Abstract
The dextran sodium sulfate (DSS) model of colitis is widely used as a result of its simplicity and reproducibility and because it mimics clinicopathological disease features. Its effectiveness depends on the mouse strain, DSS MW, and brand. Quantitative RT-PCR (qRT-PCR) is highly sensitive for analyzing cytokine mRNA expression. We analyzed an acute model of DSS treatment in Balb/c mice for the onset of colitis using qRT-PCR for the quantification of a mouse cytokine transcript. We compared differences among 1--and 2-step qRT-PCR for transcript quantification, the effect of multiple concentrations of DSS, and the use of 2 reference genes in 3 portions of the colon. A reliable and sensitive 1-step protocol for qRT-PCR was established with a modified double LiCl precipitation for RNA isolation. The variability of 2 reference genes, β-actin and eukaryotic elongation factor 2, was compared, and expression of IL-6 was analyzed in 3 segments of the colon. The RNA cleaning protocol prevented inhibition of qRT-PCR by DSS, and RNA loss was minimized. No clinical differences among the different DSS concentrations were seen on d 7, but higher concentrations resulted in the appearance of earlier symptoms. Higher efficiency and sensitivity of the 1-step qRT-PCR reaction using eukaryotic elongation factor 2 were obtained and also less variability. Although expression levels of IL-6 were high in the middle and distal colon, the middle section had consistently less variability in values. Thus, this segment is recommended for future studies. These factors influence the statistical significance of data and need to be considered to get accurate and reliable results and to improve comparisons of the published colitis experiments.
Keywords: DSS model, reference genes, IL-6 expression, inflammation
INTRODUCTION
In 1985, Ohkusa1 first described the dextran sodium sulfate (DSS) as a model of colonic inflammation in hamsters mimicking inflammatory bowel diseases (IBDs) and resembling mainly ulcerative colitis. Thereafter, the model was extrapolated to mice, and it has gained popularity as a result of its simplicity, reproducibility, controllability, and effectiveness and because it mimics some of the clinicopathological features of human IBD.2 DSS is a negatively charged, water-soluble polysaccharide that causes disruption of the epithelial barrier of the colonic tissue. Although the exact mode of action has not yet been elucidated, the most accepted mechanism is the loss of the barrier function, along with the penetration of the microbiome and the proinflammatory intestinal products. Early descriptions of the model suggested that damage was limited to the distal colon. A recent study proved that lesions occur all along the gastrointestinal tract, not being specific to the large bowel.3
Acute and chronic inflammation, colonic adenocarcinoma, and necrotizing enterocolitis models have been reported by variations on dosing, inclusion of azomethane, and animal age, respectively.4 In addition, the effectiveness of the DSS model depends on the strain and colony of mice used, as well as the DSS MW. As DSS is a polysaccharide, its MW can vary according to the number of monosaccharides attached, modifying its pharmacokinetics. Murine colitis is induced by DSS of 40–50 kDa, whereas low MW DSS causes proximal lesions, and a larger MW fails to establish colitis, as its absorption is diminished.5 Moreover, DSS preparations of the same MW made by different manufacturers are also a source of variation.6
Quantitative RT-PCR (qRT-PCR) is a highly sensitive and specific tool for analyzing mRNA expression of proinflammatory and anti-inflammatory cytokines and has been proposed as the method of choice for quantification of cytokine profiles in immune cells or inflamed tissues.7 In addition, it allows the detection of many different cytokines from relatively small sample amounts of RNA.8 There are 2 ways the qRT-PCR can be done. In the 1-step method, the RT reaction is performed in the same tube as the PCR and usually uses specific primers for cDNA synthesis. The 2-step method synthesizes cDNA from the isolated mRNA in a separate reaction, and then the PCR is performed. The 2-step method typically uses random hexamers or oligo(dT) primers, which allow RT of all mRNA into cDNA. Assay-specific primers may be used in the 2-step reaction as well, but this approach is limited by RNA amounts, as a different RT reaction must be performed for each mRNA to be analyzed. Therefore, the efficiency and sensitivity of the qRT-PCR may depend on the priming methods.9, 10
In this study, we compared the efficiency and sensitivity of these 2 qRT-PCR methods for the relative quantification of 2 reference genes that differ in their stability during DSS-induced colitis11 and IL-6 as a marker of proinflammatory cytokine expression during DSS-induced colitis. As DSS has been reported as an inhibitor of both the RT and Taq polymerase, we used a modified protocol for the elimination of DSS traces in colonic samples by double precipitation of RNA with LiCl, which increases its purity and is based on a previously described method.12 Most papers, where qRT-PCR is performed to analyze cytokine profiles during DSS-induced colitis, do not include the removal of polysaccharides, which may cause alterations in the results and thus, could interfere with the conclusions reached. The development of a sensitive method for qRT-PCR in DSS-induced colitis models is crucial for accurate results and to support the detection of significant differences in gene expression without the need to use high DSS concentrations.
Therefore, here, we established a standardized, reliable, and sensitive 1-step protocol for qRT-PCR for the DSS-induced model of colitis, considering the quality of the isolated RNA, election of the reference gene, and the portion of the colonic tissue analyzed.
MATERIALS AND METHODS
Animals
Mice were bred at the Instituto Nacional de Pediatria and cared for by the team under the supervision of Dr. Ramon Garcia, head of the animal facility. All efforts were made to minimize animal suffering. Female Balb/c mice (6–8 wk old) were housed in groups of no more than 5 animals per cage in pathogen-free conditions in a temperature-controlled room (24°C) and on a 12/12-light cycle with food and water ad libitum. Cages were cleaned every 48 h, and mice were handled with care. There was no mortality.
The Institutional Research and Ethics Committees of the Faculty of Medicine, National University of Mexico, and the Faculty of Health Sciences of Anahuac University approved all animal procedures in accordance with Mexican Official Guidelines (NOM-062-ZOO-1999; Project 116/2015) that are in strict agreement with recommendations in the Guide for the Care and Use of Laboratory Animals of the NIH National Institutes of Health (Bethesda, MD, USA).
Induction of colitis
DSS was dissolved in tap water and given as drinking water to mice at 3, 4, or 5% concentration for 7 d, fresh DSS was prepared every 48 h, and the control group received only water. On d 7, animals were euthanized by sevoflurane overdose (Svofast; Baxter International, Deerfield, IL, USA). Two experiments were performed. The first was aimed to establish the ideal concentration of DSS; 4 groups were established: control group (n = 3), 3% DSS (Affymetrix, Santa Clara, CA, USA; n = 3), 4% DSS (n = 4), and 5% DSS (n = 5). The number of animals was higher in the 4 and 5% DSS groups, because a higher mortality rate was expected. In the second experiment, a control group (n = 5) and a 3% DSS group (n = 5) were used, based on the results of the first experiment.
Clinical evaluation
The severity of colitis was evaluated daily, according to a disease activity index (DAI) modified from Chassaing et al.,13 Taylor et al.,14 and Maxwell et al.,15 according to the following parameters: 1) weight loss (<5% = 0, 5–10% = 1, 10–20% = 2, >20% = 3), 2) stool appearance (normal = 0, soft but formed = 1, very soft = 2, watery stools or incapacity to evacuate = 3), 3) presence of blood (no blood = 0, traces of blood = 1, blood on rectum = 2, blood on fur = 3), and 4) movement (normal = 0, piloerection = 1, lethargy = 2, motionless = 3). As macroscopic markers of inflammation, colonic length was measured, which indicates local edema.
RNA extraction
To analyze mRNA expression, once the colon length was registered, the cecum was removed, washed with PBS, pH 7.4, and supplemented with protease inhibitors (Complete; Roche, Indianapolis, IN, USA) to remove all traces fecal material using a straight stainless-steel oral gavage needle, gauge 22, and the remaining tissue was divided in 3 segments: proximal, middle, and distal. One centimeter of each segment was isolated and kept in Trizol reagent (Thermo Fisher Scientific, Waltham, MA, USA) at −70°C until use. The manufacturer’s protocol was used to isolate RNA. Colonic tissue was homogenized in the Trizol Reagent using a PowerGen Model 125 Homogenizer (Thermo Fisher Scientific) for 1 min at 30,000 rpm. As DSS inhibition of PCR was observed, a double 2.5 M LiCl precipitation that precipitates large RNA was added after the Trizol protocol. RNA was incubated on ice for 90 min and centrifuged at 12,000 g for 30 min at 4°C. The resulting RNA pellet was dissolved in 200 μl RNase-free water. The LiCl precipitation, incubation, and centrifugation were repeated once more. RNA was dissolved in 200 μl in 0.3 M sodium acetate and precipitated overnight at −20°C with 1 vol isopropyl alcohol. The samples were centrifuged at 12,000 g for 30 min at 4°C, the supernatant was removed, and the pellet containing the RNA was washed with 500 μl 75% ethanol. The RNA pellet was dissolved in 20–50 μl RNase-free water and kept at −70°C until use. RNA concentration was quantified with the Nano-Drop 2000 Spectrophotometer (Thermo Fisher Scientific). RNA quality was evaluated by absorbance at 260/280 nm (A260/A280) and A260/A230 ratios with values of 1.94–2.09 and 2.02–2.20, respectively. RNase-free methods, as well as tips and tubes guaranteed to be RNase free, were used throughout the procedures.
qRT-PCR
For qRT-PCR, 1-step and 2-step reactions were compared. For both assays, a Taqman gene-expression assay was performed using commercially available, predesigned primers and probes (Thermo Fisher Scientific) for β-actin (Mm00607939_s1), both primers and probe map within a single exon: exon 6, GenBank AK078935.1: location 1233, amplicon length 115 (a qPCR without the RT step was performed to confirm that genomic DNA is not amplified); eukaryotic translation elongation factor 2 (Eef2; Mm01171435_gH), probe spans exons: exon boundary 2–3, RefSeq NM_007907.2: location 316, amplicon length 62 bp; and IL-6 (Mm00446190_m1), probe spans exons: exon boundary 2–3, RefSeq NM_031168.1, location 237, amplicon length 78 bp. All reactions were performed on a LightCycler 2.0 (Roche). Probe efficiency was calculated by a 4-point curve with 10-fold dilutions. For the 1-step reaction, cDNA starting dilution was prepared from 1 μg DNA; for the 2-step reaction, RNA starting dilution was 200 ng. Triplicate amplifications for each dilution were performed.
For the 1-step protocol, RNA was diluted, and the reaction was prepared in 10 μl containing 3 μl RNA, 5 μl Luna Universal Probe One-Step Reaction Mix (New England BioLabs, Ipswich, MA, USA), 0.5 μl Luna WarmStart RT Enzyme Mix (New England BioLabs), 0.5 μl predesigned Taqman assay probe plus primers, and 1 μl RNase-free water. Gene-expression assays (Thermo Fisher Scientific) are supplied as a 20× stock containing 18 μM per primer and 5 mM Taqman probe; 1× final concentrations were 900 nM for each primer and 250 nM probe, as recommended by the manufacturer. Parameters for PCR amplification were the following: RT (55°C for 10 min), initial denaturalization (95°C for 1 min), followed by 45 cycles of denaturalization (95°C for 10 s), annealing and extension (60°C for 30 s), and detection at 72°C for 1 s, as recommended by the manufacturer.
For the 2-step protocol, first, 1 μg RNA was submitted to RT using oligo(dT)20 primers, provided with the SuperScript III First Strand Synthesis System (Thermo Fisher Scientific), according to the manufacturer’s instructions. For the RT reaction, 1 μg RNA was incubated for 5 min at 65°C with 50 μM oligo(dT)20, 10 mM deoxynucleotide, and RNase-free water in 10 μl and then placed on ice for 1 min. Next, 10 μl of a mixture of 25 mM MgCl2, 0.1 M DTT, 40 U/μl RNaseOut, 200 U/μl SuperScript III, and RT buffer, provided by the manufacturer, was added and incubated for 50 min at 50°C. The reaction was terminated at 85°C for 5 min. Finally, 1 μl RNase H was added to each tube and incubated for 20 min at 37°C to hydrolyze specifically the phosphodiester bonds of RNA hybridized to DNA. cDNA was stored at −20°C until use. cDNA concentrations were measured following the RT reaction with the Nano-Drop 2000 Spectrophotometer.
The resulting cDNA was used in a 10-μl reaction that contained 3 μl cDNA, 2 μl Taqman Universal PCR Master Mix (Roche), 0.5 μl Taqman assay probe, and 4.5 μl RNase-free water. Parameters for PCR amplification were 95°C for 10 min, followed by 45 cycles each consisting of denaturation at 95°C for 10 s, annealing at 60°C for 10 s, and extension at 72°C for 10 s. Relative quantification calculations were performed by the Pfaffl method.16 β-Actin and Eef2 were used as potential reference genes to normalize values and compare their stability.
Statistical analysis
The graphs and statistical analysis were performed using Prism 6.0 software (GraphPad Software, La Jolla, CA, USA). Statistical significance for multiple groups was calculated using Kruskal-Wallis analysis, followed by Dunn’s post hoc test. Mann-Whitney test was performed for comparison between 2 groups. All data are shown as means ± sd. A value of P < 0.05 was considered significant, and detailed significance values are shown in figure legends.
RESULTS
Optimal DSS concentration
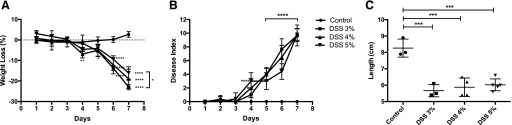
Induction of colitis with different concentrations of DSS was analyzed in the treated groups compared with the control group, and no significant differences were seen on water consumption. Based on weight loss, DAI, and colon length, all DSS-treated groups showed a significant degree of inflammation compared with the control group (Fig. 1A–C). The 3% DSS group lost significantly more weight than the 5% DSS group (Fig. 1A). Figure 1B shows the daily DAI; on d 4, a significant difference among the 3 groups was observed, as 4 and 5% DSS groups showed a higher DAI compared with the 3% DSS group (P < 0.05). However, by d 7, when necropsies were performed, all DSS-treated groups presented the same DAI. Likewise, at d 7, the colon length was reduced in the DSS-treated groups compared with the control, but no significant difference was observed among the different DSS groups (Fig. 1C).
FIGURE 1.
Analysis of clinical parameters for different concentrations of DSS. Weight loss (A), DAI (B), and colon length (C) were evaluated during 7 d of oral administration of DSS. DSS was used at a concentration of 3, 4, and 5%. Two-way ANOVA was used for weight loss and DAI analysis and 1-way ANOVA for colon length. Results are expressed as means ± sd compared with control mice; n = 3–5 mice per group. *P < 0.05, **P < 0.01, ***P = 0.001, ****P < 0.0001.
DSS inhibits RT and Taq polymerase
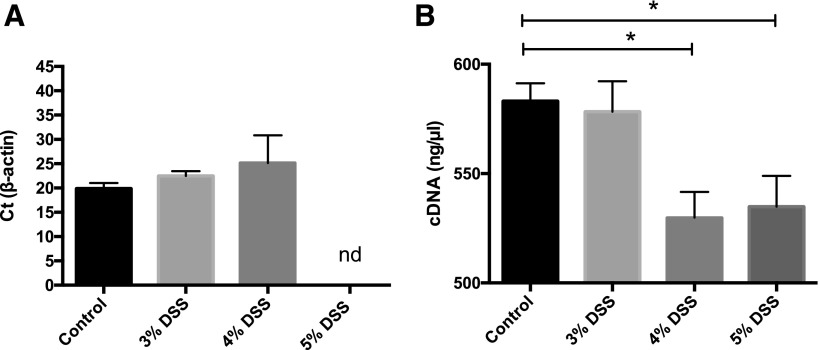
Residual DSS in colonic tissue in RNA preparations after Trizol isolation may interfere with qRT-PCR.12 Figure 2A shows the Ct value of the cDNA prepared from RNA, obtained from the middle portion of the colon of mice treated with the different concentrations of DSS. A 2-step reaction was performed. A delay in the Ct value was observed as the concentration of DSS increased, whereas in the 5% DSS group, no amplification was seen after 45 cycles. To analyze if traces of DSS in colonic RNA samples interfered with the RT step, cDNA produced from 1 μg RNA in the RT reaction was evaluated. cDNA concentrations decreased when higher concentrations of DSS were used (Fig. 2B).
FIGURE 2.
Traces of DSS inhibit PCR amplification in a 2-step reaction and cDNA synthesis using the SuperScript III First Strand Synthesis System. Ct values for β-actin of the cDNA were prepared from RNA from the colon of mice treated with the different concentrations of DSS. A delay in Ct value is observed with increasing concentrations of DSS (A). Traces of DSS in colonic RNA samples interfered with the RT reaction as lower concentrations of cDNA were obtained with increasing concentrations of DSS (B). Results are expressed as means ± sd; n = 3–5 mice per group. Kruskall-Wallis, *P < 0.05; nd, not detected.
RNA precipitation to remove residual DSS
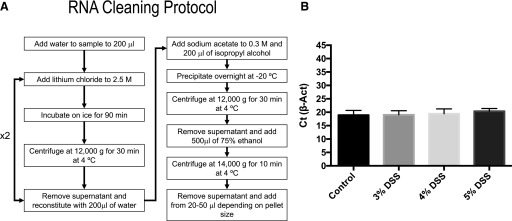
A protocol to eliminate residual DSS from colonic samples was reported by Viennois et al.12 They proposed an RNA cleaning method by a double precipitation with 0.1 vol 8 M LiCl. However, in our hands, at the concentration of LiCl used by the authors, no RNA precipitation occurred. We thus increased the LiCl final concentration to 2.5 M, followed by an incubation on ice for 90 min, substituted the final ethanol precipitation with 1 vol isopropyl alcohol in the presence of 0.3 M sodium acetate, and increased the precipitation time to overnight at −20°C to reduce the RNA loss. Figure 3A shows the whole cleaning protocol that resulted in an uncontaminated RNA. Figure 3B shows the Ct values for β-actin in the 2-step reaction; no interference of the combined RT and qPCR steps was observed, as β-actin mean Ct values were the same with all DSS concentrations used.
FIGURE 3.
DSS cleaning protocol results in efficient amplification of cDNA. Flow chart showing the main steps for double LiCl precipitation (A). Ct values for β-actin of cDNA were prepared from RNA from the colon of mice treated with the different concentrations of DSS after the cleaning protocol. Traces of DSS were efficiently removed, and amplification was no longer inhibited by traces of DSS (B).
The 1-step protocol has greater efficiency and sensitivity than the 2-step protocol
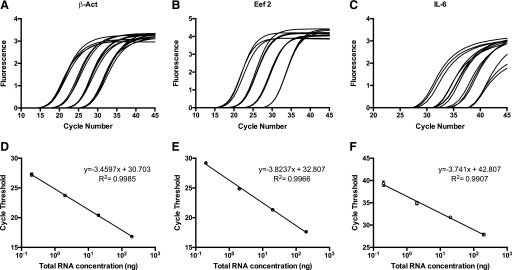
To compare the efficiency and sensitivity of 1-step and 2-step qRT-PCR, curves with 10-fold dilutions were analyzed. Representative 1-step reactions for 2 potential reference genes (β-actin and Eef2) and for IL-6 are shown in Fig. 4A–C. Each reaction was run in triplicate, and these points were plotted on a graph of the log of RNA concentration vs. Ct (Fig. 4D–F). Table 1 shows the correlation coefficient (R2) values calculated from the line of best fit, the reaction efficiencies, and the average Ct. The standard curves generated for all 3 genes produced linear results. For IL-6, no amplification was observed in the last dilution using the 2-step protocol. Efficiencies were lower in the 2-step reaction (84, 80, and 84%, respectively) compared with those obtained with the 1-step method (95, 88, and 92%, respectively).
FIGURE 4.
qRT-PCR curves for β-actin, Eef2, and IL-6 using the 1-step protocol. Real-time fluorescence using 1:10-fold dilutions starting at 200 ng total RNA from colon run in triplicate using primers/probe for β-actin (A), Eef2 (B), and IL-6 (C). B) The standard curves generated from the Ct values of each of the dilutions and the equation of the line of best fit for β-actin (D), Eef2 (E), and IL-6 (F).
TABLE 1.
Comparison of 1-step and 2-step methods for reference and IL-6 genes
| Primer | R2 2-step | R2 1-step | Efficiency 2-step, % | Efficiency 1-step, % | Average Ct 2-step | Average Ct 1-step |
|---|---|---|---|---|---|---|
| β-Actin | 0.996 | 0.998 | 84 | 95 | 17.81 ± 0.12 | 14.65 ± 0.65 |
| Eef2 | 0.990 | 0.996 | 80 | 88 | 24.40 ± 0.48 | 21.16 ± 0.48 |
| IL-6 | 0.994 | 0.991 | 84 | 92 | 38.17 ± 1.05 | 35.44 ± 0.60 |
To assess the sensitivity of both methods, we compared the Ct values for the 3 genes with each method. There was a 3-cycle difference for all 3 genes tested, with the 1-step reaction more sensitive. Table 2 shows the Ct values and descriptive statistics of the 10-fold dilutions used in the generation of the standard curves for the 1-step reactions for all 3 genes. sds for 20 ng reactions were >0.3, whereas higher Ct variations were observed with 2 and 0.2 ng, which is expected when higher Ct values are obtained. The 20 ng of RNA is within the linear dynamic range of the standard curves and was used to analyze the relative expression of IL-6 in the 1-step reactions for the colon samples from the control and the DSS-treated groups.
TABLE 2.
Descriptive performance of 1-step dilution curves for β-actin, Eef2, and IL-6 using RNA from control mice (Ct values)
| Primer | RNA, ng | Minimum | Maximum | Median | Mean | sd |
|---|---|---|---|---|---|---|
| β-Actin | 200 | 16.79 | 16.86 | 16.82 | 16.82 | 0.035 |
| 20 | 20.21 | 20.54 | 20.48 | 20.41 | 0.176 | |
| 2 | 23.67 | 23.86 | 23.68 | 23.74 | 0.107 | |
| 0.2 | 26.95 | 27.46 | 27.33 | 27.25 | 0.265 | |
| Eef2 | 200 | 17.32 | 17.96 | 17.61 | 17.63 | 0.320 |
| 20 | 21.15 | 21.43 | 21.36 | 21.31 | 0.145 | |
| 2 | 24.72 | 24.98 | 24.85 | 24.85 | 0.130 | |
| 0.2 | 29.19 | 29.21 | 29.19 | 29.20 | 0.111 | |
| IL-6 | 200 | 27.55 | 28.14 | 27.93 | 27.87 | 0.299 |
| 20 | 31.59 | 32.03 | 31.62 | 31.75 | 0.246 | |
| 2 | 34.54 | 35.23 | 34.95 | 34.91 | 0.347 | |
| 0.2 | 38.76 | 39.91 | 39.20 | 39.29 | 0.580 |
IL-6 expression varies in different colon segments
As histologic damage by DSS may vary, depending on the colon segment studied, we analyzed the relative expression of IL-6 as a marker of inflammation in the proximal, middle, and distal portions of the colon. Table 3 shows the Ct values and descriptive statistics of the 3 genes in the different colon segments in control and inflammatory conditions. Higher sds were observed in the proximal region for all 3 genes. In the distal segment, sds were high for IL-6 in both groups of mice. The middle section showed the least variability, as shown by the lower sds observed in both control and inflammatory conditions.
TABLE 3.
Descriptive performance of 1-step reactions for β-actin, Eef2, and IL-6 in the different colon segments (Ct values)
|
Gene |
Minimum |
Maximum |
Median |
Mean |
sd |
|---|---|---|---|---|---|
| Proximal colon | |||||
| β-Actin control | 19.94 | 24.31 | 22.81 | 22.77 | 1.10 |
| β-Actin DSS | 16.09 | 20.91 | 20.26 | 19.26 | 1.75 |
| Eef2 control | 20.60 | 22.61 | 21.62 | 21.58 | 0.55 |
| Eef2 DSS | 19.93 | 22.94 | 20.82 | 20.83 | 1.09 |
| IL-6 control | 35.80 | 39.22 | 36.96 | 37.07 | 1.02 |
| IL-6 DSS | 27.12 | 35.94 | 33.07 | 32.82 | 3.17 |
| Middle colon | |||||
| β-Actin control | 17.86 | 19.04 | 18.20 | 18.28 | 0.40 |
| β-Actin DSS | 17.25 | 18.57 | 18.04 | 17.98 | 0.36 |
| Eef2 control | 20.68 | 21.60 | 20.81 | 20.99 | 0.34 |
| Eef2 DSS | 20.78 | 22.51 | 21.49 | 21.65 | 0.54 |
| IL-6 control | 34.29 | 35.14 | 34.65 | 34.67 | 0.25 |
| IL-6 DSS | 26.88 | 28.73 | 27.92 | 27.83 | 0.61 |
| Distal colon | |||||
| β-Actin control | 17.38 | 18.19 | 17.66 | 17.70 | 0.24 |
| β-Actin DSS | 15.67 | 18.78 | 17.01 | 17.10 | 0.89 |
| Eef2 control | 19.76 | 21.21 | 2.30 | 20.38 | 0.49 |
| Eef2 DSS | 20.24 | 20.91 | 20.49 | 20.56 | 0.22 |
| IL-6 control | 30.74 | 35.80 | 34.15 | 33.67 | 1.65 |
| IL-6 DSS |
25.16 | 33.91 | 27.07 | 28.56 | 3.08 |
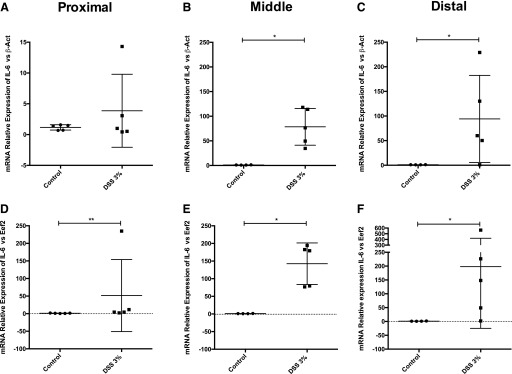
We calculated the IL-6 relative expression considering the amplification efficiencies using β-actin and Eef2 as reference genes (Fig. 5). DSS treatment resulted in a significantly higher relative expression of the proinflammatory cytokine IL-6 in all 3 colon segments as expected. IL-6 expression was higher in the middle and distal colon compared with the proximal segment when both β-actin and Eef2 were used as reference genes. However, higher relative expression levels were observed when Eef2 was used as a reference gene (Fig. 5A–F). In the proximal section, when β-actin was used for normalization, no statistically significant differences were observed in contrast to normalization with Eef2 (Fig. 5A, B).
FIGURE 5.
Variability of the proinflammatory cytokine IL-6 relative expression in different colon segments. mRNA relative expression of IL-6 in the 3 segments of the colon, proximal (A, D), middle (B, E), and distal (C, F). qRT-PCR was performed by the 1-step protocol, and data are expressed as the relative expression calculated using the Pfaffl method16 and β-actin (A–C) or Eef2 (D–F) as reference genes. Results are expressed as means ± sd; n = 5 mice per group. Mann-Whitney, *P < 0.05, **P < 0.01.
DISCUSSION
The DSS experimental model is extensively used in the literature to study IBD. However, this model shows high variations in the criteria that impacts the reproducibility of the results. Thus, urgent procedures to get reliable conclusions are needed. A recent systematic review demonstrated that failure to report all essential criteria is a cause of concern, as the reproducibility of the DSS model may be affected, and these factors have been shown to alter the severity of the disease.17
The concentration of DSS administered in the drinking water is an important source of variation, as concentrations between 1.5 and 5% are accepted for the Balb/c strain in acute colitis.18, 19 Taking into account that on d 7, no differences were found among the different DSS concentrations, 3% DSS is a better choice for this model, but when greater DAI is needed earlier in the experiment, 4 and 5% could be an option. Differences in susceptibility between sexes has been observed in many inflammatory and autoimmune diseases.20 In DSS-induced colitis, greater male susceptibility has been observed. However, both males and females develop DSS-induced colitis robustly.13 In this study, we used female mice, as males tend to have higher mortality rates and have been shown to be more prone to establish territorial and social dominance and a more aggressive behavior that may result in fighting and injury that might negatively impact the outcome of the study.21 Female mice establish social dominance, but fighting usually does not occur, and thus, the use of female mice is recommended.22 Nevertheless, Alex et al.23 showed by multivariate analysis that there are no differences in gender in the cytokine profiles in the acute model of DSS-induced colitis.
Expression of proinflammatory cytokines is upregulated in DSS-induced colitis. qRT-PCR is widely used to analyze cytokine expression as a result of its high sensitivity and reproducibility.8 However, DSS in colonic tissue inhibits qRT-PCR amplification in a dose-dependent manner. Kerr et al.24 demonstrated that DSS causes little damage to the total RNA, but DSS concentrations of 10 nM or higher inhibited amplification. They also found that 2.5% DSS administration to the mice interfered with the qRT-PCR and proposed the first protocol to clean DSS-exposed RNA by a column-based poly-A method.24 Viennois et al.,12 found that DSS inhibits both the RT and the Taq polymerase by measuring the activity of both enzymes with RNA incubated with different concentrations of DSS. They also proposed a method to remove DSS traces by a double precipitation with LiCl.12 Our results also show inhibition of both the PCR amplification and RT reaction. We modified the protocol published by Viennois et al.,12 as the concentration of LiCl proposed by the authors caused a significant loss of RNA in our hands. In our protocol, LiCl was used at a final concentration of 2.5 M, and an overnight precipitation with isopropanol was added, which resulted in higher yields of RNA and no interference in the qRT-PCR. LiCl RNA precipitation precipitates large RNAs and excludes small RNAs that remain in the supernatant but offers the advantage of not precipitating DNA, carbohydrates, and protein, eliminating inhibitors of translation or cDNA synthesis.25 In our study, the expected loss of small RNAs, as a result of LiCl precipitation, was not an issue, as microRNA expression was not analyzed. Contamination of colonic tissue with DSS has not been appropriately addressed in the majority of articles using the DSS colitis model. DSS decontamination is crucial to have accurate, reliable, and reproducible results for cytokine-expression analysis and should be included in all expression experiments for this colitis model.
The qRT-PCR chemistry used had an impact on the efficiency and sensitivity of the amplification data. We found that the 1-step method showed higher efficiencies, compared with the 2-step method. These 2 methods differ in the RT enzymes and primers used. In the 1-step protocol, the Luna WarmStart RT was used; this enzyme was designed in silico and uses specific primers. For the 2-step protocol, the SuperScript III RT and oligo(dT)20 for priming were used. The different RTs probably differ in their relative efficiency. In addition, gene expression may be affected by the priming method, as primer-specific methods tend to produce more complete copies than oligo(dT), as RNA secondary structures may form and interfere with the oligo(dT) binding. Specific primers are a more sensitive option for absolute or relative quantification. However, this primer method needs separate RT reactions when using the 2-step method, and RNA amounts are thus a limiting factor. The higher sensitivity of the 1-step method is particularly important for low-expression genes, such as cytokines, as it represents an ∼8-fold difference in detection levels, assuming 100% efficiency of the PCR. In the case of low copy mRNA, specific primers may be more efficient in generating full-length cDNA.9, 10 Thus, this variation may be attributed to the priming method and the enzyme-specific efficiencies that may change with the nature of expression of each gene. Additionally, other reagents and conditions may interact with the PCR reaction. Levesque-Sergerie et al.26 found that concentrations beyond 1 μg RNA using Superscript III resulted in equal or lower concentrations of cDNA. In our study, the higher sensitivity of the 1-step method allowed the use of 10 times less starting RNA concentration that prevented possible RNA interference in the RT reaction. The use of a small amount of starting RNA could compensate for the main disadvantage of the use of specific primers in the 1-step method, i.e., the need to use an aliquoted sample of RNA for each reaction. In addition, cDNA has been shown to degrade faster than the respective RNA.27
For the analysis of IL-6 expression, we used the 2 reference genes—β-actin and Eef2—as the normalizing reference genes. Eissa et al.11 indicated that among 13 reference genes for the model of DSS, Eef2 had the lowest variation in the DSS model. The authors showed that colonic inflammation resulted in higher Ct values in 11 of the 13 reference genes studied, including β-actin.11 However, no DSS cleaning process was added to their RNA isolation protocol, so inhibition by traces of DSS could be, at least in part, responsible for the decline in expression observed in their results. After the RNA cleaning protocol was implemented in our study, DSS-induced inflammation did not lead to low reference gene expression.
Differences in function, permeability, tight junction proteins, and microbiota composition have been reported in different regions of the colon.28, 29 These variations may explain the different IL-6 expression levels in the proximal vs. middle and distal segments found in our study. In addition, the density of commensal bacteria increases along the length of the gastrointestinal tract.30 DSS causes disruption of the intestinal barrier and more bacteria, and their byproducts may reach the lamina propria and induce higher levels of IL-6 in more distal portions of the gut. In human studies, IL-6 levels have been related to disease progression and recurrences; furthermore, local levels have been positively correlated with endoscopic severity and histologic grade. The DSS model has similarities with Crohn’s disease, which mainly has IL-6 overexpression in the terminal ileus (not addressed in this study) and distal colon.31
Likewise, histologic findings during DSS-induced colitis differ depending on the colon segment. Loss of mucosal architecture and damage was shown to be greater in the middle and distal regions.5 Our data on IL-6 expression are in accordance with the histologic findings, as expression of this proinflammatory cytokine was lower in the proximal region. However, the exact mechanisms by which DSS induces stronger mucosal inflammation in the distal colon need further studies. Different colon segments are frequently used for cytokine expression, myeloperoxidase activity, and histologic analysis, and the possible association between these parameters and cytokine expression could differ depending on the colon segment analyzed. Thus, middle and distal colon should be used for RNA extraction, but the proximal segment will underestimate the results.
In summary, as the most widely used experimental model of colitis, the DSS model has many experimental advantages, as it is administered orally and has an effect in a short period of time. However, there are certain issues that need special attention while designing the protocol to get accurate results, beginning with the use the adequate DSS brand and concentration, the inclusion of a cleaning method for RNA to avoid DSS contamination of the colonic tissue, and the selection of adequate reference genes and qRT-PCR chemistry, as well as using the same region of the colon to analyze the different parameters. By taking all of these considerations into account we could improve comparisons of the published colitis experiments and reinforce the DSS murine model as an even better tool for advancing our knowledge on IBD, identifying new therapeutic targets and testing possible new drugs to prevent or treat IBD.
ACKNOWLEDGMENTS
The authors thank Dr. Ramon Garcia, head of the animal facility at the Instituto Nacional de Pediatria, and his team for providing and taking care of the mice used in this study. This work was supported by Grant IN223816 from PAPIIT, Direccion General de Asuntos del Personal Academico (DGAPA), Universidad Nacional Autonoma de Mexico (UNAM), Mexico. The funders had no role in study design; data collection; and analysis, decision to publish, or preparation of the manuscript. The authors declare no conflicts of interest.
REFERENCES
- 1.Ohkusa T. [Production of experimental ulcerative colitis in hamsters by dextran sulfate sodium and changes in intestinal microflora]. Nippon Shokakibyo Gakkai Zasshi 1985;82:1327–1336. [PubMed] [Google Scholar]
- 2.Okayasu I, Hatakeyama S, Yamada M, Ohkusa T, Inagaki Y, Nakaya R. A novel method in the induction of reliable experimental acute and chronic ulcerative colitis in mice. Gastroenterology 1990;98:694–702. [DOI] [PubMed] [Google Scholar]
- 3.Elsheikh W, Flannigan KL, McKnight W, Ferraz JGP, Wallace JL. Dextran sulfate sodium induces pan-gastroenteritis in rodents: implications for studies of colitis. J Physiol Pharmacol. 2012;63:463–469. [PubMed] [Google Scholar]
- 4.Ginzel M, Feng X, Kuebler JF, et al. Dextran sodium sulfate (DSS) induces necrotizing enterocolitis-like lesions in neonatal mice. PLoS One 2017;12:e0182732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kitajima S, Takuma S, Morimoto M. Histological analysis of murine colitis induced by dextran sulfate sodium of different molecular weights. Exp Anim. 2000;49:9–15. [DOI] [PubMed] [Google Scholar]
- 6.Bamba S, Andoh A, Ban H, et al. The severity of dextran sodium sulfate-induced colitis can differ between dextran sodium sulfate preparations of the same molecular weight range. Dig Dis Sci. 2012;57:327–334. [DOI] [PubMed] [Google Scholar]
- 7.Giulietti A, Overbergh L, Valckx D, Decallonne B, Bouillon R, Mathieu C. An overview of real-time quantitative PCR: applications to quantify cytokine gene expression. Methods 2001;25:386–401. [DOI] [PubMed] [Google Scholar]
- 8.Amsen D, de Visser KE, Town T. Approaches to determine expression of inflammatory cytokines. Methods Mol Biol. 2009;511:107–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wacker MJ, Godard MP. Analysis of one-step and two-step real-time RT-PCR using SuperScript III. J Biomol Tech. 2005;16:266–271. [PMC free article] [PubMed] [Google Scholar]
- 10.Bustin SA, Nolan T. Pitfalls of quantitative real-time reverse-transcription polymerase chain reaction. J Biomol Tech. 2004;15:155–166. [PMC free article] [PubMed] [Google Scholar]
- 11.Eissa N, Hussein H, Wang H, Rabbi MF, Bernstein CN, Ghia JE. Stability of reference genes for messenger RNA quantification by real-time PCR in mouse dextran sodium sulfate experimental colitis. PLoS One 2016;11:e0156289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Viennois E, Chen F, Laroui H, Baker MT, Merlin D. Dextran sodium sulfate inhibits the activities of both polymerase and reverse transcriptase: lithium chloride purification, a rapid and efficient technique to purify RNA. BMC Res Notes 2013;6:360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chassaing B, Aitken JD, Malleshappa M, Vijay-Kumar M. Dextran sulfate sodium (DSS)-induced colitis in mice. Curr Protoc Immunol. 2014;104:15.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taylor BC, Zaph C, Troy AE, et al. TSLP regulates intestinal immunity and inflammation in mouse models of helminth infection and colitis. J Exp Med. 2009;206:655–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maxwell JR, Brown WA, Smith CL, Byrne FR, Viney JL. Methods of inducing inflammatory bowel disease in mice. Curr Protoc Pharmacol. 2009; Chapter 5:Unit 5. [DOI] [PubMed] [Google Scholar]
- 16.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bramhall M, Flórez-Vargas O, Stevens R, Brass A, Cruickshank S. Quality of methods reporting in animal models of colitis. Inflamm Bowel Dis. 2015;21:1248–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eichele DD, Kharbanda KK. Dextran sodium sulfate colitis murine model: an indispensable tool for advancing our understanding of inflammatory bowel diseases pathogenesis. World J Gastroenterol. 2017;23:6016–6029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wirtz S, Popp V, Kindermann M, et al. Chemically induced mouse models of acute and chronic intestinal inflammation. Nat Protoc. 2017;12:1295–1309. [DOI] [PubMed] [Google Scholar]
- 20.Ngo ST, Steyn FJ, McCombe PA. Gender differences in autoimmune disease. Front Neuroendocrinol. 2014;35:347–369. [DOI] [PubMed] [Google Scholar]
- 21.Bábíčková J, Tóthová Ľ, Lengyelová E, et al. Sex differences in experimentally induced colitis in mice: a role for estrogens. Inflammation 2015;38:1996–2006. [DOI] [PubMed] [Google Scholar]
- 22.DeVoss J, Diehl L. Murine models of inflammatory bowel disease (IBD): challenges of modeling human disease. Toxicol Pathol. 2014;42:99–110. [DOI] [PubMed] [Google Scholar]
- 23.Alex P, Zachos NC, Nguyen T, et al. Distinct cytokine patterns identified from multiplex profiles of murine DSS and TNBS-induced colitis. Inflamm Bowel Dis. 2009;15:341–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kerr TA, Ciorba MA, Matsumoto H, et al. Dextran sodium sulfate inhibition of real-time polymerase chain reaction amplification: a poly-A purification solution. Inflamm Bowel Dis. 2012;18:344–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zununi Vahed S, Barzegari A, Rahbar Saadat Y, Mohammadi S, Samadi N. A microRNA isolation method from clinical samples. Bioimpacts 2016;6:25–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Levesque-Sergerie JP, Duquette M, Thibault C, Delbecchi L, Bissonnette N. Detection limits of several commercial reverse transcriptase enzymes: impact on the low- and high-abundance transcript levels assessed by quantitative RT-PCR. BMC Mol Biol. 2007;8:93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wilkening S, Bader A. Quantitative real-time polymerase chain reaction: methodical analysis and mathematical model. J Biomol Tech. 2004;15:107–111. [PMC free article] [PubMed] [Google Scholar]
- 28.Collins FL, Rios-Arce ND, Atkinson S, et al. Temporal and regional intestinal changes in permeability, tight junction, and cytokine gene expression following ovariectomy-induced estrogen deficiency. Physiol Rep. 2017;5:e13263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fihn BM, Jodal M. Permeability of the proximal and distal rat colon crypt and surface epithelium to hydrophilic molecules. Pflugers Arch 2001;441:656–662. [DOI] [PubMed] [Google Scholar]
- 30.Tropini C, Earle KA, Huang KC, Sonnenburg JL. The gut microbiome: connecting spatial organization to function. Cell Host Microbe 2017;21:433–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Drastich P, Frolova-Brizova L, Zanvit P, Spicak J, Tlaskalova-Hogenova H. Spontaneous in vitro IL-6 production in various intestinal segments in patients with inflammatory bowel disease. Folia Microbiol (Praha) 2011;56:185–190. [DOI] [PubMed] [Google Scholar]