Abstract
Background
Circular RNA (circRNA) is a novel member of the noncoding RNA and function as efficient microRNA sponges with gene-regulatory potential, especially the circular RNA ciRS-7 (CDR1as)/tumor suppressor miRNA-7 (miR-7) signals. However, the function of CDR1as/miR-7 in non-small cell lung cancer (NSCLC) is unknown.
Methods
Normal lung tissues (n=20), adjacent non-tumor tissues (n=60), and NSCLC tissues (n=60) were collected to determine the expression and significance of CDR1as/miR-7. Lung cancer cell lines A549 and H460 were overexpressed or knocked down of CDR1as, miR-7 to determine the tumor growth etc. The CDR1as/miR-7-related pathway were analyzed.
Results
CDR1as levels was robustly increased with the development of NSCLC (P<0.001) and the NSCLC tissues harbored highest expression of CDR1as, which negatively correlated to the expression of miR-7. Patients with high expression of CDR1as had high TNM stage (P=0.004), more lymph nodes metastasis (LNM) (P=0.021) and shorted overall survival time (OS) (P=0.0135). The CDR1as level was an independent prognostic factor for the patients with NSCLC. Overexpression of CDR1as induced increased cell vitalities and growth, which could be abrogated by knockdown of CDR1as or overexpressed miR-7 to induce apoptosis and G1/S arrest. Mechanistically, CDR1as functioned as miR-7 sponges to up-regulate the key target genes of miR-7 including EGFR, CCNE1 and PIK3CD. The results in vivo further confirmed that CDR1as functioned as oncogene to inhibit the anti-tumor effects of tumor suppressor miR-7 by up-regulation of proliferation index Ki-67, EGFR, CCNE1 and PIK3CD levels.
Conclusion
Overexpressed CDR1as in NSCLC functions promotes the tumor progression via miR-7 signals.
Keywords: CDR1as, miR-7, prognosis, tumor growth, NSCLC
Introduction
Lung cancer is a global health burden and remains the leading cause of cancer-related death worldwide, accounting for 19.4% of cancer-related deaths and 1.59 million deaths annually.1 Lung cancer is divided in two main groups including non-small-cell and small-cell carcinoma and non-small-cell lung cancer (NSCLC), which accounts for over 85% of all lung cancers.2 Because the majority of patients present with locally advanced or metastatic disease during early diagnosis, the prognosis for advanced-stage disease is poor and the 5-year overall survival (OS) rate is 17% in the US.3 Therefore, early detection of resectable and potentially curable disease may reduce the overall death rate for NSCLC.
Recently, the tumor suppressor or tumorigenic roles of noncoding RNAs (ncRNAs) have been identified in different types of cancer. The ncRNAs account for 95% of the total RNA in eukaryotic transcription, and they have no protein coding capacity; they include short microRNAs (miRNA), long ncRNAs, and circular RNAs (circRNA).4,5 circRNAs are newly discovered endogenous ncRNAs that are hundreds or even thousands of bases in length with a covalently closed structure, giving them a higher tolerance to RNA exonuclease.6 Therefore, due to the features such as structural stability, high abundance, and tissue-specific expression, the circRNAs could function as biomarkers for cancer.7 The mechanisms of circRNAs in gene regulation are involved with its function of competing endogenous RNAs or microRNA sponges during various biological processes and diseases including carcinogenesis.8
As the most extensively investigated circRNA, CDR1as, also known as ciRS-7, has been reported to participate the neu-ropsychiatric disorders,9 insulin secretion,9 myocardial infarction, and cancer.10,11 CDR1as was upregulated in the mice with myocardial infarction under hypoxic treatment, and CDR1as overexpression promoted cell apoptosis, but was reversed by miR-7a overexpression.10 In mammalian brain, animals with knockout of CDR1as displayed impaired sensorimotor gating, a deficit in the ability to filter out unnecessary information associated with neuropsychiatric disorders, which was associated with miRNA-7 (miR-7) and miR-671.6 In colorectal cancer (CRC), Weng et al11 found that the expression of CDR1as was significantly upregulated in CRC tissues compared with matched normal mucosa. Overexpression of ciRS-7 in CRC cell lines inhibited the tumor suppressive effects of miR-7 and resulted in a more aggressive oncogenic phenotype via inactivation of miR-7 targets and EGFR and RAF1 oncogenes. However, the role of CDR1as in NSCLC is unclear.
As the miR-7 target, the frequency of EGFR mutations in various Asian populations increases to 22%–62% of NSCLC.12 The EGFR inhibitors are approved by the US Food and Drug Administration for the patients with advanced NSCLC with activating EGFR mutations.13 Thus, we speculated that the CDR1as/miR-7 signals were associated with the development of NSCLC. The expressions, clinical significance, and functions of CDR1as/miR-7 in NSCLC were investigated in vitro and in vivo.
Materials and methods
Patients and tissue specimens
Normal lung tissues (n=20), adjacent nontumor tissues (n=60), and NSCLC tissues (n=60) were collected, as approved by the Clinical Research Ethics Committee of Yan’an People’s Hospital. All patients and healthy volunteers provided written informed consent. The patients with primary lung cancer were diagnosed by hematoxylin and eosin staining by experienced pathologists from the Department of Pathology at Yan’an People’s Hospital. None of the patients underwent preoperative chemotherapy and/or radiotherapy. Patients with other kinds of cancer or some other autoimmune diseases (eg, rheumatoid arthritis, systemic lupus erythematous, diabetes, etc) were absolutely excluded. Besides, pregnant and lactating individuals were also excluded from this study. All these retrospective specimens were handled according to ethical and legal standards of Yan’an People’s Hospital. The tissue samples were isolated from surgically removed samples and then stored at −80°C until use. All the patients were followed up until October 15, 2017. High or low expression of CDR1as was defined by the median of its expression.
RNA isolation and qRT-PCR
Total RNA from tissues or cell lines were extracted using Trizol reagent (Invitrogen, Carlsbad, CA, USA) and quantitative real-time RT-PCR (qRT-PCR) was performed according to the manufacturer’s instructions and 2−ΔΔCt method was used to estimate relative expression changes of genes including CDR1as and miR-7. The expression levels were normalized to GAPDH and U6 for gene expression.
Cell lines and reagents
The lung cancer cell lines A549 and H460 was purchased from the cell bank of the Chinese Academy of Sciences (Shanghai, People’s Republic of China) and cultured in Dulbecco’s Modified Eagle’s Medium supplemented with 10% fetal bovine serum (Life Technologies, Carlsbad, CA, USA), ampicillin and streptomycin at 37°C and in 5% CO2. pcDNA3-CDR1as and its negative control (which had an inserted ciRS-7 sequence only but no invert repeat flanking introns into the pcDNA3.1-empty plasmid, resulting in no circular CDR1as), shRNA-CDR1as, were conducted by GenePharma (Shanghai, People’s Republic of China). miR-7 mimics or negative control was purchased from RiboBio (Guangzhou, People’s Republic of China). Anti-GAPDH, -EGFR, -CCNE1, and -PIK3CD antibodies were obtained from Cell Signaling Tech (Denver, MA, USA) and Abcam (Cambridge, MA, USA).
Cell transfection
The A549 and H460 cell lines were cultured to about 80% confluence in 12/96-well plates and then, using Lipofectamine 2000 (Invitrogen), were transfected with indicated agents according to the manufacturer’s instructions.
CCK-8 assay
The A549 and H460 cell lines were transfected with pcDNA3 or siRNAs and then were harvested, washed with PBS, and then CCK-8 kit (Kumamoto, Japan) mixed with Dulbecco’s Modified Eagle’s Medium was used for cell viability assay, and the absorbance was measured at 450 nm using a microplate reader.
Immunohistochemistry
To determine the expression of ki-67 in tissues, 2-μm-thick, formalin-fixed and paraffin-embedded specimen sections were used. After the slides were incubated in xylene for 5 min, 100% ethanol was used for 10 min, followed by 95% ethanol for 10 min. Antigen unmasking was performed, and then the slides were blocked with 3% hydrogen peroxide for 30 min at room temperature. Then, the primary antibody for ki-67 was incubated with the formalin-fixed paraffin-embedded sections at 4°C overnight; then, the goat anti-mouse secondary antibody (Zymed Laboratories Inc., South San Francisco, CA, USA) were used for the detection of ki-67. After that, the EnVision Detection System kit (DAKO, Glostrup, Denmark) was used to assess the results.
Flow cytometry assay
For the cell cycle analysis, conditional A549 and H460 cells were stained with PI staining solution (10 μg/mL RNase A and 50 μg/mL PI) at 37°C for 30 min in the dark. The cell cycle distribution was analyzed using a flow cytometry with the Cell-Quest software. For the apoptosis analysis, the cells were fixed in cold 70% ethanol at −20°C for 2 h. About 10 mg/mL RNase was added and stained with 2 μL of annexin V mixed with 2 μL of PI (eBioscience, Santa Clara, CA, USA), and the results were assessed according to the manufacturer’s instructions and analyzed using a flow cytometer.
Western blots
Conditional A549 and H460 cells were harvested and, according to the manufacturer’s instructions, the whole cell protein extracts were resolved on a 10% SDS denatured polyacrylamide gel and were then transferred onto a nitrocellulose membrane, which was then blocked with 5% BSA in TBST buffer for 1 h at room temperature. Subsequently, incubation was performed with primary antibodies overnight at 4°C. GAPDH was used as the loading control in the Western blotting. After washing with TBST buffer, the blots were then incubated with HRP-conjugated secondary antibody for 1 h at room temperature. After washing with TBST buffer, the blots were visualized using the ECL-Plus reagent (Millipore, Billerica, MA, USA).
Tumor model
To investigate the role of CDR1as in vivo, 2×106 A549 cells transfected with lentivirus vector of pcDNA3.1-CDR1as or miR-7 alone, or both, and the samples were subcutaneously injected into the rear flank of nude mice (5 per group). The tumor sizes were measured 3 days apart, and the tumor volumes were calculated using the formula V (cm3) = width2 (cm2) * length (cm)/2. The animal study was approved by institutional animal research committee of Yan’an People’s Hospital, and animals were cared for following the guidelines for use and care of laboratory animals.
Statistical analyses
The results were analyzed using the Statistical Package for Social Sciences version 16.0 (SPSS 16.0, SPSS Inc., Chicago, IL, USA) and the Prism statistical software package (Version 5.0, Graphpad Software Inc., La Jolla, CA, USA). The Mann–Whitney U-test was used the differences between more than two groups were analyzed by the Kruskal–Wallis test. Kaplan–Meier survival curves and the log-rank statistic were used to analyze the prognostic significance of CDR1as. Correlations between the expression of CDR1as and miR-7 were analyzed by Spearman’s Rho analysis. Correlations of expression of CDR1as and clinicopathological characters were analyzed by Pearson χ2. Cox proportional hazard regression was used for univariate and multivariate analysis of OS according to CDR1as expression. P<0.05 was considered statistically significant.
Results
The expression of CDR1as was upregulated in NSCLC
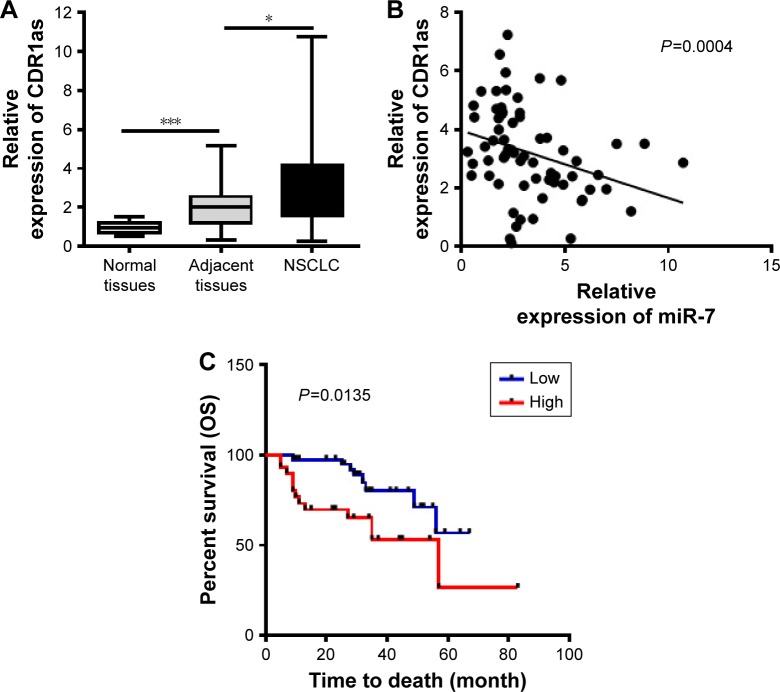
To investigate the clinical significance of CDR1as in NSCLC, the expressions of CDR1as in normal lung tissues (n=20), adjacent nontumor tissues (n=60), and NSCLC tissues (n=60) were determined by qPCR. The results showed that the CDR1as level was increased in NSCLC tissues and that the normal tissues had the lowest level of CDR1as (Figure 1A). Moreover, the statistical analysis in the three groups indicated that 19 out of 20 (95%) normal tissues showed low expression of CDR1as; this percentage was decreased to 48.3% in adjacent nontumor tissues, and the majority of NSCLC tissues showed high expression of CDR1as (76.7%) (Table 1). Because the miR-7 as tumor suppressor is often downregulated in cancer, the correlation between CDR1as and miR-7 was estimated. We found that the expression of CDR1as in tumor samples negatively correlated with the miR-7 level, which indicated that the CDR1as might inhibit miR-7 in NSCLC (Figure 1B).
Figure 1.
CDR1as is overexpressed in NSCLC and predicts a poor prognosis.
Notes: (A) The expression of CDR1as in normal lung tissues (n=20), adjacent nontumor tissues (n=60), and NSCLC tissues (n=60) samples were determined by qPCR. (B) The correlation between CDR1as and miR-7 was analyzed. (C) The OS was analyzed by Kaplan–Meier survival curves according to the three degree of CDR1as. *P<0.05, ***P<0.001, data represent the mean ± SD.
Abbreviations: NSCLC, non-small-cell lung cancer; OS, overall survival; qPCR, quantitative polymerase chain reaction.
Table 1.
CDR1as expression in patients with NSCLC
| Types | n | CDR1as expression
|
χ2 | P-values | |
|---|---|---|---|---|---|
| Low (%) | High (%) | ||||
| Normal lung tissues | 20 | 19 (95.0) | 1 (5.0) | 24.12 | 0.0001*** |
| Adjacent nontumor tissues | 60 | 29 (48.3) | 31 (51.7) | ||
| NSCLC | 60 | 19 (23.3) | 41 (76.7) | ||
Note:
P < 0.001.
Abbreviation: NSCLC, non-small-cell lung cancer.
Overexpressed CDR1as predicts poor clinical outcome of patients with NSCLC
We next analyzed the expression of CDR1as and the clinicopathological characteristics of NSCLC patients. The results showed that the expression of CDR1as was not associated with the gender, age, smoking, pT, histology, and differentiation of NSCLC patients, but the expression of CDR1as was found to be positively associated with TNM stage and lymph node metastasis (LNM) (Table 2). We also analyzed the prognostic role of CDR1as in the 60 NSCLC patients who were followed up until October 15, 2017. The OS was analyzed according to the expression of CDR1as. The patients with high expression of CDR1as had shorter OS than those with low expression of CDR1as (χ2=6.103 by Log-rank Test) (Figure 1C). Univariate and multivariate analyses were performed to analyze the prognostic role of CDR1as for NSCLC. The results revealed that gender, age, histology, TNM stage, LNM, and differentiation were not independent prognostic indicators for OS, but the pT and the expression of CDR1as were the independent prognostic factors for the OS of patients with NSCLC (Table 3). These results indicated that the upregulated CDR1as participated in the progression of NSCLC and could be a biomarker for NSCLC.
Table 2.
Relationships between CDR1as expression and clinical-pathologic characteristics in NSCLC patients
| Characteristics | n=70 | CDR1as expression
|
P-values | |
|---|---|---|---|---|
| Low (n/%) | High (n/%) | |||
| Sex | 0.155 | |||
| Male | 23 | 13 (56.5) | 20 (43.5) | |
| Female | 27 | 6 (22.2) | 21 (77.7) | |
| Age | 0.174 | |||
| <50 | 36 | 9 (25) | 27 (75) | |
| ≥50 | 24 | 10 (41.6) | 14 (58.3) | |
| Smoking | 0.919 | |||
| Smokers | 31 | 10 (32.3) | 21 (67.7) | |
| Never smokers | 29 | 9 (31) | 20 (69) | |
| pT | 0.077 | |||
| T1–2 | 31 | 13 (41.9) | 18 (58.1) | |
| T3–4 | 29 | 6 (20.7) | 23 (79.3) | |
| Histology | 0.141 | |||
| Squamous | 36 | 14 (38.9) | 22 (61.1) | |
| Adenocarcinoma | 24 | 5 (20.8) | 19 (79.2) | |
| TNM stage | 0.004** | |||
| I–II | 22 | 12 (54.5) | 10 (45.5) | |
| III–IV | 48 | 7 (14.6) | 31 (85.4) | |
| Differentiation | 0.088 | |||
| Well | 38 | 15 (39.5) | 23 (60.5) | |
| Moderate to poor | 22 | 4 (18.2) | 18 (81.8) | |
| LNM | 0.021** | |||
| No | 28 | 13 (46.4) | 15 (53.6) | |
| Yes | 32 | 6 (18.8) | 26 (81.2) | |
| Total | 70 | 19 (100) | 41 (100) | |
Note:
P < 0.001.
Abbreviations: LNM, lymph node metastasis; NSCLC, non-small-cell lung cancer.
Table 3.
Prognostic factors in the Cox proportional hazards model
| Variables | OS
|
|||||
|---|---|---|---|---|---|---|
| HR | Univariate 95% CI | Sig | HR | Multivariate 95% CI | Sig | |
| Gender | ||||||
| Male vs female | 1.232 | 0.528–1.312 | 0.782 | |||
| Age | ||||||
| <50 vs ≥50 | 0.982 | 0.785–2.201 | 0.936 | |||
| Smoking | ||||||
| No vs yes | 2.723 | 1.823–4.192 | 0.021 | 3.556 | 2.238–3.984 | 0.152 |
| pT | ||||||
| T1–2 vs T3–4 | 3.112 | 2.230–4.762 | 0.002* | 5.397 | 3.343–7.842 | 0.0001* |
| Histology | ||||||
| Squamous vs adenocarcinoma | 1.274 | 0.542–2.723 | 0.803 | |||
| TNM stage | ||||||
| I–II vs III–IV | 1.682 | 0.926–2.698 | 0.437 | |||
| Differentiation | ||||||
| Low vs moderate-high | 2.091 | 1.134–2.803 | 0.182 | |||
| LNM | ||||||
| − vs + | 1.892 | 1.117–2.437 | 0.235 | |||
| CDR1as expression | ||||||
| Low vs high | 3.583 | 1.668–4.826 | 0.006* | 6.132 | 2.923–7.556 | 0.0001* |
Note:
P < 0.05.
Abbreviations: CI, confidence interval; HR, hazard ratio; LNM, lymph node metastasis; OS, overall survival.
CDR1as inhibits the tumor suppressor miR-7 to promote cell growth in vitro
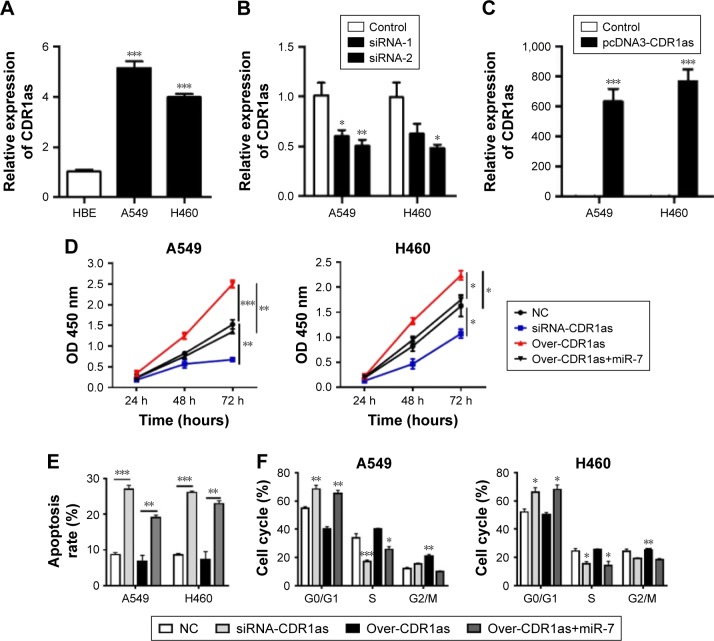
The clinical data had demonstrated that CDR1as was an unfavorable factor for the development of NSCLC; thus, we here further investigated the role of CDR1as in vitro in lung cancer cell lines A549 and H460. The expressions of CDR1as in normal lung epithelial cells HBE and lung cancer cell lines A549 and H460 were assessed, and it was found that the CDR1as level was higher in cancer cells than in normal lung epithelial cells (Figure 2A). The knockdown and overexpression experiments of CDR1as in A549 and H460 were performed (Figure 2B and C); the cell vitality was significantly impaired by the siRNA-CDR1as and improved by the overexpression of CDR1as. However, the CDR1as-induced increase of cell vitality could be inhibited by miR-7 (Figure 2D). Similarly, overexpression of CDR1as also abrogated the miR-7-induced apoptosis and G1/S arrest, which indicated that the tumor suppressive effects of miR-7 could be negatively regulated by CDR1as in NSCLC (Figure 2E and F).
Figure 2.
Overexpression of CDR1as antagonized the availability of miR-7 for tumor growth.
Notes: (A) The expression of CDR1as in normal lung epithelial cells HBE and lung cancer cell lines A549 and H460 were determined by qPCR. (B and C) The efficiency of knockdown or overexpression in A549 and H460 was determined. (D) The cell vitalities of two cell lines were estimated by CCK-8 assay. (E and F) The apoptosis and cell cycle were analyzed by flow cytometry. *P<0.05, **P<0.01, ***P<0.001, data represent the mean ± SD. Control bars are missing from part C as the level of control group was too low.
Abbreviations: qPCR, quantitative polymerase chain reaction; NC, negative control.
The cell growth-related targets of miR-7 were upregulated by CDR1as
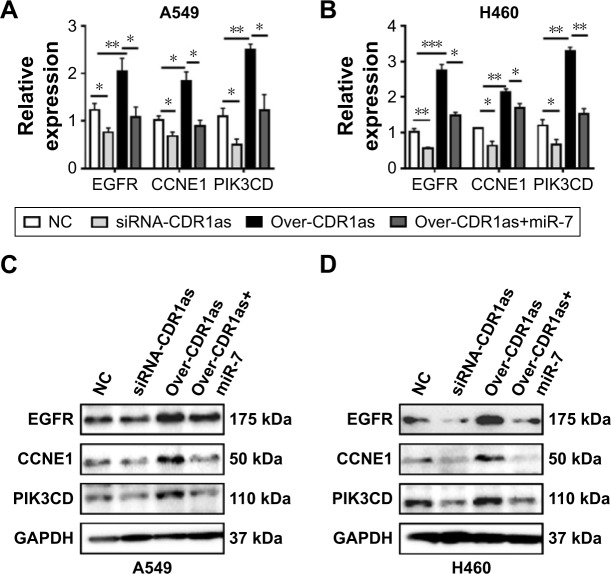
To clarify the potential mechanism of CDR1as-induced cell growth, we next investigated the miR-7 and its cell growth-related target genes, EGFR, CCNE1, and PIK3CD. We found that overexpression of CDR1as remarkably increased the gene expression of EGFR, CCNE1, and PIK3CD, which were restored by the overexpression of miR-7 in A549 (Figure 3A and C) and H460 (Figure 3B and D). These data suggested that CDR1as might promote the cell growth via the miR-7/EGFR/CCNE1/PIK3CD signals in NSCLC.
Figure 3.
The target genes of miR-7 are upregulated by CDR1as.
Notes: (A and B) The direct target genes’, EGFR, CCNE1, and PIK3CD, expression are determined by qPCR and (C and D) Western blotting. *P<0.05, **P<0.01, ***P<0.001, data represent the mean ± SD.
Abbreviations: qPCR, quantitative polymerase chain reaction; NC, negative control.
CDR1as functions as an oncogene to elevate tumor growth in vivo
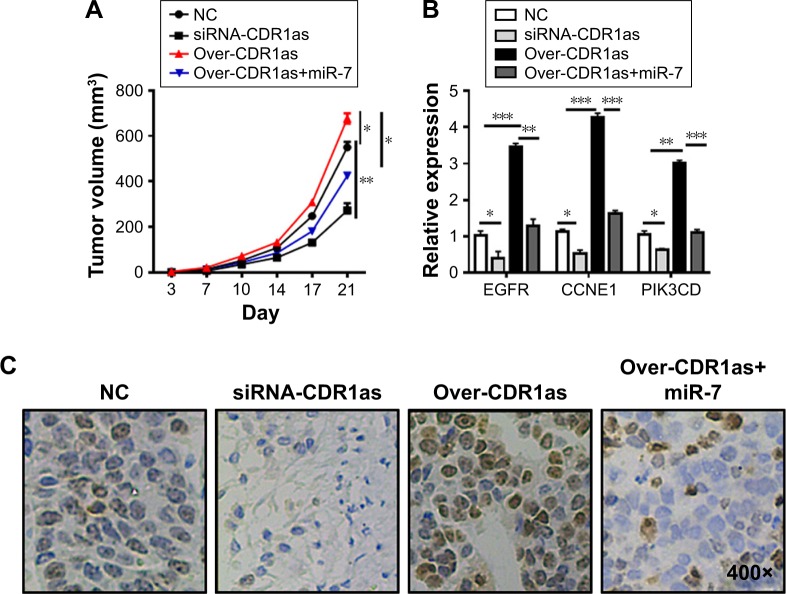
To confirm the tumorigenic role of CDR1as in vivo, the xenograft model of human A549 was established and the nude mice were administrated conditional tumor cells that were knocked down or had overexpressed CDR1as and/or miR-7. Knockdown of CDR1as significantly restricted the tumor growth with small tumor volume, whereas the tumor growth could be enhanced by the overexpression of CDR1as and inhibited by miR-7 (Figure 4A). The expression of tumor proliferation indication Ki-67 and miR-7 target genes were determined (Figure 4B), and it was found that CDR1as in vivo inhibits the function of miR-7 to upregulate the levels of EGFR, CCNE1, and PIK3CD (Figure 4C).
Figure 4.
ciRS-7 regulates miR-7 activity to promote tumor growth in vivo.
Notes: A549 cells were inoculated with lentivirus vector of pcDNA3.1-CDR1as or miR-7 alone, or both, and subcutaneously injected into nude mice. (A) The mean tumor size (mm3) was analyzed. (B) The expression levels of Ki67 in tumor tissues were estimated in different treatment groups. (C) The EGFR, CCNE1, and PIK3CD expression levels were determined by qPCR. *P<0.05, **P<0.01, ***P<0.001, data represent the mean ± SD.
Abbreviations: qPCR, quantitative polymerase chain reaction; NC, negative control.
Discussion
NSCLC accounts for over 85% of all lung cancers. Due to toxicity and/or efficacy issues, several currently available treatment options developed in the last decade for NSCLC (eg, bevacizumab, pemetrexed, and nintedanib) are not approved or are not suitable for use in patients with squamous histology.12 The investigation of diagnostic and prognostic factors for patients with NSCLC is conducive to the early detection, which also could provide novel targets for targeted therapy.10 In this study, we identified that the circRNA CDR1as functioned as an oncogene via the inhibition of tumor suppressor miR-7 and could predict poor clinical outcome of patients with NSCLC.
circRNAs are abundant, stable, conserved, and nonrandom products of RNA splicing, and mounting evidence have shed light on its function in normal cellular differentiation and tissue homeostasis as well as in disease development.14 Since circRNAs do not have susceptible 5′ and 3′ ends, they are resistant to endonuclease enzymatic degradation, which leads to more stability than linear RNAs and makes them very abundant in the cytoplasm.15 In basal cell carcinoma (BCC), Sand et al16 identified 23 upregulated (such as hsa_circ_0075828, hsa_circ_0075825, hsa_circ_0022383) and 48 downregulated (such as hsa_circ_0022392) circRNAs with 354 miRNA response elements in BCC that are potentially involved in the molecular pathogenesis of BCC.16 circRNA-Foxo3 was reported to be minimally expressed in breast cancer tissues when compared to benign samples, and this circRNA is likely to have a role in breast cancer progression.17 Shang et al18 found that there is a significant difference in the expression of hsa_circ_0005075 between hepatocellular carcinoma (HCC) and normal hepatocellular tissues, and high expression of hsa_circ_0005075 predicts high HCC tumor size. In this study, we found that CDR1as was upregulated in NSCLC tissues and the high expression of CDR1as correlated with the high stage of TNM, LNM, and shorter OS. Similarly, other circRNAs were also found to be involved in the progression of NSCLC. The expression of cir-ITCH was significantly decreased in lung cancer tissues.19 Upregulated circRNA 100876 in NSCLC was found to closely correlate with LNM and tumor stage and could be a potential prognostic biomarker for NSCLC.20
There are some structural features that render circRNAs its distinct potential functions, such as acting as miRNA sponges, or binding to RNA-associated proteins to form RNA–protein complexes that regulate gene transcription at the transcriptional or posttranscriptional level. The most studied miRNA in circRNA research is miR-7.21 The first evidence that circRNAs behave as miRNA sponges was CDR1as/miR-7 interaction. CDR1as contains over 70 binding sites for miR-7 and is densely bound by Argonaute proteins.6 cir-ITCH, derived from the ITCH gene, presents a sequence enriched with three miRNA-binding sites (miR-7, miR-17, and miR-214).22 circPVT1 is reported to function as let-7 sponge for the inhibition of cell senescence.23 In bladder cancer, circTCF25 as an miRNA sponge reduced the function of miR-103a-3p and miR-107 in cancerous tissue, leading to increased CDK6 expression and thus the proliferation of tumor cells.24 circHIPK3 was found to directly bind to miR-124 and inhibit miR-124 activity to upregulate two known proliferation-promoting targets (IL6R and DLX2) of miR-124 for cellular growth.25 In NSCLC, we found that CDR1as negatively correlated to miR-7 in tumor tissues and that overexpression of CDR1as in lung cancer cell lines inhibited miR-7-induced growth arrest apoptosis via upregulating the targets of miR-7 including EGFR/CCNE1/PIK3CD, which was consistent with the findings in CRC that CDR1as suppressed CRC cell proliferation and invasion via miR-7/EGFR/IGF-1R/RAF1 signals.11 Yu et al26 also reported that knockdown of CDR1as suppressed the HCC cell proliferation and invasion through targeting miR-7/CCNE1/PIK3CD expression. These findings emphasized the tumorigenic role of CDR1as/miR-7 in NSCLC.
Conclusion
We investigated the prognostic role of CDR1as in NSCLC patients and found that the high expression of CDR1as correlated with high TNM stage, LNM, and short survival time. Overexpression of CDR1as promoted the proliferation and inhibited apoptosis of lung cancer cells via upregulating cell growth-related target genes of miR-7 including EGFR, CCNE1, and PIK3CD in vitro and in vivo.
Footnotes
Author contributions
All authors contributed toward data analysis, drafting and revising the paper and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Socinski MA, Obasaju C, Gandara D, et al. Clinicopathologic features of advanced squamous NSCLC. J Thorac Oncol. 2016;11(9):1411–1422. doi: 10.1016/j.jtho.2016.05.024. [DOI] [PubMed] [Google Scholar]
- 2.Zheng M. Classification and pathology of lung cancer. Surg Oncol Clin N Am. 2016;25(3):447–468. doi: 10.1016/j.soc.2016.02.003. [DOI] [PubMed] [Google Scholar]
- 3.Stiles BM, Pua B, Altorki NK. Screening for lung cancer. Surg Oncol Clin N Am. 2016;25(3):469–479. doi: 10.1016/j.soc.2016.02.002. [DOI] [PubMed] [Google Scholar]
- 4.Kunej T, Obsteter J, Pogacar Z, Horvat S, Calin GA. The decalog of long non-coding RNA involvement in cancer diagnosis and monitoring. Crit Rev Clin Lab Sci. 2014;51(6):344–357. doi: 10.3109/10408363.2014.944299. [DOI] [PubMed] [Google Scholar]
- 5.Patil VS, Zhou R, Rana TM. Gene regulation by non-coding RNAs. Crit Rev Biochem Mol Biol. 2014;49(1):16–32. doi: 10.3109/10409238.2013.844092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang Z, Xie L, Han L, et al. Circular RNAs: Regulators of cancer-related signaling pathways and potential diagnostic biomarkers for human cancers. Theranostics. 2017;7(12):3106–3117. doi: 10.7150/thno.19016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Han C, Seebacher NA, Hornicek FJ, Kan Q, Duan Z. Regulation of microRNAs function by circular RNAs in human cancer. Oncotarget. 2017;8(38):64622–64637. doi: 10.18632/oncotarget.19930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meng S, Zhou H, Feng Z, et al. CircRNA: functions and properties of a novel potential biomarker for cancer. Mol Cancer. 2017;16(1):94. doi: 10.1186/s12943-017-0663-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Piwecka M, Glazar P, Hernandez-Miranda LR, et al. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science. 2017;357(6357) doi: 10.1126/science.aam8526. pii: eaam8526. [DOI] [PubMed] [Google Scholar]
- 10.Geng HH, Li R, Su YM, et al. The circular RNA Cdr1as promotes myocardial infarction by mediating the regulation of miR-7a on its target genes expression. PLoS One. 2016;11(3):e0151753. doi: 10.1371/journal.pone.0151753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weng W, Wei Q, Toden S, et al. Circular RNA ciRS-7-A promising prognostic biomarker and a potential therapeutic target in colorectal cancer. Clin Cancer Res. 2017;23(14):3918–3928. doi: 10.1158/1078-0432.CCR-16-2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Naylor EC, Desani JK, Chung PK. Targeted therapy and immunotherapy for lung cancer. Surg Oncol Clin N Am. 2016;25(3):601–609. doi: 10.1016/j.soc.2016.02.011. [DOI] [PubMed] [Google Scholar]
- 13.Shi Y, Au JS, Thongprasert S, et al. A prospective, molecular epidemiology study of EGFR mutations in Asian patients with advanced non-small-cell lung cancer of adenocarcinoma histology (PIONEER) J Thorac Oncol. 2014;9(2):154–162. doi: 10.1097/JTO.0000000000000033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Greene J, Baird AM, Brady L, et al. Circular RNAs: biogenesis, function and role in human diseases. Front Mol Biosci. 2017;4:38. doi: 10.3389/fmolb.2017.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19(2):141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sand M, Bechara FG, Sand D, et al. Circular RNA expression in basal cell carcinoma. Epigenomics. 2016;8(5):619–632. doi: 10.2217/epi-2015-0019. [DOI] [PubMed] [Google Scholar]
- 17.Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, Yang BB. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44(6):2846–2858. doi: 10.1093/nar/gkw027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shang X, Li G, Liu H, et al. Comprehensive circular RNA profiling reveals that hsa_circ_0005075, a new circular RNA biomarker, is involved in hepatocellular carcinoma development. Medicine (Baltimore) 2016;95(22):e3811. doi: 10.1097/MD.0000000000003811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wan L, Zhang L, Fan K, Cheng ZX, Sun QC, Wang JJ. Circular RNA-ITCH suppresses lung cancer proliferation via inhibiting the Wnt/beta-catenin pathway. Biomed Res Int. 2016;2016:1579490. doi: 10.1155/2016/1579490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yao JT, Zhao SH, Liu QP, et al. Over-expression of CircRNA_100876 in non-small cell lung cancer and its prognostic value. Pathol Res Pract. 2017;213(5):453–456. doi: 10.1016/j.prp.2017.02.011. [DOI] [PubMed] [Google Scholar]
- 21.Zhao J, Tao Y, Zhou Y, et al. MicroRNA-7: a promising new target in cancer therapy. Cancer Cell Int. 2015;15:103. doi: 10.1186/s12935-015-0259-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li F, Zhang L, Li W, et al. Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/beta-catenin pathway. Oncotarget. 2015;6(8):6001–6013. doi: 10.18632/oncotarget.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen J, Li Y, Zheng Q, et al. Circular RNA profile identifies circPVT1 as a proliferative factor and prognostic marker in gastric cancer. Cancer Lett. 2017;388:208–219. doi: 10.1016/j.canlet.2016.12.006. [DOI] [PubMed] [Google Scholar]
- 24.Zhong Z, Lv M, Chen J. Screening differential circular RNA expression profiles reveals the regulatory role of circTCF25-miR-103a-3p/miR-107-CDK6 pathway in bladder carcinoma. Sci Rep. 2016;6:30919. doi: 10.1038/srep30919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zheng Q, Bao C, Guo W, et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. 2016;7:11215. doi: 10.1038/ncomms11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu L, Gong X, Sun L, Zhou Q, Lu B, Zhu L. The circular RNA Cdr1as act as an oncogene in hepatocellular carcinoma through targeting miR-7 expression. PLoS One. 2016;11(7):e0158347. doi: 10.1371/journal.pone.0158347. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]