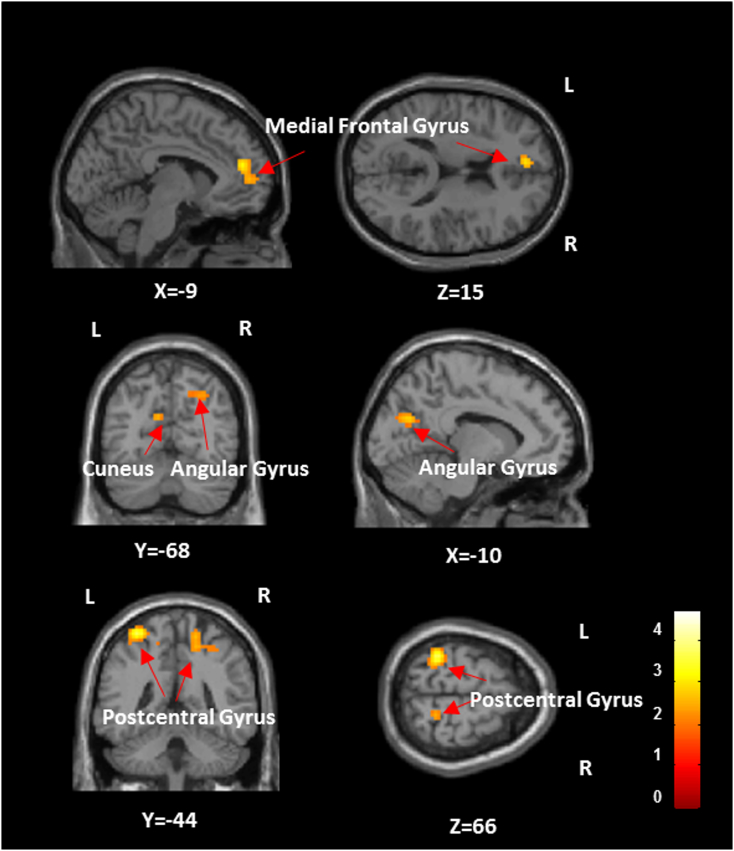
Voxels in left medial frontal gyrus, left cuneus, right angular gyrus and bilateral postcentral gyrus showed a significant interaction effect between groups and deconvolution (Fig. 7, Table S4 in supplementary material). This means that group differences between ASD and Controls in these regions would be inferred differently in DC and NDC data.
Fig. 7.
The brain regions showing significant (p-value < 0.05, FDR corrected) interaction between groups (ASD and control) and with/without applying deconvolution. Color bars represent F-values. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Finally, to assess if any global effects were impacting our findings, we compared the global signal across groups using pre-processed fMRI data. We found no significant difference between groups in derived global signal measures (p > 0.05), based on which we concluded that global signal effects did not impact any group differences observed in this study (more details in supplementary material SI-2).
4. Discussion
In this study, we tested the hypothesis that the HRF is altered in individuals with ASD as compared to controls, and that this could lead to differences in resting-state FC estimated from latent neural signal as compared with that obtained from pre-processed (but not deconvolved) BOLD fMRI data. In order to do so, we estimated the HRF at each voxel using a blind deconvolution technique and characterized significant differences in HRF characteristics such as RH, FWHM and TTP. Further, resting-state FC maps obtained from DC and NDC data had significant differences, and this impacted inferences about group differences derived from resting-state connectivity analysis. These results seem to lend credence to the fact that scenarios such as the ones shown in Fig. 1 could occur in experimental data. Therefore, in order to mitigate the uncertainty introduced by the variability of the HRF, we feel that it is desirable to perform resting-state FC analysis in the latent neural space than with BOLD data. Given the recent push to investigate FC differences as a potential imaging biomarker of ASD (Deshpande et al., 2013; Di Martino et al., 2014, Di Martino et al., 2017) as well as a metric for tracking treatment response (Levin et al., 1998), we feel that taking HRF differences in FC analysis will be critical in the clinical context.
The shape of the HRF is controlled by both non-neural and neural factors. The non-neural factors include vasculature differences, baseline cerebral blood flow, hematocrit, alcohol/caffeine/lipid ingestion, partial volume imaging of veins, global magnetic susceptibilities, slice timing differences and pulse or respiration differences (Handwerker et al., 2004) (Aguirre et al., 1998; Rangaprakash et al., 2018b). These factors induce HRF differences across both brain regions and subjects. On the other hand, systematic differences in the shape of the HRF observed between ASD and controls could at least partly be attributed to underlying neural factors that control the HRF shape.
One might wonder why specifically three HRF parameters were used. This choice was guided by prior literature, in part by Wu et al.'s findings as well as other studies that tie neurochemical mechanisms with these three HRF parameters. For example, Lindquist and Wager (2007) suggest the use of three parameters for optimal estimation of the HRF: RH, TTP, and FWHM as potential measures of response magnitude, latency, and response duration. The relationship between HRF's other morphological features and neurochemical mechanisms controlling HRF shape is yet unclear.
Concerning the choice of Wu et al.'s deconvolution technique in this study, we evaluated various methods for deconvolution and deduced that this technique is most suitable for investigating voxel-level HRF differences across subjects. For example, the “Total Activation” approach (Karahanoǧlu et al., 2013) performs impressively for finding transient and block-type co-activation patterns in the latent neural space, especially for task data. However, since it uses the same anatomical constraint across subjects, it may not fully capture the true inter-subject HRF variability. Also, HRF shape can be greatly influenced by proximity to blood vessels (Wu et al., 2013), and it might be inappropriate to impose the constraint that the HRFs within a given anatomical region must be similar.
A comprehensive account of neural factors that control the shape of the HRF is beyond the scope of this report. However, we will discuss neurochemicals that have been shown to affect the shape of the HRF and then link them with independent reports of altered neurochemistry in Autism. Fig. S1 (in supplement) shows various neurochemicals that control the coupling between neural activity and blood flow. The demand for energy due to neural activity is coupled to blood flow changes by signaling pathways controlled by various neurochemicals that directly or indirectly mediate vasodilation or vasoconstriction. The HRF is a mathematical transfer function which represents this coupling and hence could be altered as a consequence of changes in any of these neurochemicals. Specifically, glutamatergic and GABAergic interneurons impact the HRF (Bush et al., 2015; Brown et al., 2003) by releasing neuromodulators which control local cerebral blood flow (Buzsáki et al., 2007; Lozano-Soldevilla et al., 2014). In brain regions with low concentrations of GABA, taller, quicker and narrower HRFs have been observed previously (Muthukumaraswamy et al., 2012). Also, local activation of brain regions causes blood vessel dilation, which is mediated by glutamatergic actions on N-methyl-d-aspartate (NMDA) receptors (Busija et al., 2007). Attwell et al. (2010) point that GABA decreases excitability, implying that decreased GABA concentration leads to increased neuronal excitability and therefore elevated neuronal firing and higher glutamate release, which triggers the release of vasodilators, increases blood flow and modulates the HRF. However, these relationships are likely not linear. Neural activations as well as neurotransmitters could contribute to altered HRFs. Therefore, in the supplement (SI-3. Further discussion on the neurochemistry underlying our findings) we provide a detailed discussion on abnormalities in GABA, serotonin, nitric oxide (NO), glutamate and neural activations in ASD (as indicated in Fig. S1 of supplement), and ways in which such aberrations could have impacted HRF differences between groups.
Since the precuneus showed alterations in all three HRF parameters, we chose that region as the seed for FC analysis. It is noteworthy that precuneus is a core region of the Default-Mode Network (DMN) (Zhang and Li, 2012; Andrews-Hanna et al., 2014). Therefore, by using precuneus as the seed, in effect, we probed the DMN in ASD and controls using both DC and NDC datasets. Between-group differences obtained from both datasets were largely consistent with those obtained from previous studies (Di Martino et al., 2014; Lynch et al., 2013; Washington et al., 2014; Maximo et al., 2014; Uddin and Menon, 2009; Williams and Minshew, 2007; Assaf et al., 2010).
With between-group comparisons, bilateral angular gyrus and medial frontal gyrus showed higher positive connectivity in the control group compared to ASD in both DC and NDC datasets (Fig. 6, Table S3 in supplementary material). This supports the notion that under-connectivity of DMN contributes to core ASD deficits (Assaf et al., 2010; Jann et al., 2015; Ren et al., 2016). In the DC dataset, the right middle temporal gyrus exhibited hypo-connectivity with precuneus in ASD. We also detected lower negative connectivity with precuneus in superior temporal gyrus, insula and right supramarginal gyrus in ASD, with these negative correlations related to the DMN being considered as “anti-correlated” (Chang and Glover, 2010). However, in the NDC dataset, we observed only lower negative connectivity with precuneus in right superior temporal gyrus in ASD (Fig. 6, Table S3 in supplementary material).
Using repeated-measures ANOVA, we investigated whether brain regions show an interaction between group and deconvolution factors, i.e. whether significant differences between ASD and control groups were themselves significantly different between DC and NDC datasets. We found that the connectivity between the precuneus seed and the following regions – left medial frontal gyrus, left cuneus, right angular gyrus and bilateral postcentral gyrus (Fig. 7, Table S3 in supplementary material) – showed this interaction effect. This shows that FC group differences in these regions would be inferred differently in DC and NDC datasets. This is consistent with Lehmann et al. (2017) who found that the HRF to confound connectivity estimates in similar regions. This finding reinforces the point that we would be better off performing FC analysis in the latent neural space using DC data than in BOLD space using NDC so that inferences are not confounded by HRF variability.
Finally, we present some limitations of this study and point towards future directions which could address those limitations. First, we estimated the effect of HRF variability using seed-based FC using a precuneus seed. We did this since the precuneus showed alterations in all three HRF parameters. However, one could investigate the effect of voxel-wise HRF variability on voxel-wise FC differences between controls and ASD at the whole brain level, rendering a broader picture. Second, the deconvolution method chooses the threshold based on the normalized time series in each voxel of each subject. Thus, even if the ASD group has abnormal neural activations, the method only considers the relative amplitude in time series at one voxel as one event. This is nevertheless a limitation even if neural differences between the groups are captured by the latent neural time series (which is a continuous time signal that allows for amplitude variations as shown in Fig. 1) obtained from deconvolution (and not the neural event time series obtained by thresholding the BOLD data, which is binary). Third, we have discussed various neurochemical alterations in ASD and how they could have influenced the shape of the HRF. These inferences are indirect at best, since we did not directly measure the concentration of those neurochemicals. Such an endeavor, using noninvasive in vivo methods such as magnetic resonance spectroscopy in humans as well as invasive methods in animal models, could provide evidence that is more direct and further validate our observations. Fourth, all reported blind deconvolution algorithms have demonstrated their face validity using simulations (although many methods are applicable only to task data, and the number of methods capable of deconvolving resting state data are small). Therefore, in principle, we should get similar (not same) results using any blind deconvolution algorithm. However, in the absence of any reports of direct comparison of deconvolution algorithms within the same framework, it is difficult to definitively say that all of them would lead to the same conclusion. Given mounting evidence from previous literature supporting the notion that HRF variability corrupts fMRI data (Rangaprakash et al., 2017b, Rangaprakash et al., 2017c, Rangaprakash et al., 2018b; Müller et al., 2011), and given our further contribution to the understanding of HRF confound, we recommend researchers to perform deconvolution during pre-processing to minimize the confound of HRF variability.
Disclosures
The authors report no competing interests.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.nicl.2018.04.013.
Appendix A. Supplementary data
Supplementary material
References
- Abraham A., Milham M., Martino A.D., Craddock R.C., Samaras D., Thirion B., Varoquaux G. Deriving reproducible biomarkers from multi-site resting-state data: an autism-based example. NeuroImage. 2017;147:736–745. doi: 10.1016/j.neuroimage.2016.10.045. [DOI] [PubMed] [Google Scholar]
- Aguirre G.K., Zarahn E., D'esposito M. The variability of human, BOLD hemodynamic responses. NeuroImage. 1998;8(4):360–369. doi: 10.1006/nimg.1998.0369. [DOI] [PubMed] [Google Scholar]
- Alaerts K., Woolley D.G., Steyaert J., Di Martino A., Swinnen S.P., Wenderoth N. Underconnectivity of the superior temporal sulcus predicts emotion recognition deficits in autism. Soc. Cogn. Affect. Neurosci. 2013;9(10):1589–1600. doi: 10.1093/scan/nst156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews-Hanna J.R., Smallwood J., Spreng R.N. The default network and self-generated thought: component processes, dynamic control, and clinical relevance. Ann. N. Y. Acad. Sci. 2014;1316(1):29–52. doi: 10.1111/nyas.12360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assaf M., Jagannathan K., Calhoun V.D., Miller L., Stevens M.C., Sahl R., O'Boyle J.G., Schultz R.T., Pearlson G.D. Abnormal functional connectivity of default mode sub-networks in autism spectrum disorder patients. NeuroImage. 2010;53(1):247–256. doi: 10.1016/j.neuroimage.2010.05.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attwell D., Buchan A.M., Charpak S., Lauritzen M., MacVicar B.A., Newman E.A. Glial and neuronal control of brain blood flow. Nature. 2010;468(7321):232–243. doi: 10.1038/nature09613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswal B., Yetkin F.Z., Haughton V.M., Hyde J.S. Functional connectivity in the motor cortex of resting human brain using echo-planar MRI. Magn. Reson. Med. 1995;34(4):537–541. doi: 10.1002/mrm.1910340409. [DOI] [PubMed] [Google Scholar]
- Brown G.G., Eyler Zorrilla L.T., Georgy B., Kindermann S.S., Wong E.C., Buxton R.B. BOLD and perfusion response to finger-thumb apposition after acetazolamide administration: differential relationship to global perfusion. J. Cereb. Blood Flow Metab. 2003;23(7):829–837. doi: 10.1097/01.WCB.0000071887.63724.B2. [DOI] [PubMed] [Google Scholar]
- Bush K., Cisler J., Bian J., Hazaroglu G., Hazaroglu O., Kilts C. Improving the precision of fMRI BOLD signal deconvolution with implications for connectivity analysis. Magn. Reson. Imaging. 2015;33(10):1314–1323. doi: 10.1016/j.mri.2015.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busija D.W., Bari F., Domoki F., Louis T. Mechanisms involved in the cerebrovascular dilator effects of N-methyl-d-aspartate in cerebral cortex. Brain Res. Rev. 2007;56(1):89–100. doi: 10.1016/j.brainresrev.2007.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzsáki G., Kaila K., Raichle M. Inhibition and brain work. Neuron. 2007;56(5):771–783. doi: 10.1016/j.neuron.2007.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C., Glover G.H. Time-frequency dynamics of resting-state brain connectivity measured with fMRI. NeuroImage. 2010;50(1):81–98. doi: 10.1016/j.neuroimage.2009.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao-Gan Y., Yu-Feng Z. DPARSF: a MATLAB toolbox for ‘pipeline’ data analysis of resting-state fMRI. Front. Syst. Neurosci. 2010;4(May):13. doi: 10.3389/fnsys.2010.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.P., Keown C.L., Jahedi A., Nair A., Pflieger M.E., Bailey B.A., Müller R.-A. Diagnostic classification of intrinsic functional connectivity highlights somatosensory, default mode, and visual regions in autism. NeuroImage. Clin. 2015;8:238–245. doi: 10.1016/j.nicl.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng W., Rolls E.T., Gu H., Zhang J., Feng J. Autism: reduced connectivity between cortical areas involved in face expression, theory of mind, and the sense of self. Brain. 2015;138:1382–1393. doi: 10.1093/brain/awv051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordes D., Haughton V.M., Arfanakis K., Carew J.D., Turski P.A., Moritz C.H., Quigley M.A., Meyerand M.E. Frequencies contributing to functional connectivity in the cerebral cortex in ‘resting-state’ data. Am. J. Neuroradiol. 2001;22(7):1326–1333. [PMC free article] [PubMed] [Google Scholar]
- David O., Guillemain I., Saillet S., Reyt S., Deransart C., Segebarth C., Depaulis A. Identifying neural drivers with functional MRI: an electrophysiological validation. PLoS Biol. 2008;6(12) doi: 10.1371/journal.pbio.0060315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande G., Libero L.E., Sreenivasan K.R., Deshpande H.D., Kana R.K. Identification of neural connectivity signatures of autism using machine learning. Front. Hum. Neurosci. 2013;7:670. doi: 10.3389/fnhum.2013.00670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Martino A., Yan C.G., Li Q., Denio E., Castellanos F.X., Alaerts K., Anderson J.S., Assaf M., Bookheimer S.Y., Dapretto M., Deen B., Delmonte S., Dinstein I., Ertl-Wagner B., Fair D.A., Gallagher L., Kennedy D.P., Keown C.L., Keysers C., Lainhart J.E., Lord C., Luna B., Menon V., Minshew N.J., Monk C.S., Mueller S., Müller R., Nebel M.B., Nigg J.T., O'Hearn K., Pelphrey K.A., Peltier S.J., Rudie J.D., Sunaert S., Thioux M., Tyszka J.M., Uddin L.Q., Verhoeven J.S., Wenderoth N., Wiggins J.L., Mostofsky S.H., Milham M.P. The autism brain imaging data exchange: towards a large-scale evaluation of the intrinsic brain architecture in autism. Mol. Psychiatry. 2014;19(6):659–667. doi: 10.1038/mp.2013.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Martino A., O'Connor D., Chen B., Alaerts K., Anderson J.S., Assaf M., Balsters J.H., Baxter L., Beggiato A., Bernaerts S., Blanken L.M.E., Bookheimer S.Y., Braden B.B., Byrge L., Castellanos F.X., Dapretto M., Delorme R., Fair D.A., Fishman I., Fitzgerald J., Gallagher L., Keehn R.J.J., Kennedy D.P., Lainhart J.E., Luna B., Mostofsky S.H., Müller R.A., Nebel M.B., Nigg J.T., O'Hearn K., Solomon M., Toro R., Vaidya C.J., Wenderoth N., White T., Craddock R.C., Lord C., Leventhal B., Milham M.P. Enhancing studies of the connectome in autism using the autism brain imaging data exchange II. Sci. Data. 2017:4. doi: 10.1038/sdata.2017.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte J.V., Pereira J.M., Quendera B., Raimundo M., Moreno C., Gomes L., Carrilho F., Castelo-Branco M. Early disrupted neurovascular coupling and changed event level hemodynamic response function in type 2 diabetes: an fMRI study. J. Cereb. Blood Flow Metab. 2015;35(10):1671–1680. doi: 10.1038/jcbfm.2015.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatemi S.H., Reutiman T.J., Folsom T.D., Thuras P.D. GABAA receptor downregulation in brains of subjects with autism. J. Autism Dev. Disord. 2009;39(2):223–230. doi: 10.1007/s10803-008-0646-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friston K.J., Williams S., Howard R., Frackowiak R.S., Turner R. Movement-related effects in fMRI time-series. Magn. Reson. Med. 1996;35(3):346–355. doi: 10.1002/mrm.1910350312. [DOI] [PubMed] [Google Scholar]
- Friston K.J., Harrison L., Penny W. Dynamic causal modelling. NeuroImage. 2003;19(4):1273–1302. doi: 10.1016/s1053-8119(03)00202-7. [DOI] [PubMed] [Google Scholar]
- Glover G.H. Deconvolution of impulse response in event-related BOLD fMRI. NeuroImage. 1999;9(4):416–429. doi: 10.1006/nimg.1998.0419. [DOI] [PubMed] [Google Scholar]
- Grant M.M., White D., Hadley J., Hutcheson N., Shelton R., Sreenivasan K., Deshpande G. Early life trauma and directional brain connectivity within major depression. Hum. Brain Mapp. 2014;35(9):4815–4826. doi: 10.1002/hbm.22514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant M.M., Wood K., Sreenivasan K., Wheelock M., White D., Thomas J., Knight D.C., Deshpande G. Influence of early life stress on intra- and extra-amygdaloid causal connectivity. Neuropsychopharmacology. 2015;40(7):1–12. doi: 10.1038/npp.2015.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handwerker D.A., Ollinger J.M., D'Esposito M. Variation of BOLD hemodynamic responses across subjects and brain regions and their effects on statistical analyses. NeuroImage. 2004;21(4):1639–1651. doi: 10.1016/j.neuroimage.2003.11.029. [DOI] [PubMed] [Google Scholar]
- Handwerker D.A., Gonzalez-Castillo J., D'Esposito M., Bandettini P.A. The continuing challenge of understanding and modeling hemodynamic variation in fMRI. NeuroImage. 2012;62(2):1017–1023. doi: 10.1016/j.neuroimage.2012.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havlicek M., Friston K.J., Jan J., Brazdil M., Calhoun V.D. Dynamic modeling of neuronal responses in fMRI using cubature Kalman filtering. NeuroImage. 2011;56(4):2109–2128. doi: 10.1016/j.neuroimage.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes A.P., Friston K.J. Generalisability, random effects and population inference. NeuroImage. 1998;7:S754. [Google Scholar]
- Jann K., Hernandez L.M., Beck-Pancer D., Mccarron R., Smith R.X., Dapretto M., Wang D.J.J. Altered resting perfusion and functional connectivity of default mode network in youth with autism spectrum disorder. Brain Behav. 2015;9:5. doi: 10.1002/brb3.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karahanoǧlu F.I., Caballero-Gaudes C., Lazeyras F., Van De Ville D. Total activation: FMRI deconvolution through spatio-temporal regularization. NeuroImage. 2013;73:121–134. doi: 10.1016/j.neuroimage.2013.01.067. [DOI] [PubMed] [Google Scholar]
- Lehmann B.C.L., White S.R., Henson R.N., Cam-CAN, Geerligs L. Assessing dynamic functional connectivity in heterogeneous samples. NeuroImage. 2017;157:635–647. doi: 10.1016/j.neuroimage.2017.05.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin J.M., Ross M.H., Mendelson J.H., Kaufman M.J., Lange N., Maas L.C., Mello N.K., Cohen B.M., Renshaw P.F. Reduction in BOLD fMRI response to primary visual stimulation following alcohol ingestion. Psychiatry Res. 1998;82(3):135–146. doi: 10.1016/s0925-4927(98)00022-5. [DOI] [PubMed] [Google Scholar]
- Lindquist M.A., Wager T.D. Validity and power in hemodynamic response modeling: a comparison study and a new approach. Hum. Brain Mapp. 2007;28(8):764–784. doi: 10.1002/hbm.20310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe M.J., Mock B.J., Sorenson J.A. Functional connectivity in single and multislice echoplanar imaging using resting-state fluctuations. NeuroImage. 1998;7(2):119–132. doi: 10.1006/nimg.1997.0315. [DOI] [PubMed] [Google Scholar]
- Lozano-Soldevilla D., Ter Huurne N., Cools R., Jensen O. GABAergic modulation of visual gamma and alpha oscillations and its consequences for working memory performance. Curr. Biol. 2014;24(24):2878–2887. doi: 10.1016/j.cub.2014.10.017. [DOI] [PubMed] [Google Scholar]
- Lynch C.J., Uddin L.Q., Supekar K., Khouzam A., Phillips J., Menon V. Default mode network in childhood autism: posteromedial cortex heterogeneity and relationship with social deficits. Biol. Psychiatry. 2013;74(3):212–219. doi: 10.1016/j.biopsych.2012.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maximo J.O., Cadena E.J., Kana R.K. The implications of brain connectivity in the neuropsychology of autism. Neuropsychol. Rev. 2014;24(1):16–31. doi: 10.1007/s11065-014-9250-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller R.A., Shih P., Keehn B., Deyoe J.R., Leyden K.M., Shukla D.K. Underconnected, but how? A survey of functional connectivity MRI studies in autism spectrum disorders. Cereb. Cortex. 2011;21(10):2233–2243. doi: 10.1093/cercor/bhq296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthukumaraswamy S.D., Evans C.J., Edden R.A.E., Wise R.G., Singh K.D. Individual variability in the shape and amplitude of the BOLD-HRF correlates with endogenous GABAergic inhibition. Hum. Brain Mapp. Feb. 2012;33(2):455–465. doi: 10.1002/hbm.21223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen J.A., Zielinski B.A., Fletcher P., Alexander A.L., Lange N., Bigler E.D., Lainhart J.E., Anderson J.S. Abnormal lateralization of functional connectivity between language and default mode regions in autism. Mol. Autism. 2014;5(1):8. doi: 10.1186/2040-2392-5-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa S., Lee T.M. Magnetic resonance imaging of blood vessels at high fields: in vivo and in vitro measurements and image simulation. Magn. Reson. Med. 1990;16(1):9–18. doi: 10.1002/mrm.1910160103. [DOI] [PubMed] [Google Scholar]
- Penny W., Harrison L. Multivariate autoregressive models. In: Friston K., Ashburner J., Kiebel S., Nichols T., Penny W., editors. Statistical Parametric Mapping: The Analysis of Functional Brain Images. Elsevier; London: 2006. [Google Scholar]
- Press W.H., Teukolsky S.A., Vetterling W.T., Flannery B.P. The Art of Scientific Computing. 2nd ed. vol. 29. 1992. Numerical recipes in C. (no. 4) [Google Scholar]
- Rangaprakash D., Deshpande G., Daniel T.A., Goodman A.M., Robinson J.L., Salibi N., Katz J.S., Denney T.S., Dretsch M.N. Compromised hippocampus-striatum pathway as a potential imaging biomarker of mild-traumatic brain injury and posttraumatic stress disorder. Hum. Brain Mapp. 2017;38(6):2843–2864. doi: 10.1002/hbm.23551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangaprakash D., Dretsch M.N., Yan W., Katz J.S., Denney T.S., Deshpande G. Hemodynamic response function parameters obtained from resting-state functional MRI data in soldiers with trauma. Data in Brief. 2017;14 doi: 10.1016/j.dib.2017.07.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangaprakash D., Dretsch M.N., Yan W., Katz J.S., Denney T.S., Deshpande G. Hemodynamic variability in soldiers with trauma: implications for functional MRI connectivity studies. NeuroImage Clin. 2017;16:409–417. doi: 10.1016/j.nicl.2017.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangaprakash D., Dretsch M.N., Venkataraman A., Katz J.S., Denney T.S., Deshpande G. Identifying disease foci from static and dynamic effective connectivity networks: illustration in soldiers with trauma. Hum. Brain Mapp. 2018;39(1):264–287. doi: 10.1002/hbm.23841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangaprakash D., Wu G.-R., Marinazzo D., Hu X., Deshpande G. Hemodynamic response function (HRF) variability confounds resting-state fMRI functional connectivity. Magn. Reson. Med. 2018 doi: 10.1002/mrm.27146. (in press) [DOI] [PubMed] [Google Scholar]
- Rangaprakash D., Wu G.-R., Marinazzo D., Hu X., Deshpande G. Parametrized hemodynamic response function data of healthy individuals obtained from resting-state functional MRI in a 7T MRI scanner. Data in Brief. 2018;17 doi: 10.1016/j.dib.2018.01.003. 1175–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Y., Hu X., Lv J., Quo L., Han J., Liu T. Proceedings - International Symposium on Biomedical Imaging. 2016. Identifying autism biomarkers in default mode network using sparse representation of resting-state fMRI data; pp. 1278–1281. [Google Scholar]
- Reynell C., Harris J.J. The BOLD signal and neurovascular coupling in autism. Dev. Cogn. Neurosci. 2013;6:72–79. doi: 10.1016/j.dcn.2013.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan Z.Y., Vinkhuyzen A.A.E., Thompson P.M., McMahon K.L., Blokland G.A.M., de Zubicaray G.I., Calhoun V., Martin N.G., Visscher P.M., Wright M.J., Reutens D.C. Genes influence the amplitude and timing of brain hemodynamic responses. NeuroImage. 2016;124, pp:663–671. doi: 10.1016/j.neuroimage.2015.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X.-W., Dong Z.-Y., Long X.-Y., Li S.-F., Zuo X.-N., Zhu C.-Z., He Y., Yan C.-G., Zang Y.-F. REST: a toolkit for resting-state functional magnetic resonance imaging data processing. PLoS One. 2011;6(9) doi: 10.1371/journal.pone.0025031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagliazucchi E., Balenzuela P., Fraiman D., Montoya P., Chialvo D.R. Spontaneous BOLD event triggered averages for estimating functional connectivity at resting state. Neurosci. Lett. Jan. 2011;488(2):158–163. doi: 10.1016/j.neulet.2010.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagliazucchi E., Balenzuela P., Fraiman D., Chialvo D.R. Criticality in large-scale brain fmri dynamics unveiled by a novel point process analysis. Front. Physiol. 2012;3(FEB) doi: 10.3389/fphys.2012.00015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uddin L.Q., Menon V. The anterior insula in autism: under-connected and under-examined. Neurosci. Biobehav. Rev. 2009;33(8):1198–1203. doi: 10.1016/j.neubiorev.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washington S.D., Gordon E.M., Brar J., Warburton S., Sawyer A.T., Wolfe A., Mease-Ference E.R., Girton L., Hailu A., Mbwana J., Gaillard W.D., Kalbfleisch M.L., Vanmeter J.W. Dysmaturation of the default mode network in autism. Hum. Brain Mapp. 2014;35(4):1284–1296. doi: 10.1002/hbm.22252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams D.L., Minshew N.J. Understanding autism and related disorders: what has imaging taught us? Neuroimaging Clin. N. Am. 2007;17(4):495–509. doi: 10.1016/j.nic.2007.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G.R., Marinazzo D. Point-process deconvolution of fMRI BOLD signal reveals effective connectivity alterations in chronic pain patients. Brain Topogr. 2014;28(4):541–547. doi: 10.1007/s10548-014-0404-4. [DOI] [PubMed] [Google Scholar]
- Wu G.R., Liao W., Stramaglia S., Ding J.-R.J., Chen H., Marinazzo D. A blind deconvolution approach to recover effective connectivity brain networks from resting state fMRI data. Med. Image Anal. 2013;17(3):365–374. doi: 10.1016/j.media.2013.01.003. [DOI] [PubMed] [Google Scholar]
- Yan W., Rangaprakash D., Deshpande G. Hemodynamic response function parameters obtained from resting state BOLD fMRI data in subjects with autism spectrum disorder and matched healthy controls. Data in Brief. 2018 doi: 10.1016/j.dib.2018.04.126. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Li C. shan R. Functional connectivity mapping of the human precuneus by resting state fMRI. NeuroImage. 2012;59(4):3548–3562. doi: 10.1016/j.neuroimage.2011.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material