Abstract
Cwc23 is a member of the J protein family, and has been shown to interact with Ntr1, a scaffold protein that interacts with Ntr2 and Prp43 to form the NTR complex that mediates spliceosome disassembly. We show that Cwc23 is also an intrinsic component of the NTR complex, and that it interacts with the carboxyl terminus of Ntr1. Metabolic depletion of Cwc23 concurrently depleted Ntr1 and Ntr2, suggesting a role for Cwc23 in stabilizing these two proteins. Ntr1, Ntr2 and Cwc23 are stoichiometrically balanced, and form a stable heterotrimer. Depletion of Cwc23 from splicing extracts using antibodies resulted in depletion of all three proteins and accumulation of intron-lariat in the splicing reaction. Cwc23 is not required for disassembly of intron-lariat spliceosome (ILS), but facilitates disassembly of spliceosome intermediates after the actions of Prp2 and Prp16 by stabilizing the association of Ntr1 with the spliceosome. Cwc23 has a more limited effect on the association of Ntr1 with the ILS. Our data suggest that Cwc23 is important for maintaining the levels of Ntr1 and Ntr2, and that it also plays a regulatory role in targeting spliceosome intermediates for disassembly.
INTRODUCTION
Splicing is a process that removes intervening sequences from precursor mRNAs. The process is catalyzed by the spliceosome, which consists of five small nuclear RNAs (snRNAs)—U1, U2, U4, U5 and U6—in the form of ribonucleoprotein particles (RNPs), as well as many protein factors. Assembly of the spliceosome is achieved by ordered addition of the snRNPs and protein components to the pre-mRNA. Following the association of all five snRNAs, extensive remodeling of the spliceosome through release of U1 and U4, and addition of the Prp19-associated complex (NTC for NineTeen Complex) establishes the RNA catalytic center, forming the activated spliceosome (1–4). The catalytic reaction then proceeds via two steps of transesterification.
Two DEAH-box proteins, Prp2 and Prp16, are required for the catalytic reactions. In the first step, Prp2 and its cofactor Spp2 mediate the release of U2 components SF3a and SF3b to allow interaction of the branch site with the 5′ splice site (3–6). Yju2 and Cwc25 are required to stabilize the interaction in promoting the reaction (7,8). In the second step, Prp16 mediates the removal of Yju2 and Cwc25 from the spliceosome (9–11), and Slu7, Prp18 and Prp22 can then promote the reaction (12–16). After completion of the splicing reaction, the spliceosome is disassembled in two steps mediated by the DEAH-box proteins Prp22 and Prp43. Prp22 first catalyzes the release of mRNA from the spliceosome (17,18), and then Prp43 mediates disassembly of the remaining intron-lariat spliceosome (ILS) to recycle the spliceosomal components (19–25).
We have previously identified a protein complex NTR (NTC-related)—consisting of Ntr1, Ntr2 and Prp43—that is required to catalyze disassembly of the spliceosome (26). Ntr1 functions as a scaffold, interacting with Prp43 through its N-terminal G-patch domain and with Ntr2 via a middle domain (26–28). The G-patch domain of Ntr1 has been shown to activate the ATPase or helicase activity of Prp43, thereby triggering spliceosome disassembly (29–31). Prp43 can also mediate disassembly of spliceosome intermediates, but only after the actions of Prp2 and Prp16, linking its function to disposal of spliceosome intermediates in a splicing fidelity control mechanism (32–34). The Ntr1 sequence beyond its G-patch domain has been proposed to play a regulatory role in distinguishing ILS and spliceosome intermediates (35).
Cwc23 was initially identified to be a component of the Cef1/Ntc85-associated complex in a proteomic study (36), and was then demonstrated to exhibit both physical and genetic interactions with Ntr1, and to be required for pre-mRNA splicing in vivo (27,37). Recent determination of the cryo-EM structure of the ILS complex also revealed interaction of Cwc23 with Ntr1 on the spliceosome (38). Cwc23 contains a J domain, which is dispensable for cellular growth and for splicing (37). The functional role of Cwc23 in splicing has not been established.
Here, we show that Cwc23 is an intrinsic component of the NTR complex. Cwc23 associates with Ntr1 and Ntr2 to form a stable heterotrimer that can further interact with Prp43 to form a tetrameric complex. Depletion through antibodies of Cwc23 from splicing extracts resulted in depletion all three proteins, and accumulation of intron-lariat in the splicing reaction. Cwc23 interacts with Ntr1 but not with Ntr2 or Prp43. The very C-terminal segment of Ntr1 is essential and sufficient for its interaction with Cwc23. Metabolic depletion of Cwc23 led to concurrent reductions in the levels of Ntr1 and Ntr2, suggesting that Cwc23 is required for maintaining the levels of Ntr1 and Ntr2 proteins in cell extracts. Cwc23 is not required for disassembly of ILS, but it does facilitate disassembly of spliceosome intermediates by stabilizing the association of Ntr1 with the spliceosome. Our results reveal an essential role for Cwc23 in maintaining the integrity of the NTR complex, as well as a regulatory role in promoting disassembly of spliceosome intermediates.
MATERIALS AND METHODS
Yeast strains
The yeast strains used were BJ2168 (MATaprc1 prb1 pep4 leu2 trp1 ura3), YSCC23 (MATaprc1 prb1 pep4 leu2 trp1 ura3 CWC23-HA), YSCC232 (MATaprc1 prb1 pep4 leu2 trp1 CWC23::LEU2, pRS414.GAL-CWC23) and YSCC152 (MATaprc1 prb1 pep4 leu2 trp1 NTR2::LEU2, pRS414.GAL-NTR2).
Antibodies and reagents
Anti-hemagglutinin (anti-HA) monoclonal antibody 8G5F was produced by immunizing mice with a keyhole limpet hemocyanin (KLH)-conjugated HA peptide. Anti-Ntc20, anti-Cwc23, anti-Cwc25 and anti-Slu7 antibodies were produced by immunizing rabbits with corresponding full-length recombinant proteins expressed in Escherichia coli. Other antibodies were produced by immunizing rabbits with protein segments as follows: anti-Prp22, amino acids (aa) 1–484; anti-Ntr1, aa 19–367 or aa 563–708; and anti-Prp43, aa 1–101. Protein A-Sepharose (PAS) was obtained from GE Healthcare Life Sciences.
Splicing extracts and substrate
Splicing extracts were prepared according to Cheng et al. (39). Actin precursors were synthesized with SP6 RNA polymerase (Promega) using EcoRI-linearized pSP64–88 plasmid as the template.
Immunoprecipitation of spliceosomes, and immunoprecipitation and immunodepletion of components from splicing extracts
Immunoprecipitation of the spliceosome with anti-Ntc20, anti-Cwc23 or anti-Ntr1 antibody was performed according to Cheng and Abelson (40). For each 10 μl of the splicing reaction, 1 μl of anti-Ntc20, 10 μl of anti-Cwc23 and 3 μl of anti-Ntr1 antibodies were used. Splicing reaction mixtures were incubated with antibody-conjugated PAS at 4°C for 1 h. After centrifugation, the precipitates were washed four times with 1 ml of NET-2 (50 mM Tris–HCl, pH7.4, 150 mM NaCl and 0.05% NP-40), and RNA extracted for analysis. Immunoprecipitation of NTR complex from splicing extracts was performed by incubation of 100 μl of Cwc23-HA extracts with 50 μl of PAS conjugated with 100 μl monoclonal anti-HA antibody (8G5F). Immunodepletion of Cwc25, Slu7, Prp22, Ntr1, Prp43 or Cwc23 was performed by incubation of 200 μl of yeast splicing extracts with 200 μl of anti-Cwc25, 200 μl of anti-Slu7, 200 μl of anti-Prp22, 50 μl of anti-Ntr1, 80 μl of anti-Prp43 or 200 μl of anti-Cwc23 antibody, respectively, conjugated to 100 μl PAS, at 4°C for 1 h. After centrifugation, the supernatant fractions were collected as depleted extracts and stored at –80°C.
Purification of NTR complex, Ntr1–Ntr2 dimer, GST-Ntr1CT and Cwc23
The NTR complex was affinity purified from yeast as described by Tsai et al. using Ntr1-HA extracts (26). Ntr1–Ntr2 dimer was purified by co-expression of the two proteins in E. coli Rosetta strain in separate plasmids. NTR1 was cloned into plasmid vector pSUMO-2 and expressed as a HIS-SUMO-fused protein. NTR2 was cloned into plasmid vector pRSFDuet-1 and expressed as untagged protein. Ntr1–Ntr2 dimer was purified through three consecutive chromatographic steps using HisTrap, Mono-Q and Superdex-G200 columns (GE Healthcare Life Sciences). Escherichia coli cells were grown at 37°C until OD600 ∼0.6, and induced for expression of HIS-SUMO-Ntr1 and Ntr2 by addition of 0.5 mM of IPTG, and then grown at 17°C overnight. Cells harvested from each liter of the cell culture were resuspended in 8.3 ml of lysis buffer, which contains 50 mM Tris–HCl, pH 8.0, 500 mM NaCl, 20 mM imidazole and 1–2 tablets of protease inhibitor, passed through a syringe with No. 21 needle to reduce clotting, and then lysed by passing through microfludizer. Cell lysates were loaded onto a 1-ml Ni2+-charged HisTrap HP column pre-equilibrated with buffer A, which contains 50 mM Tris–HCl, pH 8.0, 500 mM NaCl and 20 mM imidazole. The column was washed with 10 column volumes of buffer A, and the bound proteins were eluted with a 15 column volumes of linear gradient of increasing amounts of imidazole to a final concentration of 500 mM. The fractions containing HIS-SUMO-Ntr1 and Ntr2 were collected, diluted five-fold with 50 mM Tris–HCl, pH 8.0, loaded onto a Mono-Q column, and eluted with a 20 column volumes of linear gradient of increasing salt concentrations from 0.1 to 1 M NaCl. After concentration, the HIS-SUMO-Ntr1 and Ntr2 dimeric complex was further purified through a Sephadex-G200 column, concentrated and added glycerol to final 20% for storage. CWC23 was cloned into plasmid vector pET-15 and expressed in E. coli Rosetta strain as HIS-tagged protein. One liter of cells were grown at 37°C until OD600 ∼0.5, and induced for expression of Cwc23 by addition of 1 mM of IPTG, and then grown at 17°C overnight. After centrifugation at 8000 rpm for 10 min, cell pellets were resuspended in 40 ml of lysis buffer (50 mM NaPO4, pH 8.0, 300 mM NaCl, 10 mM imidazole and 1 mM PMSF), and passed through a microfludizer. Cell lysates were centrifuged at 15 000 rpm for 30 min to remove cell debris, and the supernatant was applied to a 1-ml Ni-NTA affinity column. After washing with 20 column volumes of wash buffer (50 mM NaPO4, pH 8.0, 300 mM NaCl, 20 mM imidazole and 1 mM PMSF), bound proteins were eluted with 10 column volumes of elution buffer (50 mM NaPO4, pH 8.0, 300 mM NaCl, 250 mM imidazole and 1 mM PMSF). The eluent was dialyzed against 2 l of Buffer DK (20 mM HEPES-KOH, pH 7.9, 60 mM KPO4, pH 7.0, 50 mM NaCl, 0.2 mM EDTA, 0.5 mM DTT and 20% glycerol), and stored at –80°C in aliquots. The C-terminal segment of Ntr1 (Ntr1CT), representing amino acid residues 563–708, was expressed as a GST-fused protein in E. coli by cloning the corresponding DNA segment into plasmid vector pGEX-5X-3. Recombinant GST-Ntr1CT was purified by chromatography on Glutathione Sepharose (GE Healthcare Life Sciences).
Yeast two-hybrid assays
Ntr1, Ntr2, Prp43 and Ntr1 deletion mutants were fused to the LexA-DNA binding domain in plasmid pEG202, and Cwc23 fused to the Gal4-activation domain in plasmid pACT2. Ntr1 deletion mutants were constructed as described in Tsai et al. (26). Each pairs of plasmids were transformed into S. cerevisiae strain EGY48 together with the reporter plasmid pSH18–34. Interactions were assessed by the expression of β-galactosidase.
Gradient sedimentation
Sedimentation analysis of the purified NTR complex was performed in 10–30% glycerol gradients in a buffer containing 20 mM HEPES, pH 7.9, 100 mM NaCl and 0.2 mM EDTA. Gradients were centrifuged at 50 000 rpm in an SW60 rotor (Beckman Coulter) for 3 h at 4°C, and collected in 0.2 ml fractions.
Spliceosome disassembly assays
Spliceosomes were isolated by precipitation of 10 μl of the reaction mixture with 1 μl of anti-Ntc20 antibody conjugated to 10 μl PAS. The isolated spliceosomes were then incubated at 25°C for 20 min with a 30 μl buffer containing 8 mM HEPES, pH 7.9, 60 mM KPO4, pH 7.0, 20 mM NaCl, 0.08 mM EDTA, 1 mM spermidine, 2 mM ATP, 4 mM MgCl2 and 1 U/μl RNasin, together with purified Ntr1–Ntr2, Prp43 and Cwc23. After centrifugation, the supernatant was removed to a new tube, and the pellet was washed twice first with 70 μl and then 100 μl of the incubation buffer. All supernatant fractions were combined to a total volume of 200 μl. RNA was extracted from both pellet and supernatant fractions for analysis.
NTR binding assays
Splicing reactions were performed in splicing extracts depleted of various components, and the reaction mixtures were depleted of ATP by addition of 10 mM glucose and incubation at 25°C for 5 min. Following addition of Ntr1–Ntr2 dimer with or without 400 ng of recombinant His-Cwc23 and incubation at 25°C for 5 min, each 10 μl of the reaction mixture was precipitated with 3 μl of anti-Ntr1 antibody conjugated to 10 μl PAS. After removal of supernatant, the pellets were washed four times with 1 ml NET-2, and RNA extracted for analysis.
Quantification
Gels were exposed to GE Storage Phosphor Screen (GE healthcare Life Sciences). RNA bands were quantified with Typhoon™ FLA 9000 (GE healthcare Life Sciences), and analyzed with ImageQuant TL7.0 (GE healthcare Life Sciences).
RESULTS
Cwc23 is an intrinsic component of the NTR complex
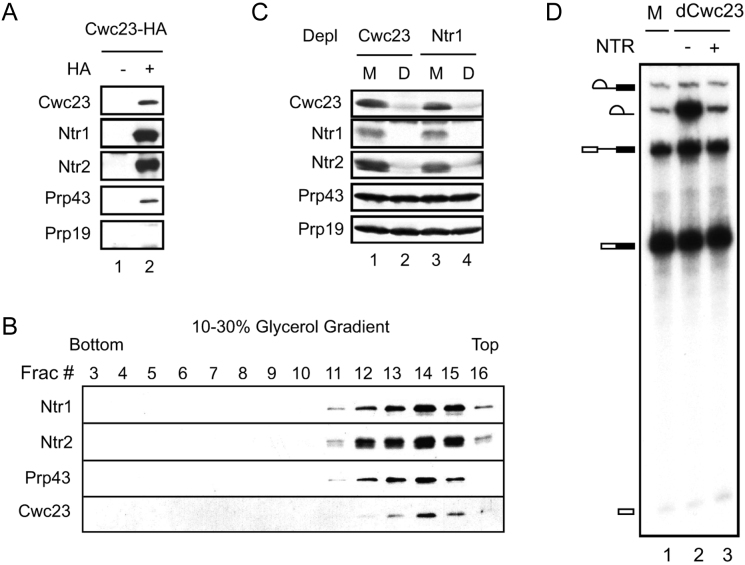
Cwc23 has previously been demonstrated to have genetic and two-hybrid interactions with Ntr1, suggesting a possible role for Cwc23 in disassembly of the spliceosome (37). To determine if Cwc23 is a component of the NTR complex, we tagged Cwc23 with HA for immunoprecipitation analysis. Splicing extracts prepared from Cwc23-HA or from an untagged strain were precipitated with anti-HA antibody and then subjected to Western blotting, probed with polyclonal anti-Ntr1, anti-Ntr2 and anti-Prp43 antibodies. As shown in Figure 1A, Ntr1, Ntr2 and Prp43 co-precipitated with Cwc23 (lane 2), but the NTC component Prp19 did not. To ascertain if Cwc23, Ntr1, Ntr2 and Prp43 form a complex, we fractionated the NTR complex purified from Ntr1-HA extracts on a 10−30% glycerol gradient, followed by Western blotting. As shown in Figure 1B, Ntr1, Ntr2, Prp43 and Cwc23 all co-sedimented in fractions 12–15, suggesting that these four proteins are associated in a complex (Figure 1B).
Figure 1.
Cwc23 is a component of the NTR complex. (A) Western blotting of Cwc23-HA extracts precipitated without (lane 1) or with (lane 2) anti-HA antibody. (B) Western blotting of gradient fractions of the NTR complex. (C) Western blotting of extracts depleted of Cwc23 (lane 2) or Ntr1 (lane 4). M, mock-depletion; D, depletion. (D) Splicing in Cwc23-depleted extracts without (lane 2) or with (lane 3) supplementation of NTR complex. M, mock-depletion; dCwc23, Cwc23-depletion.
We have previously shown that Ntr1 and Ntr2 are stoichiometrically balanced and form a stable complex that can further interact with Prp43 (26). Depletion of Ntr1 results in nearly complete depletion of Ntr2, and vice versa, whereas the amount of Prp43 remains relatively unchanged (26). To assess whether Cwc23 exhibits stoichiometric interactions with Ntr1 and Ntr2, we depleted extracts with polyclonal anti-Cwc23 or anti-Ntr1 antibody and then determined the amounts of Ntr1, Ntr2, Prp43 and Cwc23 in the depleted extracts by Western blotting. Figure 1C shows that depletion of Cwc23 resulted in nearly complete depletion of Ntr1 and Ntr2 (lane 2), and depletion of Ntr1 also resulted in complete depletion of Ntr2 and Cwc23 (lane 4), but the amounts of Prp43 or Prp19 were not significantly affected. Although Prp43 was coprecipitated with Cwc23, <10% of Prp43 was co-depleted with Cwc23 under this condition. These results indicate that the majority of Cwc23, Ntr1 and Ntr2 are associated with the NTR complex, but only a small amount of Prp43 is associated with Ntr1, Ntr2 and Cwc23. We further confirmed functional association of Cwc23 with the NTR complex through accumulation of intron-lariat when splicing was performed in extracts depleted of Cwc23 using anti-Cwc23 antibody (Figure 1D, lane 2), as well as the abolition of intron accumulation upon addition of affinity-purified NTR complex to Cwc23-depleted extracts (lane 3). Together, these results suggest that Cwc23 is associated with the NTR complex, forming a functional unit that mediates disassembly of the spliceosome.
Cwc23 interacts with the carboxyl terminus of Ntr1 and is important for maintaining levels of Ntr1 and Ntr2
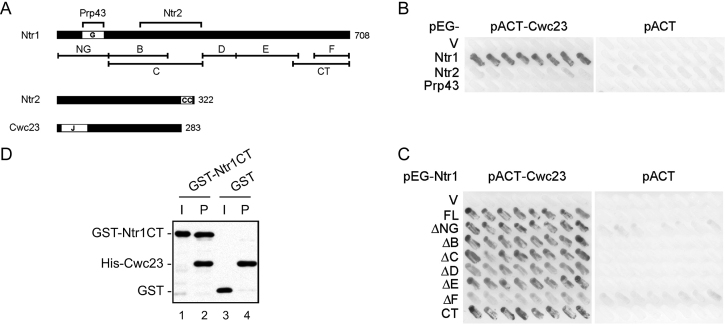
Ntr1 has previously been shown to function as a scaffold for the NTR complex through interactions with both Ntr2 and Prp43. Prp43 interacts with the G-patch domain of Ntr1, whereas Ntr2 interacts with a middle region of Ntr1 between amino acid residues 123–344 (Figure 2A) (26). We examined interactions of Cwc23 with NTR components by two-hybrid assays (Figure 2B). Ntr1, Ntr2 and Prp43 were fused to the LexA-DNA binding domain, and Cwc23 fused to the GAL4-activation domain. Interactions were measured according to β-galactosidase expression on plates. Cwc23 was found to interact with Ntr1, but not with Ntr2 or Prp43. We then performed deletion analysis to map the Cwc23-interacting domain on Ntr1. Figure 2C shows that only deletion of the C-terminal region (amino acid residues 649–708, ΔF) abolished the interaction between Ntr1 and Cwc23. In fact, the carboxyl terminal region alone (amino acid residues 564–708, CT) was sufficient for the interaction, indicating that the very C-terminal region of Ntr1 is essential and sufficient for its interaction with Cwc23. Interaction of Cwc23 with Ntr1CT was further confirmed by pull-down assays. Cwc23 tagged with 6xHis was expressed in E. coli, and conjugated with Ni-NTA. His-Cwc23-conjugated beads were then incubated with lysates expressing recombinant GST-fused Ntr1CT, and the precipitates were analyzed by Western blotting and probed with anti-GST antibody for Ntr1CT and with anti-His antibody for Cwc23. Figure 2D shows that GST-Ntr1CT (lane 2), but not GST alone (lane 4), was pulled-down by Cwc23-conjugated Ni-NTA. These results suggest that Ntr1CT is sufficient for stable interaction of Ntr1 with Cwc23.
Figure 2.
Interactions of Cwc23 with the C-terminus of Ntr1. (A) Maps of Ntr1, Ntr2 and Cwc23 proteins showing conserved motifs of the proteins and Ntr1 domains for deletion analysis. Regions responsible for Ntr1 interactions with Prp43 and Ntr2 are shown above the map. G, G-patch; CC, coiled coil; J, J-domain; CT, C-terminal fragment. (B) Two-hybrid assays for interactions of Cwc23 with NTR components. (C) Two-hybrid assays for interactions of Cwc23 with Ntr1 deletion mutants. (D) Pull-down of GST-fused Ntr1CT by His-Cwc23 probed with anti-GST and anti-Cwc23 antibodies. I, input; P, precipitates.
Fourmann et al. have shown that disassembly of purified intron-lariat spliceosome (ILS) requires only Prp43 and Ntr1, and that the latter can be replaced by the G-patch domain of Ntr1 or Pfa1, suggesting that Ntr2 is not required to mediate disassembly of purified ILS (35,41). However, Ntr2 is essential for cellular viability, and for splicing in vivo as both intron-lariat and pre-mRNA accumulate in cells metabolically depleted of Ntr2 (ΔNtr2) (26). Furthermore, when splicing was performed in ΔNtr2 extracts, in which Ntr2 was nearly completely depleted but the level of Ntr1 or Prp43 was not greatly affected, intron-lariat also accumulated, suggesting an important role of Ntr2 in spliceosome disassembly (20). Together, these results reveal a fundamental difference between disassembly of ILS in extracts and purified spliceosomes, suggesting an as yet unrevealed disassembly mechanism.
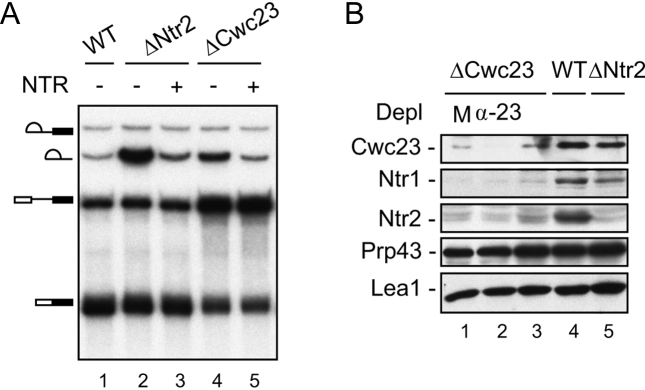
To address the role of Cwc23 in spliceosome disassembly, we constructed a CWC23-repressible yeast strain by placing CWC23 under the control of GAL1-promoter for depletion of Cwc23. Although CWC23 is an essential gene, cells of this yeast strain grew normally in glucose-supplemented media (Supplementary Figure S1). We then prepared extracts from cells grown in glucose-supplemented media and assessed the in vitro splicing reaction (Figure 3A). In contrast to ΔNtr2 extracts (lane 2) that accumulated a large amount of intron-lariat (85% of mRNA), only a smaller amount of intron-lariat (53% of mRNA) accumulated in Cwc23-metabolically-depleted extracts (ΔCwc23) (lane 4). Addition of purified NTR complex decreased the amounts of intron accumulation in both extracts (10% of mRNA in ΔNtr2 and 13% in ΔCwc23) (lanes 3 and 5). These results suggest that disassembly of the spliceosome was not as significantly impaired in ΔCwc23 extracts as in ΔNtr2 extracts. Examination of proteins in ΔCwc23 extracts by Western blotting revealed the presence of a small amount of Cwc23 (Figure 3B, lane 3), which could be further depleted using antibody against Cwc23 (lane 2), indicating that Cwc23 was not completely depleted under repressive conditions, and that the small amount of Cwc23 in ΔCwc23 extracts was sufficient for disassembly of ILS spliceosomes. Concurrently, the amounts of Ntr1 and Ntr2 were reduced to very low levels, whereas the level of Prp43 was not greatly affected. This finding suggests that Cwc23 might be required for stabilization of the Ntr1 protein, and that Ntr2 is also destabilized without Ntr1. Furthermore, the residual levels of Ntr1, Ntr2 and Cwc23 in ΔCwc23 extracts were sufficient to mediate spliceosome disassembly in the splicing reaction and to support cellular growth. In contrast, neither Ntr1 nor Cwc23 was greatly affected in Ntr2-metabolically-depleted extracts (lane 5).
Figure 3.
Metabolic depletion of Cwc23 resulted in co-depletion of Ntr1 and Ntr2. (A) Splicing assays were performed in wild-type extracts (lane 1) or extracts metabolically-depleted of Ntr2 (lanes 2 and 3) or Cwc23 (lanes 4 and 5), without (lanes 1, 2 and 4) or with (lanes 3 and 5) supplementation of purified NTR complex. (B) Western blotting of wild-type (lane 4) or Ntr2-metabolically-depleted extracts (lane 5) or untreated (lane 3), mock-treated (lane1) or anti-Cwc23 antibody-treated (lane 2) Cwc23-metabolically-depleted extracts. M, mock-treated; Depl, depletion.
Cwc23 is not required for disassembly of intron-lariat spliceosomes
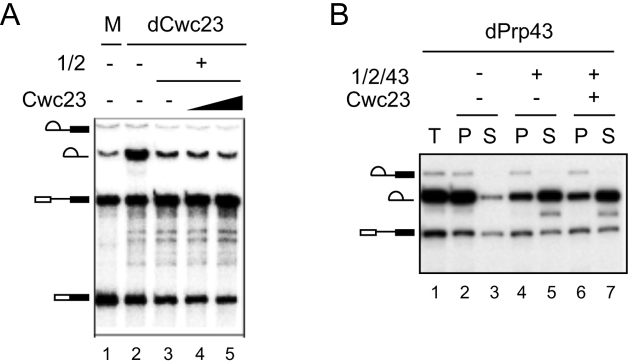
Since Cwc23 could not be completely depleted by our genetic method, we then depleted the NTR complex in vitro using antibodies and then supplemented the depleted extract with Ntr1 and Ntr2 to examine the functional role of Cwc23. Splicing extracts were depleted of NTR using anti-Cwc23 antibody. Recombinant Ntr1 and Ntr2 proteins, co-expressed and purified from E. coli (Ntr1–Ntr2), were added to the depleted extracts with or without further addition of Cwc23 prior to the reaction. Figure 4A shows accumulation of intron-lariat in the NTR-depleted extracts (lane 2). Addition of Ntr1–Ntr2 prevented intron accumulation without (lane 3) or with (lanes 4 and 5) addition of Cwc23, suggesting that Cwc23 is not required for disassembly of ILS in the extracts. We then examined whether Cwc23 is required for disassembly of the affinity-purified ILS (Figure 4B). Spliceosomes were isolated from reactions performed in Prp43-depleted extracts by precipitation with anti-Ntc20 antibody (lane 1). Around 50% of the spliceosome was recovered under this condition. The precipitated spliceosomes were then incubated with a mixture containing Ntr1–Ntr2 and Prp43 with or without Cwc23. The fractions of intron-lariat released from beads were similar in the presence (lanes 6 and 7) or absence (lanes 4 and 5) of Cwc23, suggesting that Cwc23 has no function in disassembly of purified ILS. Together, these results suggest that Cwc23 does not participate in disassembly of ILS either in extracts or on purified spliceosomes.
Figure 4.
Cwc23 is not required for disassembly of ILS. (A) Splicing was performed in Cwc23-depleted extracts (lane 2) supplemented with recombinant Ntr1–Ntr2 dimer, without (lane 3) or with the addition of 5 ng/μl (lane 4) or 40 ng/μl (lane 5) of Cwc23. (B) Splicing reactions performed in Prp43-depleted extracts were precipitated with anti-Ntc20 antibody (lane 1). The isolated spliceosomes were incubated without (lanes 2 and 3) or with (lanes 4–7) Ntr–Ntr2 dimer and Prp43, without (lanes 4 and 5) or with (lanes 6 and 7) further addition of Cwc23. Supernatant and pellet fractions were separated for analysis. M, mock; 1/2, Ntr1–Ntr2; 1/2/43, Ntr1–Ntr2 and Prp43; T, total precipitates; P, pellet; S, supernatant.
Cwc23 facilitates disassembly of spliceosome intermediates by stabilizing the association of Ntr1 with spliceosome intermediates
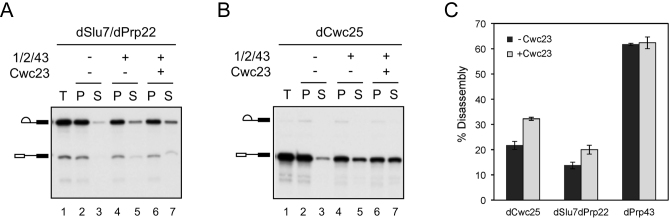
It has been shown that besides mediating disassembly of ILS, NTR also mediates disassembly of spliceosome intermediates stalled in the middle of the pathway (19,35). Spliceosomes arrested after the actions of Prp2 and Prp16 are susceptible to NTR-mediated disassembly, possibly coupling with a mechanism for splicing fidelity control (19,42). To see whether Cwc23 plays a role in mediating disassembly of spliceosome intermediates, splicing was performed in extracts depleted of Slu7 and Prp22 for spliceosomes arrested after Prp16 action, or depleted of Cwc25 for spliceosomes arrested after Prp2 action, and the spliceosomes were isolated by precipitation with anti-Ntc20 antibody. Ntr1–Ntr2 and Prp43 were added to the precipitated spliceosome with or without Cwc23, and the fractions of lariat-intron-exon 2 (Figure 5A) or pre-mRNA (5B) released from beads were analyzed. Quantification of the results revealed that in the presence of Cwc23, disassembly efficiency increased from 22% to 32% and from 14% to 20% for spliceosomes isolated from Cwc25-depleted and from Slu7 and Prp22 doubly-depleted extracts, respectively, whereas there was no significant difference observed for ILS (Figure 5C). These results suggest that Cwc23 may facilitate disassembly of spliceosome intermediates after the actions of Prp2 and Prp16.
Figure 5.
Cwc23 facilitates disassembly of spliceosome intermediates. (A) Splicing reactions performed in Slu7 and Prp22 doubly-depleted extracts were precipitated with anti-Ntc20 antibody (lane 1). The isolated spliceosomes were incubated without (lanes 2 and 3) or with (lanes 4–7) Ntr1–Ntr2 and Prp43, without (lanes 4 and 5) or with (lanes 6 and 7) further addition of Cwc23. Supernatant and pellet fractions were separated for analysis. (B) Splicing reactions performed in Cwc25-depleted extracts were precipitated with anti-Ntc20 antibody (lane 1). The isolated spliceosomes were incubated without (lanes 2 and 3) or with (lanes 4–7) Ntr1–Ntr2 dimer and Prp43, without (lanes 4 and 5) or with (lanes 6 and 7) further addition of Cwc23. Supernatant and pellet fractions were separated for analysis. (C) Quantification of disassembly efficiency in the presence or absence of Cwc23. Amounts of RNA from Figures 4B, 5A and 5B were quantified by a PhosphoImager. Data are presented as mean ± SD of three (dCwc25 and dPrp43) or four (dSlu7/dPrp22) replicates. P values for dCwc25, dSlu7/dPrp22 and dPrp43 are 0.000933, 0.002647 and 0.778647, respectively. 1/2/43, Ntr1–Ntr2 and Prp43; T, total precipitates; P, pellet; S, supernatant.
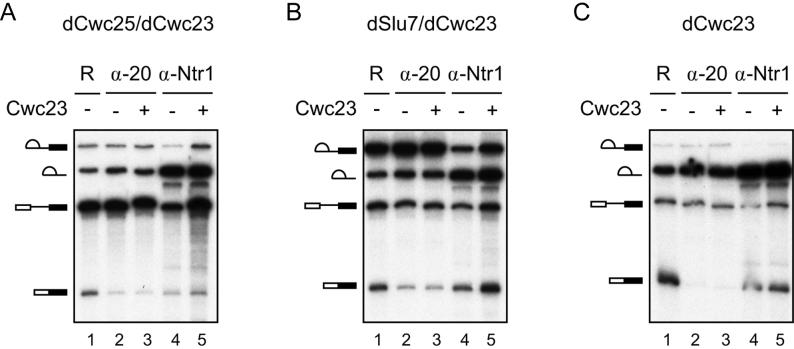
NTR has been shown to mediate disassembly of spliceosome intermediates, but with a lower efficiency than for disassembly of ILS (19), possibly due to weaker NTR interaction with spliceosome intermediates than with ILS. We have previously shown that Ntr2 might play a role in recruiting NTR to the spliceosome, and Ntr2 interacts with lower affinity for spliceosome intermediates than for ILS (19). We speculated that Cwc23 might function in stabilizing NTR on the spliceosome to facilitate disassembly of spliceosome intermediates, so we then examined the association of NTR with spliceosomes arrested after the actions of Prp2 and Prp16 in the presence or absence of Cwc23. Splicing extracts were depleted of NTR using anti-Cwc23 antibody, without (Figure 6C) or with further depletion of Cwc25 (Figure 6A) or Slu7 (Figure 6B) to block the splicing pathway after the action of Prp2 or Prp16, respectively, and then supplemented these extracts with Ntr1–Ntr2 dimer with or without addition of Cwc23. After these extracts underwent splicing, the reaction mixtures were precipitated with anti-Ntc20 or anti-Ntr1 antibody to isolate total activated and NTR-associated spliceosomes, respectively. The amounts of pre-mRNA, intron-exon 2 (IVS-E2) and excised intron (IVS) precipitated by anti-Ntr1 or anti-Ntc20 antibody from Cwc25 and Cwc23 doubly-depleted, Slu7 and Cwc23 doubly-depleted and Cwc23-depleted extracts, respectively, were measured, and the percentage of RNA precipitated by anti-Ntr1 antibody compared to that precipitated by anti-Ntc20 antibody was calculated. As shown in Table 1, in the presence of Cwc23, Ntr1 was more stably associated with all the spliceosomal complexes, i.e. those containing pre-mRNA, IVS-E2 or IVS. Nevertheless, Cwc23 stabilized 3–4-fold the Ntr1 associations with spliceosomal intermediates bur only 1.6-fold Ntr1 associations with ILS, suggesting that Cwc23 is more effective in stabilizing NTR on spliceosomal intermediates than on ILS. Figure 7 shows the steps of the splicing pathway in which Cwc23 may facilitate disassembly of stalled spliceosome intermediates.
Figure 6.
Cwc23 stabilizes the association of NTR with the spliceosome. Splicing was performed in Cwc25 and Cwc23 doubly-depleted (A), Slu7 and Cwc23 doubly-depleted (B) or Cwc23-depleted (C) extracts. Following depletion of ATP, the reaction mixtures were supplemented with Ntr1–Ntr2 dimer with or without Cwc23 and then precipitated with anti-Ntc20 (lanes 2 and 3) or anti-Ntr1 antibody (lanes 4 and 5). R, reaction mixture; α-20, anti-Ntc20 antibody. The relative amount of the reaction mixture used is 1:2.5:25 for R:α-20:α-Ntr1.
Table 1. Stabilization of NTR on the spliceosome by Cwc23.
| Extract | RNAa | % Ntr1/Ntc20b | ||
|---|---|---|---|---|
| -Cwc23 | +Cwc23 | Fold (+23/–23)c | ||
| dCwc25/dCwc23 | Pre-mRNA | 4.3 ± 0.3 | 14.7 ± 0.6 | 3.4 |
| dSlu7/dCwc23 | IVS-E2 | 1.2 ± 0.1 | 4.9 ± 0.1 | 4.1 |
| dCwc23 | IVS | 23.8 ± 2.0 | 37.7 ± 3.4 | 1.6 |
aRNA species quantified.
bData are presented as mean ± SD from three replicates. P values for dCwc25/dCwc23, dSlu7/dCwc23 and dCwc23 are 0.000011, 0.000003 and 0.003805, respectively.
cFold of Ntr1-associated RNA in the presence versus in the absence of Cwc23.
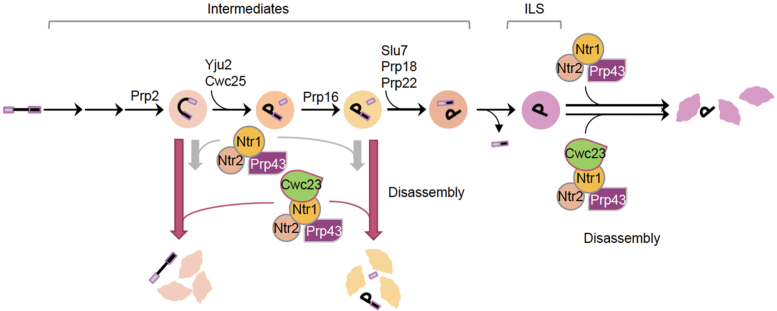
Figure 7.
Schematic for the role of Cwc23 in disassembly of spliceosome intermediates. The NTR complex mediates disassembly of ILS and spliceosome intermediates with different efficiency. The presence of Cwc23 facilitates disassembly of stalled spliceosome intermediates after the actions of Prp2 and Prp16, but does not significantly affect the disassembly of ILS. The length of arrows in the disassembly steps indicates the efficiency of disassembly.
DISCUSSION
Cwc23 was initially identified to be associated with the NTC component Cef1/Ntc85 in a proteomic study (36), and was later shown to exhibit genetic and two-hybrid interactions with the spliceosome disassembly factor Ntr1 (19,27,28). Cwc23 was also shown to be required for pre-mRNA splicing in vivo (37), but its role in the splicing reaction was unknown. Here we show that Cwc23 is an intrinsic component of the NTR complex involved in spliceosome disassembly. Cwc23, Ntr1 and Ntr2 form a stable heterotrimer, and together these proteins can further interact with Prp43 to form a functional complex that catalyzes spliceosome disassembly. In vitro depletion of Cwc23 using antibodies resulted in co-depletion of both Ntr1 and Ntr2, and depletion of Ntr1 also co-depleted Ntr2 and Cwc23, suggesting that these three proteins are present in extracts only in association with each other.
We have previously shown that Ntr2 is required for spliceosome disassembly in vitro (20). Intron-lariat accumulates during splicing in Ntr2-metabolically-depleted extracts. Stable association of Ntr1 with the spliceosome is inhibited in the absence of Ntr2, suggesting that Ntr2 may play a role in recruiting NTR or in stabilizing the association of NTR with the spliceosome (20). It has recently been reported that NTR components may also play regulatory roles in mediating spliceosome disassembly (35,41). Using affinity-purified spliceosomes, it was shown that Prp43 and the G-patch domain of Ntr1 (Ntr1GP), either as two separate components or fused as one protein, are sufficient to catalyze disassembly of ILS, suggesting that Ntr2 and the bulk of Ntr1 are dispensable for mediating disassembly. Ntr1GP has been shown to activate the ATPase or helicase activity of Prp43, triggering the disassembly reaction (29–31). Other G-patch proteins, Gno1 and Pfa1, have also been demonstrated to modulate the activity of Prp43 (35,41,43). Prp43 alone with the Ntr1GP fused to its amino terminus is sufficient to catalyze disassembly of the spliceosome after the action of Prp5, Prp28, Brr2, Prp2 or Prp22 (41), whereas full-length Ntr1 only supports Prp43 in catalyzing disassembly of ILS but not spliceosome intermediates. Amino acid sequences outside Ntr1GP were proposed to play a role in negatively regulating the function of Prp43 on spliceosome intermediates (35). Disassembly of spliceosome intermediates has been shown to link with the action of DExD/H-box proteins, several of which have been implicated in fidelity control of the splicing reaction (44–46). Thus, Ntr1 or other components of NTR may play regulatory roles in splicing fidelity control.
It is interesting that Ntr2 is required for disassembly of ILS in cell extracts, but not for affinity-purified ILS. It may be that ILS disassembly can occur via Ntr2-dependent and Ntr2-independent pathways and that the Ntr2-independent pathway is inhibited in cell extracts. Specific factors that inhibit the Ntr2-independent pathway may not be present on the affinity-purified spliceosome, so disassembly can proceed in the absence of Ntr2. This is not an unlikely scenario since Prp43 was reported to crosslink to two regions of U6 (47), and presumably may initiate disassembly by disrupting U2-U6 or U6-intron interactions. On the other hand, a recent study using Prp43-Ntr1GP fusion protein for disassembly of various purified spliceosome intermediates suggests that U2-intron interaction is a major target of Prp43 in the disassembly process (41). Whether Ntr2 is required for targeting Prp43 to U6 to initiate disassembly of the spliceosome remains to be investigated.
Since Prp43 and Ntr1GP are sufficient to catalyze disassembly of affinity-purified ILS, Cwc23 seems to be dispensable for the reaction, and we confirmed this in our analysis using spliceosomes purified from Prp43-depleted extracts. Unlike Ntr2, Cwc23 is also dispensable for catalyzing disassembly in cell extracts. However, Cwc23 does play a role in facilitating disassembly of the spliceosome intermediates that accumulate after the action of Prp2 or Prp16. In the presence of Cwc23, disassembly efficiency increased by 40–50% for spliceosome intermediates, whereas we did not observe a significant difference for ILS. This increased disassembly efficiency may be attributable to stabilization of the NTR complex on spliceosome intermediates, since the association of Ntr1 with spliceosome intermediates increased 3–4-fold in the presence of Cwc23. Although the association of Ntr1 with ILS also increased 1.6-fold in the presence of Cwc23, the presence of Cwc23 did not significantly enhance the efficiency of disassembly. Since NTR-mediated disassembly of ILS is more efficient than that of spliceosome intermediates, further stabilization of NTR by Cwc23 may not be necessary to promote disassembly.
We show that Cwc23 interacts with a C-terminal region of Ntr1. The Ntr1 region including amino acid residues 564–708 is essential and sufficient for its interaction with Cwc23. A cryo-EM structure of ILS revealed that its C-terminal region (amino acid residues 626–708) interacted with Cwc23 and Snu114 (38). Ntr1 also interacts with Ntr2 through amino acid residues 123–344 in the superhelical domain and with Prp43 through its G-patch domain (26). Cwc23, Ntr2 and Prp43 do not interact with each other. Ntr1 thus serves as a scaffold to support the NTR complex, apart from its function in activating Prp43. Ntr2 also interacts with the C-terminal domain of Brr2, which has been shown to interact with several splicing factors involved in the catalytic step, including Prp2, Prp16 and Slu7 (19,48). Brr2 has been suggested to serve as a platform for recruiting splicing factors to the spliceosome for catalytic reactions (49). Ntr2 can compete with Prp16 and Slu7 for interactions with Brr2, and it likely mediates recruitment of the NTR complex to the spliceosome through its interactions with Brr2. NTR and ILS interactions are stabilized when Ntr1 then interacts with Snu114. Alternatively, NTR can also be recruited directly through the interaction of Ntr1 with Snu114.
Cwc23 appears to play only a minor role in mediating disassembly of the spliceosome and no role in disassembly of ILS. Promoting disassembly of spliceosome intermediates is presumably important for splicing fidelity control, but it cannot account for the essentiality of CWC23 for cellular viability. However, Cwc23 is essential to maintain the integrity of the NTR complex, which is important for the recycling of spliceosomal components. Cwc23 likely protects Ntr1 from degradation by proteases by interacting with the C-terminal region of Ntr1, although we dd not find any typical motifs for ubiquitin-mediated protein degradation among sequences of the C-terminal region. Alternatively, expression of NTR1, NTR2 and CWC23 might be coordinately regulated, which is less likely since in vivo depletion of Ntr2 has little impact on the levels of Ntr1 and Cwc23.
Cwc23 is the only known splicing factor that contains a J-domain. Nevertheless, the J-domain of Cwc23 is dispensable for cellular viability and for splicing. It is also not required for the interaction of Cwc23 with Ntr1 (37), and is unlikely to play a major role in the disassembly of the spliceosome. The J-domain of Cwc23 has been shown to stimulate the ATPase activity of Hsp70 to a level similar to that by Ydj1, and changing the conserved H50 residue to Q eliminates its stimulatory ability, suggesting the Cwc23 J-domain being functional (37). Although the absence of the J domain does not compromise the interaction between Cwc23 and Ntr1, mutations in Ntr1 or Prp43 that affect the interaction between Ntr1 and Prp43 exhibit synthetic lethality with deletion of the J domain (37), suggesting that the J domain may play a role in promoting the interaction between Ntr1 and Prp43 under suboptimal conditons. On the other hand, we have observed no significant change in the levels of Ntr1 and Ntr2 in extracts with Cwc23’s J-domain deleted (data not shown), suggesting that the J domain may not contribute to maintaining the integrity of the Ntr1–Ntr2–Cwc23 trimeric complex.
Supplementary Material
ACKNOWLEDGEMENTS
We thank J. O’Brien for English editing and members of the Cheng laboratory for helpful discussions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online.
FUNDING
Academia Sinica and Ministry of Science and Technology, Taiwan: MoST105-2321-B-001-013 to S.-C.C.; MoST105-2811-B-001-051 to Y.-L.S. Funding for open access charge: MoST105-2321-B-001-013.
Conflict of interest statement. None declared.
REFERENCES
- 1. Tarn W.-Y., Hsu C.-H., Huang K.-T., Chen H.-R., Kao H.-Y., Lee K.-R., Cheng S.-C.. Functional association of essential splicing factor(s) with PRP19 in a protein complex. EMBO J. 1994; 13:2421–2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tarn W.-Y., Lee K.-R., Cheng S.-C.. Yeast precursor mRNA processing protein PRP19 associates with the spliceosome concomitant with or just after dissociation of U4 small nuclear RNA. Proc. Natl. Acad. Sci. U.S.A. 1993; 90:10821–10825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chan S.-P., Cheng S.-C.. The Prp19-associated complex is required for specifying interactions of U5 and U6 with pre-mRNA during spliceosome activation. J. Biol. Chem. 2005; 280:31190–31199. [DOI] [PubMed] [Google Scholar]
- 4. Chan S.-P., Kao D.-I., Tsai W.-Y., Cheng S.-C.. The Prp19p-associated complex in spliceosome activation. Science. 2003; 302:279–282. [DOI] [PubMed] [Google Scholar]
- 5. Kim S.-H., Lin R.-J.. Spliceosome activation by PRP2 ATPase prior to the first transesterification reaction of pre-mRNA splicing. Mol. Cell. Biol. 1996; 16:6810–6819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Warkocki Z., Schneider C., Mozaffari-Jovin S., Schmitzova J., Hobartner C., Fabrizio P., Lührmann R.. The G-patch protein Spp2 couples the spliceosome-stimulated ATPase activity of the DEAH-box protein Prp2 to catalytic activation of the spliceosome. Genes Dev. 2015; 29:94–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Liu H.-L., Cheng S.-C.. The interaction of Prp2 with a defined region of the intron is required for the first splicing reaction. Mol. Cell. Biol. 2012; 32:5056–5066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ohrt T., Prior M., Dannenberg J., Odenwalder P., Dybkov O., Rasche N., Schmitzova J., Gregor I., Fabrizio P., Enderiein J. et al. Prp2-mediated protein rearrangements at the catalytic core of the spliceosome as revealed by dcFCCS. RNA. 2012; 18:1244–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chiu Y.-F., Liu Y.-C., Chiang T.-W., Yeh T.-C., Tseng C.-K., Wu N.-Y., Cheng S.-C.. Cwc25 is a novel splicing factor required after Prp2 and Yju2 to facilitate the first catalytic reaction. Mol. Cell. Biol. 2009; 29:5671–5678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Liu Y.-C., Chen H.-C., Wu N.-Y., Cheng S.-C.. A novel splicing factor Yju2 is associated with NTC and acts after Prp2 in promoting the first catalytic reaction of pre-mRNA splicing. Mol. Cell. Biol. 2007; 27:5403–5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tseng C.-K., Liu H.-L., Cheng S.-C.. DEAH-box ATPase Prp16 has dual roles in remodeling of the spliceosome in catalytic steps. RNA. 2011; 17:145–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Horowitz D.S., Abelson J.. A U5 small nuclear ribonucleoprotein particle protein involved only in the second step of pre-mRNA splicing in Saccharomyces cerevisiae. Mol. Cell. Biol. 1993; 13:2959–2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Horowitz D.S., Abelson J.. Stages in the second reaction of pre-mRNA splicing: the final step is ATP independent. Genes Dev. 1993; 7:320–329. [DOI] [PubMed] [Google Scholar]
- 14. Ansari A., Schwer B.. SLU7 and a novel activity, SSF1, act during the PRP16-dependent step of yeast pre-mRNA splicing. EMBO J. 1995; 14:4001–4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jones M.H., Frank D.N., Guthrie C.. Characterization and functional ordering of Slu7p and Prp17p during the second step of pre-mRNA splicing in yeast. Proc. Natl. Acad. Sci. U.S.A. 1995; 92:9687–9691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schwer B., Gross C.H.. Prp22, a DExH-box RNA helicase, plays two distinct roles in yeast pre-mRNA splicing. EMBO J. 1998; 17:2086–2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Schwer B. A conformational rearrangement in the spliceosome sets the stage for Prp22-dependent mRNA release. Mol. Cell. 2008; 30:743–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wagner J.D., Jankowsky E., Company M., P. A.M., Abelson J.N.. The DEAH-box protein PRP22 is an ATPase that mediates ATP-dependent mRNA release from the spliceosome and unwinds RNA duplexes. EMBO J. 1998; 17:2926–2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chen H.-C., Tseng C.-K., Tsai R.-T., Chung C.-S., Cheng S.-C.. Link of NTR-mediated spliceosome disassembly with DEAH-box ATPases Prp2, Prp16 and Prp22. Mol. Cell. Biol. 2013; 33:514–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tsai R.-T., Tseng C.-K., Lee P.-J., Chen H.-C., Fu R.-H., Chang K.-J., Yeh F.-L., Cheng S.-C.. Dynamic interactions of Ntr1–Ntr2 with Prp43 and with U5 govern the recruitment of Prp43 to mediate spliceosome disassembly. Mol. Cell. Biol. 2007; 27:8027–8037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tseng C.-K., Chung C.-S., Chen H.-C., Cheng S.-C.. A central role of Cwc25 in spliceosome dynamics during catalytic phase of pre-mRNA splicing. RNA. 2017; 23:546–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fourmann J., Schmitzová J., Christian H., Urlaub H., Ficner R., Boon K.-L., Fabrizio P., Lührmann R.. Dissection of the factor requirements for spliceosome disassembly and the elucidation of its dissociation products using a purified splicing system. Genes Dev. 2013; 27:413–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Boon K., Auchynnikava T., Edwalds-Gilbert G., Barrass J.D., Droop A.P., Dez C., Beggs J.D.. Yeast Ntr1/Spp382 mediates Prp43 function in postspliceosomes. Mol. Cell. Biol. 2006; 26:6016–6023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Martin A., Schneider S., Schwer B.. Prp43 is an essential RNA-dependent ATPase required for release of lariat-intron from the spliceosome. J. Biol. Chem. 2002; 277:17743–17750. [DOI] [PubMed] [Google Scholar]
- 25. Arenas J.E., Abelson J.N.. Prp43: An RNA helicase-like factor involved in spliceosome disassembly. Proc. Natl. Acad. Sci. U.S.A. 1997; 94:11798–11802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tsai R.-T., Fu R.-H., Yeh F.-L., Tseng C.-K., Lin Y.-C., Huang Y.-h., Cheng S.-C.. Spliceosome disassembly catalyzed by Prp43 and its associated components Ntr1 and Ntr2. Genes Dev. 2005; 19:2991–3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pandit S., Lynn B., Rymond B.C.. Inhibition of a spliceosome turnover pathway suppresses splicing defects. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:13700–13705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pandit S., Paul S., Zhang L., Chen M., Durbin N., Harrison S.M., Rymond B.C.. Spp382p interacts with multiple yeast splicing factors, including possible regulators of Prp43 DExD/H-Box protein function. Genetics. 2009; 183:195–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tanaka N., Aronova A., Schwer B.. Ntr1 activates the Prp43 helicase to trigger release of lariat-intron from the spliceosome. Genes Dev. 2007; 21:2312–2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Christian H., Hofele R.V., Urlaub H., Ficner R.. Insights into the activation of the helicase Prp43 by biochemical studies and structural mass spectrometry. Nucleic Acids Res. 2014; 42:1162–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Robert-Paganin J., Rety S., Leulliot N.. Regulation of DEAH/RHA helicases by G-patch proteins. Biomed. Res. Int. 2015; 2015:931857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mayas R.M., Maita H., Semlow D.R., Staley J.P.. Spliceosome discards intermediates via the DEAH box ATPase Prp43p. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:10020–10025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Koodathingal P., Novak T., Piccirilli J.A., Staley J.P.. The DEAH box ATPases Prp16 and Prp43 cooperate to proofread 5' splice site cleavage during pre-mRNA splicing. Mol. Cell. 2010; 39:385–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Koodathingal P., Staley J.P.. Splicing fidelity: DEAD/H-box ATPases as molecular clocks. RNA Biol. 2013; 10:1073–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fourmann J., Tauchert M.J., Ficner R., Fabrizio P., Lührmann R.. Regulation of Prp43-mediated disassembly of spliceosomes by its cofactors Ntr1 and Ntr2. Nucleic Acids Res. 2017; 45:4068–4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ohi M.D., Link A.J., Ren L., Jennings J.L., McDonald W.H., Gould K.L.. Proteomics analysis reveals stable multiprotein complexes in both fission and budding yeasts containing Myb-related Cdc5p/Cef1p, novel pre-mRNA splicing factors, and snRNAs. Mol. Cell. Biol. 2002; 22:2011–2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sahi C., Lee T., Inada M., Pleiss J.A., Craig E.A.. Cwc23, an essential J protein critical for pre-mRNA splicing with a dispendable J domain. Mol. Cell. Biol. 2010; 30:33–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wan R., Yan C., Bai R., Lei J., Shi Y.. Structure of an intron lariat spliceosome from Saccharomyces cerevisiae. Cell. 2017; 171:120–132. [DOI] [PubMed] [Google Scholar]
- 39. Cheng S.-C., Newman A., Lin R.-J., McFarland G.D., Abelson J.N.. Preparation and fractionation of yeast splicing extract. Methods Enzymol. 1990; 181:89–96. [DOI] [PubMed] [Google Scholar]
- 40. Cheng S.-C., Abelson J.. Fractionation and characterization of a yeast mRNA splicing extract. Proc. Natl. Acad. Sci. U.S.A. 1986; 83:2387–2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Fourmann J., Dybkov O., Agafonov D.E., Tauchert M.J., Urlaub H., Ficner R., Fabrizio P., Lührmann R.. The target of the DEAH-box NTP triphosphatase Prp43 in Saccharomyces cerevisiae spliceosomes is the U2 snRNP-intron interaction. eLife. 2016; 5:e15564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mayas R.M., Maita H., Semlow D.R., Staley J.P.. Spliceosome discards intermediates via the DEAH box ATPase Prp43p. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:10020–10025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lebaron S., Papin C., Capeyrou R., Chen Y.-L., Fromont C., Monsarrat B., Caizergues-Ferrer M., Grigoriev M., Henry Y.. The ATPase and helicase activities of Prp43p are stimulated by the G-patch protein Pfa1p during yeast ribosome biogenesis. EMBO J. 2009; 28:3808–3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mayas R.M., Maita H., Staley J.P.. Exon ligation is proofread by the DExD/H-box ATPase Prp22p. Nat. Struct. Mol. Biol. 2006; 13:482–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Xu Y.-Z., Query C.C.. Competition obetween the ATPase Prp5 and branch region-U2 snRNA pairing modulates the fidelity of spliceosome assembly. Mol. Cell. 2007; 28:838–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Burgess S.M., Guthrie C.. A mechanism to enhance mRNA splicing fidelity: the RNA-dependent ATPase Prp16 governs usage of a discard pathway for aberrant lariat intermediates. Cell. 1993; 73:1377–1392. [DOI] [PubMed] [Google Scholar]
- 47. Bohnsack M.T., Martin R., Granneman S., Ruprecht M., Schleiff E., Tollervey D.. Prp43 bound at different sites on the pre-rRNA performs distinct functions in ribosome synthesis. Mol. Cell. 2009; 36:583–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. van Nues R.W., Beggs J.D.. Functional contacts with a range of splicing proteins suggest a centraal role for Brr2p in the dynamic control of the order of events in spliceosomes of Saccharomyces cerevisiae. Genetics. 2001; 157:1451–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhang L., Xu T., Maeder C., Bud L., Shanks J., Nix J., Guthrie C., Pleiss J., Zhao R.. Structural evidence for consecutive Hel308-like modules in the spliceosomal ATPase Brr2. Nat. Struct. Mol. Biol. 2009; 16:731–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.