Summary
CTLA-4 immune checkpoint blockade is clinically effective in a subset of patients with metastatic melanoma. We identify a subcluster of MAGE-A cancer-germline antigens, located within a narrow 75 kb region of chromosome Xq28, that predicts resistance uniquely to blockade of CTLA-4, but not PD-1. We validate this gene expression signature in an independent anti-CTLA-4-treated cohort and show its specificity to the CTLA-4 pathway with two independent anti-PD-1-treated cohorts. Autophagy, a process critical for optimal anti-cancer immunity, has been previously shown to be suppressed by the MAGE-TRIM28 ubiquitin ligase in vitro. We now show that the expression of the key autophagosome component LC3B and other activators of autophagy are negatively associated with MAGE-A protein levels in human melanomas, including samples from patients with resistance to CTLA-4 blockade. Our findings implicate autophagy suppression in resistance to CTLA-4 blockade in melanoma, suggesting exploitation of autophagy induction for potential therapeutic synergy with CTLA-4 inhibitors.
Keywords: Melanoma, ipilimumab, CTLA-4, cancer-germline antigen, MAGE-A, autophagy
ITI
Increased expression of a subcluster of MAGE-A cancer-germline antigens predicts resistance specific to CTLA4, but not PD1, blockade, and its association with autophagy suppression implicates the role autophagy in regulating primary resistance to anti-CTLA4 therapy in melanoma patients

Introduction
Antibodies targeting the cytotoxic T lymphocyte–associated antigen-4 (CTLA-4) pathway in advanced melanoma have yielded lasting clinical responses in a subset of patients (Hodi et al., 2010). Combining CTLA-4 blockade with antagonists of an alternative immune checkpoint pathway, the programmed death (PD-1) pathway, increases response rates in metastatic melanoma compared with either agent alone and suggests the potential benefit of combining CTLA-4 blockade with other immunotherapeutics (Wolchok et al., 2013). However, robust determinants of response and resistance to CTLA-4 blockade remain elusive, hindering efforts to rationally incorporate it in combinatorial strategies and precisely pair it with patients most likely to respond.
While several investigators have identified genomic markers of clinical outcome to CTLA-4 blockade (Gao et al., 2016; Riaz et al., 2016; Snyder et al., 2014; Van Allen et al., 2015), discovery of robust transcriptional signatures for response to immune checkpoint therapy has been limited by small sample sizes and lack of validation cohorts. Along with important genomic markers that have highlighted resistance pathways common to various checkpoint inhibitors (Gao et al., 2016; Zaretsky et al., 2016), discovery of biomarkers specific to a given immune checkpoint may illuminate the processes distinguishing these inhibitory pathways. To interrogate and identify transcriptional determinants of clinical outcome specific to CTLA-4 blockade in advanced melanoma, we analyzed RNA sequencing (RNA-seq) data from 146 melanoma biopsies representing patients from three previously reported clinical studies comprising four clinical treatment cohorts (Hugo et al., 2016; Van Allen et al., 2015; Weber et al., 2016) as well as both transcriptomic and DNA methylation data from The Cancer Genome Atlas (TCGA) (Cancer Genome Atlas, 2015). These analyses allowed us to discover an unexpected role for cancer-germline antigens in primary resistance to CTLA-4 but not PD-1 blockade and to conduct studies implicating the involvement of autophagy dysregulation in clinical outcome to ipilimumab, setting the stage for future investigations in larger, prospective trials.
Results
A specific cluster of cancer-germline antigens predicts resistance to CTLA-4 blockade
From a previously reported cohort of 110 melanoma patients treated with ipilimumab, we analyzed the 40 with associated tumor-derived RNA-seq data from pre-therapy samples (Van Allen et al., 2015). Thirty-five of these 40 patients could be unambiguously classified into the clinical benefit (CB) and no benefit (NB) designations maintained from the original report (see STAR Methods). A differential gene expression analysis comparing NB (n=22) and CB (n=13) groups identified 457 genes upregulated in NB samples (Fig. 1A and Tables S1, S2). Strikingly, 7 of the top 12 genes overexpressed in NB samples were clustered within a 75 kb region of chromosome Xq28 (median 67-fold increase, range 34–188; Figure 1B, top). An additional gene, MAGEA3, was also located within this narrow genomic region and was similarly upregulated in NB samples (24-fold increase). All 8 genes (MAGEA3, CSAG3, CSAG2, MAGEA2, MAGEA2B, CSAG1, MAGEA12, MAGEA6) encoded cancer-germline antigens (CGAs), a large family of genes notable for their restricted expression in testis and placenta during normal development but re-expression across many tumor types. These 8 genes constitute one of four subclusters within the MAGE-A family and are coordinately regulated, independent of the other three MAGE-A subclusters (Bredenbeck et al., 2008). 16 of 22 NB samples showed upregulation of at least one of these 8 CGAs, which we refer to as ‘anti-CTLA-4 Resistance associated MAGE-A (CRMA)’ genes, compared to only 2 of 13 CB samples (Fisher’s exact test, p=0.002; Figure S1A).
Figure 1. A specific cluster of cancer-germline antigens predicts resistance to CTLA-4 blockade.
(A) Volcano plot depicting genes enriched in CB and NB tumors (n=457 and 326 respectively; fold change>2, one-sided Wilcoxon test p-value<0.05). Relative positions of CRMA and immune-related genes (pink) are shown. Inset: Computational purity estimates by the ABSOLUTE algorithm are comparable between the CB and NB groups (Carter et al., 2012). (B) 75 kb region within the CRMA locus containing the 8 CGAs is shown; CSAG3 and MAGEA2 are duplicated genes. Box plots depicting the individual RNA-seq expression value for each MAGEA and CSAG gene within this locus in the discovery cohort stratified by CB and NB status (Van Allen et al., 2015). (C) Volcano plot depicting genes enriched in ‘no progressive disease’ (No PD) and ‘progressive disease’ (PD) groups at week 13 in the ipilimumab-nivolumab arm of the CheckMate 064 trial (Weber et al., 2016) (validation cohort). (D) Box plots depicting RNA-seq expression for CRMA MAGEA genes between the No PD and PD groups in validation cohort. (E) IHC staining analysis of MAGE-A protein expression in pre-ipilimumab melanoma biopsies. (F) Examples of MAGE-A protein expression from a patient in the CB (left) and NB (right) groups. Magnification, x1000. See also Figure S1, Table S1 and Table S2.
We performed several confirmatory assessments to evaluate the validity of the CRMA signature. First, we ruled out potential sequencing-related artifacts by confirming expression of the CRMA genes in the original tumor RNA from the discovery set by gene-specific RT-qPCR (Figure S1B). Second, we found similar tumor purity estimates in both patient groups (see STAR Methods), suggesting that relative enrichment of cancer cells in the NB group was unlikely to explain our finding (Figure 1A-inset). Finally, given the signature’s genomic localization to the X chromosome and potential modification by DNA damaging agents, we ensured that neither gender nor prior exposure to cytotoxic therapy (i.e. dacarbazine/temozolomide) was associated with clinical outcome (Figure S1C–S1D).
While our discovery cohort was generated from formalin-fixed samples from an observational, retrospective study, we could validate our findings in an independent RNA-seq data set generated from cryopreserved tumors from a prospective, randomized trial using pre-treatment patient samples derived from the CheckMate 064 trial (Weber et al., 2016). This cohort comprised 41 patients, divided into ‘progressive disease’ (PD; n=12) or ‘no progressive disease’ (no PD; n=29) groups (see STAR Methods for details). Again, the CRMA genes were amongst the most significantly upregulated genes (Figure 1C). Because overall survival data attributable to ipilimumab monotherapy was not available given the subsequent administration of nivolumab, we re-classified our discovery cohort based on response assessments used for the CheckMate 064 trial with “progressive disease (PD)” and “no PD” groups. RNA-seq expression values from the validation cohort were available for 5 of 8 genes in the CRMA locus (MAGEA3, MAGEA2, MAGEA2B, MAGEA12, and MAGEA6), and we observed consistent increases in all of these genes in patients with PD in both discovery (median 57-fold increase, range 24–159; maximum p-value = 0.075) and validation cohorts (median 108-fold increase, range 5–236; maximum p-value = 0.044; Figures 1C–D; Figure S1E–F.)
To assess protein-level expression of the CRMA genes in relationship to clinical response, we performed immunohistochemistry (IHC) using a MAGE-A antibody (clone 6C1) broadly reactive for gene products from the MAGE-A family (i.e. recognizing MAGEA1, A2, A3, A4, A6, A10 and A12) on 55 melanoma samples from the original discovery cohort (Van Allen et al., 2015). Forty-eight of these 55 patients could be unambiguously classified into CB and NB cohorts (see STAR Methods). Consistent with the RNA-seq analysis, the NB cohort (n=33) comprised a higher proportion of MAGE-A+ tumors compared to the CB cohort (n=15) (Figures 1E–F, 76% vs 40%, p = 0.024), further confirming the strong association of baseline transcript and protein expression of a specific cluster of MAGE-A genes with primary resistance to ipilimumab.
DNA methylation patterns in resistant tumors and CRMA-high TCGA samples
To define the transcriptome-wide effects of CRMA expression, we analyzed RNA-seq data from 465 melanomas from TCGA (Cancer Genome Atlas, 2015). Genes overexpressed in NB samples overlapped with genes positively associated with CRMA expression in TCGA melanomas (p < 2.2 × 10−16) but not with genes negatively associated with CRMA expression (Fig. 2A; Tables S3, S4; see STAR Methods). These results suggest that primary resistance to ipilumumab is associated with transcriptional programs inherent to high CRMA-expressing tumors.
Figure 2. DNA methylation patterns in resistant tumors and CRMA-high TCGA samples.
(A) Genes associated with high CRMA expression in TCGA significantly overlap with genes associated with NB tumors; hypergeometric test. (B) Methylation of MAGEA3 and MAGEA6 promoters in NB patients (orange; n=3) compared to CB patients (blue; n=3) as validated by bisulfite PCR. The plot highlights the local regression (solid line) of the mean methylation for every CpG (dots) along the MAGEA3 and MAGEA6 promoters in CB versus NB patients. The standard deviation is indicated by the shaded area. Both promoter sequences are identical within the analyzed amplicon span. (C) Volcano plot of differentially methylated probes (FDR = 0.05) across the genome between CRMA-low and CRMA-high expression groups in the TCGA melanoma cohort. Table shows PANTHER pathways (Mi et al., 2016) enriched in genes corresponding to 65,467 probes relatively hypomethylated in the CRMA-high group. Highlighted in red are two of only three PANTHER pathways enriched in NB samples. See also Figure S2, Table S2, Table S3 and Table S4.
To investigate potential mechanisms underlying the transcriptional enrichment of CRMA genes in resistant tumors, we analyzed copy number variation and DNA methylation at this locus in the clinical trial samples. We observed no copy number alterations of this region based on analysis of matched whole-exome sequencing (WES) data (Figure S2A; see STAR Methods for details). On the other hand, locus-specific methylation analysis of the MAGEA3 and MAGEA6 promoters revealed decreased DNA methylation throughout these promoters in NB samples (Figure 2B; p=3×10−6) and a slight to moderate decrease in unique methylation sites within the gene bodies of MAGEA6, MAGEA3, and MAGEA12 (Figure S2B), consistent with known epigenetic regulation of CRMA genes (Simpson et al., 2005).
We further investigated DNA methylation patterns associated with CRMA expression by querying methylation data from TCGA melanoma samples. Differential methylation analysis of 485,577 probes between samples with high (n=116) and low expression (n=117) of the CRMA locus (see STAR Methods for details) revealed 47 probes relatively hypermethylated in the “CRMA-high” group compared to 65,467 in the “CRMA-low” group (Figure 2C-top). These 65,467 probes mapped across the genome, suggesting global hypomethylation in melanoma samples with high CRMA expression (Figure S2C). Gene set enrichment analysis for genes corresponding to these 65,467 probes using PANTHER (Mi et al., 2016) revealed four pathways as significantly demethylated in the CRMA-high melanoma TCGA group (Figure 2C-bottom). Of these, cadherin signaling and Wnt pathway signaling were also two of only three PANTHER pathways enriched in genes upregulated in NB samples (Bonferroni adjusted p<0.01 and p<0.05 respectively), further implicating epigenomic dysregulation in primary resistance to anti-CTLA-4 therapy.
In addition to the CRMA cluster, we also found increased expression of additional CGAs in NB samples, though none were as highly expressed as those at the CRMA locus (Table S2). Moreover, CGAs previously described to elicit humoral and cellular responses linked to clinical outcome, such as NY-ESO-1 (Yuan et al., 2011), were not differentially expressed, nor were melanoma differentiation antigens (e.g. TYR, TYRP1, PMEL, and MLANA) (Figure S3A). Additional gene families upregulated in NB samples included those related to epithelial-to-mesenchymal transition (Kalluri and Weinberg, 2009) (i.e. CLDN1, CLDN2, EGF, EYA1, FGF2, SNAI1, WNT3) and embryonic development (HOXA2, HOXA3, HOXA5, HOXD10, HOXD11, HOXD13) (Table S2). Moreover, multiple subunits of the GABA-A receptor, which has been implicated in mediating suppression of inflammatory macrophages and anti-tumor T cells, were also enriched in NB samples (fold change: 2.7–110; 6 of 19 family members, hypergeometric test, p < 5.7 × 10−6) (Pu et al., 2016). Recent work has also implicated GABRA3, a GABA-A receptor subunit, in mediating cancer invasion and metastasis, potentially through increasing stem cell populations (Gumireddy et al., 2016). Consistent with previous reports, immune subsets were found to be significantly upregulated only in CB transcriptomes (Figure S3B–C). Altogether, the primary resistance phenotype to CTLA-4 blockade appears to be marked by CRMA overexpression and may comprise epigenomic dysregulation coupled with activation of additional programs including EMT, embryonic development, and invasion.
CRMA signature predicts clinical outcome to CTLA-4 but not PD-1 blockade
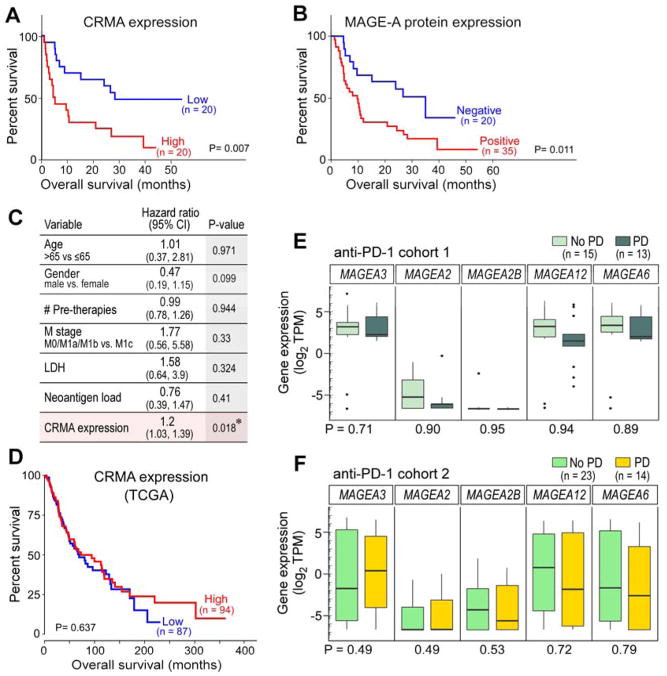
Within the full cohort from Van Allen et al (Van Allen et al., 2015) a Kaplan-Meier analysis of overall survival on ipilimumab therapy demonstrated that samples with high CRMA expression had poorer overall and progression-free survival than those with low CRMA expression (log-rank p=0.007 and p=0.006, respectively, n=40) (Figure 3A, Figure S3D). Similarly, detectable MAGE-A protein expression associated with inferior overall and progression-free survival on ipilimumab therapy (log-rank p=0.011 and p=0.032, respectively, n=55) (Figure 3B, Figure S3E). In a multivariable analysis of these 40 patients that included evaluation of neoantigen load, CRMA expression emerged as the sole independent risk factor for poor outcome after ipilimumab therapy (Cox proportional-hazards model, p=0.018; Figure 3C).
Figure 3. CRMA molecular signatures correlate with clinical outcome to CTLA-4 but not PD-1 blockade.
(A, B) Kaplan-Meier overall survival analysis comparing patients from discovery cohort classified by (A) expression of genes within the CRMA locus (median split) or (B) expression of MAGE-A protein. (C) Cox proportional hazards model of risk factors for outcomes on ipilimumab therapy. (D) Kaplan-Meier overall survival analysis of TCGA melanoma samples with high or low expression of CRMA genes. (E) Box plots of RNA-seq expression values for each MAGEA gene in the CRMA locus within no progressive disease (“No PD”; light green) (n=15) and progressive disease (“PD”; dark green) tumors (n=13) from anti-PD-1 cohort 1(Hugo et al., 2016). (F) Box plots of RNA-seq expression values for each MAGEA gene in the CRMA locus within no PD (blue; n=23) and PD (orange; n=14) pre-nivolumab melanoma tumors from CheckMate 064 (Weber et al., 2016). See also Figure S3 and Table S1.
To investigate any bias resulting from inclusion of patients with stable disease, we removed patients with stable disease from the expression analysis and re-classified the cohort into patients with objective responses (complete [CR] or partial response [PR]) and to those with progressive disease (PD). A differential expression analysis of CRMA genes between these groups confirmed the association of CRMA expression with primary resistance (median 99-fold increase, range 37–196; maximum p-value = 0.073; Figure S3F), leading us to ask whether there is any prognostic value to the signature. We then demonstrated that CRMA expression did not discriminate overall survival in the untreated TCGA melanoma cohort (Figure 3D). These findings support the predictive, rather than prognostic, nature of the CRMA signature for CTLA-4 blockade. Taken together, these data suggest CRMA expression as a transcriptomic determinant of clinical outcome to CTLA-4 blockade.
Because the CTLA-4 and PD-1 pathways drive markedly different immunobiologic processes, we hypothesized that transcriptional signatures of clinical outcomes to their antagonists would be unique to each pathway. Indeed, when we interrogated pre-treatment CRMA expression in a recently reported set of transcriptomic data from pre-PD-1 blockade treated melanomas, we found none of these genes to associate with clinical outcome to PD-1 blockade, suggesting the specificity of the CRMA signature to ipilimumab outcome (Figure 3E) (Hugo et al., 2016). We confirmed similar results in an analysis of transcriptome data from an independent nivolumab-treated cohort in a parallel arm of the CheckMate 064 trial (Table S1; Figure 3F; see STAR Methods for details), further supporting the notion that the CRMA signature is predictive for CTLA-4 blockade rather than prognostic of aggressive natural history. Collectively, these results are consistent with the notion that the CTLA-4 and PD-1 pathways occupy biologically and clinically distinct niches.
MAGE-A protein is associated with decreased expression of regulators of autophagy in melanoma
Although MAGE proteins have often been studied as immunotherapeutic targets bound to HLA molecules on the cell surface (Van Der Bruggen et al., 2002), recent studies have attributed them with key oncogenic capacities. Critical to the oncogenic functions of MAGEs may be their ability to bind to and potentiate the activity of various E3 ubiquitin ligases (Lee and Potts, 2017). MAGEA2, MAGEA3, and MAGEA6 all share specific binding to the TRIM28 ubiquitin ligase (Doyle et al., 2010). Extensive, in vitro mechanistic characterization of diverse cancer cell lines has revealed autophagy to be a prime target of the MAGE-TRIM28 complex (Pineda et al., 2015). Several other studies have demonstrated the potential for autophagy to induce potent immune responses by cytokine-mediated priming of antigen-specific IFN-γ producing T cells (Ghiringhelli et al., 2009) and stimulation of immunogenic cell death (Michaud et al., 2011). A role for MAGE-A genes in autophagy regulation in vivo in human tumors, however, has not been previously demonstrated.
We hypothesized that autophagy would be suppressed in human melanomas with elevated MAGE-A protein levels to mediate MAGE-A-driven primary resistance to CTLA-4 blockade, and hence sought to investigate whether CRMA expression was negatively associated with markers of autophagy. To explore this hypothesis, we first evaluated the potential targets of the MAGE-TRIM28 complex by examining the list of proteins negatively correlated with CRMA proteins in human melanomas from The Cancer Protein Atlas (TCPA) (see STAR Methods for details). In this unbiased analysis, the only significantly downregulated protein in tumors with high expression of the CRMA locus was 14-3-3 protein zeta (14-3-3ζ), a known activator of autophagy that acts through the AMPK pathway (Weerasekara et al., 2014) (Figure 4A, t-test, p = 0.0002, FDR=0.05).
Figure 4. MAGE-A proteins may degrade regulators of autophagy.
(A) 14-3-3ζ is downregulated in melanoma tumors with high CRMA expression. (B) In vitro screen for MAGE-TRIM28 ubiquitination substrates identifies HMGB1 (p=0.04). AMPKα1 was used as a positive control (Pineda et al., 2015). (C) Immunofluorescence staining (IF) for MAGE-A and HMGB1 in a melanoma tissue microarray (TMA) shows a negative association between the two proteins in individual cells. (D) Examples from the TMA of mutually exclusive expression of MAGE-A and HMGB1 in cells from the same tumor. Magnification x200. (E) A higher proportion of MAGE− tumors are positive for LC3B, a marker of autophagy, by IHC staining compared to MAGE+ tumors in the melanoma TMA (p=0.004) (top). Examples of LC3B+ and LC3B− tumors, magnification x400 (bottom). (F) Immunofluorescence staining for MAGE-A and HMGB1 shows mutual exclusion in five patient samples from the discovery cohort in addition to a human xenograft melanoma. Magnification x400. (G) MAGE-TRIM28 complex drives primary resistance to CTLA-4 blockade by degradation of regulators of autophagy. See also Figure S3.
We also considered the results of a recent in vitro ubiquitination screen of >9,000 recombinant proteins to identify protein targets of the MAGE-TRIM28 complex (Pineda et al., 2015). In addition to known targets such as AMPK, high-mobility group box 1 (HMGB1), a protein with well-described roles in both autophagy and immunogenic cell death that is required for dendritic cell-mediated priming of an adaptive immune response (Apetoh et al., 2007; Tang et al., 2010) was identified as a lead target (Figure 4B). To investigate whether HMGB1 was a potential target of the MAGE-TRIM28 complex in melanoma tumors in vivo, we enumerated on a cell-by-cell basis the relative expression of HMGB1 and MAGE-A in tumor sections of a tissue microarray (TMA) comprising 100 samples (9 benign nevi tumors, 91 primary and metastatic melanomas) through immunofluorescence (IF) staining. We found that the fraction of HMGB1 positive cells was comparable in MAGE-A− cells from both benign nevi and malignant tumors but was significantly reduced in MAGE-A+ cells (26% and 31% vs 8%, Chi-square test p < 2.2×10−16, Figure 4C). We also observed mutual exclusivity of HMGB1 and MAGE-A proteins within the same tumor (Figure 4D).
In order to directly explore the association of MAGE-A protein levels and autophagy, we used an available subset of melanoma samples on the TMA (n=58) for staining with microtubule-associated protein 1A/1B-light chain 3 (LC3A/B), a well-established marker of autophagosomal membranes (Schlafli et al., 2015). MAGE-A− tumors were commonly LC3B-expressing (71%, 34 of 48) compared to MAGE-A+ tumors (20%, 2 of 10, Fisher’s test, p=0.004), indicating an in vivo attenuation of autophagy in samples with high MAGE-A levels (Figure 4E). Finally, IF staining of the clinical samples from the discovery cohort for expression of HMGB1 and MAGE-A proteins revealed ubiquitous expression of HMGB1 protein in 3 of 3 MAGE-A negative tumors and absent HMGB1 expression in 2 of 2 MAGE-A positive tumors and the MAGE-A+ A375 human xenograft (Figure 4F). Taken together, these results suggest that the MAGE-A proteins are associated with suppression of autophagy in vivo in melanoma, potentially disrupting the development of optimal anticancer immunity and driving primary resistance to CTLA-4 blockade (Figure 4G).
Discussion
MAGE family members were first identified as targets of anti-tumor T cells in melanoma, and their restricted expression in immune-privileged gonadal tissues and various tumor types highlighted them as immunogenic targets (Coulie et al., 2014; De Plaen et al., 1994; Simpson et al., 2005; van der Bruggen et al., 1991). Therefore, our findings of a specific subcluster of MAGE-A genes overexpressed in melanomas resistant to CTLA-4 blockade were unexpected. However, clinical efforts to immunotherapeutically target these proteins have yielded mixed results, suggesting their in vivo immunogenicity should not be assumed (Vansteenkiste et al., 2016). Indeed, many groups have demonstrated the association of CGAs and especially the MAGE family with poor prognostic features in melanoma such as ulceration, thickness, metastases and progression in contrast to the positive prognosis afforded by immune infiltration (Azimi et al., 2012; Barrow et al., 2006; Roeder et al., 2005).
One possible explanation for our findings is that reduced CRMA expression in responding tumors is a manifestation of effective anti-MAGE-A immune activity. Responding melanoma samples are characterized by immune infiltrates that may have already selected against tumor cells expressing high levels of CRMA genes. However, we observed that other CGAs previously demonstrated to elicit cellular and humoral responses, such as NY-ESO-1, and various differentiation antigens, showed no evidence of selection in our analysis (Figure S3A). To our knowledge, this particular MAGE-A subfamily has not been shown to provoke stronger immune responses than other cancer-germline or melanoma-associated antigens; further investigation of in situ immune responses should be pursued to rule out this possibility.
An alternative explanation is that these particular CRMA genes induce immune resistance. Recently described cell-intrinsic functions for MAGE-A3/A6 have implicated these proteins in oncogene addiction and the repression of autophagy in vitro (Doyle et al., 2010; Pineda et al., 2015). We also observed a mutually exclusive expression pattern between MAGE-A and the damage-associated molecular pattern protein and alarmin HMGB1. In addition, autophagy also plays a role in upregulation of class I and II presentation through non-canonical pathways (Bronietzki et al., 2015; Tey and Khanna, 2012). Consistent with our hypothesis that MAGE-TRIM28 driven dysregulation of autophagy contributes to primary resistance to anti-CTLA-4 therapy, we observed a strong, negative association in vivo between MAGE-A protein and LC3B, a well-established marker for autophagy (Adams et al., 2016), in melanoma patient samples. We also observed a mutually exclusive expression pattern between MAGE-A and the damage-associated molecular pattern protein HMGB1, which has well-known roles in mediating the highly related processes of cellular autophagy and immunogenic cell death (Apetoh et al., 2007; Tang et al., 2010). Nevertheless, further mechanistic studies are needed to carefully dissect the role of CRMA gene-mediated suppression of autophagy in conferring resistance to CTLA-4 blockade. These findings raise the notion of synergistic potency that may emerge from combining autophagy activation with CTLA-4 blockade and should explored for clinical therapeutic development. Indeed, evidence from preclinical models suggests that activation of cancer cell autophagy enhances responses to immunogenic therapies (Di Biase et al., 2016; Pietrocola et al., 2016).
The possibility that the CRMA genes mediate resistance uniquely to CTLA-4, and not PD-1, blockade through impairment of immune priming may be initially puzzling because immune effector function is usually thought to be contingent on previous T cell priming. However, other factors could explain why CRMA expression is a marker of resistance for anti-CTLA-4 but not anti-PD-1 therapy. Given the importance of autophagy for T cell priming, which is restrained by the CTLA-4 pathway, autophagy suppression by the MAGE-TRIM28 complex could directly explain the relevance of the MAGE-A signature to anti-CTLA-4 therapy. However, unlike the restricted expression of CTLA-4 to the T cell compartment, the promiscuous expression of PD-1 in various cell types contributes to diverse and heterogeneous mechanisms underlying responses to PD-1 pathway blockade – which operate through both cell-autonomous. (i.e. melanoma cells expressing the PD-1 receptor and driven by intrinsic PD-1 pathway signaling (Kleffel et al., 2015)) and non-autonomous (e.g. reversal of T cell exhaustion (Barber et al., 2006)) processes. Furthermore, tumor biopsies (such as used in this study) provide only a snapshot in time; if MAGE-A gene expression has occurred later in tumor development after an anti-cancer effector immune response has already developed, then a PD-1 inhibitor may be able to exploit a pre-existing reservoir of intratumoral lymphocytes whereas a CTLA-4 inhibitor is still restrained by an upstream defect in priming. The classical assumption that effector immunity generally follows intact priming has been challenged by the results of the CheckMate 064 trial which found that patients treated with CTLA-4 blockade (priming manipulation) after PD-1 blockade (effector manipulation) had a significantly improved overall survival compared to the reverse sequence (Weber et al., 2016).
Although we relaxed our statistical stringency because of our small discovery cohort, we could validate our finding of CRMA gene upregulation in primary resistance to CTLA-4 blockade through confirmation in a prospective, independent cohort and technical verification by qPCR and immunohistochemistry. Because both CTLA-4 blockade and cancer vaccines impact immune priming and memory formation, our findings may also explain the long history of unsuccessful cancer vaccination efforts targeting MAGE-A3 and MAGE-A6 (Palucka and Banchereau, 2014; Pedicord et al., 2011; Saiag et al., 2016; Vansteenkiste et al., 2016). Our results further suggest that mechanisms of response and resistance to immune priming (e.g. CTLA-4 blockade) may differ substantially from those relevant to clinical manipulation of effector immunity (e.g. PD-1/PD-L1 blockade). As immunotherapeutic combinations are increasingly evaluated, understanding these mechanisms will be important for precisely pairing patients with appropriate combinations to avoid toxicity and ensure efficacy. Nevertheless, our findings should be investigated in larger, prospective cohorts to evaluate these signatures as potential biomarkers of outcome, and studied in preclinical models as potential therapeutic targets to sensitize to or combine with CTLA-4 blockade.
STAR Methods
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Catherine J. Wu (cwu@partners.org).
For the validation cohorts from the CheckMate 064 trial, we were given permission from Bristol-Myers Squibb (BMS) only to interrogate CRMA gene expression. Expression data for the CRMA genes will be provided upon request. We understand that the full gene expression data from this study will be published as part of a study that is currently under review.
Experimental Model and Subject Details
Human subjects
For data from the discovery cohort, all patients were consented to an Institutional Review Board protocol that allows research molecular characterization of tumor and germline samples (University Hospital Essen 12-4961-BO) (Van Allen et al., 2015).
For data from the CheckMate 064 study, all patients provided written informed consent, and the protocol, amendments, and patient informed consent were approved by the institutional review board (IRB) or independent ethics committee (IEC) of the participating center before initiation of the study at the site (Weber et al., 2016).
For both cohorts, gender identity and age for all human subjects are provided in Table S1.
Animals
RAG2−/− mice were obtained from the Jackson Laboratory, and bred and maintained at Harvard Medical School. For transplantation of A375 melanoma cells, female mice aged 4–6 weeks were injected subcutaneously with 2 × 106 A375 melanoma cells. Subcutaneous tumors were harvested, fixed with 10% formalin and used for immunofluorescent staining. Mice were maintained and experiments performed in accordance with IACUC approved experimental protocols.
Cell lines
A375 melanoma cell line (human; sex: female; Cat# CRL-1619, RRID:CVCL_0132) was cultured according to the manufacturer’s recommendations. Briefly, cells were grown in ATCC-formulated Dulbecco’s Modified Eagle’s Medium, Catalog No. 30-2002 supplemented with fetal bovine serum to a final concentration of 10% and pencillin/streptomycin to a final concentration of 1%. For cell collection for murine injection, culture media was removed, and cells were treated with Trypsin-0.53 mM EDTA solution for 5 minutes at 37 degrees to facilitate detachment after which time cells were spun down and resuspended in phosphate-buffered saline for injection.
Method Details
Quantitative real-time RT-PCR
We validated expression of target genes in the discovery cohort (Van Allen et al., 2015) from RNA that was extracted for RNA-sequencing. We used the following TaqMan gene expression assays (Applied Biosystems, Foster City, CA): MAGE-A3 (Hs00366532_m1), MAGE-A6 (Hs04190523_gH), MAGE-A2 (Hs01032164_g1), MAGE-A12 (Hs00855175_s1), CSAG2 (Hs01680099_gH), CSAG1 (Hs01564427_g1), CSAG3 (Hs00383358_g1), HPRT1 (Hs02800695_m1), PGK1 (Hs00943178_g1), and GAPDH (Hs02758991_g1). We performed cDNA amplification using the TaqMan Gene Expression Master Mix (Applied Biosystems) on an Applied Biosystems 7500HT Fast real-time polymerase chain reaction (PCR) System (10-minute enzyme activation and 40 cycles of 15 s at 95°C, 1 minute at 60°C). We measured samples in duplicate; “undetermined” values were assigned a cycle threshold (Ct) of 40. HPRT1, GAPDH, and PGK1 were used as housekeeping genes to calculate relative expression values according to the delta-Ct method.
Amplicon methylation analysis
We bisulfite-treated (EZ DNA Methylation-Gold™ Kit, Zymo Research) genomic DNA samples of male patients and amplified individual amplicons via PCR (using TaKaRa EpiTaq™ HS, Clonetech) and the following program: one cycle at 98°C for 10 sec followed by 40 cycles at 98°C for 10 s, 55°C for 30 s, and 72°C for 30 s. The following primer pairs were used: MAGEA3, MAGEA6 and MAGEA12 gene body, Forward Primer: GATTGTGTTTTTGAGGAGAAAATTT, Reverse Primer: CTCCCACTAACCCTAACTACAACTC. MAGEA3 and MAGEA6 gene promoter, Forward Primer: AATTTTAGGATTTTGAGGGATGAT, Reverse Primer: AAACCCTCTATCTAAAATAAAACCC. PCR products were subcloned (One Shot® TOP10 Chemically Competent E. coli, NEB) and individual colonies were sequenced for subsequent methylation analysis. For the local regression analysis (R package ‘msir’) the span smoothing parameter for loess was set to 0.4.
Tissue Microarray (TMA)
A total of 100 cases of melanocytic lesions including benign nevi (n=18), primary cutaneous (n=54) and metastatic (n=28) melanomas were selected from a tissue microarray previously constructed from the Archives of Pathology Department, Brigham and Women’s Hospital of Harvard Medical School. The hematoxylin and eosin stained sections, prior diagnoses, and reported prognostic attributes were independently reviewed and confirmed by board-certified dermatopathologist (C.G.L.). The study protocol was approved by the Institutional Review Board at Brigham and Women’s Hospital (Boston, MA). A subset of these comprising 58 melanomas (primary cutaneous, n=36; metastatic, n=22) was also available for LC3B staining.
Staining
All specimens were evaluated by conventional histopathology. Antibodies used for IHC and IF included mouse anti-MAGE antibody (6C1; Santa Cruz Biotechnology, San Diego, CA USA) and rabbit anti-HMGB1 antibody (ab18256; Abcam, Cambridge, MA). Immunohistochemistry was performed with pressure cooker heat-induced epitope retrieval on 4-mm-thick sections prepared from formalin-fixed, paraffin-embedded tissues. In addition to detection of biomarker antibodies by use of chromogen vector NovaRed peroxidase substrate (Vector laboratory, Burlingame, CA, USA), selected samples were evaluated by a dual labeling approach by combining NovaRed with a blue chromogen vector Blue AP substrate (Vector laboratory). Positive and negative tissue controls and isotype-specific irrelevant antibody controls were used to ensure specificity. Consistent with other reports of IHC for MAGE-A protein, nuclear and/or cytoplasmic staining was interpreted as a positive staining pattern; staining in any cancer cells, irrespective of percentage of positive cells or intensity, was regarded as positive.
Immunohistochemistry for LC3B (Novus Biologicals, Zug, Switzerland, rabbit polyclonal, #NB600-1384, 1:4’000, tris buffer, 95°C, 30 minutes) was performed using an automated immunostainer Leica Bond RX (Leica Biosystems, Heerbrugg, Switzerland) and the appropriate positive and negative controls as described before (Schlafli et al., 2015). Visualization was done with the Bond Polymer Refine Detection kit (Leica Biosystems, Muttenz, Switzerland, DS9800) following the manufacturer’s instructions. Staining results were evaluated by an experienced pathologist (RL) as follows: LC3B dot-like immunohistochemical staining was scored from 0 to 3: score 0 - no dots visible or barely dots visible in < 5% of the tumor cells, score 1 - detectable dots in 5–25% of the tumor cells, score 2 - detectable dots in 25–75% of the tumor cells, score 3 - dots visible in > 75% of the tumor cells. Score 1–3 were considered positive for LC3B staining.
Dual-labeling immunofluorescence was performed to complement immunohistochemistry as a means of two-channel identification of epitopes co-expressed in similar or overlapping sub-cellular locations. Briefly, 4-mm-thick paraffin sections were incubated with 1:100 mouse anti-MAGE antibody + 1:1000 rabbit anti-HMGB1 antibody at 4°C overnight and then incubated with 1:2000 Alexa Fluor 594-conjugated anti-mouse IgG and Alexa Fluor 488-conjugated anti-rabbit IgG (Invitrogen,) at room temperature for 1 hour. The sections were cover slipped with ProLong Gold anti-fade with DAPI (Invitrogen). Sections were analyzed with a BX51/BX52 microscope (Olympus America, Melville, NY, USA), and images were captured using the CytoVision 3.6 software (Applied Imaging, San Jose, CA, USA). Single label immunofluorescence was also performed using isotype-specific irrelevant primary antibodies and with switching of the secondary antibodies to ensure specificity and exclude cross reactivity.
In vitro ubiquitination screen
Briefly, ProtoArray containing > 9,000 GST-tagged recombinant proteins purified from SF9 insect cells was purchased from Invitrogen and previously performed in (Pineda et al 2015).
Quantification and Statistical Analysis
Study Design
We analyzed a previously reported RNA-seq dataset of pre-therapy samples collected from a study cohort of 40 melanoma patients treated with ipilimumab (Van Allen et al., 2015). In this study, RNA and genomic DNA were extracted from formalin-fixed, paraffin-embedded (FFPE) tumor blocks, and Illumina’s TruSeq Stranded Total RNA Sample Prep Kit was used to generate RNA-seq libraries. Patient classification was maintained from the original report (Table S1). The “clinical benefit” (CB) group (n=13) was defined as patients who achieved complete or partial response by RECIST criteria, or stable disease by RECIST criteria with overall survival greater than one year. The “no benefit” (NB) group (n=22) was defined as patients who had progressive disease by RECIST criteria or stable disease with overall survival less than 1 year. A third group of five patients was described with early progression on ipilimumab (progression-free survival <6 months) but overall survival exceeding 2 years (“NB with long-term survival”). To identify genes associated with clinical benefit and no benefit, we performed the differential expression analysis between the CB and NB groups only. The association of CRMA expression with survival outcome was evaluated in the entire cohort. Genes were identified as differentially expressed when their median expression differed by more than two-fold with a nominal one-sided p-value ≤ 0.05 (Wilcoxon test). We obtained sectioned slides from the tumors of 55 patients (15 CB, 33 NB, and 7 NB with long-term survival) from the original cohort used in Van Allen et al Science 2015 for immunohistochemistry staining. Of these, 28 patients (9 CB, 14 NB, and 5 NB with long term survival) had RNA-seq data (i.e. from the discovery cohort).
An independent, validation cohort comprised 41 patients from the CheckMate 064 trial (Weber et al., 2016) treated with ipilimumab followed by nivolumab (Table S1). The trial also studied in parallel a cohort comprising patients treated with the reverse sequence of nivolumab followed by ipilimumab. Overall survival could not be assessed in this crossover design. Response assessments, collected at week 13 before the planned switch, were used to classify patients into either no progressive disease (“No PD”; comprising stable disease, complete response and partial response, n=12) or progressive disease (“PD”; n=29) from each arm. Tumor samples were cryopreserved in RNALater. RNA-seq libraries were generated using the Stranded TruSeq method, and 75bp paired-end reads for duplexed samples were sequenced per lane (Expression Analysis, Inc; Morrisville, NC). RNA-Seq and associated clinical data were available for the following CRMA genes: MAGEA3, MAGEA2, MAGEA2B, MAGEA12, MAGEA6.
CRMA expression was evaluated in patients from two different anti-PD-1-treated cohorts. Anti-PD-1 cohort 1 comprised 28 pre-anti-PD-1-treated tumors (Hugo et al., 2016); anti-PD-1 cohort 2 comprised 37 pre-treatment tumors from the nivolumab followed by ipilimumab arm of the CheckMate064 trial (Hugo et al., 2016; Weber et al., 2016) (Table S1).
465 melanoma samples from TCGA (Cancer Genome Atlas, 2015) were used to further investigate identified gene expression and methylation signatures.
Processing and analysis of sequencing data
RNA sequencing data from the discovery cohort was aligned to the reference human genome with STAR (Dobin et al., 2013), followed by removal of duplicates and quantification with RSEM (Li and Dewey, 2011). RNA sequencing data from the CheckMate064 trial was first aligned using STAR (Dobin et al., 2013) followed by removal of duplicate reads. Gene level quantification of the reads was performed with the htseq-count tool (Anders et al., 2015).
We also obtained whole exome data for 110 patients from the discovery cohort (including the 40 with transcriptomic data) (Van Allen et al., 2015) and Infinium 450K methylation chip data for 476 samples from TCGA (Cancer Genome Atlas, 2015). Tumor purities were estimated by the ABSOLUTE algorithm (Carter et al., 2012).
TCGA gene expression analysis
We defined a metagene comprising the following CRMA genes: MAGEA2, MAGEA3, MAGEA6, MAGEA12, CSAG1, CSAG2, and CSAG3 (MAGEA2B was not quantified by TCGA). The expression of this metagene was defined as the geometric mean of its components and was computed for each of 465 TCGA melanoma samples. TCGA samples with expression values in the bottom and top quartiles for this metagene were classified into “CRMA-low” (n=117) and “CRMA-high” (n=116) groups respectively. An unbiased gene expression analysis between these two groups was performed using one-sided Wilcoxon tests with a false discovery rate (FDR) threshold of 0.05 and two-fold change threshold.
Differential expression and methylation analyses within TCGA samples were performed using a false discovery rate (Benjamini-Hochberg) of 0.05. Hypergeometric tests were used to evaluate overlap of differentially expressed genes between the clinical and TCGA cohorts. Enrichment of immune gene sets (Angelova et al., 2015) in the discovery cohort were evaluated based on the number of overlapping genes between a given gene set and genes upregulated in the CB or NB groups as assessed by the hypergeometric test. Overlaps of gene lists with pathways in the PANTHER (Protein ANalysis THrough Evolutionary Relationships) database (containing 177 pathways) were evaluated with the overrepresentation test using the Bonferroni correction (Mi et al., 2016).
TCGA methylation analysis
We retrieved Level3 Infinium 450K methylation chip data from TCGA for “CRMA-low” (n=117) and “CRMA-high” (n=116) groups. We performed a probe-level comparison between the two groups using Wilcoxon tests with an FDR of 0.05 for all 485,577 CpG probes. A probe with higher median beta values in one group was considered relatively hypermethylated in that group compared to the other.
The Cancer Protein Atlas analysis
We retrieved Level 4 (L4) normalized protein expression data for 354 proteins for TCGA melanoma samples from The Cancer Protein Atlas. We performed a differential expression analysis between the “CRMA-low” (n=88) and “CRMA-high” (n=86) groups using a two-sided t-test and FDR=5%.
Copy number analysis
We tested the clinical benefit (CB) and no benefit (NB) groups for variations in germline and somatic CNVs in Xq28 locus using GATK 4 target coverage denoising and ACNV pipelines. We collected raw coverage on whole exome Agilent targets for 110 normal and tumor samples and corrected for GC bias. We set aside the 40 samples with RNA-seq data used in this study and used the remaining 70 samples to learn the target coverage bias profile (“panel of normals”). We then denoised and normalized the coverage profile of samples used in this study using the obtained panel of normals. We detected tumor samples with anomalously low signal-to-noise ratio and normal samples with significant contamination and excluded them from the analysis. We calculated empirical distributions of raw copy ratios on all 16 Agilent targets in the Xq28 locus using an agnostic prior distribution and copy ratio likelihoods for each sample. We estimated absolute copy ratios with respect to diploid by performing allelic CNV analysis, detecting copy neutral autosomal intervals, and normalizing the raw copy ratios accordingly. We tested the two groups for germline and somatic copy number variations in the Xq28 locus using the two-sample KS test. We performed the test separately for each target, and for the copy ratio average on all 16 targets in the Xq28 locus. We found the copy ratio distributions in each case via empirical bootstrap.
Survival analysis
The association of CRMA expression with overall survival was evaluated using the Kaplan-Meier method. Patients with long-term survival with early tumor progression (n=5) in the discovery cohort were also included in the analysis (Van Allen et al., 2015). In the discovery cohort, patients with CRMA expression values above the median were considered “high” and below the median were considered “low”. The effect of CRMA expression on overall survival adjusting for age, gender, number of pre-therapies, M-stage, LDH and neoantigen load was assessed using the Cox proportional hazards model (R package ‘coxph’). For testing the association of MAGE-A protein expression with overall survival, all 55 samples with available IHC data were included.
Supplementary Material
(A) Heatmap showing relative expression of CRMA genes for CB and NB patients in the discovery cohort with annotations for gender, purity and RECIST response. (B) RT-qPCR validation of CRMA genes using three different housekeeping genes: HPRT1 (top), GAPDH (middle) and PGK1 (bottom). P-values using the Wilcoxon test and median fold changes are indicated. (C) Gender does not affect outcome after ipilimumab. (D) DTIC/temozolamide treatment history does not affect outcome after ipilimumab. Patients were included into “DTIC” group if DTIC or temozolamide were used as treatment at any timepoint prior to ipilimumab. (E) Heatmap showing relative expression of CRMA genes in the validation set (CheckMate 064) with annotations for gender and RECIST response. (F) Box plots depicting the individual fold changes for each MAGEA gene within the CRMA locus are shown for patients with no progressive (“no PD”) or progressive disease (“PD”) in the discovery cohort; p-values using the Wilcoxon test.
(A) Copy number analysis of CRMA region in CB/NB patients. Neither the locus average of copy ratios, nor copy ratios on individual targets showed a statistically significant germline or somatic variation between the two groups at 5% level. (B) Bisulfite PCR of unique methylation sites within the gene bodies of MAGEA3/A6/A12 genes highlights a slight to moderate decrease in methylation in NB patients (n=4, orange) vs. CB patients (n=4, blue). The position of 3 PCR amplicons are highlighted, and the plots highlight the mean methylation for each CpG within the amplicon region. (C) Chromosomal locations of 65,467 hypomethylated (top) and 47 hypermethylated (bottom) probes in CRMA-high TCGA melanoma samples.
(A) Heatmap showing relative gene expression of NB-enriched biological categories (see Table S2) along with lack of enrichment of NY-ESO-1 and melanoma differentiation antigens. Annotations of gender, purity and RECIST response included. (B) Percentage of differentially expressed genes related to immune response, in both NB (3.5%) and CB (56%) tumors. (C) Immune gene sets enriched in CB and NB tumors showing p-value of enrichment (signed according to whether the gene set was enriched in NB (+) or CB (−) tumors). Dashed line represents p=0.05. (D, E) CRMA gene and MAGE-A protein expression correlate with progression-free survival after CTLA-4 blockade. Progression-free survival analyses comparing patients from the discovery cohort classified by (D) CRMA gene expression or (E) MAGE-A protein expression. (F) Box plots depicting the individual RNA-seq expression value for each MAGEA and CSAG gene within this locus in the discovery cohort stratified by CR/PR and PD status (Van Allen et al., 2015).
Table S1. Clinical characteristics of discovery and validation cohorts, Related to Figures 1 and 3.
Table S3. Genes enriched in TCGA CRMA-high/low expression samples, Related to Figure 2.
Table S4. Overlapping genes between No Benefit/Clinical Benefit groups and TCGA CRMA-high/low subsets, Related to Figure 2.
Highlights.
Increased expression of a MAGE-A subcluster predicts resistance to CTLA4 blockade
This MAGE-A subcluster marks a distinct, epigenetically-defined subset of melanomas
This gene signature is specific to resistance to CTLA4, but not PD1, blockade
Autophagy is implicated in clinical resistance to CTLA4 blockade
Acknowledgments
C.J.W. is a Scholar of the Leukemia and Lymphoma Society and acknowledges support from the Blavatnik Family Foundation, National Heart, Lung, and Blood Institute (NHLBI) (1RO1HL103532-01) and National Cancer Institute (NCI) (1R01CA155010-01A1). S.A.S. acknowledges support by the NCI (R50 RCA211482A). P.B. acknowledges support by the Damon Runyon Physician-Scientist Training Award and the Conquer Cancer Foundation Young Investigator Award. The work was funded in part by the New York Stem Cell Foundation, NIH (R01DA036898) and DoD (BC134001P1). A.M. is a New York Stem Cell Foundation Robertson Investigator. D.N. is supported by Dana-Farber/Harvard Cancer Center Support Grant 5P30 CA006516. D.M. is supported by the Howard Hughes Medical Institute. This work has made extensive use of data generated by TCGA, a project of the National Cancer Institute and National Human Genome Research Institute. We thank Patrick Ott for stimulating scientific discussions; Zachary Cartun for excellent technical assistance; Samuel Lee and David Benjamin for method development for copy number analysis and Christine Horak, Donald Jackson, and Han Chang for facilitating access to RNA sequencing data from the CheckMate 064 trial.
Footnotes
Author Contributions
S.A.S. and P.B. conceived the study, designed the experiments, interpreted the results and wrote the manuscript. D.G., E.M.V.A., D.M., P.R.P. contributed data and performed analysis. M.B. and K.C. performed analysis. C.G., Q.Z., P.C.L., Z.J.C., D.B.K., A.M., A.Meissner, A.G., G.F.M., C.G.L., C.B., R. L., Y.H. performed experiments and interpreted results. B.S., D.S., L.A.W, E.M.V.A., A.S., T.M., and J.D.W. provided tissue reagents. F.S.H. and J.S.W. provided validation cohort data and interpreted results. D.N. and N.H. provided feedback on experimental design and interpretation, and edited the manuscript. C.J.W. edited the manuscript and supervised the study. All authors have read the paper.
Declaration of interests
C.J.W. and N.H. are co-founders and scientific advisory board members of Neon Therapeutics. D.S. and B.S. are on the advisory board or have received honoraria from Novartis, Roche, Bristol-Myers Squibb (BMS) and MSD Sharp & Dohme, research funding from Bristol-Myers Squibb and MSD Sharp & Dohme and travel support from Novartis, Roche, BMS and AMGEN. S.A.S. and P.B. report equity in 152 Therapeutics. S.A.S., P.B., and C.J.W. have a patent application filed based on the discoveries presented. F.S.H. reports consultant/advisory fees from Merck, Novartis, Genentech, Amgen, and EMD Serono; and research support from BMS.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams O, Dislich B, Berezowska S, Schlafli AM, Seiler CA, Kroll D, Tschan MP, Langer R. Prognostic relevance of autophagy markers LC3B and p62 in esophageal adenocarcinomas. Oncotarget. 2016;7:39241–39255. doi: 10.18632/oncotarget.9649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics (Oxford, England) 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelova M, Charoentong P, Hackl H, Fischer ML, Snajder R, Krogsdam AM, Waldner MJ, Bindea G, Mlecnik B, Galon J, et al. Characterization of the immunophenotypes and antigenomes of colorectal cancers reveals distinct tumor escape mechanisms and novel targets for immunotherapy. Genome biology. 2015;16:64. doi: 10.1186/s13059-015-0620-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apetoh L, Ghiringhelli F, Tesniere A, Obeid M, Ortiz C, Criollo A, Mignot G, Maiuri MC, Ullrich E, Saulnier P, et al. Toll-like receptor 4-dependent contribution of the immune system to anticancer chemotherapy and radiotherapy. Nat Med. 2007;13:1050–1059. doi: 10.1038/nm1622. [DOI] [PubMed] [Google Scholar]
- Azimi F, Scolyer RA, Rumcheva P, Moncrieff M, Murali R, McCarthy SW, Saw RP, Thompson JF. Tumor-infiltrating lymphocyte grade is an independent predictor of sentinel lymph node status and survival in patients with cutaneous melanoma. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2012;30:2678–2683. doi: 10.1200/JCO.2011.37.8539. [DOI] [PubMed] [Google Scholar]
- Barber DL, Wherry EJ, Masopust D, Zhu B, Allison JP, Sharpe AH, Freeman GJ, Ahmed R. Restoring function in exhausted CD8 T cells during chronic viral infection. Nature. 2006;439:682–687. doi: 10.1038/nature04444. [DOI] [PubMed] [Google Scholar]
- Barrow C, Browning J, MacGregor D, Davis ID, Sturrock S, Jungbluth AA, Cebon J. Tumor antigen expression in melanoma varies according to antigen and stage. Clin Cancer Res. 2006;12:764–771. doi: 10.1158/1078-0432.CCR-05-1544. [DOI] [PubMed] [Google Scholar]
- Bredenbeck A, Hollstein VM, Trefzer U, Sterry W, Walden P, Losch FO. Coordinated expression of clustered cancer/testis genes encoded in a large inverted repeat DNA structure. Gene. 2008;415:68–73. doi: 10.1016/j.gene.2008.02.019. [DOI] [PubMed] [Google Scholar]
- Bronietzki AW, Schuster M, Schmitz I. Autophagy in T-cell development, activation and differentiation. Immunology and cell biology. 2015;93:25–34. doi: 10.1038/icb.2014.81. [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas N. Genomic Classification of Cutaneous Melanoma. Cell. 2015;161:1681–1696. doi: 10.1016/j.cell.2015.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter SL, Cibulskis K, Helman E, McKenna A, Shen H, Zack T, Laird PW, Onofrio RC, Winckler W, Weir BA, et al. Absolute quantification of somatic DNA alterations in human cancer. Nature biotechnology. 2012;30:413–421. doi: 10.1038/nbt.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulie PG, Van den Eynde BJ, van der Bruggen P, Boon T. Tumour antigens recognized by T lymphocytes: at the core of cancer immunotherapy. Nature reviews Cancer. 2014;14:135–146. doi: 10.1038/nrc3670. [DOI] [PubMed] [Google Scholar]
- De Plaen E, Arden K, Traversari C, Gaforio JJ, Szikora JP, De Smet C, Brasseur F, van der Bruggen P, Lethe B, Lurquin C, et al. Structure, chromosomal localization, and expression of 12 genes of the MAGE family. Immunogenetics. 1994;40:360–369. doi: 10.1007/BF01246677. [DOI] [PubMed] [Google Scholar]
- Di Biase S, Lee C, Brandhorst S, Manes B, Buono R, Cheng CW, Cacciottolo M, Martin-Montalvo A, de Cabo R, Wei M, et al. Fasting-Mimicking Diet Reduces HO-1 to Promote T Cell-Mediated Tumor Cytotoxicity. Cancer Cell. 2016;30:136–146. doi: 10.1016/j.ccell.2016.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics (Oxford, England) 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle JM, Gao J, Wang J, Yang M, Potts PR. MAGE-RING protein complexes comprise a family of E3 ubiquitin ligases. Molecular cell. 2010;39:963–974. doi: 10.1016/j.molcel.2010.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Shi LZ, Zhao H, Chen J, Xiong L, He Q, Chen T, Roszik J, Bernatchez C, Woodman SE, et al. Loss of IFN-gamma Pathway Genes in Tumor Cells as a Mechanism of Resistance to Anti-CTLA-4 Therapy. Cell. 2016;167:397–404e399. doi: 10.1016/j.cell.2016.08.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghiringhelli F, Apetoh L, Tesniere A, Aymeric L, Ma Y, Ortiz C, Vermaelen K, Panaretakis T, Mignot G, Ullrich E, et al. Activation of the NLRP3 inflammasome in dendritic cells induces IL-1beta-dependent adaptive immunity against tumors. Nat Med. 2009;15:1170–1178. doi: 10.1038/nm.2028. [DOI] [PubMed] [Google Scholar]
- Gumireddy K, Li A, Kossenkov AV, Sakurai M, Yan J, Li Y, Xu H, Wang J, Zhang PJ, Zhang L, et al. The mRNA-edited form of GABRA3 suppresses GABRA3-mediated Akt activation and breast cancer metastasis. Nature communications. 2016;7:10715. doi: 10.1038/ncomms10715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodi FS, O’Day SJ, McDermott DF, Weber RW, Sosman JA, Haanen JB, Gonzalez R, Robert C, Schadendorf D, Hassel JC, et al. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med. 2010;363:711–723. doi: 10.1056/NEJMoa1003466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugo W, Zaretsky JM, Sun L, Song C, Moreno BH, Hu-Lieskovan S, Berent-Maoz B, Pang J, Chmielowski B, Cherry G, et al. Genomic and Transcriptomic Features of Response to Anti-PD-1 Therapy in Metastatic Melanoma. Cell. 2016;165:35–44. doi: 10.1016/j.cell.2016.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119:1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleffel S, Posch C, Barthel SR, Mueller H, Schlapbach C, Guenova E, Elco CP, Lee N, Juneja VR, Zhan Q, et al. Melanoma Cell-Intrinsic PD-1 Receptor Functions Promote Tumor Growth. Cell. 2015;162:1242–1256. doi: 10.1016/j.cell.2015.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AK, Potts PR. A comprehensive guide to the MAGE family of ubiquitin ligases. Journal of molecular biology. 2017 doi: 10.1016/j.jmb.2017.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Poudel S, Muruganujan A, Casagrande JT, Thomas PD. PANTHER version 10: expanded protein families and functions, and analysis tools. Nucleic acids research. 2016;44:D336–342. doi: 10.1093/nar/gkv1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaud M, Martins I, Sukkurwala AQ, Adjemian S, Ma Y, Pellegatti P, Shen S, Kepp O, Scoazec M, Mignot G, et al. Autophagy-dependent anticancer immune responses induced by chemotherapeutic agents in mice. Science (New York, NY) 2011;334:1573–1577. doi: 10.1126/science.1208347. [DOI] [PubMed] [Google Scholar]
- Palucka K, Banchereau J. SnapShot: Cancer Vaccines. Cell. 2014;157:516–516e511. doi: 10.1016/j.cell.2014.03.044. [DOI] [PubMed] [Google Scholar]
- Pedicord VA, Montalvo W, Leiner IM, Allison JP. Single dose of anti-CTLA-4 enhances CD8+ T-cell memory formation, function, and maintenance. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:266–271. doi: 10.1073/pnas.1016791108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietrocola F, Pol J, Vacchelli E, Rao S, Enot DP, Baracco EE, Levesque S, Castoldi F, Jacquelot N, Yamazaki T, et al. Caloric Restriction Mimetics Enhance Anticancer Immunosurveillance. Cancer Cell. 2016;30:147–160. doi: 10.1016/j.ccell.2016.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pineda CT, Ramanathan S, Fon Tacer K, Weon JL, Potts MB, Ou YH, White MA, Potts PR. Degradation of AMPK by a cancer-specific ubiquitin ligase. Cell. 2015;160:715–728. doi: 10.1016/j.cell.2015.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu Y, Xu M, Liang Y, Yang K, Guo Y, Yang X, Fu YX. Androgen receptor antagonists compromise T cell response against prostate cancer leading to early tumor relapse. Science translational medicine. 2016;8:333ra347. doi: 10.1126/scitranslmed.aad5659. [DOI] [PubMed] [Google Scholar]
- Riaz N, Havel JJ, Kendall SM, Makarov V, Walsh LA, Desrichard A, Weinhold N, Chan TA. Recurrent SERPINB3 and SERPINB4 mutations in patients who respond to anti-CTLA-4 immunotherapy. Nature genetics. 2016;48:1327–1329. doi: 10.1038/ng.3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder C, Schuler-Thurner B, Berchtold S, Vieth G, Driesch P, Schuler G, Luftl M. MAGE-A3 is a frequent tumor antigen of metastasized melanoma. Archives of dermatological research. 2005;296:314–319. doi: 10.1007/s00403-004-0527-7. [DOI] [PubMed] [Google Scholar]
- Saiag P, Gutzmer R, Ascierto PA, Maio M, Grob JJ, Murawa P, Dreno B, Ross M, Weber J, Hauschild A, et al. Prospective assessment of a gene signature potentially predictive of clinical benefit in metastatic melanoma patients following MAGE-A3 immunotherapeutic (PREDICT) Annals of oncology: official journal of the European Society for Medical Oncology/ESMO. 2016;27:1947–1953. doi: 10.1093/annonc/mdw291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlafli AM, Berezowska S, Adams O, Langer R, Tschan MP. Reliable LC3 and p62 autophagy marker detection in formalin fixed paraffin embedded human tissue by immunohistochemistry. Eur J Histochem. 2015;59:2481. doi: 10.4081/ejh.2015.2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson AJ, Caballero OL, Jungbluth A, Chen YT, Old LJ. Cancer/testis antigens, gametogenesis and cancer. Nature reviews Cancer. 2005;5:615–625. doi: 10.1038/nrc1669. [DOI] [PubMed] [Google Scholar]
- Snyder A, Makarov V, Merghoub T, Yuan J, Zaretsky JM, Desrichard A, Walsh LA, Postow MA, Wong P, Ho TS, et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. The New England journal of medicine. 2014;371:2189–2199. doi: 10.1056/NEJMoa1406498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D, Kang R, Livesey KM, Cheh CW, Farkas A, Loughran P, Hoppe G, Bianchi ME, Tracey KJ, Zeh HJ, 3rd, et al. Endogenous HMGB1 regulates autophagy. J Cell Biol. 2010;190:881–892. doi: 10.1083/jcb.200911078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tey SK, Khanna R. Autophagy mediates transporter associated with antigen processing-independent presentation of viral epitopes through MHC class I pathway. Blood. 2012;120:994–1004. doi: 10.1182/blood-2012-01-402404. [DOI] [PubMed] [Google Scholar]
- Van Allen EM, Miao D, Schilling B, Shukla SA, Blank C, Zimmer L, Sucker A, Hillen U, Foppen MH, Goldinger SM, et al. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science (New York, NY) 2015;350:207–211. doi: 10.1126/science.aad0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Bruggen P, Traversari C, Chomez P, Lurquin C, De Plaen E, Van den Eynde B, Knuth A, Boon T. A gene encoding an antigen recognized by cytolytic T lymphocytes on a human melanoma. Science. 1991;254:1643–1647. doi: 10.1126/science.1840703. [DOI] [PubMed] [Google Scholar]
- Van Der Bruggen P, Zhang Y, Chaux P, Stroobant V, Panichelli C, Schultz ES, Chapiro J, Van Den Eynde BJ, Brasseur F, Boon T. Tumor-specific shared antigenic peptides recognized by human T cells. Immunological reviews. 2002;188:51–64. doi: 10.1034/j.1600-065x.2002.18806.x. [DOI] [PubMed] [Google Scholar]
- Vansteenkiste JF, Cho BC, Vanakesa T, De Pas T, Zielinski M, Kim MS, Jassem J, Yoshimura M, Dahabreh J, Nakayama H, et al. Efficacy of the MAGE-A3 cancer immunotherapeutic as adjuvant therapy in patients with resected MAGE-A3-positive non-small-cell lung cancer (MAGRIT): a randomised, double-blind, placebo-controlled, phase 3 trial. The lancet oncology. 2016;17:822–835. doi: 10.1016/S1470-2045(16)00099-1. [DOI] [PubMed] [Google Scholar]
- Weber JS, Gibney G, Sullivan RJ, Sosman JA, Slingluff CL, Jr, Lawrence DP, Logan TF, Schuchter LM, Nair S, Fecher L, et al. Sequential administration of nivolumab and ipilimumab with a planned switch in patients with advanced melanoma (CheckMate 064): an open-label, randomised, phase 2 trial. Lancet Oncol. 2016;17:943–955. doi: 10.1016/S1470-2045(16)30126-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weerasekara VK, Panek DJ, Broadbent DG, Mortenson JB, Mathis AD, Logan GN, Prince JT, Thomson DM, Thompson JW, Andersen JL. Metabolic-stress-induced rearrangement of the 14-3-3zeta interactome promotes autophagy via a ULK1- and AMPK-regulated 14-3-3zeta interaction with phosphorylated Atg9. Mol Cell Biol. 2014;34:4379–4388. doi: 10.1128/MCB.00740-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolchok JD, Kluger H, Callahan MK, Postow MA, Rizvi NA, Lesokhin AM, Segal NH, Ariyan CE, Gordon RA, Reed K, et al. Nivolumab plus ipilimumab in advanced melanoma. N Engl J Med. 2013;369:122–133. doi: 10.1056/NEJMoa1302369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Adamow M, Ginsberg BA, Rasalan TS, Ritter E, Gallardo HF, Xu Y, Pogoriler E, Terzulli SL, Kuk D, et al. Integrated NY-ESO-1 antibody and CD8+ T-cell responses correlate with clinical benefit in advanced melanoma patients treated with ipilimumab. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:16723–16728. doi: 10.1073/pnas.1110814108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaretsky JM, Garcia-Diaz A, Shin DS, Escuin-Ordinas H, Hugo W, Hu-Lieskovan S, Torrejon DY, Abril-Rodriguez G, Sandoval S, Barthly L, et al. Mutations Associated with Acquired Resistance to PD-1 Blockade in Melanoma. N Engl J Med. 2016;375:819–829. doi: 10.1056/NEJMoa1604958. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Heatmap showing relative expression of CRMA genes for CB and NB patients in the discovery cohort with annotations for gender, purity and RECIST response. (B) RT-qPCR validation of CRMA genes using three different housekeeping genes: HPRT1 (top), GAPDH (middle) and PGK1 (bottom). P-values using the Wilcoxon test and median fold changes are indicated. (C) Gender does not affect outcome after ipilimumab. (D) DTIC/temozolamide treatment history does not affect outcome after ipilimumab. Patients were included into “DTIC” group if DTIC or temozolamide were used as treatment at any timepoint prior to ipilimumab. (E) Heatmap showing relative expression of CRMA genes in the validation set (CheckMate 064) with annotations for gender and RECIST response. (F) Box plots depicting the individual fold changes for each MAGEA gene within the CRMA locus are shown for patients with no progressive (“no PD”) or progressive disease (“PD”) in the discovery cohort; p-values using the Wilcoxon test.
(A) Copy number analysis of CRMA region in CB/NB patients. Neither the locus average of copy ratios, nor copy ratios on individual targets showed a statistically significant germline or somatic variation between the two groups at 5% level. (B) Bisulfite PCR of unique methylation sites within the gene bodies of MAGEA3/A6/A12 genes highlights a slight to moderate decrease in methylation in NB patients (n=4, orange) vs. CB patients (n=4, blue). The position of 3 PCR amplicons are highlighted, and the plots highlight the mean methylation for each CpG within the amplicon region. (C) Chromosomal locations of 65,467 hypomethylated (top) and 47 hypermethylated (bottom) probes in CRMA-high TCGA melanoma samples.
(A) Heatmap showing relative gene expression of NB-enriched biological categories (see Table S2) along with lack of enrichment of NY-ESO-1 and melanoma differentiation antigens. Annotations of gender, purity and RECIST response included. (B) Percentage of differentially expressed genes related to immune response, in both NB (3.5%) and CB (56%) tumors. (C) Immune gene sets enriched in CB and NB tumors showing p-value of enrichment (signed according to whether the gene set was enriched in NB (+) or CB (−) tumors). Dashed line represents p=0.05. (D, E) CRMA gene and MAGE-A protein expression correlate with progression-free survival after CTLA-4 blockade. Progression-free survival analyses comparing patients from the discovery cohort classified by (D) CRMA gene expression or (E) MAGE-A protein expression. (F) Box plots depicting the individual RNA-seq expression value for each MAGEA and CSAG gene within this locus in the discovery cohort stratified by CR/PR and PD status (Van Allen et al., 2015).
Table S1. Clinical characteristics of discovery and validation cohorts, Related to Figures 1 and 3.
Table S3. Genes enriched in TCGA CRMA-high/low expression samples, Related to Figure 2.
Table S4. Overlapping genes between No Benefit/Clinical Benefit groups and TCGA CRMA-high/low subsets, Related to Figure 2.