Abstract
DNA methylation is an essential epigenetic mark crucial for normal mammalian development. This modification controls the expression of a unique class of genes, designated as imprinted, which are expressed monoallelically and in a parent-of-origin-specific manner. Proper parental allele-specific DNA methylation at imprinting control regions (ICRs) is necessary for appropriate imprinting. Processes that deregulate DNA methylation of imprinted loci cause disease in humans. DNA methylation patterns dramatically change during mammalian development: first, the majority of the genome, with the exception of ICRs, is demethylated after fertilization, and subsequently undergoes genome-wide de novo DNA methylation. Secondly, after primordial germ cells are specified in the embryo, another wave of demethylation occurs, with ICR demethylation occurring late in the process. Lastly, ICRs reacquire DNA methylation imprints in developing germ cells. We describe the past discoveries and current literature defining these crucial dynamics in relation to imprinted genes and the rest of the genome.
Keywords: genomic imprinting, imprinted genes, reprogramming, DNA methylation, epigenetics, primordial germ cells, oocyte, sperm, zygote
This review summarizes the changes in DNA methylation at imprinting control regions, which regulate monoallelic imprinted gene expression, from fertilization, embryogenesis, and PGC development, which are essential for proper development.
Introduction
All cells within an organism contain the same genetic information, yet the phenotypes of these cells vary drastically across tissues and stages of development. Epigenetic control of gene expression allows for the distinct usage and expression of genes, ultimately giving rise to the diverse set of cells and functions within the body. Epigenetics broadly encompasses heritable mechanisms that lead to changes in gene expression without altering the underlying DNA sequence, such as nucleosome positioning and composition, histone post-translational modifications, noncoding RNAs, and DNA methylation. DNA methylation, one of the best studied epigenetic modifications, involves the covalent addition of a methyl group to the carbon 5 position on cytosine (5mC), typically within the context of a cytosine guanine dinucleotide, connected by a phosphodiester bond (CpG). This epigenetic mark is involved in a variety of functions in the mammalian genome, including X chromosome inactivation, gene silencing, genomic stability, cellular identity, and genomic imprinting. While DNA methylation is a stable and heritable epigenetic mark, this modification is also highly dynamic, particularly during mammalian development. The global changes in DNA methylation after fertilization and in primordial germ cells (PGCs) are central to embryonic epigenetic reprogramming.
In this review, we summarize the findings from the available literature focusing on epigenetic reprogramming in mammals with a particular emphasis on imprinted genes. As existing methodologies have become more sensitive, and new techniques developed, we have attained a more complete understanding of the processes and significance of epigenetic reprogramming. Here, we present the genes and proteins involved in regulating imprinted DNA methylation (see Tables 1 and 2 for examples of a paternally methylated locus, H19, and for a maternally methylated locus, Igf2r), the kinetics and levels of DNA methylation changes, as well as how different regions of the genome are affected and targeted during these processes. Lastly, we discuss remaining questions regarding reprogramming of imprints.
Table 1.
Genetic models of imprinted expression and DNA methylation regulators at the H19 locus in mice.
| Genotype | Tissue | Allelic expression | H19 ICR DNA methylation | Reference |
|---|---|---|---|---|
| Dnmt1n/n(hypomorph) | E10.5 embryos | H19 biallelic; Igf2 repressed | ND | [18] |
| Dnmt1s/s (replication foci targeting domain mutation) | E9.5 concepti | H19 biallelic | ND | [109] |
| Dnmt1n/n | E9.5 concepti | H19 biallelic; Igf2 repressed | ND | [110] |
| Dnmt1+/n | E9.5 concepti | *H19 biallelic | ND | |
| Dnmt1+/c(c = null) | E9.5 concepti | H 19 = | ND | |
| Dnmt1mat–/+ | E3.5 blastocysts | ND | *H19 ↓ | [81] |
| Dnmt1mat–/− | E3.5 blastocysts | ND | H19 ↓ | |
| Uhrf1−/− | ESCs | ND | H19 ↓ | [111] |
| Uhrf1−/− | E9.5 embryos | H19 biallelic; Igf2 repressed | ND | |
| Dnmt3amat–/−; Dnmt3b+/− | E9.5 embryos | ND | H 19 = | [112] |
| Dnmt3a2lox/1lox, TNAP–Cre x WT male (maternal deletion only) | E10.5 embryos | ND | H 19 = | [113] |
| Dnmt3a2lox/1lox, TNAP–Cre–/– males | Spermatogonia (P11 Testis) | ND | H19 ↓ | |
| Paternal germline deletion Dnmt3b | Pups# | ND | H 19 = | |
| Maternal germline deletion Dnmt3b | Pups# | ND | H 19 = | |
| Dnmt3a−/−; Dnmt3b−/− DKO (Maternal-zygotic null) | E9.5 embryos | H 19 = | H 19 = | [81] |
| Dnmt3l−/− | Spermatogonia (P11 Testis) | ND | H19 ↓ | [113] |
| Dnmt3l−/− | ESCs | ND | Igf2 DMR2 ↓ | [112] |
| Dnmt3lmat–/− | E9.5 embryos | ND | H 19 = | |
| Dnmt3lmat–/+ | Embryos# | H 19 = ; Igf2 = | H 19 = | [114] |
| Dppa3mat–/+ | PN5 zygotes | ND | H19 ↓ | [71] |
| Dppa3−/− | E12.5 PGCs | ND | H 19 = | [115] |
| Zfp57−/− (zygotic only) | E11.5-E13.5 embryos | ND | H 19 = | [76] |
| Zfp57mat–/− (maternal-zygotic) | E11.5-E13.5 embryos | ND | H 19 = | |
| Zfp57+/− | Sperm | ND | H 19 = | |
| Zfp57−/− | Sperm | ND | H 19 = | |
| Trim28chatwo/chatwo (zygotic hypomorph) | E8.5 embryos | *H19 biallelic; Igf2 repressed | *H19 ↓ | [82] |
| Trim28L–/L– (zygotic null) | E7.5 embryos | Igf2 repressed | *H19 ↓ | |
| Hypomorphic maternal-zygotic Trim28 | E7.5 embryos | H19 biallelic | ND | |
| Trim28mat–/+(maternal null) | 8-cell stage embryos | ND | *H19 ↓ | [83] |
| Trim28mat–/+ (maternal null) | E12.5 embryos | ND | *H19 ↓; * H19 secondary DMR ↓ | [116] |
| E4.5 embryos | ND | *H19 ↓ | ||
| [Tet1−/−;Tet2−/−] female x WT male | P1–2 pups | ND | *H19 ↑ | [101] |
| [Tet1−/+;Tet2−/+] female x [Tet1−/−;Tet2-/−] male | P1–2 pups | ND | *H19 ↑ |
Table Key: *(partially penetrant); ND (no data); = (no change); ↓ (hypomethylated); ↑ (hypermethylated); # (age not explicitly stated); P (postnatal day); PN (pronuclear stage); ESCs (embryonic stem cells); PGCs (primordial germ cells).
Table 2.
Genetic models of imprinted expression and DNA methylation regulators at the Igf2r locus in mice.
| Genotype | Tissue | Allelic expression | Igf2r ICR DNA methylation | Reference |
|---|---|---|---|---|
| Dnmt1n/n(hypomorph) | E10.5 embryos | Igf2r = | *Igf2r ↓ | [18] |
| Dnmt1s/s (replication foci targeting domain mutation) | E9.5 embryos | Igf2r repressed | ND | |
| Dnmtn/c(c = null) | E9.5 embryos | Igf2r repressed | Igf2r ↓ | |
| Dnmt3amat–/−;Dnmt3b+/− | E9.5 embryos | ND | Igf2r ↓ | [112] |
| Dnmt3a2lox/1lox, TNAP–Cre × WT male (maternal deletion only) | E10.5 embryos | Igf2r repressed | Igf2r ↓ | [113] |
| Dnmt3l−/− | ESCs | ND | Igf2r = | [112] |
| Dnmt3lmat–/− | E9.5 embryos | ND | Igf2r ↓ | |
| Zfp57mat–/− (maternal-zygotic) | E11.5-E13.5 embryos | ND | * Igf2r ↓ | [76] |
| Zfp57−/− (zygotic null) | E11.5-E13.5 embryos | ND | * Igf2r ↓ | |
| Zfp57+/− | Oocytes | ND | * Igf2r ↓ | |
| Zfp57−/− | Oocytes | ND | * Igf2r ↓ | |
| Trim28chatwo/chatwo (zygotic hypomorph) | E8.5 embryos | * Airn biallelic | ND | [82] |
| Trim28mat–/+(maternal null) | 8-cell stage embryos | ND | * Igf2r ↓ | [83] |
| Tet1Gt/Gt (catalytic domain removed) | E9.5 embryos | * Airn repressed | * Igf2r ↑ | [96] |
Table Key: *(partially penetrant); ND (no data); = (no change); ↓ (hypomethylated); ↑ (hypermethylated); # (age not explicitly stated) P (postnatal day); PN (pronuclear stage); ESCs (embryonic stem cells); PGCs (primordial germ cells).
Epigenetic reprogramming
Reprogramming in mammals consists of two main waves: the first occurs in the zygote and preimplantation embryo, and the second, in the developing germ line. In both cases, these waves include dramatic loss of DNA methylation, followed by subsequent de novo methylation. Although not the focus of this review, histone modifications are also dynamically remodeled during these two waves of reprogramming and there may be mechanistic links between these two epigenetic processes [1, 2]. Thus, epigenetic reprogramming refers to broad changes in epigenetic modifications, such as DNA methylation and histone modifications, that lead to changes in gene expression and cell potency.
The concept of epigenetic reprogramming was initially recognized by Art Riggs when he proposed a role for DNA methylation in facilitating the process of X inactivation in female mammals [3]. In the 1980s, Jähner and Jaenisch observed that changes in gene expression were correlated with DNA methylation and postulated that DNA methylation “may be a condition for ‘resetting’ the genome” [4]. Later, Monk et al. found that while sperm DNA is highly methylated, blastocysts had very low methylation. Gains of DNA methylation were observed from the blastocyst stage to embryonic day (E)6.5 in the epiblast. By E12.5 and E14.5, PGCs exhibited low levels of DNA methylation, while the somatic DNA methylation levels remained similar to E6.5 epiblasts. Monk and colleagues also observed methylation increases in certain repetitive sequences in male germ cells at E16.5 but not in female germ cells [5]. This study demonstrated that the early embryo and the germline were likely undergoing dynamic DNA methylation changes.
Why would these reprogramming events be necessary for early mammalian development? In mammals, germ cells are specified from the epiblast [6, 7]. Therefore, reprogramming of DNA methylation is required to erase the epiblast-specific pattern of DNA methylation enabling the subsequent acquisition of sperm- or egg-specific epigenetic marks [5]. This is also true for the zygote, which must erase the cell-type specific DNA methylation marks that define the sperm and oocyte in order to facilitate DNA methylation patterns characteristic of somatic cells. DNA methylation erasure is also postulated to ensure that abnormal epigenetic marks are not transmitted to the next generation [8, 9]. Lastly, erasure of DNA methylation in the fetal germ line provides a blank slate so that parental-specific imprinting marks can be properly established according to the sex of the developing embryo [10].
Genomic imprinting
Genomic imprinting is a phenomenon where a subset of genes in the mammalian genome is expressed from a single parental allele. We contrast this definition of imprinted genes with genes that show allelically biased expression, as the mechanism governing biased expression is unclear. Currently, approximately 150 imprinted genes in mice and about 100 in humans have been identified, many of which are imprinted in both species. Imprinted genes tend to be found in clusters, and this allows for their coordinated regulation by a cis-acting regulatory element called an imprinting control region (ICR). These elements are characterized by parental-allele-specific DNA methylation, which regulates their unique expression pattern (reviewed in [11]).
The inequivalence of the paternal and maternal genomes was known well before the identification of imprinted genes. Nuclear transfer experiments in mice demonstrated that both maternal and paternal contributions were necessary for viable pups, whereas uniparental embryos failed to develop to term. Additionally, it was shown that uniparental disomies in specific genomic regions were detrimental or gave rise to phenotypes dependent upon the parent of origin, in both mouse and humans. Lastly, a subset of transgenic mouse lines exhibited parent-of-origin specific expression of the transgene, suggesting that some genomic sequences could be differentially modified in the germline [11].
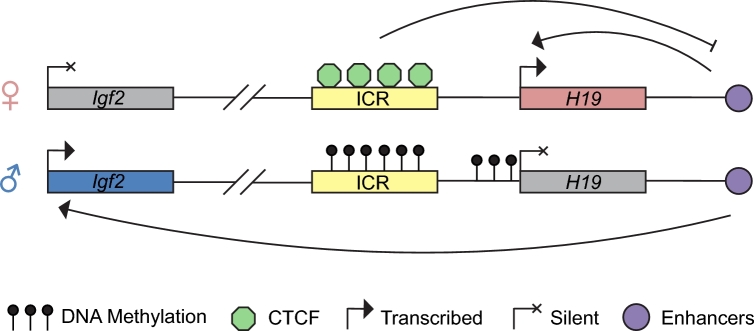
The first three imprinted genes were described in the early 1990s: insulin growth factor receptor 2 (Igf2r) [12], H19 [13], and insulin growth factor 2 (Igf2) [14]. Two of these genes (H19 and Igf2) are linked (Figure 1), which prompted the original suggestion that imprinted genes are clustered. Once imprinted genes were identified, the question of how this unique expression pattern is conferred was pursued. It was hypothesized that a specific sequence could be marked epigenetically, or a sequence could be recognized by a trans-acting epigenetic regulatory protein. Given the suggestion that imprints must be set in the germline, maintained through fertilization, and erased in embryonic germ cells, it was speculated that DNA methylation may be the epigenetic mark that fit these criteria [14]. Consequently, rigorous searches for differential DNA methylation were undertaken. For example, at the H19 locus, a region of paternal-specific methylation was found in a 7–9 kilobase region encompassing part of the H19 gene itself, as well as a 5΄ region [15]. This region was further refined to a 2 kilobase region 5΄ of H19 that was found to be DNA methylated in sperm but not oocytes, and this differential methylation was maintained during development [16, 17]. Definitive evidence that DNA methylation regulates imprinted gene expression came from a mouse knockout model of the DNA methyltransferase 1 (Dnmt1) gene, which resulted in DNA hypomethylation and loss of imprinted gene expression (Tables 1 and 2) [18]. More recently, genome-wide analyses using F1 hybrid animals have confirmed existing regions as well as uncovered new differentially methylated regions (DMRs) in mice [19].
Figure 1.
Schematic of the H19 imprinted locus in mice. Arrows show enhancer activation of transcription. The blunt-ended arrow indicates the enhancer-blocker function of the insulator formed by the CTCF-bound unmethylated maternal ICR. The details of imprinted regulation at this locus are provided in the text.
Ultimately, the test of whether a DMR conferred imprinted expression of one or multiple genes was undertaken using genetic deletion in mice. Loss of imprinted gene expression upon deletion of a DMR provided evidence that the sequence was causal in conferring imprinted gene expression—these regions are designated as ICRs. In addition to regulation of monoallelic gene expression, ICRs have other functions. The H19 ICR, for example, is DNA methylated exclusively on the paternal allele [17]. CCCTC binding factor (CTCF), a multifunctional, methylation-sensitive architectural protein, binds to the unmethylated maternal ICR and forms a functional insulator blocking the access of downstream enhancers from interacting with the upstream Igf2 promoter [20]. On the paternal allele, DNA methylation at the ICR prevents CTCF binding, allowing the downstream enhancers to interact with the Igf2 promoter and promote transcription. The DNA methylation from the ICR spreads into the H19 promoter, silencing H19 expression [21] (Figure 1). Other ICRs serve as promoters for noncoding RNAs. For example, the paternally unmethylated Igf2r ICR is the promoter for the paternally expressed Airn, the KvDMR serves as the promoter for the paternally expressed Kcnq1ot1, and the Snrpn ICR is the promoter for Ube3a-ats [11].
The importance of proper monoallelic expression of imprinted genes is exemplified by their misregulation in human imprinting disorders and the abnormal phenotypes described in various genetic mouse models where imprinted gene dosage or ICR mutations have been constructed. For example, in patients where there is an abnormal gain of methylation at the H19 ICR, H19 is silenced and IGF2 is biallelically expressed, resulting in an overgrowth disorder known as Beckwith-Wiedemann syndrome [22].
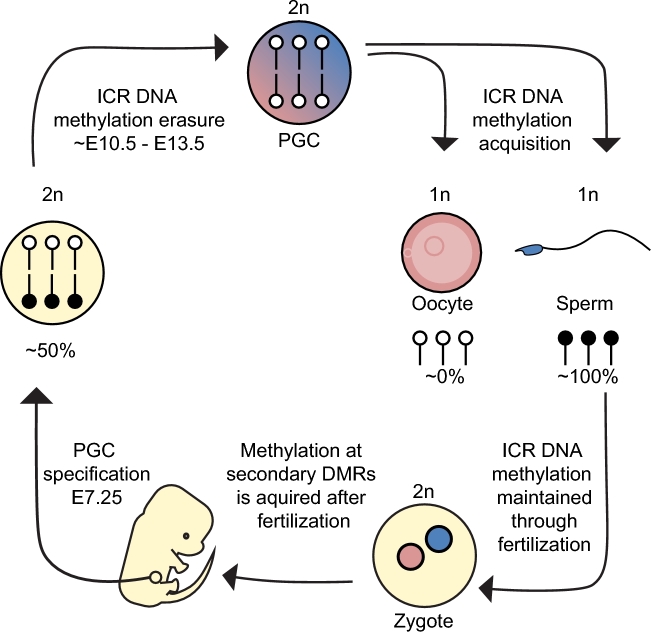
Intriguingly, these parent-specific DNA methylation marks at ICRs are dynamically regulated during development (Figure 2). DNA methylation imprints are first set in the germline and maintained through fertilization and preimplantation development, despite a nearly complete demethylation of the genome. DNA methylation imprints are then erased in the developing PGCs of the embryo, which allows the establishment of sex-specific marks in the gametes (as reviewed in [23]). Below, we describe the regulators of DNA methylation, observations of changing ICR DNA methylation levels beginning with maintenance at fertilization, erasure of imprints following the specification of PGCs, and the acquisition of imprints in the developing germ cells. ICR methylation dynamics are described in the context of concurrent genome-wide changes in DNA methylation.
Figure 2.
The life cycle of a DNA methylation imprint. ICRs obtain their parent-of origin-specific DNA methylation in the haploid genomes of germ cells. This differential methylation is maintained through fertilization. In contrast, secondary DMRs acquire DNA methylation after fertilization. PGCs are specified from the epiblast and thus need to erase imprinted methylation as PGCs develop, allowing a ground state for the acquisition of imprinted methylation in the germline.
How is DNA methylation regulated?
DNA methyltransferases
DNA methylation is a catalytic reaction carried out through the use of the methyl donor, S-adenosylmethionine. This covalent modification of cytosine residues typically occurs in a CpG context, although non-CpG methylation has been observed, particularly in the brain and in oocytes [24, 25]. Collectively, the DNA methyltransferase enzymes (DNMTs) add and maintain levels of DNA methylation throughout the genome.
The first identified family member of DNMTs was DNMT1. Dnmt1 homozygous null animals die during midgestation, indicating the importance of DNA methylation during development. DNMT1 maintains DNA methylation by copying existing methylation patterns onto newly replicated DNA strands. Evidence suggests that DNMT1 can methylate cytosines in a de novo fashion [26], although the enzyme's preferred substrate is hemimethylated DNA [27]. The maintenance function of DNMT1 is accomplished by binding to ubiquitin-like with PHD and ring finger domains 1 (UHRF1, also known as nuclear protein, 95 kDa (NP95)) [28]. UHRF1 binds to proliferating cell nuclear antigen, the sliding clamp of the DNA replication fork [29]. UHRF1 additionally is recruited to the replication fork by DNA ligase 1 (LIG1) [30]. Thus, DNMT1 is targeted to the replication machinery and explains the mechanism behind its maintenance function.
Evidence for additional DNMTs became apparent when residual DNA methylation was observed in Dnmt1 null embryonic stem cells (ESCs) and mice [31]. Through homology searches, two additional methyltransferases were discovered, called DNMT3A and DNMT3B [32]. These de novo methyltransferases can add DNA methylation with equal affinity for nonmethylated and hemimethylated DNA substrates and have also been shown to have some maintenance functions as well [32, 33]. Dnmt3a null mice die approximately four weeks after birth, whereas Dnmt3b homozygous knockout animals die after E9.5 [32, 34].
Lastly, the DNMT3L protein lacks a catalytic domain but binds to both of the de novo methyltransferases and acts as a stimulatory cofactor. DNMT3L contains a plant homeodomain-like domain (PHD) that recognizes unmethylated histone 3 lysine 4 (H3K4) residues and therefore is important for targeting the enzymes to chromatin. Dnmt3l homozygous knockout animals also die around E9.5 (reviewed in [35, 36]). Interestingly, mouse ESCs can tolerate the combined deletion of DNTM1, DNMT3A, and DNMT3B despite the near complete abolishment of DNA methylation. However, these cells have compromised differentiation, emphasizing the critical role of DNA methylation in development [37].
Evidence for active DNA demethylation
Breaking a carbon-carbon bond between the methyl group and the cytosine ring was thought to be impossible given the extreme thermodynamic input required in a physiological setting. However, evidence that DNA methylation could be removed was first demonstrated by Gjerset and Martin, who described demethylation in nuclear extracts from erythroleukemia cells where 5mC was replaced with an unmodified cytosine. This demethylation occurred in the absence of DNA synthesis, was proportional to the amount of protein added to the reaction, and was abolished by the addition of both proteinase K and heat inactivation, indicating an enzymatic activity [38]. This demethylation was specific to DNA methylated CpGs [39, 40]. Likewise, HeLa cell extract exhibited such an activity and additionally demonstrated newly generated abasic sites in the template, indicating glycosylase involvement [41]. Lastly, it was shown that fusion of somatic cells with germ cells caused extensive DNA demethylation of somatic nuclei [8]. This early evidence pointed to an active process whereby 5mC is replaced without DNA replication. While many candidate proteins and pathways have been described as the origin of a demethylating activity (for further information, see [42–44]), one of the most promising discoveries involves a family of proteins called the Ten-eleven translocation (TET) family, as described below.
Oxidative demethylation by TET1, TET2, and TET3
In 2002, a fusion protein containing part of the mixed lineage leukemia (MLL) histone methyltransferase and a previously uncharacterized protein, called leukemia-associated protein with a CxxC domain (LCX), was described in patients with MLL [45]. However, it was not until 2009 that the relevance of this new protein became apparent [46, 47]. A newly described cytosine modification, 5-hydroxymethylcytosine (5hmC), was an oxidation product of 5mC and was reported in Purkinje neurons and mouse ESC DNA [46, 47]. Crucially, Tahiliani et al. demonstrated that LCX, now known as ten-eleven translocation methylcytosine dioxygenase 1 (TET1), was responsible for generating 5hmC and that this activity depended on a functional catalytic domain as well as Fe(II) and alpha-ketoglutarate. The two other TET family members, TET2 and TET3, were also shown to catalyze the 5hmC reaction, and all three family members can further oxidize 5hmC to 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC) [48–50]. The use of Dnmt3a/3b null mice revealed a reduction in 5fC, suggesting that 5fC accumulation was likely dependent on 5mC first being oxidized by TET to 5hmC [51]. The structure of the TET2 catalytic domain revealed that the enzyme works by using a base-flipping mechanism. The methyl group of cytosine does not participate in the DNA–enzyme interaction and thus the active site can accommodate the larger oxidized cytosine bases [52]. Together, these important studies supported the idea that the TET family of proteins is responsible for 5mC oxidation and thus led to the prevailing view that TETs are a major regulator of DNA demethylation.
TDG and base excision repair
Thymine DNA glycosylase (TDG) was originally described for its DNA glycosylase activity on G/T mismatches. Nevertheless, there had been hints that this enzyme was also involved in DNA demethylation [41, 53, 54]. It is now appreciated that TDG cleaves 5fC and 5caC rapidly in vitro, whereas the enzyme shows no activity on 5hmC [50]. This cleavage results in an abasic site that is subsequently repaired by base excision repair. Indeed, inhibition of base excision repair proteins downstream of TDG such as apurinic/apyrimidinic endonuclease 1 or poly(ADP-ribose) polymerase 1 (PARP1) [55–57] in cultured mouse zygotes leads to accumulations of 5mC in zygotes [57]. Other experiments that inhibited PARP pharmacologically in pregnant dams also led to increases in DNA methylation at certain imprinted genes in fetal PGCs. TDG null embryos die around E12.5 [58, 59], further underscoring this protein's role in a critical developmental pathway.
Preimplantation DNA methylation reprogramming
Genome-wide DNA demethylation
After fertilization, the paternal pronucleus undergoes rapid demethylation before the onset of DNA replication whereas the maternal genome demethylates more slowly over multiple cell divisions. These kinetics indicate that the paternal genome is actively demethylated whereas the maternal genome is demethylated in a passive, replication-dependent manner [60]. Multiple groups have described TET3 activity as well as the accumulation of all three oxidized cytosine bases in both parental pronuclei [61, 62]. This accumulation depends on TET3 and TET1 as depletion of TET3 in the oocyte leads to an impairment of 5hmC accumulation, whereas depletion of both TET1 and TET3 leads to a complete absence of 5hmC at the eight-cell stage [63–65]. It is likely that 5hmC, 5fC, and 5caC are removed by passive dilution as these bases are only found on one half of the paternally derived chromatids [66, 67].
Interestingly, the oxidation of 5mC in the zygote may not be necessary for the global demethylation as previously thought. Shen and colleagues demonstrated that demethylation still occurred in paternal pronuclei despite a conditional oocyte-specific Tet3 knockout, suggesting that zygotic TET3 was partially responsible for demethylating the paternal genome and that only certain regions were dependent on TET3 for 5mC oxidation. When wild-type zygotes were treated with the replication inhibitor aphidicolin, 5mC levels did not decrease, despite the continued activity of TET3 as evidenced by the presence of the oxidized bases 5hmC, 5fC, and 5caC [61]. Further evidence that 5hmC accumulation does not drive DNA demethylation was demonstrated by precise staging of zygotes and detection of 5mC and 5hmC using antibody staining [68]. This experiment demonstrated that the global wave of DNA demethylation is complete by pronuclear stage (PN)3, which is before the accumulation of 5hmC. It was further demonstrated that the accumulation of 5hmC was dependent on previously underappreciated activities of both maternally inherited DNMT3A and DNMT1 [68]. Thus, the role of TET3 in the zygote may not be connected to the initial DNA demethylation of the genomes after fertilization, but may be serving to protect normally unmethylated regions from inappropriate acquisition of DNA methylation [68].
Imprinted regions escape preimplantation DNA methylation reprogramming
Even before the discovery of imprinted genes, it was appreciated that the functional differences between maternal and paternal genomes remained intact during genome-wide demethylation that occurred in the zygote [69]. Now, it is clear that a generalized feature of ICRs is the maintenance of DNA methylation and simultaneous protection from demethylation after fertilization. How are ICRs protected? Developmental pluripotency-associated 3 (DPPA3) is a highly expressed protein in oocytes, PGCs, and both pronuclei in the zygote [70]. When DPPA3 is deleted, both maternal and paternal genomes lose methylation. This is also apparent for ICRs. Dppa3 maternal-null zygotes lose imprinted DNA methylation at the H19 (Table 1), Peg1, Peg3, Peg10, and Rasgrf1 ICRs. In contrast, the IG-DMR, Snrpn, and Peg5 ICRs remained methylated, indicating that DPPA3 is partially responsible for the maintenance of DNA methylation at a subset of ICRs [71]. It has been suggested that DPPA3 exerts a maintenance function by binding to and inhibiting the activity of the C-terminal catalytic domain of TET2 and TET3 [72]. However, these results should be interpreted with caution as the co-immunoprecipitations were conducted in human embryonic kidney 293T cells [72], which express low levels of TET proteins endogenously [73, 74]. Also in 293T cells, approximately 60% of TET3 chromatin immunoprecipitation followed by deep sequencing (ChIP-seq) peaks overlapped with DPPA3 peaks, and vice versa. Analysis of the DNA sequence bound by DPPA3 indicated a motif preference found in the ICRs of Peg1, Peg3, Peg10, and H19. [72]. Additionally, DPPA3 can bind to H3K9me2, which is enriched on the methylated allele of imprinted genes, further explaining the targeting and protection of ICRs afforded by DPPA3 binding [75].
Perhaps a more compelling factor that is involved in the protection of ICR methylation during preimplantation development is zinc finger protein 57 (ZFP57) [76]. Null embryos from Zfp57 heterozygous matings show a partial lethality phenotype [76]. Maternal-zygotic null zygotes exhibit changes in total expression of imprinted genes regulated by the IG-DMR and loss of ICR DNA methylation, but the ICR DNA methylation and imprinted gene expression at the H19/Igf2 locus are unaffected (Table 1). The maternal-zygotic Zfp57 mutants also lose methylation at the Snrpn, Peg1, Peg3, and Peg5/Nnat ICRs [76]. Interestingly, ZFP57 binds only to the methylated ICR and this methylation is necessary for its binding [77, 78]. The allele-specific binding is neither observed at secondary DMRs (DMRs that gain methylation in the somatic lineage as opposed to in the germline, Figure 2) nor at DMRs unrelated to imprinted genes in the germline [78]. ChIP-seq experiments demonstrated that a six-base pair motif, TGCCGC, is found at almost all of the known ICRs. This motif is sufficient to maintain imprinted methylation at the Snrpn ICR when it is integrated away from its endogenous locus, but mutations of the motif cause a loss of methylation maintenance [79].
The ability of ZFP57 to serve an ICR methylation maintenance function lies in its interacting partners. ZFP57 binds tripartite motif-containing 28 (TRIM28, also known as KAP1 or TIF1-beta), which is a corepressor that recruits repressive histone modifiers like histone deacetylases, the histone methyltransferase SETDB1, and DNA methyltransferases to chromatin [80]. The recruitment of maternal DNMT1 is essential for imprint maintenance as maternal-zygotic Dnmt1 mutation also leads to demethylation at ICRs in zygotes [81]. Trim28 hypomorphic zygotic mutants maintain imprinted DNA methylation at the IG-DMR, but loss-of-function zygotic mutants lose DNA methylation at IG-DMR in a partially penetrant manner, indicating the amount of TRIM28 is important for ICR protection. Hypomorphic maternal-zygotic Trim28 mutants exhibit biallelic expression of H19 (Table 1), Gtl2, and Snrpn [82]. Additionally, single cell methylation analysis of six ICRs (H19, IG-DMR, Igf2r, Snrpn, Peg3, Nnat), demonstrated that maternal loss of Trim28 resulted in highly variable demethylation of these ICRs within the same blastomere (Tables 1 and 2). This result suggests that maternal Trim28 deficiency leads to an incompletely penetrant ICR demethylation phenotype [83].
Peri-implantation de novo methylation of the genome
Following zygotic epigenetic reprogramming, the blastocyst is hypomethylated with the exception of imprinted genes, intracisternal A particle elements (IAPs), and a subset of gene promoters, including genes enriched in functions such as gamete generation and sexual reproduction [84, 85]. Beginning at the early blastocyst stage and culminating by E6.5, the genome gains DNA methylation globally while certain CpG islands remain hypomethylated [24, 85]. PGCs are specified from cells with this aforementioned methylated state, which then must be subsequently erased, as discussed in the following section.
PGC DNA methylation reprogramming
PGCs are specified at E7.25 in the mouse. After specification, PGCs proliferate and migrate from the epiblast toward the genital ridge. PGCs then undergo a second wave of demethylation to erase the epiblast cell fate and facilitate germ-cell fate. Here, DNA is demethylated at ICRs [2]. First, levels of genome-wide methylation decrease as early as E8.0, while some regions maintain DNA methylation. A later, second wave targets these initially resistant regions, leading to the lowest levels of methylation in PGCs by E13.5. This two-step demethylation process may be important for suppressing premature differentiation of the germline [86].
Bulk genome-wide demethylation in PGCs: the early wave
Numerous studies have demonstrated genome-wide demethylation in PGCs from E7.25 to E13.5. Immunofluorescence staining of 5mC showed that PGCs are globally demethylated starting as early as E8.0, concomitant with the onset of PGC migration. Genome-wide profiling confirmed these observations [87–89]. Demethylation is accompanied by global loss of H3K9me [90], downregulation of DNMT3A [90], and cytoplasmic localization of DNMT3B. DNMT1 is expressed and localized to the nucleus from E10.5 to E13.5 [91]. In PGCs, Uhrf1 mRNA is also consistently downregulated after E7.25, and UHRF1 protein expression is undetectable between E8.5 and E11.5 [92], indicating impaired targeting for DNMT1 to the replication fork [93]. Passive dilution of DNA methylation was further demonstrated using hairpin bisulfite sequencing, where strand-specific DNA methylation can be determined. In PGCs, hemimethylated DNA strands significantly increased between E10.5 and E11.5 at long-interspersed nuclear elements-1 [92]. Overall, these observations indicate that a variety of mechanisms facilitate DNA demethylation, including downregulation and nuclear exclusion of de novo and maintenance DNA methylation machinery and absence of the targeting factors.
Does the oxidation of 5mC by TET proteins contribute to global DNA demethylation in PGCs? Tet1 and Tet2 mRNA and protein are detectable in PGCs between approximately E9.5 and E12.5. Notably, Tet3 mRNA and protein levels are undetectable, indicating 5hmC is most likely generated by TET1 and TET2 in PGCs [57, 94, 95]. PGCs assayed for 5mC and 5hmC with mass spectrometry showed increases in 5hmC starting at E8.75 through E12.5. However, immunostaining did not reveal changes in the levels of 5fC and 5caC, indicating that further oxidation of 5hmC is not involved in the bulk wave of DNA demethylation [95, 96]. Consistently, in chromosome spreads, 5hmC was only detected on one sister chromatid suggesting that dilution of the oxidized base through passive replication is responsible for demethylation in PGCs [96]. To determine the role of TET1 catalytic activity on DNA demethylation in PGCs, an allele without the catalytic domain of TET1 was generated (Tet1Gt) and methylation was profiled in Tet1Gt E13.5 PGCs. Overall, global levels of methylation were unchanged, suggesting that TET1 does not contribute to genome-wide demethylation but may play roles in locus-specific demethylation instead [94].
It is notable that while overall levels of DNA methylation decrease beginning at E8.0 in PGCs, and reach a minimum at E13.5, not all parts of the genome follow these demethylation kinetics. Certain regions are initially resistant to this wave of DNA methylation, indicating DNA methylation is maintained. These regions include ICRs, a subset of repetitive elements, and CpG islands on the X-chromosome [88, 91, 97]. In order for DNA demethylation-resistant loci to maintain DNA methylation, two processes are required. First, the loci must be resistant to demethylation, possibly through the use of proteins that protect DNA from demethylation. Second, methylation must be maintained during cell division. DNMT1, despite the low levels of UHRF1 in PGCs [90], methylates the H19 ICR, Snrpn ICR, and IAP elements. This suggests that DNMT1 can localize to specific regions including IAPs and ICRs to maintain DNA methylation [86].
ICR demethylation in PGCs
As stated above, ICRs retain their methylation in PGCs until approximately E10.5 and then are demethylated in a gene-specific stereotypical pattern. This result was first demonstrated by assaying DNA methylation at individual imprinted genes. Early profiling studies in F1 hybrid mice demonstrated that the H19 ICR was considerably demethylated on both the maternal and paternal allele by E13.5 in PGCs [21]. Lee et al. generated embryos from PGC clones and used the imprinting status of these embryos as proxies for the imprinted status in the parental PGCs. ICRs in embryos derived from E11.5 PGCs had very different levels of methylation, indicating this demethylation of ICRs occurs somewhat stochastically across a population of PGCs. This result also suggested that each ICR had its own demethylation timing: the Nnat ICR was one of the earliest to demethylate, the H19 ICR was “intermediate,” whereas the Peg10 ICR was the slowest and thus the most resistant to DNA demethylation. The timing of DNA methylation in PGCs and PGC clones had good concordance [98]. Additionally, locus-specific bisulfite sequencing of PGCs revealed that the Peg3, Lit1, Snrpn, and H19 ICRs were demethylated between E11.5 and E12.5, and this demethylation persisted until E13.5 [91]. Sato and colleagues sorted GFP transgenic PGCs and found Igf2r ICR methylation at E10.5, but demethylation initiated at E11.5 [99].
Simultaneous profiling of all ICR methylation in smaller cell numbers was greatly enhanced by whole-genome bisulfite sequencing. Using this technique, at E10.5, ICRs were approximately 40% methylated. At E13.5, in both males and female PGCs, none of the paternally methylated ICRs showed appreciable DNA methylation, confirming the results from earlier locus-specific approaches [97]. Hackett et al. also found that demethylation timing depended on the imprinted gene in question. Whereas Peg10 and Peg3 ICRs were slow to demethylate, Igf2r ICR and the KvDMR exhibited faster demethylation kinetics [95]. Thus, how long DNA methylation is retained depends on the ICR.
TET-mediated demethylation of ICRs in the second wave of PGC demethylation
The second wave of demethylation includes ICR demethylation. The expression of TET1 and TET2 in PGCs prompted investigators to examine whether these enzymes participate in ICR demethylation. Multiple lines of evidence were consistent with this idea. Fusion of embryonic germ cells derived from E12.5 PGCs with B cells resulted in ICR demethylation, which coincided with a rapid accumulation of 5hmC [100]. Tet1 depletion using short-hairpin RNA interference in this system caused a loss of 5hmC accumulation at the H19 ICR, and the failure to demethylate, despite the presence of TET2 [100]. Physiological experiments directly showed a role for TETs in ICR demethylation. Using glucosyltransferase-quantitative polymerase chain reaction (Glu-qPCR) to measure locus-specific 5hmC, levels of DNA methylation loss correlated with 5hmC gains at the KvDMR, Peg10, Igf2r, and Peg3 ICRs in PGCs [95]. 9.5- and 10.5-day embryos generated from a homozygous Tet1Gtmouse mated to wild-type mouse [94], or 13.5-day embryos generated from Tet1; Tet2 double knockout mice mated to either Tet1; Tet2 double heterozygous mice or to wild-type mice [101], showed hypermethylation at ICRs as well as dysregulated total levels of imprinted gene expression (Tables 1 and 2) [96, 101]. Together, evidence strongly suggests that TET1 may be a prominent mediator of ICR erasure.
Despite the accumulation of 5hmC in wild-type PGCs, dependent upon the catalytic activity of at least TET1, the further processing of this oxidized base may occur by replication-dependent dilution. Multiple studies report a decrease in 5hmC levels to be consistent with the predicted replication rate of PGCs [95, 102]. Indeed, levels of Tdg mRNA, the protein responsible for cleaving 5fC and 5caC from the genome, drop from E9.5 to E13.5 in PGCs [102]. Therefore, evidence for active processing of 5hmC at ICRs is still wanting.
Reacquisition of DNA methylation at ICRs
Once PGCs have completed demethylation at E13.5, genome-wide and locus-specific remethylation initiates. This remethylation is essential for ICRs to acquire their parental-specific imprints during germ cell development. Male germ cells acquire DNA methylation at ICRs shortly after DNA demethylation ceases, and DNA methylation is completed mostly before birth. In contrast, maternal-specific imprints are acquired predominantly after birth during oocyte growth [103, 104].
While our understanding of how ICRs are targeted for remethylation in the developing germ cells is incomplete, well-validated observations have been made. First, DNA methylation imprints are acquired when the histone modifications H3K4me2/3 are low, levels of the histone demethylases KDM1A and KDM1B are high, and H3K36me3 is high [105]. Additionally, transcription likely plays a major role in the timing of imprinting acquisition, although transcription is not sufficient to explain this timing [103, 106, 107]. A unique case is found at the Rasgrf1 imprinted locus where piwi-interacting RNAs have been proposed to play a role in DNA methylation establishment in the male germline [108]. While DNMT3B, DNMT3A, and DNMT3L are required for methylating the Rasgrf1 ICR, other ICRs, such as the H19 and Igf2r ICRs, are targeted by DNMT3A and DNMT3L (Tables 1 and 2) [103]. Interestingly, the timing of acquisition of imprinted marks is not the same on the previously paternal and previously maternal alleles, indicating a non-equivalence of the underlying chromatin state, despite the lack of DNA methylation in PGCs [21, 104]. Thus, a complex interplay between DNA methylation regulators, chromatin regulators, transcription, and long-noncoding RNAs together functions in a complex manner to methylate the regions crucial for imprinted expression in the next generation.
Remaining questions and future directions
The fields of genomic imprinting and epigenetic reprogramming have greatly expanded over the past decades. Advances in the discovery of key genes and proteins, application of novel, sensitive, locus-specific and genome-wide techniques, and use of genetic knockout models have elucidated many aspects of dynamic DNA methylation genome wide and at ICRs in the early embryo and in PGCs. However, many questions remain. The observation that a subset of ICRs is sensitive to the loss of protective proteins (such as DPPA3 and ZFP57, see Tables 1 and 2) shows us that our understanding of DNA demethylation escape in the zygote remains incomplete. Additionally, in PGCs, the relative contributions of TET oxidation and other proposed demethylation pathways remain to be determined. The use of conditional alleles may be useful in deciphering these roles. It is also still unclear how ICRs are specifically targeted for demethylation. This remains an intriguing question and may relate to intricate communication between chromatin modifications, trans factors, and epigenetic regulatory proteins. Similarly, mechanisms leading to the reacquisition of imprinted DNA methylation warrant further investigation. Given these exciting questions, we eagerly anticipate forthcoming discoveries.
Acknowledgments
We dedicate this manuscript to the memory of Denise Barlow, who made pioneering contributions to the study of imprinting mechanisms. We would like to thank members of the Bartolomei lab for critical reading and editing of the manuscript, particularly Frances Xin and Joanne Thorvaldsen. We apologize to the researchers whose primary work could not be cited due to space constraints.
Footnotes
Grant support: This work was supported by the National Institutes of Health [R37GM051279 to MSB, T32GM008216 to JMS, F31GM119271 to JMS]
References
- 1. Hajkova P, Ancelin K, Waldmann T, Lacoste N, Lange UC, Cesari F, Lee C, Almouzni G, Schneider R, Surani MA. Chromatin dynamics during epigenetic reprogramming in the mouse germ line. Nature 2008; 452(7189):877–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Saitou M, Yamaji M. Primordial germ cells in mice. Cold Spring Harb Perspect Biol 2012; 4(11):a008375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Riggs AD. X inactivation, differentiation, and DNA methylation. Cytogenet Cell Genet 1975; 14(1):9–25. [DOI] [PubMed] [Google Scholar]
- 4. Jähner D, Jaenisch R. DNA methylation in early mammalian development. In: Cedar H, Razin A, Riggs AD (eds.), Dna Methylation, Biochemistry, And Biological Significance. New York: Springer; 1984:189–219. [Google Scholar]
- 5. Monk M, Boubelik M, Lehnert S. Temporal and regional changes in DNA methylation in the embryonic, extraembryonic and germ cell lineages during mouse embryo development. Development 1987; 99:371–382. [DOI] [PubMed] [Google Scholar]
- 6. Anderson R, Copeland TK, Schöler H, Heasman J, Wylie C. The onset of germ cell migration in the mouse embryo. Mech Dev 2000; 91(1–2):61–68. [DOI] [PubMed] [Google Scholar]
- 7. Ginsburg M, Snow MH, McLaren A. Primordial germ cells in the mouse embryo during gastrulation. Development 1990; 110:521–528. [DOI] [PubMed] [Google Scholar]
- 8. Surani MA. Reprogramming a somatic nucleus by trans-modification activity in germ cells. Semin Cell Dev Biol 1999; 10(3):273–277. [DOI] [PubMed] [Google Scholar]
- 9. Heard E, Martienssen RA. Transgenerational epigenetic inheritance: myths and mechanisms. Cell 2014; 157(1):95–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tada S, Tada T, Lefebvre L, Barton SC, Surani MA. Embryonic germ cells induce epigenetic reprogramming of somatic nucleus in hybrid cells. EMBO J 1997; 16(21):6510–6520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Barlow DP, Bartolomei MS. Genomic imprinting in mammals. Cold Spring Harb Perspect Biol 2014; 37:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Barlow DP, Stöger R, Herrmann BG, Saito K, Schweifer N. The mouse insulin-like growth factor type-2 receptor is imprinted and closely linked to the Tme locus. Nature 1991; 349(6304):84–87. [DOI] [PubMed] [Google Scholar]
- 13. Bartolomei MS, Zemel S, Tilghman SM. Parental imprinting of the mouse H19 gene. Nature 1991; 351(6322):153–155. [DOI] [PubMed] [Google Scholar]
- 14. DeChiara TM, Robertson EJ, Efstratiadis A. Parental imprinting of the mouse insulin-like growth factor II gene. Cell 1991; 64(4):849–859. [DOI] [PubMed] [Google Scholar]
- 15. Bartolomei MS, Webber AL, Brunkow ME. Epigenetic mechanisms underlying the imprinting of the mouse H19 gene. Genes Dev 1993; 7(9):1663–1673. [DOI] [PubMed] [Google Scholar]
- 16. Tremblay KD, Saam JR, Ingram RS, Tilghman SM, Bartolomei MS. A paternal–specific methylation imprint marks the alleles of the mouse H19 gene. Nat Genet 1995; 9(4):407–413. [DOI] [PubMed] [Google Scholar]
- 17. Tremblay KD, Duran KL, Bartolomei MS. A 5' 2-kilobase-pair region of the imprinted mouse H19 gene exhibits exclusive paternal methylation throughout development. Mol Cell Biol 1997; 17(8):4322–4329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li E, Beard C, Jaenisch R. Role for DNA methylation in genomic imprinting. Nature 1993; 366(6453):362–365. [DOI] [PubMed] [Google Scholar]
- 19. Xie W, Barr CL, Kim A, Yue F, Lee AY, Eubanks J, Dempster EL, Ren B. Base-resolution analyses of sequence and parent-of-origin dependent DNA methylation in the mouse genome. Cell 2012;148(4):816–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Singh P, Lee DH, Szabó PE. More than insulator: multiple roles of CTCF at the H19-Igf2 imprinted domain. Front Gene 2012; 3:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Davis TL, Yang GJ, McCarrey JR, Bartolomei MS. The H19 methylation imprint is erased and re-established differentially on the parental alleles during male germ cell development. Hum Mol Genet 2000; 9(19):2885–2894. [DOI] [PubMed] [Google Scholar]
- 22. Cooper WN, Luharia A, Evans GA, Raza H, Haire AC, Grundy R, Bowdin SC, Riccio A, Sebastio G, Bliek J, Schofield PN, Reik W et al. Molecular subtypes and phenotypic expression of Beckwith–Wiedemann syndrome. Eur J Hum Genet 2005; 13(9):1025–1032. [DOI] [PubMed] [Google Scholar]
- 23. Macdonald WA, Mann MRW. Epigenetic regulation of genomic imprinting from germ line to preimplantation. Mol Reprod Dev 2014; 81(2):126–140. [DOI] [PubMed] [Google Scholar]
- 24. Smith ZD, Chan MM, Mikkelsen TS, Gu H, Gnirke A, Regev A, Meissner A. A unique regulatory phase of DNA methylation in the early mammalian embryo. Nature 2012; 484(7394):339–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Guo JU, Su Y, Shin JH, Shin J, Li H, Xie B, Zhong C, Hu S, Le T, Fan G, Zhu H, Chang Q et al. Distribution, recognition and regulation of non-CpG methylation in the adult mammalian brain. Nat Neurosci 2014; 17(2):215–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Vertino PM, Chiu Yen R-W, Gao J, Baylin SB. De novo methylation of CpG island sequences in human fibroblasts overexpressing DNA (cytosine-5-)-methyltransferase. Mol Cell Biol 1996; 16(8):4555–4565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fatemi M, Hermann A, Pradhan S, Jeltsch A. The activity of the murine DNA methyltransferase Dnmt1 is controlled by interaction of the catalytic domain with the N-terminal part of the enzyme leading to an allosteric activation of the enzyme after binding to methylated DNA. J Mol Biol 2001; 309(5):1189–1199. [DOI] [PubMed] [Google Scholar]
- 28. Bostick M, Kim JK, Esteve P-O, Clark A, Pradhan S, Jacobsen SE. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science 2007; 317(5845):1760–1764. [DOI] [PubMed] [Google Scholar]
- 29. Uemura T, Kubo E, Kanari Y, Ikemura T, Tatsumi K, Muto M. Temporal and spatial localization of novel nuclear protein NP95 in mitotic and meiotic cells. Cell Struct Funct 2000; 25(3):149–159. [DOI] [PubMed] [Google Scholar]
- 30. Ferry L, Fournier A, Tsusaka T, Arita K, Ferry L, Fournier A, Tsusaka T, Adelmant G, Shimazu T, Matano S. Methylation of DNA ligase 1 by G9a/GLP recruits UHRF1 to replicating DNA and regulates DNA methylation. Mol Cell 2017; 67(4):550–565.e5. [DOI] [PubMed] [Google Scholar]
- 31. Li E, Beard C, Forster ACC, Bestor TH, Jaenisch R. DNA methylation, genomic imprinting, and mammalian development. Cold Spring Harb Symp Quant Biol 1993; 58:297–305. [DOI] [PubMed] [Google Scholar]
- 32. Okano M, Xie S, Li E. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat Am Inc 1998; 19:219–220. [DOI] [PubMed] [Google Scholar]
- 33. Rhee I, Jair K-W, Yen R-WC, Lengauer C, Herman JG, Kinzler KW, Vogelstein B, Baylin SB, Schuebel K. CpG methylation is maintained in human cancer cells lacking DNMT1. Nature 2000; 404(6781):1003–1007. [DOI] [PubMed] [Google Scholar]
- 34. Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 1999; 99(3):247–257. [DOI] [PubMed] [Google Scholar]
- 35. Jurkowska RZ, Jeltsch A. Mechanisms and Biological Roles of DNA Methyltransferases and DNA Methylation: From Past Achievements to Future Challenges. DNA Methyltransferases - Role and Function. Switzerland: Springer International Publishing; 2016:1–17. [DOI] [PubMed] [Google Scholar]
- 36. Dan J, Chen T. Genetic Studies on Mammalian DNA Methyltransferases. DNA Methyltransferases - Role and Function, vol. 945 Switzerland:Springer International Publishing; 2016:123–150. [DOI] [PubMed] [Google Scholar]
- 37. Tsumura A, Hayakawa T, Kumaki Y, Takebayashi SI, Sakaue M, Matsuoka C, Shimotohno K, Ishikawa F, Li E, Ueda HR, Nakayama JI, Okano M. Maintenance of self-renewal ability of mouse embryonic stem cells in the absence of DNA methyltransferases Dnmt1, Dnmt3a and Dnmt3b. Genes Cells 2006; 11(7):805–814. [DOI] [PubMed] [Google Scholar]
- 38. Gjerset RA, Martin DW. Presence of a DNA demethylating activity in the nucleus of murine erythroleukemic cells. J Biol Chem 1982; 257:8581–8583. [PubMed] [Google Scholar]
- 39. Razin A, Szyf M, Kafri T, Roll M, Giloh H, Scarpa S, Carotti D, Cantoni GL. Replacement of 5-methylcytosine by cytosine: a possible mechanism for transient DNA demethylation during differentiation. Proc Natl Acad Sci USA 1986; 83(9):2827–2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Razin A, Levine A, Kafri TAL, Agostinit S, Gomi T, Cantoni GL. Relationship between transient DNA hypomethylation and erythroid differentiation of murine erythroleukemia cells. Proc Natl Acad Sci USA 1988; 85(23):9003–9006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Vairapandi M, Duker NJ. Enzymic removal of 5-methylcytosine from DNA by a human DNA-glycosylase. Nucleic Acids Res 1993; 21(23):5323–5327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ooi SKT, Bestor TH. The colorful history of active DNA demethylation. Cell 2008; 133(7):1145–1148. [DOI] [PubMed] [Google Scholar]
- 43. Dean W. Pathways of DNA Demethylation. DNA Methyltransferases—Role and Function, vol. 945 Switzerland: Springer International Publishing; 2016:247–274. [Google Scholar]
- 44. Bochtler M, Kolano A, Xu GL. DNA demethylation pathways: additional players and regulators. Bioessays 2017; 39(1):1–13. [DOI] [PubMed] [Google Scholar]
- 45. Ono R, Taki T, Taketani T, Taniwaki M, Kobayashi H, Hayashi Y. LCX, leukemia-associated protein with a CXXC domain, is fused to MLL in acute myeloid leukemia with trilineage dysplasia having t(10;11)(q22;q23). Cancer Res 2002; 62:4075–4080. [PubMed] [Google Scholar]
- 46. Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, Rao A. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 2009; 324(5929):930–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in purkinje neurons and the brain. Science 2009; 324(5929):929–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ito S, D’Alessio AC, Taranova O V, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature 2010; 466(7310):1129–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 2011; 333(6047):1300–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. He Y-F, Li B-Z, Li Z, Liu P, Wang Y, Tang Q, Ding J, Jia Y, Chen Z, Li L, Sun Y, Li X et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science 2011; 333(6047):1303–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Pfaffeneder T, Hackner B, Truß M, Münzel M, Müller M, Deiml CA, Hagemeier C, Carell T. The discovery of 5-formylcytosine in embryonic stem cell DNA. Angew Chem Int Ed 2011; 50(31):7008–7012. [DOI] [PubMed] [Google Scholar]
- 52. Hu L, Li Z, Cheng J, Rao Q, Gong W, Liu M, Shi YG, Zhu J, Wang P, Xu Y. Crystal structure of TET2-DNA complex: insight into TET-mediated 5mC oxidation. Cell 2013; 155(7):1545–1555. [DOI] [PubMed] [Google Scholar]
- 53. Jost JP, Oakeley EJ, Zhu B, Benjamin D, Thiry S, Siegmann M, Jost YC. 5-Methylcytosine DNA glycosylase participates in the genome-wide loss of DNA methylation occurring during mouse myoblast differentiation. Nucleic Acids Res 2001; 29(21):4452–4461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zhu B, Zheng Y, Hess D, Angliker H, Schwarz S, Siegmann M, Thiry S, Jost JP. 5-methylcytosine-DNA glycosylase activity is present in a cloned G/T mismatch DNA glycosylase associated with the chicken embryo DNA demethylation complex. Proc Natl Acad Sci USA 2000; 97(10):5135–5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ciccarone F, Klinger FG, Catizone A, Calabrese R, Zampieri M, Bacalini MG, de Felici M, Caiafa P. Poly(ADP-ribosyl)ation acts in the DNA demethylation of mouse primordial germ cells also with DNA damage-independent roles. PLoS One 2012; 7(10):e46927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kawasaki Y, Lee J, Matsuzawa A, Kohda T, Kaneko-Ishino T, Ishino F. Active DNA demethylation is required for complete imprint erasure in primordial germ cells. Sci Rep 2015; 4:3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hajkova P, Jeffries SJ, Lee C, Miller N, Jackson SP, Surani MA. Genome-wide reprogramming in the mouse germ line entails the base excision repair pathway. Science 2010; 329:78–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Cortázar D, Kunz C, Selfridge J, Lettieri T, Saito Y, MacDougall E, Wirz A, Schuermann D, Jacobs AL, Siegrist F, Steinacher R, Jiricny J et al. Embryonic lethal phenotype reveals a function of TDG in maintaining epigenetic stability. Nature 2011; 470:419–423. [DOI] [PubMed] [Google Scholar]
- 59. Cortellino S, Xu J, Sannai M, Moore R, Caretti E, Cigliano A, Le Coz M, Devarajan K, Wessels A, Soprano D, Abramowitz LK, Bartolomei MS et al. Thymine DNA glycosylase is essential for active DNA demethylation by linked deamination-base excision repair. Cell 2011; 146:67–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Seah MKY, Messerschmidt DM. From germline to soma: epigenetic dynamics in the mouse preimplantation embryo. In: Plusa B, Hadjantonakis AK. (eds.), Current Topics in Developmental Biology. Academic Press; 2018; 128:203–235. [DOI] [PubMed] [Google Scholar]
- 61. Shen L, Inoue A, Lu F, Zhang Correspondence Y, He J, Liu Y, Zhang Y, Lu F, Zhang Y. Tet3 and DNA replication mediate demethylation of both the maternal and paternal genomes in mouse zygotes. Cell Stem Cell 2014; 15:459–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Tsukada Y-I, Akiyama T, Nakayama KI. Maternal TET3 is dispensable for embryonic development but is required for neonatal growth. Sci Rep 2015; 5:15876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Gu TP, Guo F, Yang H, Wu HP, Xu GF, Liu W, Xie ZG, Shi L, He X, Jin SG, Iqbal K, Shi YG et al. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature 2011; 477:606–610. [DOI] [PubMed] [Google Scholar]
- 64. Wossidlo M, Nakamura T, Lepikhov K, Marques CJ, Zakhartchenko V, Boiani M, Arand J, Nakano T, Reik W, Walter J. 5-Hydroxymethylcytosine in the mammalian zygote is linked with epigenetic reprogramming. Nat Comms 2011; 2:241. [DOI] [PubMed] [Google Scholar]
- 65. Kang J, Lienhard M, Pastor WA, Chawla A, Novotny M, Tsagaratou A, Lasken RS, Thompson EC, Surani MA, Koralov SB, Kalantry S, Chavez L et al. Simultaneous deletion of the methylcytosine oxidases Tet1 and Tet3 increases transcriptome variability in early embryogenesis. Proc Natl Acad Sci USA 2015; 112:E4236–E4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Inoue A, Zhang Y. Replication-dependent loss of 5-hydroxymethylcytosine in mouse preimplantation embryos. Science 2011; 334:194–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Inoue A, Shen L, Dai Q, He C, Zhang Y. Generation and replication-dependent dilution of 5fC and 5caC during mouse preimplantation development. Cell Res 2011; 21:1670–1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Amouroux R, Nashun B, Shirane K, Nakagawa S, Hill PWS, D’Souza Z, Nakayama M, Matsuda M, Turp A, Ndjetehe E, Encheva V, Kudo NR et al. De novo DNA methylation drives 5hmC accumulation in mouse zygotes. Nat Cell Biol 2016; 18:225–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Surani MAH, Barton SC, Norris ML, Surani L-L, Barton SC, Norris ML. Nuclear transplantation in the mouse: Heritable differences between parental genomes after activation of the embryonic genome. Cell 1986; 45:127–136. [DOI] [PubMed] [Google Scholar]
- 70. Sato M, Kimura T, Kurokawa K, Fujita Y, Abe K, Masuhara M, Yasunaga T, Ryo A, Yamamoto M, Nakano T. Identification of PGC7, a new gene expressed specifically in preimplantation embryos and germ cells. Mech Dev 2002; 113:91–94. [DOI] [PubMed] [Google Scholar]
- 71. Nakamura T, Arai Y, Umehara H, Masuhara M, Kimura T, Taniguchi H, Sekimoto T, Ikawa M, Yoneda Y, Okabe M, Tanaka S, Shiota K et al. PGC7/Stella protects against DNA demethylation in early embryogenesis. Nat Cell Biol 2007; 9:64–71. [DOI] [PubMed] [Google Scholar]
- 72. Bian C, Yu X. PGC7 suppresses TET3 for protecting DNA methylation. Nucleic Acids Res 2014; 42:2893–2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Grosser C, Wagner N, Grothaus K, Horsthemke B. Altering TET dioxygenase levels within physiological range affects DNA methylation dynamics of HEK293 cells. Epigenetics 2015; 10:819–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Wu H, Zhang Y. Mechanisms and functions of Tet protein-mediated 5-methylcytosine oxidation. Genes Dev 2011; 25:2436–2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Nakamura T, Liu Y-J, Nakashima H, Umehara H, Inoue K, Matoba S, Tachibana M, Ogura A, Shinkai Y, Nakano T. PGC7 binds histone H3K9me2 to protect against conversion of 5mC to 5hmC in early embryos. Nature 2012; 486:415–419. [DOI] [PubMed] [Google Scholar]
- 76. Li X, Ito M, Zhou F, Youngson N, Zuo X, Leder P, Ferguson-Smith AC. A maternal-zygotic effect gene, Zfp57, maintains both maternal and paternal imprints. Dev Cell 2008; 15:547–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Quenneville S, Verde G, Corsinotti A, Kapopoulou A, Jakobsson J, Offner S, Baglivo I, Pedone PV, Grimaldi G, Riccio A, Trono D. In embryonic stem cells, ZFP57/KAP1 recognize a methylated hexanucleotide to affect chromatin and DNA methylation of imprinting control regions. Mol Cell 2011; 44:361–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Strogantsev R, Krueger F, Yamazawa K, Shi H, Gould P, Goldman-Roberts M, McEwen K, Sun B, Pedersen R, Ferguson-Smith AC. Allele-specific binding of ZFP57 in the epigenetic regulation of imprinted and non-imprinted monoallelic expression. Genome Biol 2015; 16:112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Anvar Z, Cammisa M, Riso V, Baglivo I, Kukreja H, Sparago A, Girardot M, Lad S, De Feis I, Cerrato F, Angelini C, Feil R et al. ZFP57 recognizes multiple and closely spaced sequence motif variants to maintain repressive epigenetic marks in mouse embryonic stem cells. Nucleic Acids Res 2016; 44:1118–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Messerschmidt DM. Should I stay or should I go: protection and maintenance of DNA methylation at imprinted genes. Epigenetics 2012; 7:969–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Hirasawa R, Chiba H, Kaneda M, Tajima S, Li E, Jaenisch R, Sasaki H. Maternal and zygotic Dnmt1 are necessary and sufficient for the maintenance of DNA methylation imprints during preimplantation development. Genes Dev 2008; 22:1607–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Alexander KA, Wang X, Shibata M, Clark AG, García-García MJ. TRIM28 controls genomic imprinting through distinct mechanisms during and after early genome-wide reprogramming. Cell Rep 2015; 13:1194–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Lorthongpanich C, Cheow LF, Balu S, Quake SR, Knowles BB, Burkholder WF, Solter D, Messerschmidt DM. Single-cell DNA-methylation analysis reveals epigenetic chimerism in preimplantation embryos. Science 2013; 341:1110–1112. [DOI] [PubMed] [Google Scholar]
- 84. Kim S-H, Kang Y-K, Koo D-B, Kang M-J, Moon S-J, Lee K-K, Differential DNA methylation reprogramming of various repetitive sequences in mouse preimplantation embryos. Biochem Biophys Res Commun 2004; 324:58–63. [DOI] [PubMed] [Google Scholar]
- 85. Saitou M, Kagiwada S, Kurimoto K. Epigenetic reprogramming in mouse pre-implantation development and primordial germ cells. Development 2012; 139:15–31. [DOI] [PubMed] [Google Scholar]
- 86. Hargan-Calvopina J, Taylor S, Cook H, Chiang Y-SS, Chen P-YY, Correspondence ATC, Hu Z, Lee SA, Yen M-RR, Clark AT, Chiang Y-SS, Chen P-YY et al. Stage-specific demethylation in primordial germ cells safeguards against precocious differentiation. Dev Cell 2016; 39:75–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Popp C, Dean W, Feng S, Cokus SJ, Andrews S, Pellegrini M, Jacobsen SE, Reik W. Genome-wide erasure of DNA methylation in mouse primordial germ cells is affected by AID deficiency. Nature 2010; 463:1101–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Seisenberger S, Andrews S, Krueger F, Arand J, Walter J, Santos F, Popp C, Thienpont B, Dean W, Reik W. The dynamics of genome-wide DNA methylation reprogramming in mouse primordial germ cells. Mol Cell 2012; 48:849–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Guibert S, Forné T, Weber M. Global profiling of DNA methylation erasure in mouse primordial germ cells. Genome Res 2012; 22:633–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Seki Y, Hayashi K, Itoh K, Mizugaki M, Saitou M, Matsui Y. Extensive and orderly reprogramming of genome-wide chromatin modifications associated with specification and early development of germ cells in mice. Dev Biol 2005; 278:440–458. [DOI] [PubMed] [Google Scholar]
- 91. Hajkova P, Erhardt S, Lane N, Haaf T, El-Maarri O, Reik W, Walter J, Surani MA. Epigenetic reprogramming in mouse primordial germ cells. Mech Dev 2002; 117:15–23. [DOI] [PubMed] [Google Scholar]
- 92. Ohno R, Nakayama M, Naruse C, Okashita N, Takano O, Tachibana M, Asano M, Saitou M, Seki Y. A replication-dependent passive mechanism modulates DNA demethylation in mouse primordial germ cells. Development 2013; 140:2892–2903. [DOI] [PubMed] [Google Scholar]
- 93. Kurimoto K, Yabuta Y, Ohinata Y, Shigeta M, Yamanaka K, Saitou M. Complex genome-wide transcription dynamics orchestrated by Blimp1 for the specification of the germ cell lineage in mice. Genes Dev 2008; 22:1617–1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Yamaguchi S, Hong K, Liu R, Shen L, Inoue A, Diep D, Zhang K, Zhang Y. Tet1 controls meiosis by regulating meiotic gene expression. Nature 2012; 492:443–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Hackett JA, Sengupta R, Zylicz JJ, Murakami K, Lee C, Down TA, Surani MA. Germline DNA demethylation dynamics and imprint erasure through 5-hydroxymethylcytosine. Science 2013; 339:448–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Yamaguchi S, Shen L, Liu Y, Sendler D, Zhang Y. Role of Tet1 in erasure of genomic imprinting. Nature 2013; 504:460–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Kobayashi H, Sakurai T, Miura F, Imai M, Mochiduki K, Yanagisawa E, Sakashita A, Wakai T, Suzuki Y, Ito T, Matsui Y, Kono T. High-resolution DNA methylome analysis of primordial germ cells identifies gender-specific reprogramming in mice. Genome Res 2013; 23:616–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Lee J, Inoue K, Ono R, Ogonuki N, Kohda T, Kaneko-Ishino T, Ogura A, Ishino F. Erasing genomic imprinting memory in mouse clone embryos produced from day 11.5 primordial germ cells. Development 2002; 129:1807–1817. [DOI] [PubMed] [Google Scholar]
- 99. Sato S, Yoshimizu T, Sato E, Matsui Y. Erasure of methylation imprinting ofIgf2r during mouse primordial germ-cell development. Mol Reprod Dev 2003; 65:41–50. [DOI] [PubMed] [Google Scholar]
- 100. Piccolo FM, Bagci H, Brown KE, Landeira D, Soza-Ried J, Feytout A, Mooijman D, Hajkova P, Leitch HG, Tada T, Kriaucionis S, Dawlaty MM et al. Different roles for Tet1 and Tet2 proteins in reprogramming-mediated erasure of imprints induced by EGC fusion. Mol Cell 2013; 49:1023–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Dawlaty MM, Breiling A, Le T, Raddatz G, Barrasa MI, Cheng AW, Gao Q, Powell BE, Li Z, Xu M, Faull KF, Lyko F et al. Combined deficiency of Tet1 and Tet2 causes epigenetic abnormalities but is compatible with postnatal development. Dev Cell 2013; 24:310–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Kagiwada S, Kurimoto K, Hirota T, Yamaji M, Saitou M. Replication-coupled passive DNA demethylation for the erasure of genome imprints in mice. EMBO J 2013; 32:340–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Stewart KR, Veselovska L, Kelsey G. Establishment and functions of DNA methylation in the germline. Epigenomics 2016; 8:1399–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Lucifero D, Mann MRW, Bartolomei MS, Trasler JM. Gene-specific timing and epigenetic memory in oocyte imprinting. Hum Mol Genet 2004; 13:839–849. [DOI] [PubMed] [Google Scholar]
- 105. Gahurova L, Tomizawa S, Smallwood SA, Stewart-Morgan KR, Saadeh H, Kim J, Andrews SR, Chen T, Kelsey G. Transcription and chromatin determinants of de novo DNA methylation timing in oocytes. Epigenetics Chromatin 2017; 10:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Stewart KR, Veselovska L, Kim J, Huang J, Saadeh H, Tomizawa SI, Smallwood SA, Chen T, Kelsey G. Dynamic changes in histone modifications precede de novo DNA methylation in oocytes. Genes Dev 2015; 29:2449–2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Henckel A, Chebli K, Kota SK, Arnaud P, Feil R. Transcription and histone methylation changes correlate with imprint acquisition in male germ cells. EMBO J 2012; 31:606–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Watanabe T, Tomizawa S, Mitsuya K, Totoki Y, Yamamoto Y, Kuramochi-Miyagawa S, Iida N, Hoki Y, Murphy PJ, Toyoda A, Gotoh K, Hiura H et al. Role for piRNAs and noncoding RNA in de novo DNA methylation of the imprinted mouse Rasgrf1 locus. Science 2011; 332:848–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Caspary T, Cleary MA, Baker CC, Guan XJ, Tilghman SM. Multiple mechanisms regulate imprinting chromosome 7 gene cluster. Dev Biol 1998; 198:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Weaver JR, Sarkisian G, Krapp C, Mager J, Mann MRW, Bartolomei MS. Domain-specific response of imprinted genes to reduced DNMT1. Mol Cell Biol 2010; 30:3916–3928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Sharif J, Muto M, Takebayashi S-I, Suetake I, Iwamatsu A, Endo TA, Shinga J, Mizutani-Koseki Y, Toyoda T, Okamura K, Tajima S, Mitsuya K et al. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature 2007; 450:908–912. [DOI] [PubMed] [Google Scholar]
- 112. Hata K, Okano M, Lei H, Li E. Dnmt3L cooperates with the Dnmt3 family of de novo DNA methyltransferases to establish maternal imprints in mice. Development 2002; 129:1983–1993. [DOI] [PubMed] [Google Scholar]
- 113. Kaneda M, Okano M, Hata K, Sado T, Tsujimoto H, Li E, Sasaki H, Tsujimoto N, Li E, Sasaki H. Essential role for de novo DNA methyltransferase Dnmt3a in paternal and maternal imprinting. Nature 2004; 429:900–903. [DOI] [PubMed] [Google Scholar]
- 114. Bourc’his D, Xu G-L, Lin C-S, Bollman B, Bestor TH. Dnmt3L and the establishment of maternal genomic imprints. Science 2001; 294:2536–2539. [DOI] [PubMed] [Google Scholar]
- 115. Nakashima H, Kimura T, Kaga Y, Nakatani T, Seki Y, Nakamura T, Nakano T. Effects of Dppa3 on DNA methylation dynamics during primordial germ cell development in mice. Biol Reprod 2013; 88:125. [DOI] [PubMed] [Google Scholar]
- 116. Messerschmidt DM, de Vries W, Ito M, Solter D, Ferguson-Smith A, Knowles BB, Trim28 is required for epigenetic stability during mouse oocyte to embryo transition. Science 2012; 335:1499–1502. [DOI] [PubMed] [Google Scholar]