Abstract
Motivation
Large genomic datasets combining genotype and sequence data, such as for expression quantitative trait loci (eQTL) detection, require perfect matching between both data types.
Results
We described here MBV (Match BAM to VCF); a method to quickly solve sample mislabeling and detect cross-sample contamination and PCR amplification bias.
Availability and Implementation
MBV is implemented in C ++ as an independent component of the QTLtools software package, the binary and source codes are freely available at https://qtltools.github.io/qtltools/.
Supplementary information
Supplementary data are available at Bioinformatics online.
Introduction
Current genetic studies routinely combine genotypes at polymorphic sites with various molecular phenotypes measured through next generation sequencing (e.g. RNA-seq or ChIP-seq). Ensuring the exact correspondence between molecular phenotypes and genotypes for each studied individual as well as detecting technical issues, such as cross-sample contaminations or PCR-generated amplification bias, are critical for maximizing the discovery power in population genomic studies. To achieve this on large datasets, typically including thousands of individuals, we propose here MBV, a method that rapidly and accurately measures allelic consistency between genotype and any type of sequence data reducing the analytical noise. Our approach extends those published earlier; VerifyBamID (Jun et al., 2012) and IDCheck (Huang et al., 2013) performing this quality control on large datasets within reasonable running times.
2 Materials and methods
MBV takes as input a VCF file containing the genotype data for multiple samples and a BAM file with the mapped sequences (from either single or paired-end sequencing run) of a molecular assay (e.g. RNA-seq, CAGE, ChIP-seq). It first piles up sequencing reads at each single-nucleotide-variant (SNV) site in the VCF file. It then discards poorly covered SNVs (as defined by a minimal-coverage parameter) and measures, for each individual in the VCF, the proportions of heterozygous and homozygous genotypes for which both alleles are captured by the sequencing reads (BAM file; Supplementary Methods S1). Finally, the two resulting concordance measures are reported for each individual in the output file (Supplementary Methods S2), together with other secondary statistics. In order to rapidly identify which individual matches the sequence data, the two concordance measures can be visualized on a scatter plot (similarly to Hoen et al., 2013): a ‘match’ appears as a point close to 100% concordance for both measures whereas all mismatches appear as a distant cluster of points (Fig. 1A). As described later, unexpected intermediate positions allow the user to detect sample cross-contaminations or PCR amplification bias. In addition, since MBV reports concordance metrics for all genotyped individuals, it can detect swapped and contaminating samples if included in the input VCF.
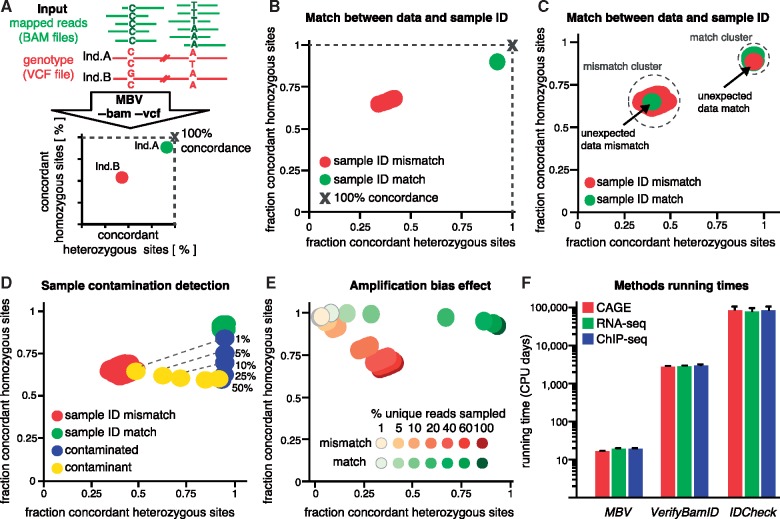
Fig. 1.
(A) Schematic representation of the MBV method. (B–E) Scatter plots of the concordance at heterozygous genotypes (X-axis) versus concordance at homozygous genotypes (Y-axis). Results for a typical CAGE BAM file (individual GM12814), ‘X’ indicating the 100% concordance point, are shown on (B) Aggregated results for 21 RNA-seq BAM files are shown on (C) Mislabeling scenarios are indicated with black arrows. (D) The results for simulation of cross-samples contamination (blue) with a known contaminant (yellow). Percentage of contamination is indicated. (E) The results for simulated amplification bias in sequence data. (F) The estimated running time to process 1000 individuals across multiple molecular assays (Supplementary Methods S6)
To test and characterize the various properties of this new tool, we use data for 21 1000-Genomes individuals (The 1000 Genomes Project Consortium, 2015) for which we have: (I) genotype data produced with whole genome sequencing, (II) gene expression data from RNA-seq (Waszak et al., 2015) and a tagging technology CAGE (Cap Analysis of Gene Expression, Takahashi et al., 2012, unpublished data from A.F.), (III) chromatin-binding profile for the second largest subunit of RNA polymerase II and the transcription factors PU.1 (Waszak et al., 2015) as well as (IV) genome wide distribution of 3 histone modifications (H3K4me1, H3K4me3 and H3K27ac; Waszak et al., 2015; Supplementary Materials). For all the 147 corresponding BAM files, mapped with BWA (Li and Durbin, 2009), we run MBV and investigated the outcome as follows. First, we computed the number of variant sites as a function of the minimal sequencing coverage required and conclude that a minimal coverage of 10 reads provides enough variants for reasonable estimates of the two concordance measures across all molecular assays (Supplementary Fig. S1). We then produced 147 scatter plots (heterozygous versus homozygous concordance), one for each BAM file, showing consistent pattern (Fig. 1B): a cluster of points corresponding to all genotyped individuals not matching to the sequence data (in red) and a point close to the 100% concordance (in green), which corresponds to a match between the genotype and the sequence data. Combining the scatter plots for the 21 individuals, we obtained two well-defined clusters, one for the matches and the other for the mismatches (Fig. 1C). Remarkably, the relative positions of both clusters remain stable regardless of the source of the genotypes (sequencing or imputation; Supplementary Methods S3; Supplementary Fig. S2) or the molecular phenotype assayed (Supplementary Fig. S3).
Next, we investigated how the position of additional samples relative to these two clusters can be informative of mislabeling or technical issues. First, a sample that is unexpectedly part of the mismatch cluster involves sample mislabeling; the genotype and sequence data belong to distinct individuals (Fig. 1C;unexpected data mismatch). Second, a sample that is unexpectedly part of the match cluster also implies sample mislabeling; the genotype and sequence data belong to the same individual but are incorrectly labeled with distinct IDs (Fig. 1C;unexpected data match). Finally, as shown by our simulations (Supplementary Methods S4–5), samples located in neither of the two clusters should be controlled for technical bias during sample or library preparation. We observed that (I) increasing cross-sample contaminations leads to decreased homozygous concordance values with no change in heterozygous concordance (Fig. 1D, in accordance with Castel et al., 2015) while (II) increasing amplification bias leads to decreased heterozygous concordance with no change in homozygous concordance (Fig. 1E).
In terms of running time MBV is at least two orders of magnitude faster than other methods to match the genotype and sequencing data. Specifically, we estimated that MBV requires 19 CPU days to perform all the pairwise comparisons (n = 106) required for 1000 samples with RNA-seq and genotype data, while VerifyBamID and IDCheck need 2,807 and 77,652 CPU days respectively (Fig. 1F;Supplementary Methods S6).
3 Discussion
We described here a new software, MBV, to rapidly ensure genome-wide matching between genotype and sequencing data. This method can be applied to a single or a collection of samples to detect a variety of issues involving mismatches between sequences and genotypes such as sample mislabeling, cross-sample contaminations and amplification bias introduced at library preparation steps. All this can be achieved for thousands of individuals in reasonable running times, therefore making MBV suitable for eQTL studies.
Supplementary Material
Acknowledgements
The computations were performed at the Vital-IT (http://www.vital-it.ch) Center for High-Performance Computing of the Swiss Institute of Bioinformatics.
Funding
This work was supported by a Louis Jeantet Foundation [E.T.D.]; Swiss National Science Foundation [31003A_149984 to E.T.D.]; Swiss National Science Foundation SystemsX [3826 to E.T.D.]; National Institute of Health NIMH GTEx [MH101814 to E.T.D.]; Swiss National Science Foundation Ambizione [PZ00P3–154728 to A.F.]; Swiss National Science Foundation [163180 to S.E.A.].
Conflict of Interest: none declared.
References
- Castel S.E. et al. (2015) Tools and best practice for data processing in allelic expression analysis. Genome Biol., 16, 195.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoen P.A.C’t. et al. (2013) Reproducibility of high-throughput mRNA and small RNA sequencing across laboratories. Nat. Biotechnol., 31, 1015–1022. [DOI] [PubMed] [Google Scholar]
- Huang J. et al. (2013) A tool for RNA sequencing sample identity check. Bioinformatics, 29, 1463–1464. [DOI] [PubMed] [Google Scholar]
- Jun G. et al. (2012) Detecting and estimating contamination of human DNA samples in sequencing and array-based genotype data. AJHG, 91, 839–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Durbin R. (2009) Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics, 25, 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H. et al. (2012) 5’ end-centered expression profiling using Cap-analysis gene expression and next-generation sequencing. Nat. Protoc., 7, 542–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The 1000 Genomes Project Consortium. (2015) A global reference for human genetic variation. Nature, 526, 68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waszak S.M. et al. (2015) Population variation and genetic control of modular chromatin architecture in humans. Cell, 162, 1039–1050. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.