Abstract
Cas9-assisted genome editing was used to construct an engineered glucose-phosphorylation-negative S. cerevisiae strain, expressing the Lactobacillus plantaruml-arabinose pathway and the Penicillium chrysogenum transporter PcAraT. This strain, which showed a growth rate of 0.26 h−1 on l-arabinose in aerobic batch cultures, was subsequently evolved for anaerobic growth on l-arabinose in the presence of d-glucose and d-xylose. In four strains isolated from two independent evolution experiments the galactose-transporter gene GAL2 had been duplicated, with all alleles encoding Gal2N376T or Gal2N376I substitutions. In one strain, a single GAL2 allele additionally encoded a Gal2T89I substitution, which was subsequently also detected in the independently evolved strain IMS0010. In 14C-sugar-transport assays, Gal2N376S, Gal2N376T and Gal2N376I substitutions showed a much lower glucose sensitivity of l-arabinose transport and a much higher Km for d-glucose transport than wild-type Gal2. Introduction of the Gal2N376I substitution in a non-evolved strain enabled growth on l-arabinose in the presence of d-glucose. Gal2N376T, T89I and Gal2T89I variants showed a lower Km for l-arabinose and a higher Km for d-glucose than wild-type Gal2, while reverting Gal2N376T, T89I to Gal2N376 in an evolved strain negatively affected anaerobic growth on l-arabinose. This study indicates that optimal conversion of mixed-sugar feedstocks may require complex ‘transporter landscapes’, consisting of sugar transporters with complementary kinetic and regulatory properties.
Keywords: yeast, pentose fermentation, l-arabinose, transporter engineering, laboratory evolution, bioethanol, gene duplication
This research describes the construction, laboratory evolution and characterization of l-arabinose fermenting Saccharomyces cerevisiae strains that are able to consume specifically l-arabinose in the presence of d-glucose and d-xylose.
INTRODUCTION
Saccharomyces cerevisiae is used on a massive scale for industrial production of ethanol from cane sugar and hydrolysed corn starch, in which fermentable sugars predominantly occur as hexoses or hexose dimers (Renewable Fuel Association 2017). In contrast, hydrolysis of lignocellulosic feedstocks such as the agricultural residues corn stover, corn cobs and wheat straw, yields mixtures of pentose and hexose sugars (Lynd 1996; van Maris et al.2006). Industrial ethanol production from lignocellulosic feedstocks therefore requires efficient conversion of pentose and hexose sugars into ethanol (Lynd 1996). In most lignocellulosic hydrolysates, d-xylose represents up to 25% of the total sugar monomers while another pentose, l-arabinose, typically accounts for up to 3% of the sugar content. In some industrially relevant hydrolysates, such as those derived from corn-fibre hydrolysates and sugar-beet pulp, l-arabinose can account for up to 26% of the sugar content (Grohmann and Bothast 1994; Grohmann and Bothast 1997; van Maris et al.2006).
Since wild-type S. cerevisiae strains cannot ferment pentoses, metabolic engineering is required to enable utilization of these substrates. Strategies to engineer S. cerevisiae for d-xylose fermentation are either based on heterologous expression of a fungal d-xylose reductase and xylitol dehydrogenase or on expression of a heterologous d-xylose isomerase (Jeffries 1983; Kuyper et al.2003; Kuyper et al.2005). Current l-arabinose-fermenting S. cerevisiae strains are based on functional expression of genes encoding l-arabinose isomerase (AraA), l-ribulokinase (AraB) and l-ribulose-5-phosphate-4-epimerase (AraD) from bacteria such as Escherichia coli, Bacillus subtilis or Lactobacillus plantarum (Sedlak and Ho 2001; Becker and Boles 2003; Wisselink et al.2007). Unlike the fungal l-arabinose pathway (Richard et al.2002), the bacterial pathway enables conversion of l-arabinose to d-xylulose-5-phosphate via redox-cofactor-independent reactions (Becker and Boles 2003; Wisselink et al.2007). As previously shown for engineered d-xylose-consuming strains (Kuyper et al.2005), overexpression of the S. cerevisiae genes encoding xylulokinase (Xks1, EC 2.7.1.17), ribulose 5-phosphate epimerase (Rpe1, EC 5.3.1.1), ribulose 5-phosphate isomerase (Rki1, EC 5.3.1.6), transketolase (Tkl1, EC 2.2.1.1) and transaldolase (Tal1, EC 2.2.1.2) enabled efficient coupling of the heterologously expressed l-arabinose pathway to glycolysis and alcoholic fermentation (Wisselink et al.2007). Subsequent laboratory evolution yielded an efficient l-arabinose fermenting strain, which combined a high ethanol yield (0.43 g g−1) with an ethanol production rate of 0.29 g h−1 [g biomass] −1 and an arabinose consumption rate of 0.70 g h−1 [g biomass]−1) (Wisselink et al.2007). Transcriptome studies identified that increased expression of the genes encoding ‘secondary’ isoenzymes of transaldolase (NQM1) and transketolase (TKL2) contributed to improved l-arabinose fermentation of this laboratory-evolved strain (Wisselink et al.2010). Improvements in l-arabinose fermentation were also achieved by codon optimization of bacterial l-arabinose genes to match the codon preference of highly expressed glycolytic yeast genes (Wiedemann and Boles 2008).
Transport of pentose sugars in S. cerevisiae occurs by facilitated diffusion, mediated by native HXT hexose transporters (Hamacher et al.2002; Becker and Boles 2003). Transport of l-arabinose predominantly occurs via the hexose transporter Gal2 which, however, exhibits a much lower affinity for l-arabinose than for d-glucose or galactose (Kou, Christensen and Cirillo 1970; Becker and Boles 2003; Subtil and Boles 2011; Subtil and Boles 2012). Of the other S. cerevisiae HXT transporters, only Hxt9p and Hxt10p show low rates of l-arabinose transport (Subtil and Boles 2011). In practice, expression of GAL2 has been found to be essential for l-arabinose fermentation by strains that do not express heterologous transporters (Becker and Boles 2003; Wisselink et al.2010; Subtil and Boles 2011; Wang et al.2017). Recently, an in silico model of the three-dimensional structure of Gal2, based on the crystal structure of the E. coli xylose permease XylEp, was used to predict mutations in GAL2 that improve l-arabinose docking in Gal2. Based on this theoretical analysis, single-amino-acid substitutions were introduced at position 85 of Gal2 and shown to significantly increase l-arabinose transport activity (Wang et al.2017).
Since transcription of GAL2 is both induced by d-galactose and repressed by d-glucose, design of l-arabinose-fermenting S. cerevisiae strains often includes expression of GAL2 behind a strong constitutive promoter (Tani et al.2016; Wang et al.2017). Alternatively, laboratory evolution can result in upregulation of GAL2 (Wisselink et al.2009; Wisselink et al.2010). However, l-arabinose transport by Gal2 is also subject to strong competitive inhibition by d-glucose (Subtil and Boles 2012; Knoshaug et al.2015), which precludes simultaneous utilization of l-arabinose and d-glucose in batch cultures grown on sugar mixtures (Wisselink et al.2009; Wisselink et al.2010).
To improve the kinetics of l-arabinose transport, transporter genes from other yeasts, filamentous fungi and plants have been functionally expressed in l-arabinose-metabolizing S. cerevisiae strains (Subtil and Boles 2011; Verho, Penttilä and Richard 2011; Knoshaug et al.2015; Li et al.2015; Bracher et al.2018). Km values of these heterologous transporters for l-arabinose ranged from 0.13 to 263 mM, while reported Vmax values ranged from 0.4 to 171 [nmol l-arabinose (mg biomass)−1 min−1]. Several heterologous transporters, including the high-affinity l-arabinose proton symporter PcAraT from P. chrysogenum (Bracher et al.2018) allowed for l-arabinose uptake in the presence of d-glucose (Subtil and Boles 2011; Knoshaug et al.2015; Li et al.2015; Bracher et al.2018).
Laboratory evolution of pentose-fermenting S. cerevisiae strains in which d-glucose phosphorylation was abolished has been successfully used to select strains in which l-arabinose or d-xylose uptake is less sensitive to d-glucose inhibition (Farwick et al.2014; Nijland et al.2014; Wisselink et al.2015). Single-amino-acid substitutions, found at corresponding positions in the hexose transporters Hxt7 (N370) and Gal2 (N376), as well as in a chimera of Hxt3 and Hxt6 (N367), were shown to substantially decrease inhibition of d-xylose transport by d-glucose (Farwick et al.2014; Nijland et al.2014).
The aim of the present study was to identify mutations that enable anaerobic growth of engineered, l-arabinose-fermenting S. cerevisiae strains on l-arabinose in the presence of d-glucose and d-xylose, with a focus on mutations that affect uptake of l-arabinose. To this end, we constructed a glucose-phosphorylation-negative, l-arabinose-fermenting S. cerevisiae strain by Cas9-assisted genome editing. This strain was then subjected to prolonged laboratory evolution in sequential batch bioreactor cultures, grown on mixtures of l-arabinose, d-xylose and d-glucose. Subsequently, causal mutations for glucose-insensitive growth on l-arabinose were identified by whole-genome sequencing and functional analysis of mutant alleles in non-evolved strain backgrounds.
MATERIALS AND METHODS
Strains and maintenance
The S. cerevisiae strains used and constructed in this study (Table 1) were derived from the CEN.PK lineage (Entian and Kötter 2007; Nijkamp et al.2012). All stock cultures were grown in shake flasks on synthetic medium (SM, see below) or, in case of auxotrophic strains, on yeast-extract/peptone (YP) medium (10 g L−1 Bacto yeast extract (Becton Dickinson, Franklin Lakes, NJ) and 20 g L−1 Bacto Peptone (Becton Dickinson)) (Verduyn et al.1992). These media were either supplemented with 20 g L−1d-glucose or, in case of the constructed glucose-phosphorylation-negative strains, 20 g L−1l-arabinose. For storage and in pre-cultures, single-colony isolates obtained after laboratory evolution were grown in medium containing 20 g L−1 of each d-glucose, d-xylose and l-arabinose. After adding glycerol (30% vol/vol) to these cultures, 1 mL aliquots were stored at −80°C.
Table 1.
Saccharomyces cerevisiae strains used in this study.
| Strain | Relevant genotype | Reference |
|---|---|---|
| CEN.PK 113–7D | MATa MAL2–8c SUC2 | Nijkamp et al.2012 |
| IMX080 | MATa ura3–52 his3–1 leu2–3112 MAL2–8c SUC2 glk1::Sphis5, hxk1::KlLEU2 | Solis-Escalante et al.2015 |
| IMX486 | MATa ura3–52 his3–1 leu2–3112 MAL2–8c SUC2 glk1::Sphis5, hxk1::KlLEU2 gal1::cas9-amdS | This study |
| IMX604 | MATa ura3–52 his3–1 leu2–3112 MAL2–8c SUC2 glk1::Sphis5, hxk1::KlLEU2 gal1::cas9- amdS gre3::pTDH3_RPE1 pPGK1_TKL1, pTEF1_TAL1 pPGI1_NQM1 pTPI1_RKI1 pPYK1_TKL2 | This study |
| IMX658 | MATa ura3–52 his3–1 leu2–3112 MAL2–8c SUC2 glk1::Sphis5, hxk1::KlLEU2 gal1::cas9- amdS gre3::pTDH3_RPE1 pPGK1_TKL1, pTEF1_TAL1 pPGI1_NQM1 pTPI1_RKI1 pPYK1_TKL2 gal80::(pTPI_araA_tCYC)*9 pPYK-araB-tPGI1 pPGK-araD-tTDH3 | This study |
| IMX660 | MATa ura3–52 his3–1 leu2–3112 MAL2–8c SUC2 glk1::Sphis5, hxk1::KlLEU2 gal1::cas9- amdS gre3::pTDH3_RPE1 pPGK1_TKL1, pTEF1_TAL1 pPGI1_NQM1 pTPI1_RKI1 pPYK1_TKL2 gal80::(pTPI_araA_tCYC)*9 pPYK-araB-tPGI1 pPGK-araD-tTDH3 hxk2::KlURA3 | This study |
| IMX728 | MATa ura3–52 his3–1 leu2–3112 MAL2–8c SUC2 glk1::Sphis5, hxk1::KlLEU2 gal1::cas9- amdS gre3::pTDH3_RPE1 pPGK1_TKL1, pTEF1_TAL1 pPGI1_NQM1 pTPI1_RKI1 pPYK1_TKL2 gal80::(pTPI_araA_tCYC)*9 pPYK-araB-tPGI1 pPGK-araD-tTDH3 hxk2:: PcaraT | This study |
| IMX1106 | MATa ura3–52 his3–1 leu2–3112 MAL2–8c SUC2 glk1::Sphis5, hxk1::KlLEU2 gal1::cas9- amdS gre3::pTDH3_RPE1 pPGK1_TKL1, pTEF1_TAL1 pPGI1_NQM1 pTPI1_RKI1 pPYK1_TKL2 gal80::(pTPI_araA_tCYC)*9 pPYK-araB-tPGI1 pPGK-araD-tTDH3 hxk2::KlURA3 gal2::gal2N376I | This study |
| IMX1386 | MATa ura3–52 his3–1 leu2–3112 MAL2–8c SUC2 glk1::Sphis5, hxk1::KlLEU2 gal1::cas9- amdS gre3::pTDH3_RPE1 pPGK1_TKL1, pTEF1_TAL1 pPGI1_NQM1 pTPI1_RKI1 pPYK1_TKL2 gal80::(pTPI_araA_tCYC)*9 pPYK-araB-tPGI1 pPGK-araD-tTDH3 hxk2::KlURA3 CAN1::pGAL2_GAL2_tGAL2 | This study |
| IMS0514 | IMX728 that has undergone laboratory evolution on a mixture of arabinose and glucose under aerobic conditions | This study |
| IMS0520 | IMX728 that has undergone laboratory evolution on mixture of arabinose, glucose and xylose under anaerobic conditions | This study |
| IMS0521 | IMX728 that has undergone laboratory evolution on mixture of arabinose, glucose and xylose under anaerobic conditions | This study |
| IMS0522 | IMX728 that has undergone laboratory evolution on mixture of arabinose, glucose and xylose under anaerobic conditions | This study |
| IMS0523 | IMX728 that has undergone laboratory evolution on mixture of arabinose, glucose and xylose under anaerobic conditions | This study |
| IMW088 | As IMS0522; PcAraTΔ | This study |
| IMW091 | As IMS0522; gal2I89T | This study |
| DS68616 | Mat a, ura3–52, leu2–112, gre3::loxP, loxP-Ptpi:TAL1, loxP-Ptpi::RKI1, loxP-Ptpi-TKL1, loxP-Ptpi-RPE1, delta::Padh1XKS1Tcyc1-LEU2, delta::URA3-Ptpi-xylA-Tcyc1 | DSM, The Netherlands |
| DS68625 | DS68616, his3::loxP, hxt2::loxP-kanMX-loxP, hxt367::loxP-hphMX-loxP, hxt145::loxP-natMX-loxP, gal2::loxP-zeoMX-loxP | Nijland et al.2014 |
| DS68625-Gal2 | DS68616, his3::loxP, hxt2::loxP-kanMX-loxP, hxt367::loxP-hphMX-loxP, hxt145::loxP-natMX-loxP, gal2::loxP-zeoMX-loxP | This study |
| DS68625-Gal2N376I | DS68616, his3::loxP, hxt2::loxP-kanMX-loxP, hxt367::loxP-hphMX-loxP, hxt145::loxP-natMX-loxP, gal2::loxP-zeoMX-loxP | This study |
| DS68625-Gal2N376S | DS68616, his3::loxP, hxt2::loxP-kanMX-loxP, hxt367::loxP-hphMX-loxP, hxt145::loxP-natMX-loxP, gal2::loxP-zeoMX-loxP | This study |
| DS68625-Gal2N376T | DS68616, his3::loxP, hxt2::loxP-kanMX-loxP, hxt367::loxP-hphMX-loxP, hxt145::loxP-natMX-loxP, gal2::loxP-zeoMX-loxP | This study |
| DS68625-Gal2N376T/T89I | DS68616, his3::loxP, hxt2::loxP-kanMX-loxP, hxt367::loxP-hphMX-loxP, hxt145::loxP-natMX-loxP, gal2::loxP-zeoMX-loxP | This study |
| DS68625-Gal2T89I | DS68616, his3::loxP, hxt2::loxP-kanMX-loxP, hxt367::loxP-hphMX-loxP, hxt145::loxP-natMX-loxP, gal2::loxP-zeoMX-loxP | This study |
| DS68625-mcs | DS68616, his3::loxP, hxt2::loxP-kanMX-loxP, hxt367::loxP-hphMX-loxP, hxt145::loxP-natMX-loxP, gal2::loxP-zeoMX-loxP | This study |
Cultivation and media
All strain characterization studies were performed in SM prepared as described previously (Verduyn et al.1992). Carbon source and vitamin solutions were added after autoclaving the medium for 20 min at 121 °C. Concentrated solutions (50% w/w) of d-glucose, d-xylose and l-arabinose were autoclaved separately at 110 °C for 20 min. Prior to inoculation 20 g L−1l-arabinose (SMA), 20 g L−1d-glucose (SMG), 20 g L−1l-arabinose and 20 g L−1d-glucose (SMAG) or 20 g L−1l-arabinose, 20 g L−1d-glucose and 20 g L−1d-xylose (SMAGX) were added to SM as carbon sources. Aerobic shake-flask cultures were grown in an orbital shaker at 200 rpm set at 30 °C, using 500-ml flasks containing 100 ml medium. For plates 20 g L−1 agar (BD) was added prior to autoclaving at 121 °C for 20 min. Aerobic shake-flask cultures used as pre-cultures for anaerobic cultures were inoculated with frozen stocks and, in late-exponential phase, a 1 mL sample was used to start a second aerobic pre-culture. Anaerobic batch cultures used for characterization were inoculated from these cultures to obtain an initial OD660 of 0.5. Anaerobic shake-flask cultures were incubated at 30°C in an Innova anaerobic chamber (5% H2, 6% CO2 and 89% N2, New Brunswick Scientific, Edison, NJ) in 50 mL shake flasks placed on an orbital shaker set at 200 rpm. Bioreactor batch cultures were performed at 30°C in 2-L laboratory bioreactors (Applikon, Delft, The Netherlands) with a working volume of 1 L. Culture pH was controlled at 5.0 by automatic addition of 2 M KOH and cultures were stirred at 800 rpm. Anaerobic bioreactors were equipped with Viton O-rings and Norprene tubing (Cole Palmer Instrument Company, Vernon Hills, IL) to minimize oxygen diffusion and continuously sparged with nitrogen gas (< 10 ppm oxygen) at 0.5 L min−1. After autoclaving, synthetic media were supplemented with 0.2 g L−1 antifoam C (Sigma-Aldrich, St. Louis, MO). Furthermore, the anaerobic growth factors Tween 80 (420 mg L−1) and ergosterol (10 mg L−1) were added to the medium as described previously (Verduyn et al.1990). Laboratory evolution experiments were performed in sequential batch reactors (SBRs). On-line measurement of CO2 concentrations in the off gas of SBRs was used as input for a control routine programmed in MFCS/win 3.0 (Sartorius AG, Göttingen, Germany). An empty-refill cycle was automatically initiated when the CO2 concentration in the off gas had first exceeded a threshold value of 0.2% and subsequently declined below 0.1% as fermentable sugars were depleted. After the emptying phase, when approximately 7% of the initial culture volume was left in the reactor, the reactor was automatically refilled with fresh medium from a 20-L reservoir, which was continuously sparged with nitrogen gas. After ca. 450 generations of selective growth, single colony isolates were obtained by plating culture samples on SMAGX agar, supplemented with Tween 80 (420 mg L−1) and ergosterol (10 mg L−1). Plates were incubated in an anaerobic chamber for 6 d and restreaked three times to obtain single-cell lines.
Plasmid and strain construction
The plasmids constructed and used in this study are shown in Additional File 1. The open reading frames of araA [Genbank: ODO63149.1, encoding l-arabinose isomerase], araB [Genbank: ODO63147.1, encoding l-ribulose kinase] and araD [Genbank: ODO63147.1, encoding l-ribulose-5-phosphate epimerase] were codon optimized based on the codon usage in glycolytic genes in S. cerevisiae (Wiedemann and Boles 2008), synthesized behind the promoter of TPI1, PGK1 and PYK1 originating from CEN.PK113–7D and cloned in pMK-RQ based vectors by GenArt GmbH (Regensburg, Germany). These plasmids were named pUD354-pUD356 and transformed into E. coli DH5a cells. Plasmid DNA was isolated from E. coli cultures using a GenElute Plasmid kit (Sigma-Aldrich, St. Louis, MO). PCR amplification of expression cassettes and plasmid fragments was performed using Phusion High Fidelity DNA Polymerase (Thermo Scientific, Waltham, MA). S. cerevisiae strains were transformed as described by Gietz and Woods (Gietz et al.1995). To facilitate CRISPR/Cas9-mediated genome editing in strain S. cerevisiae IMX080, a Cas9 expression cassette PCR-amplified from p414-TEF1p-cas9-CYCt using primers 4653–5981 as well as a second fragment containing the amdSYM marker cassette, PCR amplified from pUG-amdSYM with primers 3093–1678 (Solis-Escalante et al.2013), were in vivo co-assembled and integrated (Kuijpers et al.2013) into the GAL1 locus through a double cross over mediated by homologous recombination (Additional File 3a). After each transformation round, the cells were restreaked thrice and correct integration of all the fragments was confirmed by diagnostic PCR. Transformants were selected on solid SM with acetamide (Solis-Escalante et al.2013) as sole nitrogen source and checked by diagnostic PCR using DreamTaq polymerase (Thermo Scientific), according to the manufacturer's protocol. The resulting strain IMX486 was subsequently transformed with 200 pmol each of eight DNA fragments containing expression cassettes for the genes encoding the enzymes of the non-oxidative branch of the pentose phosphate pathway, which were PCR amplified from pUD344–346 and two 500bp fragments containing flanking regions of GRE3 amplified from CEN.PK113–7D genomic DNA, using oligonucleotide primers shown in Additional File 2. These fragments were co-transformed with 500 ng of plasmid pUDE335 to induce a double strand break in the GRE3 locus using CRISPR-Cas9 (Additional File 3b). The cells were plated on SMD plates and correct assembly of all six fragments in the GRE3 locus was verified by diagnostic colony PCR. Plasmid pUDE335 was counter selected on YP with 20 g L−1d-glucose (YPD) agar with 5-fluoroorotic acid (5-FOA) as described previously (Mans et al.2015). The resulting strain IMX604 was then transformed with nine DNA fragments that carried expression cassettes for araA and single fragments for araB and araD amplified from pUD354–356. The primers used for PCR amplification of these fragments added homologous regions required for in-vivo assembly and integration into the GAL80 locus (Additional File 3c). The fragments were co-transformed with gRNA-plasmid pUDE348, which was constructed as described previously (Verhoeven et al.2017). After verification of the correct assembly and integration of the fragments by diagnostic PCR, plasmid pUDE348 was counter selected with 5-FOA, yielding strain IMX658. HXK2 was deleted by inducing a double strand break using the gRNA plasmid pUDE327, and using an expression cassette for the PcaraT gene, obtained by PCR amplification from plasmid Pwt118, as the repair fragment (Additional File 3d). After counter selection of pUDE327 with 5-FOA, a DNA fragment carrying the URA3 gene from CEN.PK113–7D was amplified using primers 2641–1522 and transformed to restore a wild-type URA3 gene, yielding strain IMX728. Strain IMX660 was constructed from IMX658 by transforming a KlURA3 based knock-out cassette targeting the HXK2 locus.
Strain IMX1386 was obtained by co-transforming IMX728 with plasmid pROS13 and a fragment containing the promoter region, ORF and terminator of GAL2, PCR amplified from genomic DNA of strain CEN.PK113–7D with primer pair 7285–7286. Transformants were selected on YP with 20 g L−1l-arabinose (YPA) supplemented with G418. After verifying correct integration of the GAL2 expression cassette in the CAN1 locus, the gRNA plasmid was counter selected by serial plating on YPA as described previously (Mans et al.2015). The gRNA plasmids pUDR172 and pUDR187 were constructed using pROS13 as template by Gibson assembly according to the method described by Mans et al. (Mans et al.2015). The single nucleotide polymorphism (SNP) at position 1127 in GAL2 was inserted by co-transforming strain IMX660 with pUDR172 and a repair fragment containing the single nucleotide change resulting in the amino acid substitution from N to I at position 376. Expression cassettes of the GAL2, GAL2N376I and GAL2N376S variants were amplified using genomic DNA from CEN.PK113–7D, IMS0520 and IMS0514 as template using the primers F_Gal2_XbaI and R_Gal2_Cfr9I (Supplemental Table S2) and subsequently cloned into plasmid pRS313-P7T7. To obtain the GAL2N376T, GAL2N376T/T89I and GAL2T89I mutants, fragments were PCR amplified from CEN.PK113–7D genomic DNA using the primers shown in Supplemental Table S2. Subsequently, overlap PCR amplifications were done to obtain the full-length genes encoding for Gal2N376T, Gal2N376T/T89I and Gal2T89I. All genes were cloned into pRS313-P7T7 and subsequently confirmed by Sanger sequencing (Baseclear).
The Gal2 variants and the pRS313-P7T7-mcs plasmid (as an empty plasmid/control) were transformed to the hexose transporter deletion strain DS68625 and positive colonies were named DS68625-Gal2, DS68625-Gal2 N376I, DS68625-Gal2 N376S, DS68625-Gal2 N376T, DS68625-Gal2 N376T/T89I, DS68625-Gal2 T89I and DS68625-mcs. To revert the altered amino acid position 89 in Gal2 from isoleucine to threonine, pUDR187 and a repair fragment containing the original coding sequence were co-transformed. Inactivation of PcAraT in IMS0522 was achieved by in-vivo assembling of the pROS13 backbone and the gRNA expression cassette, both PCR amplified from pROS13 using the primers shown in Additional File 2 and providing a repair oligo for HXK2. After verifying correct transformants the plasmids were removed by counter selection on YPAGX plates and the resulting strains were named IMW091 and IMW088, respectively.
Analytical methods
To monitor growth of batch cultures, a Libra S11 spectrometer (Biochrom, Cambridge, United Kingdom) was used for optical density measurements at 660 nm. To calculate specific growth rates, biomass dry weight measurements were performed on at least six samples taken during the exponential growth phase. Dry weight measurements were performed by filtering 10 mL culture samples over pre-weighed nitrocellulose filter (pore size, 0.45 μm; Gelman Laboratory, Ann Arbor, MI). The filter was washed with demineralized water and dried in a microwave oven (Bosch, Stuttgart, Germany) for 20 min at 360 W. The correlation between these CDW measurements and the corresponding OD data was used to estimate CDW in samples for which no direct CDW measurements were done. Metabolite concentrations in culture supernatants, obtained by centrifugation, were determined by high-performance liquid chromatography (HPLC) on an Agilent 1260 HPLC (Agilent Technologies, Santa Clara, CA) equipped with a Bio-Rad HPX 87 H column (Bio-Rad, Hercules, CA). The column was eluted at 60 °C with 0.5 g L−1 H2SO4 at a flow rate of 0.6 ml min−1. Detection was by means of an Agilent G1362A refractive-index detector and an Agilent G1314F VWD detector. CO2 and O2 concentrations were measured in bioreactor off gas using an NGA 2000 analyzer (Rosemount Analytical, Orrville, OH) after it was first cooled by a condenser (2 °C) and dried with a Permapure MD-110–48P-4 dryer (Permapure, Toms River, NJ). Correction for ethanol evaporation was done for all bioreactor experiments as described previously (Guadalupe Medina et al.2010).
DNA sequence analysis
Genomic DNA for sequencing was isolated using the QIAGEN Blood & Cell Culture Kit With 100/G Genomic-tips (QIAGEN, Valencia, CA) according to the manufacturer's protocol. DNA libraries were prepared using the Nextera XT DNA library Preparation Kit (Illumina, San Diego, CA), yielding 300bp fragments. The Libraries were sequenced using the Illumina MiSeq platform (Illumina, San Diego, CA, USA). Data were aligned and mapped to the CEN.PK113–7D genome using the Burrows–Wheeler alignment tool (Li and Durbin 2009). Variant calling by Pilon (Walker et al.2014) was checked with the Integrated Genomics Viewer (Thorvaldsdóttir, Robinson and Mesirov 2013). The Poisson mixture model based algorithm Magnolya (Nijkamp et al.2012) was used to detect and quantify chromosomal copy number variations (CNV). Copy numbers of AraA, AraB, AraD and PcAraT were estimated as described previously using the average read depth of the chromosomes that did not contain any duplication according to the CNV analysis done with Magnolya (Verhoeven et al.2017). Genome sequence data of strains IMS0520, IMS0521, IMS0522, IMS0523, IMS514 and parental strains IMX728 and IMX660 have been deposited at the NCBI Sequence Read archive (www.ncbi.nlm.nih.gov/sra) with the corresponding BioProject ID PRJNA414371. GAL2 ORFs from strains IMS0002 and IMS0010 and the nucleotides surrounding position T89 in IMS0003 and IMS0007 were PCR amplified from genomic DNA using primers as listed in Additional File 2 and sanger sequenced (Baseclear).
Sugar transport assays
Strains expressing GAL2 alleles in the DS68625 strain background were pre-grown in aerobic shake flasks on SM with 20 g L−1d-xylose, after which cells were collected by centrifugation (3000 g, 3 min), washed and re-suspended in SM without sugar. Uptake experiments were initiated by adding [14C] l-arabinose or [14C] d-glucose (ARC St. Louis, MO, USA) to the cell suspension at concentrations ranging from 0.2 to 500 mM. [14C] l-arabinose and [14C] d-glucose (50–60 mCi mmol−1) were added at concentrations of 0.1 mCi ml−1. At set time points, uptake was arrested by adding 5 mL of ice-cold 0.1 M LiCl, filtration over 0.45-μm HV membrane filters (Millipore, France) and washing with 5 mL ice-cold 0.1 M LiCl. Radioactivity on the filters was then counted using a Liquid Scintillation Counter (PerkinElmer, Waltham, MA) in Ultima Gold MV Scintillation cocktail (PerkinElmer). d-Glucose inhibition experiments were measured using 50 mM [14C] l-arabinose with [14C] d-glucose added at concentrations between 50 and 500 mM.
RESULTS
Construction of a S. cerevisiael-arabinose specialist strain
To investigate uptake and growth on l-arabinose in the presence of d-glucose, an l-arabinose ‘specialist strain’ was constructed by Cas9-assisted genome editing. First, a Cas9 expression cassette (DiCarlo et al.2013) and the counter-selectable marker cassette amdSYM (Solis-Escalante et al.2013) were in vivo assembled and integrated at the GAL1 locus in S. cerevisiae IMX080 (hxk1Δ glk1Δ) (Additional File 3a). In the resulting strain IMX486 (hxk1Δ glk1Δ gal1Δ::{Spcas9-amdSYM}, HXK2 was the sole remaining functional hexose kinase. Subsequently, cassettes for constitutive expression of RPE1, RKI1, TKL1, TKL2, TAL1 and NQM1, which encode (iso)enzymes of the non-oxidative pentose-phosphate pathway (NPPP), were in vivo assembled and integrated at the GRE3 locus of strain IMX486, yielding strain IMX658 (hxk1Δ glk1Δ gal1Δ::{Spcas9-amdSYM} gre3Δ::{NPPP}) (Additional File 3b). Inactivation of GRE3 prevents xylitol formation by the Gre3 non-specific aldose reductase (Träff et al.2001; Kuyper et al.2005). To introduce a functional bacterial l-arabinose pathway (ARAP) (Wisselink et al.2007), the Lactobacillus plantarum genes encoding l-arabinose isomerase (AraA), ribulokinase (AraB) and ribulose-5P epimerase (AraD) were codon optimized and placed under the control of the strong constitutive promoters of TPI1, PGI1 and TDH3, respectively. For high expression of AraA, we used the multi-copy tandem integration strategy previously described for high-level expression of xylose isomerase which, in laboratory evolution experiments, facilitates rapid adaptation of copy number by homologous recombination (Verhoeven et al.2017). Using this approach, nine copies of the AraA cassette and single copies of the AraB and AraD cassettes were integrated at the GAL80 locus by in vivo assembly and Cas9-mediated integration (Additional File 3c). Inactivation of GAL80 eliminates the need for galactose induction of GAL2 expression (Torchia et al.1984; Ostergaard et al.2001; Tani et al.2016). Expression of the high-affinity l-arabinose transporter PcAraT from P. chrysogenum enables l-arabinose uptake in the presence of glucose (Bracher et al.2018). To simultaneously abolish the ability to metabolize d-glucose, an expression cassette for PcAraT (Bracher et al.2018) was integrated into the HXK2 locus. Subsequent introduction of a functional URA3 allele from S. cerevisiae CEN.PK113–7D (Additional File 3d) yielded the prototrophic ‘arabinose specialist’ S. cerevisiae strain IMX728 (hxk1Δ glk1Δ gal1Δ::{Spcas9-amdSYM} gre3Δ::{NPPP} hxk2Δ::PcaraT gal80Δ::{ARAP}).
In aerobic shake flasks on SMA, strain IMX728 exhibited a specific growth rate of 0.26 h−1 (Fig. 1). As anticipated after deletion of all four genes encoding glucose-phosphorylating enzymes (Farwick et al.2014; Nijland et al.2014; Wisselink et al.2015), no growth was observed on SMGD. Consistent with the incomplete inhibition of PcAraT by glucose (Bracher et al.2018), strain IMX728 grew at a specific growth rate of 0.04 h−1 in aerobic batch cultures on SMAGX (Fig. 1).
Figure 1.
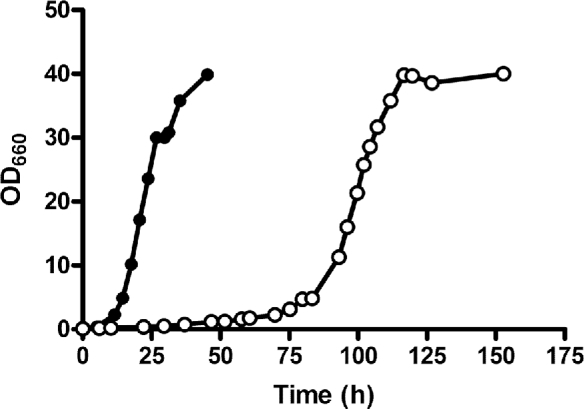
Growth curves of S. cerevisiae strain IMX728 (engineered, non-evolved arabinose-consuming, glucose-phosphorylation-negative, expressing PcaraT) in aerobic batch cultures on SM containing 20 g L−1l-arabinose (SMA) as sole carbon and energy source (•) and in SM with 20 g L−1l-arabinose, 20 g L−1d-glucose and 20 g L−1d-xylose added ( ). Data shown in the figure represent from a single growth experiment, data from a replicate experiment are shown in Additional File 4. Derived biomass-specific conversion rates in the replicate experiments differed by less than 5%.
). Data shown in the figure represent from a single growth experiment, data from a replicate experiment are shown in Additional File 4. Derived biomass-specific conversion rates in the replicate experiments differed by less than 5%.
Anaerobic laboratory evolution of an engineered l-arabinose specialist in the presence of d-glucose
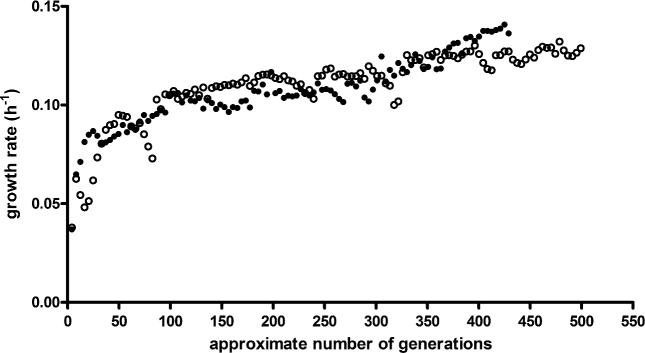
The ability of strain IMX728 to grow anaerobically on l-arabinose as sole carbon source was investigated in anaerobic bioreactors. In contrast to the fast and instantaneous growth in aerobic shake flasks (Fig. 1), slow anaerobic growth on l-arabinose was observed only after approximately 150 h (Additional File 5). To select for spontaneous mutants that combined an increased anaerobic growth rate on l-arabinose with a decreased sensitivity to growth inhibition by d-glucose and d-xylose, strain IMX728 was grown in duplicate anaerobic SBRs on SMAGX. Continuous measurements of CO2 concentrations in the off gas of the SBRs were used to automatically initiate empty-refill cycles and monitor the increase of specific growth rate over time (Additional File 6). After an initial cultivation cycle under aerobic conditions, the first anaerobic cycle required a 17d period before growth was observed. Only then, within 24 h, both replicate cultures showed an exponential increase in CO2 production, corresponding to an estimated specific growth rate of 0.07 h−1. Both SBRs were operated for 254 d (Fig. 2), after which specific growth rates had increased to above 0.13 h−1. Specific growth rates on SMAGX of 31 single-colony isolates obtained from the SBR cultures were then measured in anaerobic shake flasks (Additional File 7). Four selected isolates, named IMS0520 to IMS0523, showed specific growth rates of 0.13 to 0.17 h−1 in anaerobic bioreactor batch cultures on SMAGX, which corresponded closely to the growth rates estimated from on-line CO2 analysis of the evolving SBR cultures (Additional File 8).
Figure 2.
Specific growth rate (h−1) estimated from CO2 production profiles in two parallel laboratory evolution experiments in anaerobic SBRs SBR 1 ( ) and SBR 2 (•). Both cultures were inoculated with the l-arabinose consuming, glucose-phosphorylation-negative S. cerevisiae strain IMX728 and grown on SM with 20 g L−1l-arabinose, 20 g L−1d-glucose and 20 g L−1d-xylose. The first data point for each experiment corresponds to the initial aerobic batch cultivation, all subsequent data points represent estimated growth rates in anaerobic SBR cycles.
) and SBR 2 (•). Both cultures were inoculated with the l-arabinose consuming, glucose-phosphorylation-negative S. cerevisiae strain IMX728 and grown on SM with 20 g L−1l-arabinose, 20 g L−1d-glucose and 20 g L−1d-xylose. The first data point for each experiment corresponds to the initial aerobic batch cultivation, all subsequent data points represent estimated growth rates in anaerobic SBR cycles.
Evolved strains show mutations, duplication and allelic variation in GAL2
To identify the mutations that enabled growth of laboratory-evolved isolates IMS0520-IMS0523 on l-arabinose in the presence of d-glucose and d-xylose, their genomes were sequenced and compared to that of their common parental strain IMX728. Variant screening for non-synonymous single-nucleotide mutants and insertion/deletions revealed that, together, the four strains carried 9 single-nucleotide mutations in open reading frames (Table 2). All four isolates carried mutations in GAL2 that changed the asparagine in position 376 of Gal2, into an isoleucine (Gal2N376I) or a threonine (Gal2N376T) in isolates originating from SBR 1 and 2, respectively. Isolate IMS0522 contained an additional single-nucleotide mutation in GAL2 resulting in a Gal2T89I substitution, which, however, was observed only in 57% of the reads already suggesting a copy number increase of GAL2 locus.
Table 2.
Single-nucleotide mutations in l-arabinose-metabolizing, glucose-phosphorylation-negative S. cerevisiae strains IMS0520-IMS0523, evolved for anaerobic growth on l-arabinose in the presence of d-xylose and d-glucose in biological duplicate (reactors 1 and 2). Mutants were identified alignment of the whole-genome sequence data to S. cerevisiae IMX728. Read coverage observed for each SNP was higher than 99% unless stated otherwise in the table. Descriptions of gene functions are derived from the Saccharomyces Genome Database (as of 19–11-2017).
| Gene and strain | Nucleotide change | Amino acid change | Description |
|---|---|---|---|
| GAL2 | Galactose permease; required for utilization of galactose and also able to transport glucose (Kou, Christensen and Cirillo 1970) | ||
| IMS0520 (reactor 1) | A1127T | N376I | |
| IMS0521 (reactor 1) | A1127T | N376I | |
| IMS0522 (reactor 2) | A1127C | N376T | |
| IMS0523 (reactor 2) | C266T (57%) | T89I | |
| A1127C | N376T | ||
| BMH1 | 14–3-3 protein, major isoform; controls, involved in regulation of exocytosis, vesicle transport, Ras/MAPK and rapamycin-sensitive signalling, aggresome formation, spindle position checkpoint | ||
| IMS0520 (reactor 1) | G383T | G128V | |
| IMS0522 (reactor 2) | G64T | E22* | |
| IMS0523 (reactor 2) | G64T | E22* | |
| DCK1 | Dock family protein (Dedicator Of CytoKinesis), homolog of human DOCK1; interacts with Ino4p; cytoplasmic protein that relocates to mitochondria under oxidative stress | ||
| IMS0522 (reactor 2) | T2890C (42%) | Y964H | |
| IPT1 | Inositolphosphotransferase; involved in synthesis of mannose-(inositol-P)2-ceramide (M(IP)2C) sphingolipid; can mutate to resistance to the antifungals syringomycin E and DmAMP1 and to K. lactis zymocin | ||
| IMS0522 (reactor 2) | C938T | S313F | |
| UPC2 | Sterol regulatory element binding protein; induces sterol biosynthetic genes, upon sterol depletion; acts as a sterol sensor, binding ergosterol in sterol rich conditions; | ||
| IMS0523(reactor 2) | A2648G | Y883C | |
| RPL6B | Ribosomal 60S subunit protein L6B; binds 5.8S rRNA; homologous to mammalian ribosomal protein L6, | ||
| IMS0520 (reactor 1) | T784A | E261G |
In addition to mutations in GAL2, single-nucleotide changes were identified in DCK1, IPT1 UPC2 and RPL6B, which each occurred in only one of the four isolates. IPT1 encodes an inositol phosphotransferase involved in sphingolipid production (Dickson et al.1997), while UPC2 plays a key role in regulation of sterol metabolism and uptake (Crowley et al.1998). Both mutations may have affected membrane composition and, thereby, the uptake of l-arabinose. Mutations in BMH1 were observed in strains IMS0520, IMS0522 and IMS0523. In view of the pleiotropic phenotype of bmh1 mutants (van Heusden et al.1992; Roberts, Mösch and Fink 1997; Van Hemert, van Heusden and Steensma 2001; Wang et al.2009), we decided to not investigate the impact of these mutations on l-arabinose fermentation. Since, GAL2 was the only transporter gene whose coding region was found to be mutated in strain IMS0521, further analysis was focused on the GAL2 mutations found in the four strains.
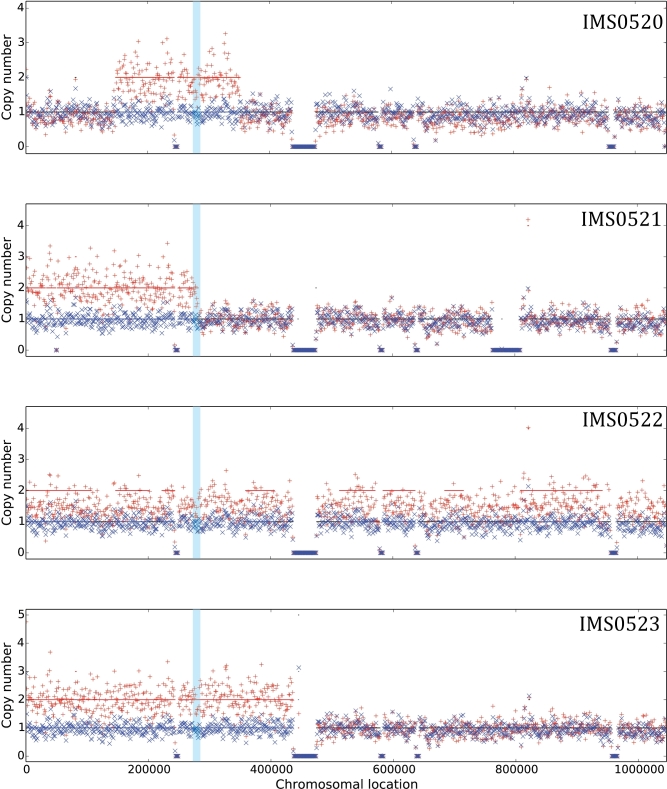
As frequently observed in laboratory evolution experiments with S. cerevisiae (Oud et al.2012; de Vries, Pronk and Daran 2017), read depth analysis revealed changes in copy number of specific regions in the genomes of the evolved strains (Additional File 9). Notably, regions of Chromosome XII, which harbors GAL2, were found to be duplicated in all four isolates, with strain IMS0522 showing a probable duplication of this entire chromosome (Fig. 3). To test the physiological significance of the resulting duplication of GAL2, an additional copy of the wild-type GAL2 allele was integrated in the genome of the parental ‘arabinose specialist’ IMX728, yielding strain IMX1386. Similar to S. cerevisiae IMX728, the strain with two copies of GAL2 required 120h before growth on l-arabinose was observed in anaerobic batch cultures (Additional File 10), indicating that duplication of GAL2 was not, in itself, sufficient to enable instantaneous anaerobic growth on l-arabinose.
Figure 3.
Chromosomal copy number estimations for chromosome 12 in the evolved anaerobically l-arabinose-growing, glucose-tolerant S. cerevisiae strains IMS0520 to IMS0523. Chromosomal copy numbers were estimated using the Magnolya algorithm (Nijkamp et al.2012). Reads of the four strains (red crosses) show duplications relative to the corresponding chromosome 12 sequences in the common parental strain IMX728 (blue crosses). The light blue region corresponds to the location of GAL2.
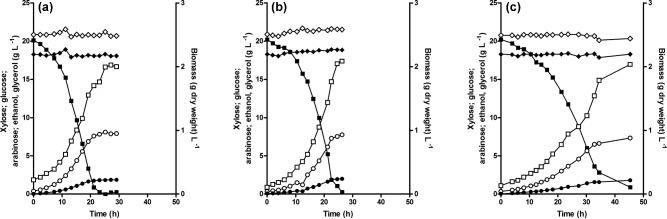
Read-depth comparisons of the genes encoding the enzymes of the bacterial l-arabinose pathway (araA, araB, araD) revealed some differences in copy number of araA and araB between evolved strains and the parental strain (Table 3). However, these differences were not consistent across the four evolved strains. Read-depth analysis of the PcaraT transporter gene revealed no copy number changes in any of the four evolved isolates. To examine the contribution of PcaraT to the physiology of the evolved strains, it was deleted in evolved strain IMS0522, resulting in strain IMW088. In duplicate anaerobic bioreactor cultures of strains IMS0522 and IMW088 on SMAGX, both completely ferment the l-arabinose in the medium. The strain lacking PcaraT (IMW088) did, however, exhibit a lower specific growth rate than strain IMS0522 (0.10 h−1 and 0.12h−1, respectively; Table 4) and, consequently, required substantially more time for complete conversion of l-arabinose in the anaerobic batch cultures (Fig. 4).
Table 3.
Estimated copy numbers of the heterologously expressed Lactobacillus plantarum genes araA, araB and araD and Penicillium chrysogenum PcaraT in the evolved, anaerobically growing and glucose-tolerant ‘arabinose specialist’ strains S. cerevisiae IMS0520-IMS0523, obtained from two independent evolution experiments in SBRs (1 and 2), and in the parental strain IMX728. Copy numbers were estimated by comparing gene read depths to the average of all chromosomes.
| Reference | Reactor 1 | Reactor 2 | |||
|---|---|---|---|---|---|
| Gene | IMX728 | IMS0520 | IMS0521 | IMS0522 | IMS0523 |
| araA | 17.5 | 8.7 | 19.3 | 19.5 | 16.4 |
| araB | 0.8 | 0.9 | 3.1 | 2.2 | 1.2 |
| araD | 0.9 | 1.0 | 1.2 | 1.0 | 1.1 |
| PcaraT | 0.9 | 1.1 | 1.0 | 1.1 | 1.2 |
| 1 Excluded chromosomes: | 3, 10, 12, 16 | 2, 8, 12 | 12 | 12, 16 | |
1Chromosomes excluded from the average read depth, based on duplication of chromosomal regions identified with the Magnolya algorithm (Nijkamp et al.2012).
Table 4.
Specific growth rates, biomass and product yields and carbon recoveries in anaerobic bioreactor batch cultures of S. cerevisiae strains IMS0522 (d-glucose-phosphorylation-negative strain evolved for l-arabinose fermentation in presence of d-xylose and d-glucose), IMW088 (IMS0522 with PcaraTΔ) and IMW091 (IMS0522 with Gal289I reverted to Gal289T) in SM with 20 g L−1l-arabinose, 20 g L−1d-glucose and 20 g L−1d-xylose. The values are average and mean deviation of data from two independent cultures of each strain.
| Yield (g g−1 L-arabinose consumed) | Carbon recovery | |||||
|---|---|---|---|---|---|---|
| μmax (h−1) | Biomass | Ethanol | Glycerol | Acetate | (%) | |
| IMS0522 | 0.121 ± 0.001 | 0.075 ± 0.002 | 0.384 ± 0.004 | 0.090 ± 0.004 | 0.018 ± 0.001 | 96.9 ± 0.1 |
| IMW088 | 0.102 ± 0.01 | 0.079 ± 0.005 | 0.381 ± 0.006 | 0.094 ± 0.002 | 0.019 ± 0.002 | 99.3 ± 2.1 |
| IMW091 | 0.069 ± 0.01 | 0.060 ± 0.005 | 0.394 ± 0.004 | 0.088 ± 0.001 | 0.020 ± 0.001 | 103.4 ± 1.4 |
Figure 4.
Growth and extracellular metabolite concentrations in anaerobic cultures of the evolved l-arabinose consuming, glucose-phosphorylation negative S. cerevisiae strain IMS0522 and two derived strains with specific genetic modifications. Cultures were grown in bioreactors containing SM with 20 g L−1l-arabinose, 20 g L−1d-glucose and 20 g L−1d-xylose. (a) IMS0522, (b) IMW088 (PcaraTΔ), (c) IMW091 (Gal289I restored to Gal289T).  l-arabinose;
l-arabinose;  biomass dry weight; • glycerol;
biomass dry weight; • glycerol;  ethanol; ⧫ d-glucose;
ethanol; ⧫ d-glucose;  d-xylose. Data shown in the figure represent from a single growth experiment for each strain, data from independent replicate experiments are shown in Additional File 11. Derived biomass-specific conversion rates in the replicate experiments differed by less than 5%.
d-xylose. Data shown in the figure represent from a single growth experiment for each strain, data from independent replicate experiments are shown in Additional File 11. Derived biomass-specific conversion rates in the replicate experiments differed by less than 5%.
Amino acid substitutions in Gal2 at position N376 enable l-arabinose uptake in the presence of d-glucose and d-xylose
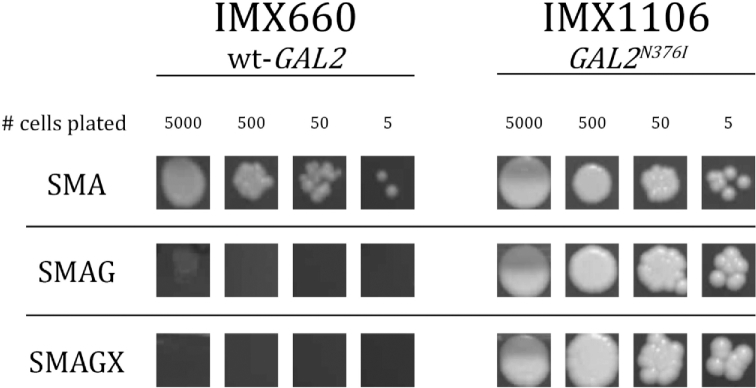
A previous laboratory evolution study with d-xylose-fermenting, glucose-phosphorylation-deficient S. cerevisiae strains also identified mutations that led to a single-amino-acid change in Gal2 at position 376, which enabled growth on d-xylose in the presence of d-glucose (Farwick et al.2014). In the present study, the parental strain IMX728 could not only import l-arabinose via Gal2, but also via the fungal l-arabinose transporter PcAraT, which is much less sensitive to glucose inhibition than Gal2 (Bracher et al.2018). To investigate whether the observed mutations in GAL2 were also required for growth on l-arabinose in the presence of d-glucose in a strain background without PcAraT, a short laboratory evolution experiment was conducted with the ‘arabinose specialist’ strain IMX660, which differed from strain IMX728 by the absence of PcAraT. After approximately 100 h incubation in an aerobic shake flask on 20 g L−1l-arabinose and d-glucose, growth was observed and strain IMS0514 was obtained as a single colony isolate and confirmed to have a stable phenotype for growth on SMAG (data not shown). As in strains IMS0520-IMS0523, whole-genome sequencing of strain IMS0514 revealed a single mutation in GAL2 at position 1128, in this case resulting in a Gal2N376S substitution. Additionally, introduction of the Gal2N376I substitution in both GAL2 alleles of strain IMS0520 enabled immediate growth on SMAG of the arabinose specialist strain lacking PcAraT (IMX1106, Fig. 5).
Figure 5.
Growth of engineered glucose-phosphorylation-negative S. cerevisiae strains on l-arabinose in the presence and absence of d-glucose and d-xylose. An approximate set numbers of cells of strain S. cerevisiae IMX1106, expressing Gal2 with amino acid substitution N376 and parental strain IMX660 were plated on solid SM with l-arabinose (SMA), l-arabinose and d-glucose (SMAG) or l-arabinose, d-glucose and d-xylose (SMAGX). Plates were incubated aerobically at 30°C for 4 d.
Sugar transport kinetics of Gal2 variants obtained by laboratory evolution
To examine inhibition by d-glucose of l-arabinose transport by different Gal2 variants found in evolved strains, the corresponding evolved GAL2 alleles were expressed in S. cerevisiae DS68625. In this strain, which does not express the pathway required for arabinose metabolism, the hexose-transporter genes HXT1 to HXT7 and GAL2 have been disrupted (Nijland et al.2014). To examine the extent to which d-glucose inhibited l-arabinose uptake by the different Gal2 variants, [14C]-l-arabinose-uptake experiments were performed at different concentrations of d-glucose (Fig. 6). Consistent with earlier reports (Subtil and Boles 2011; Farwick et al.2014), uptake of l-arabinose (50 mM) by wild-type Gal2 was inhibited by ca. 80% in the presence of 100 mM d-glucose (Fig. 6). d-glucose inhibition was not significantly affected by the Gal2T89I substitution but, under the same conditions, l-arabinose uptake by Gal2N376I, Gal2N376S and Gal2N376T was inhibited by only 20–30% (Fig. 6 and Additional File 12). The Gal2N376I, Gal2N376S and Gal2N376T substitutions yielded 25–60% lower Km values for l-arabinose than wild-type Gal2, while their Km values for d-glucose were one to two orders of magnitude higher than those of wild-type Gal2 (Table 5). As a result, the ratios of the Km values for l-arabinose vs. d-glucose was two orders of magnitude lower for the strains expression Gal2 variants with substitutions at position 376. For Gal2 variants that only carried an amino acid substitution at this position, transport capacities (Vmax) for l-arabinose and d-glucose differed by less than two-fold from those of wild-type Gal2 (Table 5). These changes in transport kinetics are consistent with a strongly reduced competitive inhibition of l-arabinose transport by d-glucose.
Figure 6.
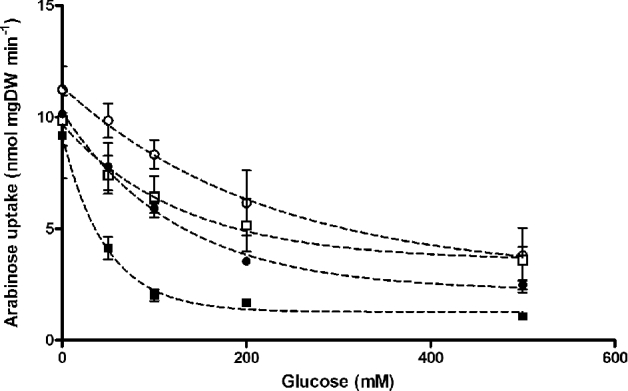
Effect of d-glucose on specific rates of L-arabinose uptake by different Gal2 variants. Uptake experiments were performed with 50 mM [14C-] l-arabinose in the presence of different d-glucose concentrations. Symbols indicate uptake rates observed with strain S. cerevisiae DS68625 expressing Gal2N376T (○), Gal2N376S (•), Gal2N376I ( ) and wild-type Gal2 (
) and wild-type Gal2 ( ).
).
Table 5.
Km and Vmax values for l-arabinose and d-glucose for Gal2 variants with amino acid substitutions at positions N376 and T89. Transport kinetics were measured by uptake studies with radioactive sugars after expression of GAL2 alleles in S. cerevisiae DS68625. Values are average and mean deviation of two independent sets of uptake experiments. The detection limit for d-glucose uptake (Vmax) was 1.8 nmol−1 (mg biomass)−1 min−1. Data used to calculate Km and Vmax values are shown in Additional File 13.
| Km (mM) | Km ratio | Vmax (nmol (mg biomass)−1 min−1) | |||
|---|---|---|---|---|---|
| Gal2 variant | l-arabinose | d-glucose | Ara/Glc | l-arabinose | d-glucose |
| Gal21 | 335 ± 21 | 1.9 ± 0.2 | 176 | 75 ± 5 | 26 ± 1 |
| Gal2N376I | 117 ± 16 | 101 ± 47 | 1 | 39 ± 3 | 32 ± 18 |
| Gal2N376S | 186 ± 33 | 38 ± 1 | 5 | 64 ± 2 | 28 ± 1 |
| Gal2N376T | 171 ± 17 | 57 ± 1 | 3 | 65 ± 2 | 17 ± 4 |
| Gal2T89I + N376T | 103 ± 40 | - | - | 30 ± 2 | <1.8 |
| Gal2T89I | 99 ± 18 | 7 ± 0.2 | 15 | 22 ± 3 | 13 ± 0.1 |
1kinetic data for wild-type Gal2 were derived from Bracher et al. (2018).
A single Gal2T89I substitution increased the Km for d-glucose transport from 1.9 to 7 mM, while it decreased the Km for l-arabinose from 335 to 99 mM. For both sugars, the Gal2T89I substitution caused a 2–3 fold reduction of Vmax (Table 5). These kinetic properties suggest that Gal2T89I may, by itself, confer a selective advantage at low extracellular l-arabinose concentrations. Remarkably, a Gal2 variant that harboured both the Gal2N376T and Gal2T89I substitutions no longer transported d-glucose, while Km and Vmax values for l-arabinose were similar to those of Gal2T89I (Table 5).
The Gal2T89I substitution also occurs in a previously evolved pentose-fermenting strain
Strain S. cerevisiae IMS0010 is an engineered pentose-fermenting strain that was previously evolved for improved anaerobic fermentation kinetics on mixtures of d-glucose, d-xylose and l-arabinose (Wisselink et al.2009). Sequence analysis of GAL2 in IMS0010 showed that this strain contains the same Gal2T89I substitution found in one of the two GAL2 alleles in the independently evolved glucose-phosphorylation-negative strain IMS0522. Analysis of GAL2 sequences in two intermediate strains in the construction and laboratory evolution of strain IMS0010 (strains IMS0002 and IMS0007; (Wisselink et al.2009)) showed that this mutation was acquired during the evolution for improved kinetics of mixed-substrate utilization.
Reverting the GalT89I substitution negatively affects l-arabinose consumption
CO2 production profiles of anaerobic bioreactor batch cultures (Additional File 8) indicated that strain IMS0522, in which one of the two GAL2 alleles encodes both the Gal2N376T and Gal2T89I substitutions, grew faster on l-arabinose in the presence of d-glucose and d-xylose than strain IMS0523, in which both copies of GAL2 only encode the Gal2N376T substitution. To investigate whether the additional Gal2T89I substitution contributed to this phenotype, the allele encoding this substitution was restored to ‘Gal2N376T only’. Anaerobic batch cultures of the resulting strain IMW091 on SMAGX showed a substantially reduced specific growth rate relative to its parental strain IMS0522 (0.069 h−1 and 0.12 h−1, respectively, Table 4). As a consequence, the time needed to completely ferment l-arabinose in the presence of d-glucose and d-xylose was much shorter for the parental strain IMS0522 (Gal2N376T/Gal2N376T T89I) than for the strain IMW091 (Gal2N376T/Gal2N376T T); 25 and 45 h, respectively) (Fig. 4).
DISCUSSION
The S. cerevisiae strain used for this evolutionary engineering study combined genetic modifications that were previously shown to enable or stimulate growth on l-arabinose. Combined expression of a bacterial l-arabinose pathway (Sedlak and Ho 2001; Becker and Boles 2003; Wisselink et al.2007), overexpression of native non-oxidative pentose-phosphate-pathway enzymes (Kuyper et al.2005; Wisselink et al.2007), deletion of the non-specific aldose reductase gene GRE3 (Träff et al.2001), deregulated expression of the Gal2 galactose/l-arabinose transporter (Tani et al.2016; Wang et al.2017), here accomplished by deletion of GAL80 (Torchia et al.1984; Ostergaard et al.2001; Tani et al.2016) and expression of the fungal l-arabinose transporter PcAraT (Bracher et al.2018) yielded a strain that, in aerobic shake-flask cultures, exhibited a specific growth rate on l-arabinose of 0.26 h−1. Moreover, deletion of all four genes encoding enzymes with glucose-phosphorylating activity rendered this ‘arabinose-specialist’ strain S. cerevisiae IMX728 unable to grow on glucose. Construction of this strain was completed within 3 months, which illustrates the power of Cas9-assisted genome editing combined with in vivo and in vitro DNA assembly methods (DiCarlo et al.2013; Kuijpers et al.2013; Mans et al.2015; Tsai et al.2015).
The specific growth rate of strain IMX728 on l-arabinose in aerobic batch cultures, which was achieved without evolutionary engineering or random mutagenesis, corresponded to 65% of the specific growth rate on glucose of the parental strain CEN.PK113–7D (Solis-Escalante et al.2015) and is the highest growth rate on l-arabinose hitherto reported for an engineered S. cerevisiae strain (Wisselink et al.2007; Wiedemann and Boles 2008; Wang et al.2013). However, the genetic modifications in this strain did not enable anaerobic growth on l-arabinose. Anaerobic growth on l-arabinose requires that the biomass-specific l-arabinose consumption rate (qarabinose) is sufficiently high to meet the ATP requirement for cellular maintenance (ca. 1 mmol ATP (g biomass)−1 h−1, (Boender et al.2009)). If l-arabinose uptake occurs via Gal2-mediated facilitated diffusion, anaerobic, fermentative metabolism of l-arabinose yields 1.67 ATP, while transport of l-arabinose exclusively via PcAraT-mediated proton symport would result in a 60% lower ATP yield (Weusthuis et al.1994). Even if l-arabinose metabolism in the aerobic batch cultures of strain IMX728 would occur exclusively via the energetically much more favorable process of respiratory dissimilation (Bakker et al.2001; Verhoeven et al.2017), its specific growth rate of 0.26 h−1 would still correspond to a qarabinose of at least 3 mmol g−1 h−1. This rate of l-arabinose metabolism should be sufficient to meet anaerobic maintenance requirements via fermentative metabolism. The inability of strain IMX728 to instantaneously grow on l-arabinose in anaerobic cultures therefore suggests that it cannot achieve the same qarabinose under those conditions, which might reflect suboptimal expression, folding, membrane environment and/or stability of wild-type Gal2 in anaerobic cultures.
The observation that the two GAL2 copies in individual strains evolved for anaerobic growth in l-arabinose in the presence of d-glucose and d-xylose encoded the same GalN376 substitution, indicated that these substitutions most probably preceded gene duplication. The importance of these substitutions is further underlined by the observation that introduction of a single copy of the GALN376I allele enabled growth of an ‘arabinose specialist’ strain on l-arabinose in the presence of glucose and/or l-arabinose under aerobic conditions (Fig. 5). GAL genes are transcriptionally regulated via pathways involving Mig1 and Gal80 (Ostergaard et al.2001). In the present study, Gal80-mediated repression was eliminated by deletion of its encoding gene. The observed growth of glucose-phosphorylation-negative strains expressing an evolved GAL2N376 allele on l-arabinose in glucose-containing media indicates that any Mig1-mediated glucose repression of GAL2 was incomplete. In the glucose-phosphorylation-negative strains, absence of Hxk2, which is involved in Mig1-dependent glucose repression (Ahuatzi et al.2007) is likely to have glucose derepression of GAL genes. Indeed, deletion of HXK2 has been reported to stimulate co-consumption of glucose and galactose (Raamsdonk et al.2001). Additionally, in the same S. cerevisiae genetic background, combined deletion of MIG1, GAL6 and GAL80 was shown not to significantly affect GAL2 transcript levels (Bro et al.2005).
Two previous studies used d-glucose-phosphorylation-negative d-xylose-fermenting strains to evolve for d-glucose tolerance of d-xylose utilization (Farwick et al.2014; Nijland et al.2014). These studies identified mutations in hexose transporters that reduced d-glucose sensitivity of d-xylose transport. In Gal2, which also transports d-xylose (Farwick et al.2014) and in a chimera of Hxt3 and Hxt6 (Hxt36) (Nijland et al.2014), these mutations caused amino acid substitutions at the corresponding positions N376 and N367, respectively. Using in silico models of the three-dimensional structure of Gal2 and Hxt36, based on the crystal structure of the E. coli XylE d-xylose/H+ symporter, changes of the amino acid at these positions were predicted to confine the space in the substrate binding pocket for the hydroxymethyl group of d-glucose (Sun et al.2012; Farwick et al.2014; Nijland et al.2014). Both studies showed that size, charge, polarity and presence of hydrophobic side chains for the substituted amino acid strongly influenced relative transport activities with d-glucose and d-xylose. The present study shows that modifications of Gal2 at this position also enable transport of l-arabinose in the presence of d-glucose. Of the Gal2N376 substitutions identified and kinetically analyzed in our study, Gal2N376T showed the highest Km and lowest Vmax for d-glucose. Moreover, this substitution decreased the Km for l-arabinose without affecting Vmax for uptake of this pentose.
Of the four d-glucose-tolerant evolved strains isolated from the evolution experiments, strain IMS0522 showed significantly better l-arabinose fermentation performance than the other strains. Its l-arabinose consumption rate of 1.6 ± 0.08 g (g biomass)−1 h−1 and ethanol production rate of 0.61 ± 0.01 g (g biomass)−1 h−1 in anaerobic cultures are the highest reported to date for an engineered S. cerevisiae strain (Wisselink et al.2007; Wiedemann and Boles 2008; Wang et al.2013). In addition to a Gal2N376T substitution, one of the GAL2 copies in this strain encoded an additional Gal2T89I substitution, whose restoration to wild type resulted in a fermentation performance similar to that of the other strains. The physiological relevance of the mutation was further supported by its identification in a strain that was previously evolved for mixed-sugar fermentation (Wisselink et al.2009). By itself, the Gal2T89I showed a reduced Km for l-arabinose and an increased Km for d-glucose, while the Vmax for both sugars was decreased relative to that of wild-type Gal2. The Gal2T89I variant was also investigated in a recent study that systematically investigated amino acid substitutions at positions within or close to the predicted l-arabinose binding pocket of Gal2 (Wang et al.2017). Subsequent expression of the Gal2T89I substitution did not show a significant effect on the growth rate on l-arabinose (Wang et al.2017). However, our results suggest that, by itself, this substitution is advantageous for l-arabinose uptake in the presence of glucose and/or at low L-arabinose concentrations. Additionally, in the evolved strain IMS0010, in which Gal2 contained only this substitution, the mutations may have contributed to anaerobic co-consumption of l-arabinose and d-xylose (Wisselink et al.2009).
Transport assays with the Gal2N376T,T89I variant showed complete loss of d-glucose transport capacity, combined with a reduced Km and Vmax for l-arabinose. In the context of the SBR protocol in which this variant evolved, these kinetic properties are particularly relevant during the growth phase in which the l-arabinose concentration declined, while d-glucose in still present at 20 g L−1. The complete loss of glucose transport activity of the Gal2N376T,T89I, which was encoded by only one of two copies of GAL2 in the laboratory-evolved strain originating from a duplication of Chromosome XII, underline the importance of chromosomal CNV in laboratory evolution and neofunctionalization of duplicated genes (Gorter de Vries, Pronk and Daran 2017). These results from a 500-generation laboratory evolution experiment illustrate how, during evolution in dynamic natural environments, similar duplication and neofunctionalization events, may have contributed to the diverse kinetic characteristics and substrate specificities of the Hxt transporter family in S. cerevisiae (Reifenberger, Boles and Ciriacy 1997; Biswas et al.2013; Jordan et al.2016).
In addition to Gal2, the parental strain used in the evolution experiments expressed the fungal l-arabinose transporter PcAraT, which upon expression in S. cerevisiae mediates high-affinity, l-arabinose-proton symport (Km = 0.13 mM) and is much less sensitive to glucose inhibition than wild-type Gal2 (Bracher et al.2018). During the evolution experiments, no loss of function mutations was observed in PcAraT. This observation indicates that its presence did not confer a strong selective disadvantage, for example as a result of the activity of a futile cycle caused by simultaneous facilitated diffusion and proton symport of l-arabinose. Moreover, the observations that deletion of PcAraT resulted in decreased specific growth rate on l-arabinose and that presence of PcAraT coincided with a faster consumption of l-arabinose towards the end of anaerobic batch cultures (Fig. 4b), are in line with its reported low Vmax and low Km for l-arabinose upon expression in S. cerevisiae (Bracher et al.2018). These results demonstrate the potential of high-affinity l-arabinose transporters to efficiently convert low concentrations of l-arabinose towards the end of fermentation processes, thereby preventing prolonged ‘tailing’ of industrial fermentation processes.
CONCLUSIONS
Laboratory evolution of engineered, l-arabinose-metabolizing and glucose-phosphorylation-negative S. cerevisiae yielded evolved strains that anaerobically fermented l-arabinose in the presence of d-glucose and d-xylose. Amino acid substitutions in Gal2 that affected the kinetics of l-arabinose and d-glucose transport played a key role in this evolution. The best performing evolved strain contained two different mutated alleles of GAL2, encoding Gal2 variants with distinct kinetic properties. This result demonstrates the importance of engineering ‘transporter landscapes’ for uptake of individual sugars, consisting of transporters with complementary kinetic and/or regulatory properties, for efficient sugar conversion in dynamic, mixed-sugar fermentation processes.
SUPPLEMENTARY DATA
Supplementary data are available at FEMSYR online.
Acknowledgements
This work was performed within the BE-Basic R&D Program (http://www.be-basic.org/), which is financially supported by an EOS Long Term grant from the Dutch Ministry of Economic Affairs, Agriculture and Innovation (EL&I). The authors thank Marcel van den Broek (TUD), Pilar de la Torre (TUD), Ioannis Papapetridis (TUD), Laura Valk (TUD), Renzo Rozenbroek (TUD), Paul de Waal (DSM), Hans de Bruijn (DSM) and Paul Klaassen (DSM) for their input in this project.
AUTHORS’ CONTRIBUTIONS
MDV, JMD, AJAvM, JMB, JN, AD and JTP together designed this study; MDV, JMB, JN and JB designed and performed all wet-lab experiments; MDV and JTP wrote the manuscript. All authors read and commented a draft version of the manuscript and approved the submitted version.
AVAILABILITY OF SUPPORTING DATA
The genome sequence data of strains IMS0520, IMS0521, IMS0522, IMS0523, IMS514 and parental strains IMX728 and IMX660 have been deposited in Genbank (http://www.ncbi.nlm.nih.gov/), BioProject ID (PRJNA414371).
FUNDING AND COMPETING INTERESTS
This work was performed within the BE-Basic R&D Program (http://www.be-basic.org/), which is financially supported by an EOS Long Term grant from the Dutch Ministry of Economic Affairs, Agriculture and Innovation (EL&I). MDV, AJA vM and JTP are inventors on patent applications that include elements of this study. DSM markets technology for biofuels production from lignocellulosic feedstocks, holds IP positions in this field and co-funded the research described in this publication.
Conflicts of interest. None declared.
REFERENCES
- Ahuatzi D, Riera A, Peláez R et al. Hxk2 regulates the phosphorylation state of Mig1 and therefore its nucleocytoplasmic distribution. J Biol Chem 2007;282:4485–93. [DOI] [PubMed] [Google Scholar]
- Bakker BM, Overkamp KM, van Maris AJ et al. Stoichiometry and compartmentation of NADH metabolism in Saccharomyces cerevisiae. FEMS Microbiol Rev 2001;25:15–37. [DOI] [PubMed] [Google Scholar]
- Becker J, Boles E. A modified Saccharomyces cerevisiae strain that consumes L-arabinose and produces ethanol. Appl Environ Microbiol 2003;69:4144–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas C, Djordjevic JT, Zuo X et al. Functional characterization of the hexose transporter Hxt13p: an efflux pump that mediates resistance to miltefosine in yeast. Fungal Gene Biol 2013;61:23–32. [DOI] [PubMed] [Google Scholar]
- Boender LG, de Hulster EA, van Maris AJ et al. Quantitative physiology of Saccharomyces cerevisiae at near-zero specific growth rates. Appl Environ Microbiol 2009;75:5607–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracher JM, Verhoeven MD, Wisselink HW et al. The Penicillium chrysogenum transporter PcAraT enables high-affinity, glucose-insensitive l-arabinose transport in Saccharomyces cerevisiae. Biotechnol Biofuels 2018;11:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bro C, Knudsen S, Regenberg B et al. Improvement of galactose uptake in Saccharomyces cerevisiae through overexpression of phosphoglucomutase: example of transcript analysis as a tool in inverse metabolic engineering. Appl Environ Microbiol 2005;71:6465–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowley JH, Leak FW, Shianna KV et al. A mutation in a purported regulatory gene affects control of sterol uptake in Saccharomyces cerevisiae. J Bacteriol 1998;180:4177–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiCarlo JE, Norville JE, Mali P et al. Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res 2013;41:4336–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson RC, Nagiec EE, Wells GB et al. Synthesis of mannose-(inositol-P) 2-ceramide, the major sphingolipid in Saccharomyces cerevisiae, requires the IPT1 (YDR072c) gene. J Biol Chem 1997;272:29620–5. [DOI] [PubMed] [Google Scholar]
- Entian K-D, Kötter P. 25 Yeast genetic strain and plasmid collections. Method Microbiol 2007;36:629–66. [Google Scholar]
- Farwick A, Bruder S, Schadeweg V et al. Engineering of yeast hexose transporters to transport D-xylose without inhibition by D-glucose. Proc Natl Acad Sci 2014;111:5159–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz RD, Schiestl RH, Willems AR et al. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast 1995;11:355–360. [DOI] [PubMed] [Google Scholar]
- Gorter de Vries AR, Pronk JT, Daran J-MG. Industrial relevance of chromosomal copy number variation in Saccharomyces yeasts. Appl Environ Microbiol 2017;83:e03206–03216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grohmann K, Bothast RJ. Pectin-rich residues generated by processing of citrus fruits, apples, and sugar beets. ACS Symposium Series 1994;566:372390. 1994;566:372–390. [Google Scholar]
- Grohmann K, Bothast RJ. Saccharification of corn fibre by combined treatment with dilute sulphuric acid and enzymes. Process Biochem 1997;32:405–15. [Google Scholar]
- Guadalupe Medina V, Almering MJ, van Maris AJ et al. Elimination of glycerol production in anaerobic cultures of a Saccharomyces cerevisiae strain engineered to use acetic acid as an electron acceptor. Appl Environ Microbiol 2010;76:190–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamacher T, Becker J, Gárdonyi M et al. Characterization of the xylose-transporting properties of yeast hexose transporters and their influence on xylose utilization. Microbiology 2002;148:2783–8. [DOI] [PubMed] [Google Scholar]
- Jeffries TW. Utilization of xylose by bacteria, yeasts, and fungi. Pentoses and Lignin, Springer, Berlin, Heidelberg, 1983; pp. 1–32. [DOI] [PubMed] [Google Scholar]
- Jordan P, Choe J-Y, Boles E et al. Hxt13, Hxt15, Hxt16 and Hxt17 from Saccharomyces cerevisiae represent a novel type of polyol transporters. Sci Rep 2016:6:23502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoshaug EP, Vidgren V, Magalhães F et al. Novel transporters from Kluyveromyces marxianus and Pichia guilliermondii expressed in Saccharomyces cerevisiae enable growth on l-arabinose and d-xylose. Yeast 2015;32:615–28. [DOI] [PubMed] [Google Scholar]
- Kou S-C, Christensen MS, Cirillo VP. Galactose transport in Saccharomyces cerevisiae II. Characteristics of galactose uptake and exchange in galactokinaseless cells. J Bacteriol 1970;103:671–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuijpers NG, Chroumpi S, Vos T et al. One-step assembly and targeted integration of multigene constructs assisted by the I-SceI meganuclease in Saccharomyces cerevisiae. FEMS Yeast Res 2013;13:769–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuyper M, Hartog MM, Toirkens MJ et al. Metabolic engineering of a xylose-isomerase-expressing strain for rapid anaerobic xylose fermentation. FEMS Yeast Res 2005;5:399–409. [DOI] [PubMed] [Google Scholar]
- Kuyper M, Harhangi HR, Stave AK et al. High-level functional expression of a fungal xylose isomerase: the key to efficient ethanolic fermentation of xylose by Saccharomyces cerevisiae. FEMS Yeast Res 2003;4:69–78. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009;25:1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Xu J, Cai P et al. Functional analysis of two L-arabinose transporters from filamentous fungi reveals promising characteristics for improved pentose utilization in Saccharomyces cerevisiae. Appl Environ Microbiol 2015;81:4062–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynd LR. Overview and evaluation of fuel ethanol from cellulosic biomass: technology, economics, the environment, and policy. Annu Rev Energy Environ 1996;21:403–65. [Google Scholar]
- Mans R, van Rossum HM, Wijsman M et al. CRISPR/Cas9: a molecular Swiss army knife for simultaneous introduction of multiple genetic modifications in Saccharomyces cerevisiae. FEMS Yeast Res 2015;15:fov004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijkamp JF, van den Broek MA, Geertman JMA et al. De novo detection of copy number variation by co-assembly. Bioinformatics 2012;28:3195–202. [DOI] [PubMed] [Google Scholar]
- Nijkamp JF, van den Broek M, Datema E et al. De novo sequencing, assembly and analysis of the genome of the laboratory strain Saccharomyces cerevisiae CEN.PK113-7D, a model for modern industrial biotechnology. Microb Cell Fact 2012;11:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijland JG, Shin HY, de Jong RM et al. Engineering of an endogenous hexose transporter into a specific D-xylose transporter facilitates glucose-xylose co-consumption in Saccharomyces cerevisiae. Biotechnol Biofuels 2014;7:168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostergaard S, Walløe KO, Gomes CS et al. The impact of GAL6, GAL80, and MIG1 on glucose control of the GAL system in Saccharomyces cerevisiae. FEMS Yeast Res 2001;1:47–55. [DOI] [PubMed] [Google Scholar]
- Oud B, van Maris AJ, Daran J-M et al. Genome-wide analytical approaches for reverse metabolic engineering of industrially relevant phenotypes in yeast. FEMS Yeast Res 2012;12:183–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raamsdonk LM, Diderich JA, Kuiper A et al. Co-consumption of sugars or ethanol and glucose in a Saccharomyces cerevisiae strain deleted in the HXK2 gene. Yeast 2001;18:1023–33. [DOI] [PubMed] [Google Scholar]
- Reifenberger E, Boles E, Ciriacy M. Kinetic characterization of individual hexose transporters of Saccharomyces cerevisiae and their relation to the triggering mechanisms of glucose repression. FEBS J 1997;245:324–33. [DOI] [PubMed] [Google Scholar]
- Renewable Fuels Association World fuel ethanol, 2017; production. available at: http://ethanolrfa.org/resources/industry/statistics/.
- Richard P, Putkonen M, Väänänen R et al. The missing link in the fungal L-arabinose catabolic pathway, identification of the L-xylulose reductase gene. Biochemistry 2002;41:6432–7. [DOI] [PubMed] [Google Scholar]
- Roberts RL, Mösch H-U, Fink GR. 14-3-3 proteins are essential for RAS/MAPK cascade signaling during pseudohyphal development in S. cerevisiae. Cell 1997;89:1055–65. [DOI] [PubMed] [Google Scholar]
- Sedlak M, Ho NW. Expression of E. coli araBAD operon encoding enzymes for metabolizing L-arabinose in Saccharomyces cerevisiae. Enzyme Microb Technol 2001;28:16–24. [DOI] [PubMed] [Google Scholar]
- Solis-Escalante D, Kuijpers NG, Barrajon-Simancas N et al. A minimal set of glycolytic genes reveals strong redundancies in Saccharomyces cerevisiae central metabolism. Eukaryotic Cell 2015;14:804–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solis-Escalante D, Kuijpers NG, Bongaerts N et al. amdSYM, a new dominant recyclable marker cassette for Saccharomyces cerevisiae. FEMS Yeast Res 2013;13:126–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subtil T, Boles E. Improving L-arabinose utilization of pentose fermenting Saccharomyces cerevisiae cells by heterologous expression of L-arabinose transporting sugar transporters. Biotechnol Biofuels 2011;4:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subtil T, Boles E. Competition between pentoses and glucose during uptake and catabolism in recombinant Saccharomyces cerevisiae. Biotechnol Biofuels 2012;5:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Zeng X, Yan C et al. Crystal structure of a bacterial homologue of glucose transporters GLUT1-4. Nature 2012;490:361–6. [DOI] [PubMed] [Google Scholar]
- Tani T, Taguchi H, Fujimori KE et al. Isolation and characterization of xylitol-assimilating mutants of recombinant Saccharomyces cerevisiae. J Biosci Bioeng 2016;122:446–55. [DOI] [PubMed] [Google Scholar]
- Thorvaldsdóttir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Briefings in Bioinformatics 2013;14:178–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torchia T, Hamilton R, Cano C et al. Disruption of regulatory gene GAL80 in Saccharomyces cerevisiae: effects on carbon-controlled regulation of the galactose/melibiose pathway genes. Mol Cell Biol 1984;4:1521–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Träff K, Cordero RO, Van Zyl W et al. Deletion of the GRE3 aldose reductase gene and its influence on xylose metabolism in recombinant strains of Saccharomyces cerevisiae expressing the xylA and XKS1 genes. Appl Environ Microbiol 2001;67:5668–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai C-S, Kong II, Lesmana A et al. Rapid and marker-free refactoring of xylose-fermenting yeast strains with Cas9/CRISPR. Biotechnol Bioeng 2015;112:2406–11. [DOI] [PubMed] [Google Scholar]
- Van Hemert M, van Heusden G, Steensma H. Yeast 14-3-3 proteins. Yeast 2001;18:889–95. [DOI] [PubMed] [Google Scholar]
- van Heusden GPH, Wenzel TJ, Lagendijk EL et al. Characterization of the yeast BMH1 gene encoding a putative protein homologous to mammalian protein kinase II activators and protein kinase C inhibitors. FEBS Lett 1992;302:145–50. [DOI] [PubMed] [Google Scholar]
- van Maris AJ, Abbott DA, Bellissimi E et al. Alcoholic fermentation of carbon sources in biomass hydrolysates by Saccharomyces cerevisiae: current status. Antonie van Leeuwenhoek 2006;90:391–418. [DOI] [PubMed] [Google Scholar]
- Verduyn C, Postma E, Scheffers WA et al. Physiology of Saccharomyces cerevisiae in Anaerobic Glucose-Limited Chemostat Cultures. Microbiology 1990;136:395–403. [DOI] [PubMed] [Google Scholar]
- Verduyn C, Postma E, Scheffers WA et al. Effect of benzoic acid on metabolic fluxes in yeasts: A continuous-culture study on the regulation of respiration and alcoholic fermentation. Yeast 1992;8:501–17. [DOI] [PubMed] [Google Scholar]
- Verho R, Penttilä M, Richard P. Cloning of two genes (LAT1,2) encoding specific l-arabinose transporters of the l-arabinose fermenting yeast Ambrosiozyma monospora. Appl Biochem Biotechnol 2011;164:604–11. [DOI] [PubMed] [Google Scholar]
- Verhoeven MD, Lee M, Kamoen L et al. Mutations in PMR1 stimulate xylose isomerase activity and anaerobic growth on xylose of engineered Saccharomyces cerevisiae by influencing manganese homeostasis. Sci Rep 2017;7:46155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker BJ, Abeel T, Shea T et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 2014;9:e112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Skinner C, Easlon E et al. Deleting the 14-3-3 protein Bmh1 extends life span in Saccharomyces cerevisiae by increasing stress response. Genetics 2009;183:1373–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Shen Y, Zhang Y et al. Improvement of L-arabinose fermentation by modifying the metabolic pathway and transport in Saccharomyces cerevisiae. BioMed Research International 2013;2013:461204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Li Y, Qiu C et al. Identification of important amino acids in Gal2p for improving the l-arabinose transport and metabolism in Saccharomyces cerevisiae. Front Microbiol 2017;8:1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weusthuis RA, Pronk JT, Van Den Broek P et al. Chemostat cultivation as a tool for studies on sugar transport in yeasts. Microbiol Rev 1994;58:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedemann B, Boles E. Codon-optimized bacterial genes improve L-arabinose fermentation in recombinant Saccharomyces cerevisiae. Appl Environ Microbiol 2008;74:2043–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisselink HW, Toirkens MJ, Wu Q et al. Novel evolutionary engineering approach for accelerated utilization of glucose, xylose, and arabinose mixtures by engineered Saccharomyces cerevisiae strains. Appl Environ Microbiol 2009;75:907–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisselink HW, Van Maris AJA, Pronk JT et al. Polypeptides with permease activity. US Patent 9034608 B2, 2015.
- Wisselink HW, Toirkens MJ, Del Rosario Franco Berriel M et al. Engineering of Saccharomyces cerevisiae for efficient anaerobic alcoholic fermentation of l-arabinose. Appl Environ Microbiol 2007;73:4881–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisselink HW, Cipollina C, Oud B et al. Metabolome, transcriptome and metabolic flux analysis of arabinose fermentation by engineered Saccharomyces cerevisiae. Metab Eng 2010;12:537–51. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.