Abstract
Laboratory insect colonies are an essential part of experimental insect science. Formalized naming of laboratory stocks is standard practice in model organisms such as mice and fruit flies, but crucial details such as colony origin and standard names are often lacking in nonmodel systems. For institutions involved in rearing multiple nonmodel species, effective monitoring requires standardized naming and nomenclature, from establishment to production, distribution, and publication. Insect rearing has been the cornerstone of the Insect Production and Quarantine Laboratories (IPQL) at the Great Lakes Forestry Centre for over 70 yr, but the histories of the insect colonies in this facility have not been adequately documented and formal, standardized names do not exist. We propose a standardized naming framework that we applied to the eight species reared at the IPQL to rectify these deficiencies. We also present the origin and history of each colony, essential information that is challenging to obtain post hoc. We suggest that other research institutions consider developing similar standards, so they can accurately document, communicate, and track laboratory insect their within the facilities and through the scientific literature.
Keywords: quarantine, forest entomology, biological control, insect rearing, invasive species
Laboratory colonies are a vital component of scientific research on animals. When Thomas Hunt Morgan established colonies of the dipteran Drosophila melanogaster Meigen (Diptera: Drosophilidae) in 1907, this species quickly became a model organism, revolutionizing our understanding of genes, heredity, and inheritance (Rubin and Lewis 2000, Jennings 2011). Other major scientific advances occurred in developmental biology, genomics, embryology, behavioral ecology, and disease biology with the development of laboratory colonies of nematode Caenorhabditis elegans (Maupas) (Rhabditida: Rhabditidae) (Ankeny 2001), darkling beetles (Wang et al. 2007), zebrafish (Howe et al. 2013), and mice (Phifer-Rixey and Nachman 2015).
Laboratory colonies have a number of advantages over wild-caught organisms. First, laboratory colonies are largely uniform. Field-collected material can be highly variable in age, nutritional condition, health, and genetic diversity. These differences can confound experimental results. Laboratory colonies provide disease-free organisms with a known rearing history, thereby reducing experimental variability. Second, laboratory colonies can provide research organisms all year long. In nature, most organisms are active for limited periods throughout the year, creating a narrow window during which researchers can sample wild populations. Specific life stages are also transient or unavailable for large portions of the year, further limiting research opportunities. Ultimately, laboratory colonies are positioned to provide high-quality organisms that display consistent performance, assuming nutrition and disease are effectively managed. They facilitate and accelerate research and are integral to many facets of biological research.
Despite their importance, laboratory colonies often lack crucial details such as a unique name and a detailed history of their establishment. Appropriate naming is essential for accurate identification. Names need to be unique and follow a set of standardized nomenclatural rules to be effective. Unique names allow researchers to precisely document the use of laboratory colonies and communicate this information to the research community. Precise identification of laboratory colonies can be essential (Kuno 2010), particularly if specific colonies or strains are used as industry or regulatory standards (McGuire et al. 1997, Macoris et al. 2005). When new colonies are established, the originating labs rarely publish the details of where and when founding individuals were collected. As a result, source populations and the geographic origin of laboratory colonies are often unknown, buried in unpublished archives, or restricted to a limited set of knowledgeable personnel. Although these details may not seem important at the time, their value may arise unexpectedly in the future. It is difficult, if not impossible, to unearth this information post hoc (Kuno 2010). Therefore, it is essential that institutions clearly document colony history and ensure that they adequately name colonies as they are established.
The Insect Production and Quarantine Laboratories (IPQL) is a multispecies rearing and quarantine facility at the Great Lakes Forestry Centre (GLFC, Natural Resources Canada, Canadian Forest Service) in Sault Ste. Marie, Ontario, Canada (http://www.nrcan.gc.ca/forests/research-centres/glfc/13467). This facility contains both clean rearing facilities for native insects, as well as a Level 2 Plant Pest Containment (PPC2) quarantine facility (Canadian Food Inspection Agency compliance no. PC-2013-034). The facility produces insects to facilitate pest management research within the Canadian Forest Service. The IPQL (also previously known as the Insect Production Unit) has maintained a wide range of pest insects in laboratory colonies since the 1940s. Initial rearing efforts were aimed at producing large numbers of forest insect pests to act as bioincubators for viral products (van Frankenhuyzen et al. 2016), and since that time, the IPQL has reared as many as 51 different species of insects (AD Roe unpublished, Ebling 2013).
The IPQL was the originating institution for many laboratory colonies of forest insect pests. The originating institution is the institution that establishes a species of insect in colony. To date, no formal nomenclature exists for the laboratory colonies reared within IPQL. This lack of formalized nomenclature has led to sporadic and inconsistent documentation of their use in the literature. Without consistent documentation, researchers cannot accurately communicate their use of IPQL colonies. Formal naming not only benefits IPQL in terms of quality and production control, but will also allow researchers to track colony use in the literature and document their impact on forestry research. We propose rules for a formal standardized nomenclature that we apply to existing (and future) IPQL colonies. We will also describe the history of colony establishment using historical and contemporary knowledge of each insect colony. One overarching purpose of this article is to highlight the importance of capturing the history of laboratory colonies. With the standardized approach we propose, we wish to encourage other institutions to follow our lead and document the history of their own insect colonies.
Methods
Nomenclature
Standardized nomenclature exists for many strains of vertebrate laboratory animals. Mouse strain nomenclature dates back to the early 1940s (Snell 1941) and is guided by the International Committee on Standardized Genetic Nomenclature for Mice (http://www.informatics.jax.org/nomen/inc.shtml). Other model systems have adopted these established rules to ensure community-wide standardization (e.g., rat and Drosophila fruit flies). We base our proposed nomenclature on naming conventions outlined by Festing (1993) and established for Drosophila (http://rice.bio.indiana.edu:7082/docs/nomenclature/lk/nomenclature.html#Introduction) and mice (http://www.informatics.jax.org/mgihome/nomen/strains.shtml#oacc). In Fig. 1, we deconstruct our proposed standard code for one of our Asian longhorned beetle stocks and define each component. Each code will have the following: 1) laboratory code, 2) a stock code, 3) species code, 4) geographic code (optional), and 5) family number. First, the laboratory code represents to institution where the insect stocks are maintained; in this case, the GLFC. We have registered the Glfc laboratory code with the Institute for Laboratory Animal Research (http://dels.nas.edu/global/ilar/Lab-Codes), the official registry for laboratories with stocks and strains of laboratory animals. Second, the stock code respresents the research unit that maintains the insect colonies. Here, the Insect Production and Quarantine Laboratories maintain all insect colonies; hence, we apply IPQL as the stock code. All insect colonies within the facility will bear the same laboratory and stock code; although if other colonies are established at the GLFC, new stock codes may be created. Third, species codes represent the scientific name of the insect. They are four letters long, with the first capital letter the first letter of the genus name, and the remaining letters are the first three letters of the specific epithet (e.g., Anoplophora glabripennis Motschulsky (Coleoptera: Cerambycidae), henceforth, Agla). If conflicting species codes arise, then an alphanumeric value (e.g., Agla2) could be added to distinguish between two different species. Fourth, geographic origin is represented by three capitalized letters (e.g., WMA = Worcester, MA, USA). This component is optional and we include it when the colony has a known provenance and has not experienced introduction of genetic material from multiple geographic locations. Fifth, family number denotes the distinct breeding unit reared through successive generations.
Fig. 1.
Deconstruction of proposed standard code for the IPQL insect stocks.
Origin
We provide a history of each insect colony including information on geographic origin or the source institution. We were able to find provenance details on our recently established colonies, but many details were missing for our older colonies.
Genetics
There is a clear distinction between laboratory strains and laboratory stocks (Festing 1993). A strain is an inbred population bred using a defined breeding scheme for +20 generations to create a population with a homozygous genetic background. A strain has a defined genetic history and all members of the strain can be traced back to a single mating pair. A stock, on the other hand, is a heterozygous outbred population with a stabilized genetic composition. Outbred stocks are genetically undefined, meaning that the genetic composition of an individual is unknown. Closed colonies represent stocks where all matings are among members of the same colony and new genetic material is not introduced from generation to generation. We can name stocks after four generations of closed outbreeding. From this point on in the article, we will refer to the IPQL colonies as stocks.
Breeding
Outbred stocks must maintain a stable genetic composition over many generations. Defined breeding systems minimize inbreeding and avoid artificially selecting specific phenotypes, leading to long-term health and viability of laboratory stocks. Festing (1993) summarizes several breeding systems used for rearing laboratory stocks: maximum avoidance of inbreeding, rotational mating, and chance mating. IPQL selects breeding systems based on colony size, mate selection, and mating chamber design.
Characteristics
We highlight rearing conditions for each stock such as diet, rearing temperature, and diapause conditions.
Additional Information
Here we include any additional information that may be pertinent to each stock. For example, we describe bottleneck events (e.g., population crashes due to disease) or if a genetic infusion event occurred.
Results
Overview
The IPQL currently rears eight insect species. Table 1 provides a summary of these species, their origins, and their new standard names. All IPQL stocks described below are closed colonies of outbred stocks. When IPQL needs to expand its stocks, it splits healthy existing families in a process termed fractionation. IPQL generally does not infuse laboratory stocks with wild-caught populations. Genetic infusion increases the risks of introducing pathogens to clean, healthy laboratory stocks. If IPQL wants to develop a new clean laboratory stock, they rear wild or diseased insects separately through a multigenerational rearing process (van Frankenhuyzen et al. 2004). We only integrate these new families into the clean facility once they have passed stringent quality control standards. We will designate these new families as official stocks once we have reared them in a closed system for at least four generations.
Table 1.
Insect species reared by the Insect Production and Quarantine Laboratories
| Scientific name | Common name | New standard code | Year | Origin |
|---|---|---|---|---|
| LEPIDOPTERA | ||||
| Tortricidae | ||||
| C. fumiferana | Spruce budworm | Glfc:IPQL:Cfum01 to Cfum16 | 1961 | Multiple sites, ON |
| C. occidentalis | Western spruce budworm | Glfc:IPQL:Cocc01 to Cocc06 | 1980 | Kamloops, BC; Merritt, BC; Berkeley X #Coc-FS-01, Corvallis OR |
| Erebidae | ||||
| O. leucostigma | Whitemarked tussock moth | Glfc:IPQL:OleuSSM01 to OleuSSM04 | 1963 | Sault Ste. Marie, ON |
| Noctuidae | ||||
| T. ni | Cabbage looper | Glfc:IPQL:Tni01 to Tni04 | 1980 | AAFC, Harrow ON |
| COLEOPTERA | ||||
| Buprestidae | ||||
| A. planipennis | Emerald ash borer | |||
| PON | Glfc:IPQL:AplaPON01 | 2014 | Parkhill, ON, Canada | |
| EON | Glfc:IPQL:AplaEON02 | 2015 | Exeter, ON, Canada | |
| Glfc:IPQL:Apla03 | 2016 | Mixed, five sites in ON and QC | ||
| Glfc:IPQL:Apla04 | 2017 | Mixed, two sites in southern ON | ||
| Cerambycidae | ||||
| A. glabripennis | Asian longhorned beetle | |||
| UIC | Glfc:IPQL:AglaUIC01 | 2010 | Ravenswood, IL, USA | |
| WMA | Glfc:IPQL:AglaWMA01 | 2010 | Worcester, MA, USA | |
| EBCL | Glfc:IPQL:AglaEBCL01 | 2016 | Multiple sites: Hohhot, Inner Mongolia China, Langfang, Hebei Province, China, five sites in France | |
| T. fuscum | Brown spruce longhorned beetle | Glfc:IPQL:TfuscBNS01 | 2011 | Bedford, NS |
| HYMENOPTERA | ||||
| Eulophidae | ||||
| T. planipennisi | Glfc:IPQL:Tpla01 | 2017 | China | |
New standard codes are shown for each family. Common name, year of establishment, and origin are also included.
In IPQL, we use two breeding regimes to maintain outbred stocks and to minimize the risk of inbreeding. We select the breeding system for each organism based on the organism’s biology and our desired stock size. Regardless of the breeding system, we start each new generation for every family with at least 25 males. This allows us to use either rotational or chance breeding. In chance breeding systems, we select mates at random from among all members of a family. For these families, the population is large enough that the risk of inbreeding is low. In rotational breeding, we know the pedigree of parental material and we avoid mating with close relatives. This ensures more genetically diverse offspring than would occur by chance.
Stock Naming and History
Spruce Budworm (SBW—Choristoneura fumiferana Clem.) (Lepidoptera: Tortricidae)
Stock: Great Lakes Forestry Centre Insect Production and Quarantine Laboratories C. fumiferana colony (Originating Institution); 16 families of mixed geographic origin.
Symbol: Glfc:IPQL:Cfum01 thru Glfc:IPQL:Cfum16
Origin: Mixed source, wild caught from multiple Ontario, Canada populations.
Genetics: Closed colony, more than 20 generations since establishment in 1961, 21 generations since quality control (QC) screening.
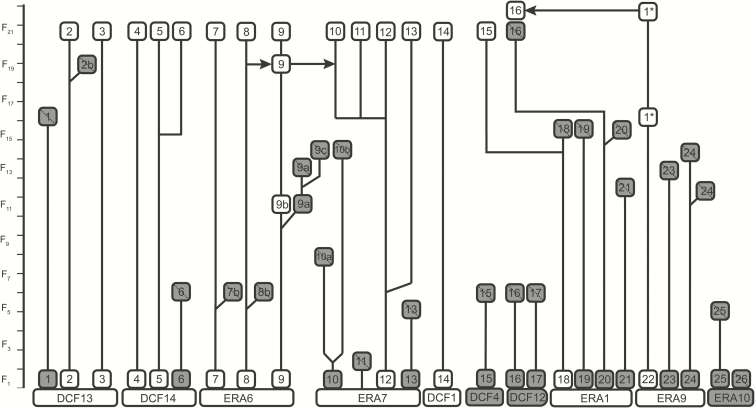
Breeding: We initiate each new generation via chance mating within mating chambers (i.e., large polyethylene bags). Each mating chamber is set up with 100 males and 100 females chosen at random from within each family. We set up a maximum of 10 mating chambers per family. IPQL maintains 16 families for each generation (Fig. 2).
Fig. 2.
Relationships among families of C. fumiferana (SBW) reared at the IPQL. Families were derived from single pair matings taken from subcolonies of SBW (DCF and ERA, meaning unknown). We have documented SBW family history since 2006 when a quality control program was initiated. White boxes are lineages that currently exist in IPQL and grey boxes indicate terminated lineages. Fractionation (i.e., splitting events) and interfamily infusions (arrows) are shown.
Characteristics: Obligate diapause. Adults show strong flight capabilities. Larvae are reared on McMorran diet (McMorran 1965) at 23 ± 3°C, 55 ± 10% RH, and 16:8 (L:D) h. Second instar larvae are stored at 23 ± 3°C, 55 ± 10% RH, and 16:8 (L:D) h for 2 wk of prediapause prior to a 20- to 34-wk diapause at 4°C (24 wk is optimal).
Additional Information: Our colony was established on artificial diet in 1961. Wild-caught adults were collected to initiate the colony and disease-free wild-caught stock were regularly infused into the colony for at least 20 yr (Grisdale 1970, 1984). Genetic infusion ceased in the 1990s, but the exact timing is unknown. In 2001, a severe microsporidian infection was detected within the colony. This coincided with a severe reduction in progeny production (van Frankenhuyzen et al. 2004). Disease-free colonies were recovered using 332 clean families derived from 10 subcolonies. Offspring from these matings were split into 26 families to provide continuous biweekly colony production throughout the year (Fig. 2). Over time, we have reduced these 26 families to 16 families to meet changing research needs and to eliminate diseased lineages. Fractionation events and mergers are shown in the family tree. Rearing procedures are available (http://cfs.nrcan.gc.ca/pubwarehouse/pdfs/35687.pdf).
Western Spruce Budworm (WSBW—Choristoneura occidentalis Freeman) (Lepidoptera: Tortricidae)
Stock: Great Lakes Forestry Centre Insect Production and Quarantine Laboratories C. occidentalis stock; six families of mixed geographic origin (Originating Institution).
Symbol: Glfc:IPQL:Cocc01 to Glfc:IPQL:Cocc06
Origin: Mixed source, Kamloops, BC and Merritt, BC, Canada.
Genetics: Closed colony, six generations since genetic infusion in 2009.
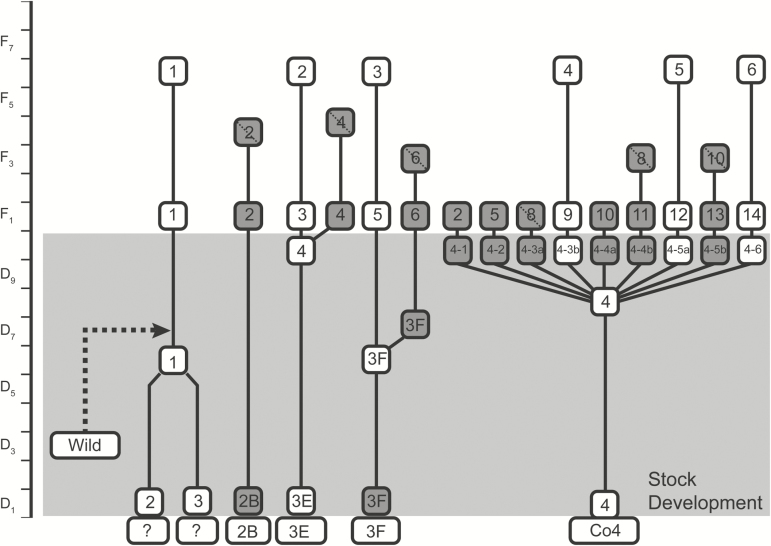
Breeding: We use chance mating for each successive generation. Each mating chamber is set up with 100 males and 100 females chosen at random from each family, with a maximum of 10 mating chambers per family. IPQL currently maintains six families of WSBW (Fig. 3).
Fig. 3.
Relationships among families of C. occidentalis (WSBW) reared at IPQL. Families were derived from single-pair matings taken from subcolonies of WSBW. Stock development phase (grey) represents the period of time when the stocks were being cleaned of pathogens. In 2009, family 1 was accidentally infused with wild stock (dashed line). Boxes and lines as in Fig. 2.
Characteristics: Obligate diapause. Adults show strong flight capabilities. Larvae are reared on McMorran diet (McMorran 1965) at 23 ± 3°C, 55 ± 10% RH, and 16:8 (L:D) h. Second instar larvae are stored at 23 ± 3°C, 55 ± 10% RH, and 16:8 (L:D) h for 2 wk of prediapause prior to a 20- to 34-wk diapause at 4°C (24 wk is optimal).
Additional Information: The Kamloops, BC stock was originally established in the 1980s, but crashed due to a microsporidial infection in 2001 (van Frankenhuyzen et al. 2004). The stock was regrown using clean families as described for SBW. Concurrently, we established a new stock with field-collected insects from Merritt, BC in 2008. We kept the two colonies separate for three generations, but these were accidentally merged in 2009 (Fig. 3). The field-collected material was still contaminated with microsporidia, so family 1 was treated Fumigil-B (van Frankenhuyzen et al. 2004) to control residual microsporidial infections. We expanded the colony into full production in 2012 after it was determined to be disease-free. Detailed rearing procedures are available (http://cfs.nrcan.gc.ca/pubwarehouse/pdfs/35690.pdf).
White Marked Tussock Moth (WMTM—Orgyia leucostigma J.E. Smith) (Lepidoptera: Erebidae)
Stock: Great Lakes Forestry Centre Insect Production and Quarantine Laboratories O. leucostigma stock (Originating Institution); four geographically defined families.
Symbol: Glfc:IPQL:OleuSSM01 to Glfc:IPQL:OleuSSM04
Origin: Sault Ste. Marie, ON, Canada (SSM). Colony was initiated on artificial diet in 1963.
Genetics: Closed colony, more than 20 generations; 15 generations since QC screening.
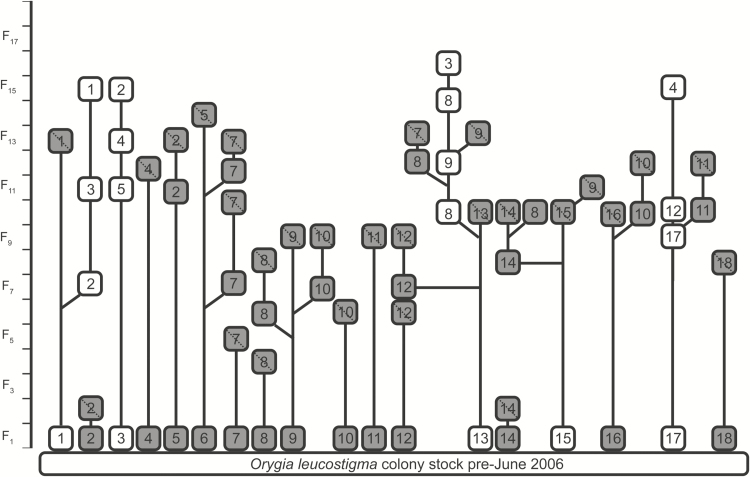
Breeding: We use chance mating for each successive generation. Each mating chamber (i.e., a large plastic box) is set up with 50 females and 50 males. We select individuals at random from each family, with a maximum of five mating chambers per family. IPQL currently maintains four families of WMTM (Fig. 4).
Fig. 4.
Relationships among families of O. leucostigma (WMTM) reared at IPQL. We have documented WMTM family history since 2006 when we initiated a quality control program. Box colors and lines as in Fig. 2.
Characteristics: Larvae are reared on Bell diet (Bell et al. 1981) at 22 ± 3°C, 50 ± 10% RH, and 12:12 (L:D) h. Eggs are stored for 1 mo in prechill conditions (13 ± 3°C, 16:8 (L:D) h, n/a RH) before they are transferred to a 2.5 ± 1°C cold room for 20- to 32-wk cold storage. All stages of the insect have urticating hairs that can cause skin rashes, requiring appropriate safety precautions when handling. Detailed rearing procedures are available (http://cfs.nrcan.gc.ca/pubwarehouse/pdfs/35692.pdf).
Cabbage Looper (Tni—Trichoplusia ni Hübner) (Lepidoptera: Noctuidae)
Stock: Great Lakes Forestry Centre Insect Production and Quarantine Laboratories stock (Receiving Institution).
Symbol: Glfc:IPQL:Tni01 to Glfc:IPQL:Tni04
Origin: We inherited the Tni colony from the Harrow Research and Development Centre (Harrow, ON, Canada; Agriculture and Agri-Food Canada). Geographic origin is unknown.
Genetics: Closed colony, more than 20 generations; 190 generations since QC screening.
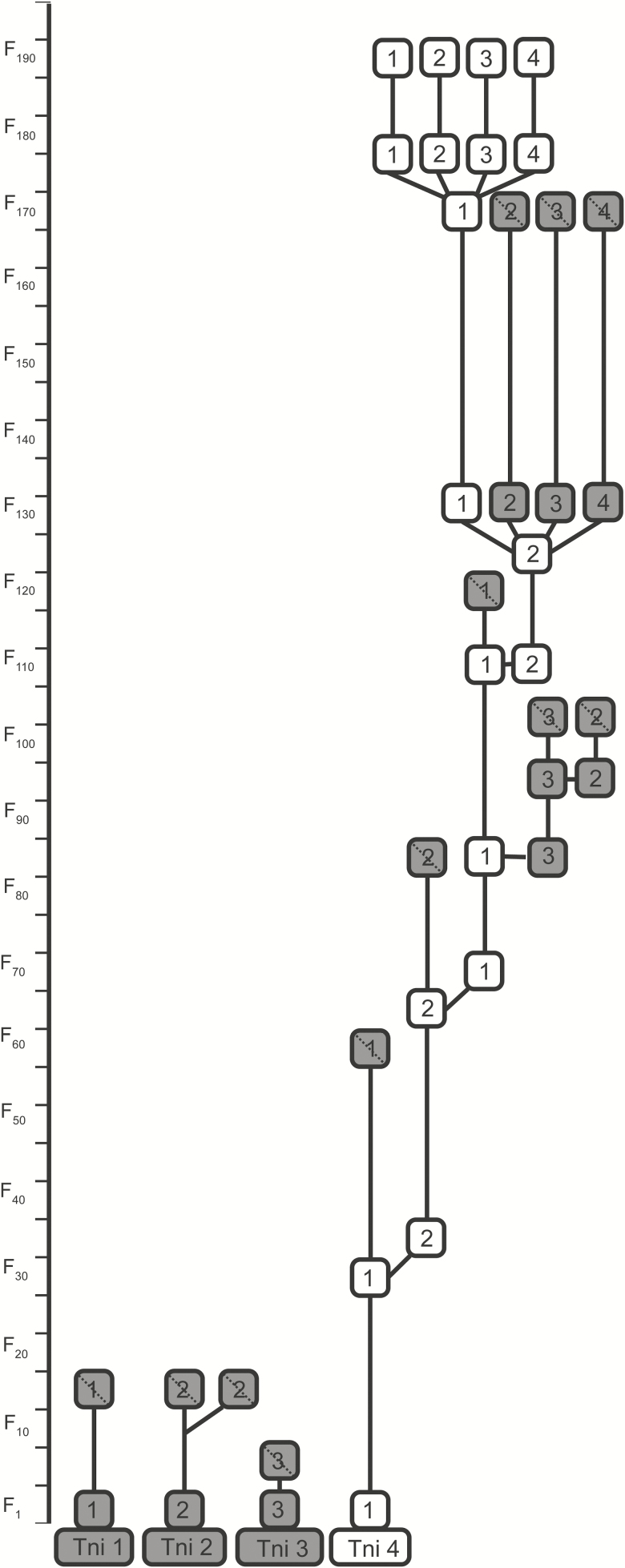
Breeding: We use chance mating for each successive generation. Each mating chamber (i.e., a large plastic box) is set up with 100 males and 100 females chosen at random from each family, with a maximum of two mating chambers per family. IPQL currently maintains four families of Tni (Fig. 5).
Fig. 5.
Relationships among families of T. ni (Tni) reared at IPQL. We have documented Tni family history since 2006 when we initiated a quality control program. Box colors and lines as in Fig. 2.
Characteristics: Larvae are reared on McMorran diet (McMorran 1965) at 27 ± 3°C, 55 ± 10% RH, and 16:8 (L:D) h. This species does not require diapause. Eggs are stored at 4°C for up to 10 d. Detailed rearing procedures are available (http://cfs.nrcan.gc.ca/pubwarehouse/pdfs/35686.pdf).
Asian Longhorned Beetle (ALB—Anoplophora glabripennis Motschulsky) (Coleoptera: Cerambycidae)
Stock: Great Lakes Forestry Centre Insect Production and Quarantine Laboratories stock; three geographically defined families (Receiving Institution).
Symbol: Glfc:IPQL:AglaUIC01
Origin: Ravenswood, Illinois, USA (UIC). Dr. Melody Keena (USDA Forest Service Quarantine Facility, Ansonia CT) established the original UIC stock from approximately 1,400 individuals collected in 1999 from infested wood harvested in Ravenswood, IL (41.58 N, −87.42 W). Adults, pupae, and larvae were collected off infested maple (Acer platanoides L., Acer saccharinum L.) and ash (Fraxinus sp.). In 2012, we initiated our stock using 125 larvae donated to IPQL from Dr. Keena.
Genetics: Closed colony; 26th generation since establishment; 8th generation at IPQL.
Symbol: Glfc:IPQL:AglaWMA01
Origin: Worcester, Massachusetts, USA (WMA). Dr. Melody Keena (USDA Forest Service Quarantine Facility, Ansonia CT) established the original WMA stock from 68 individuals collected in late 2008 and early 2009 from infested wood harvested in Worcester, MA (42.31 N, −71.81 W; 42.31 N, −71.80 W). Adults and larvae were collected off maple (Acer rubrum L.) and willow (Salix discolor Muhl.). In 2012, we received six larvae to initiate our colony at the IPQL from Dr. Keena. This colony has now been discontinued at the USDA facility.
Genetics: Closed colony; 15th generation since establishment in colony; eighth generation at IPQL.
Symbol: Glfc:IPQL:AglaEBCL01
Origin: We received 100 larvae from the European Biological Control Laboratory (EBCL; USDA, ARS) based in Saint-Gély-du-Fesc Cedex, France, which was the originating institution. The stock was established by EBCL from 400 ALB larvae imported from two sites in China: Hohhot, Inner Mongolia (198 larvae collected on 10–14 April 2002 on Populus × beijingensis W.Y. Hsu and Populus × canadensis Moench); Langfang, Hebei Province (200 larvae collected on 8–10 April 2002 on Salix matsudana f. umbraculifera (synonym of Salix babylonica L.)). Five genetic infusion events occurred with small numbers of ALB individuals collected from invasive populations in Europe (Gien, FR in 2003; Sainte-Anne-sur-Brivet, FR in 2004, Corbetta, IT in 2007; Strasbourg, FR in 2008 and 2010).
Genetics: Closed colony, >10 generations; first generation at IPQL.
Breeding: We use a rotational breeding system for our ALB colony. Individual mating chambers (i.e., large glass pickle jars plus host plant sticks) are set up with one male and one female. We maintain detailed pedigree records and select mating pairs from unrelated individuals to ensure that full or half siblings are not interbred.
Characteristics: We maintain one family from each geographic location. We rear our stocks using modified protocols provided by M. Keena (unpublished). We rear larvae on artificial diet as described by Keena (2005) at 23°C, 60% RH in darkness for 10 wk. After 10 wk, larvae are stored at 7°C for a minimum of 12 wk. After 12 wk in the chill, we return larvae to rearing conditions (23°C, 60% RH) until pupation. Following pupation, we provide adults striped maple (A. pensylvanicum L.) for maturation feeding prior to mating. After 10 d of maturation feeding, we set up mating pairs with striped maple oviposition logs. We remove eggs by peeling logs and we place neonates onto diet immediately after hatching.
Additional Information: ALB is a regulated plant pest and we rear our stocks within our quarantine facility (PPC-2, CFIA written authorization WA-2013–017).
Brown Spruce Longhorned Beetle (BSLB—Tetropium fuscum Fabricius) (Coleoptera: Cerambycidae)
Stock: Great Lakes Forestry Centre Insect Production and Quarantine Laboratories stock (Receiving Institution).
Symbol: Glfc:IPQL:TfusBNS01
Origin: Bedford, Nova Scotia, Canada (44.750293N, -66.679157W).
Genetics: Closed colony, six generations at IPQL.
Breeding: Mating chambers are set up with five pairs of insect selected at random from each family. We set up six mating chambers per family per generation. IPQL maintains one family of BSLB.
Characteristics: We rear our colony on white spruce (Picea glauca (Moench) Voss) log bolts. We set up mating cages with five pairs of BSLB which are allowed to mate and oviposit on waxed log bolts. Log bolts are incubated at 23°C, 15% RH, 16:8 (L:D) h for 8 wk, and then the infested bolts are chilled at 7°C for 12 wk. Following the chill period, we return the bolts to 23°C and adults are allowed to emerge.
Additional Information: BSLB is a regulated plant pest and we rear our stock within our quarantine facility (CFIA written authorization WA-2013–017).
Emerald Ash Borer (EAB—Agrilus planipennis Fairmarie) (Coleoptera: Buprestidae)
Stock: Great Lakes Forestry Centre Insect Production and Quarantine Laboratories stock; two geographically defined families, two mixed families (Originating Institution).
Symbol: Glfc:IPQL:AplaPON01
Origin: Initiated in 2014 near Parkhill, ON, Canada (PON). We collected insects from three species of ash trees (Fraxinus pennsylvanica Marshall, Fraxinus americana Marsh., and Fraxinus nigra Pott) harvested in woodlots owned by Ausable Bayfield Conservation Authority. We harvested infested bolts on 29–31 October at the Wright Thompson Tract (43.21364 N, −81.64276 W) on Mooseville Drive north of Parkhill, ON.
Genetics: Closed colony, five generations.
Symbol: Glfc:IPQL:AplaEON02
Origin: Initiated in 2015 near Exeter, ON, Canada (EON). We collected insects from three species of ash trees (F. pennsylvanica, F. americana, and F. nigra) harvested on 22 October 2014 at Johnson Management Area (43.34573 N, −81.55772 W) on Parr Line west of Exeter, ON.
Genetics: Closed colony, four generations.
Symbol: Glfc:IPQL:Apla03
Origin: Initiated in 2016 from trees harvested in 2015 from five sites in eastern Canada: private woodlot 404693 Beaconfield Road, south of Woodstock, ON (43.02486 N, −80.74046 W), Middleton McConkey Tract (Long Point Region Conservation Authority, F. americana) southeast of Tillsonburg, ON (42.81081 N, −80.62296 W), Gatineau Park, QC (National Capital Commission, F. pennsylvanica) (45.63985 N, −75.94650 W), Conroy Road Ottawa, ON (National Capital Commission, F. pennsylvanica) (45.34207 N, −75.60870 W), and Wildwood Conservation Area (Upper Thames River Conservation Authority, F. pennsylvanica) east of St. Marys, ON (43.24596 N, −81.05746 W).
Genetics: Closed colony, three generations.
Symbol: Glfc:IPQL:Apla04
Origin: Initiated in 2017. We collected trees in October 2016 from two sites in southern Ontario: Private woodlot 95 7th Line North Oro-Medonte Twp (44.48283 N, −79.53720 W) and Middleton McConkey Tract (Long Point Region CA property, F. americana) (42.81081 N, −80.62296 W).
Genetics: Closed colony, two generations.
Breeding: Individual mating chambers (i.e., large plastic cup with foliage) are set up with three pairs of insects selected at random from a family. Every family has between 400 and 500 mating cups set up per generation.
Characteristics: We rear our stock on ash mini-bolts using a rearing method developed at GLFC (Roe et al. in prep) with guidance from the USDA Biological Control and Production Facility (Brighton, MI). Eggs are embedded in white (F. americana) and green ash (F. pennsylvanica) mini-bolts and reared at 27°C, 70% RH for 10–12 wk. We store logs at 15°C for 2 wk of prediapause and then store the infested mini-bolts at 4–8°C for a minimum 90-d diapause. After diapause, we return to rearing temperatures and collect the adults. We feed adults fresh ash foliage from ever-bearing ash (F. uhdei (Wenz.) Lingelsh), set up mating chambers, and harvest eggs for the next generation.
Additional Information:We maintain four families of EAB, although only two families (PON, EON) have been in colony for at least four generations (see above). We recently initiated two more families (Apla03, 04), although these have not yet been through four generations in the laboratory. We chose to describe these two families in order to document their history and establish their standard name.
We also use these EAB stocks to rear a parasitoid wasp (Tetrastichus planipennisi Yang (Hymenoptera: Eulophidae)) used as a biological control agent. We describe the parasitoid stock below. Prior to 2010, EAB was considered a regulated pest within the Sault Ste. Marie, ON area and rearing was conducted within our quarantine facility. After EAB was discovered in our area, we no longer needed to maintain the colony within the quarantine facility.
Tetrastichus planipennisi Yang (Hymenoptera: Eulophidae)
Stock: Great Lakes Forestry Centre Insect Production and Quarantine Laboratories T. planipennisi stock (Receiving Institution).
Symbol: Glfc:IPQL:Tpla01
Origin: Liaoning Province, China.
Genetics: Closed colony, +10 generations; two generation at IPQL. We initiated our stock from roughly 5,000 females which were donated in 2017 from the USDA Biological Control and Production Facility (Brighton, MI). The Brighton colony was originally derived from a colony initiated at the Beneficial Insect Introduction Research Laboratory (Newark, Delaware). This original colony was established in 2008 from parasitized emerald ash borer larvae harvested in the Liaoning province in northeastern China (Duan et al. 2011, Duan and Oppel 2012).
Breeding: Mating chambers are set up with 100 females and 30 males selected at random. Between 30 and 50 mating chambers are set up using all available adults. Breeding occurs prior to exposure to EAB-infested logs.
Characteristics: We rear our colony on our EAB larval stock developing in ash mini-bolts using the method described in the EAB section. Once female parasitoids have been mated, 10–12 females plus a few males are provided EAB-infested white ash bolts as an oviposition substrate. Bolts are incubated at 27°C 80% RH 16:8 (L:D) h. A chill period is optional and bolts can be stored up to 5 mo at 4°C 90% RH to synchronize emergence with research needs.
Additional Information: We use this stock as a biological control agent for EAB (Duan et al. 2013) and each year we release a proportion of our colony into the wild. In 2017, we reintroduced a small number of wild-caught T. planipennisi into the laboratory stock. We will use these for future releases, but not for stock maintenance. In the future, we will keep breeding stock separate to maintain a closed colony.
Discussion
The IPQL maintains a state of the art rearing facility that rears stocks of multiple insect species year round. Researchers have used insects from the IPQL stocks to answer a wide range of research questions, many of which would have been impossible without access to high-quality laboratory stocks. For example, our stocks have been used to understand insect pathology and develop a range of microbial control products such as Bacillus thuringiensis, Abietiv, Disparvirus, Lecontvirus, Gypchek, and Virtuss (reviewed in van Frankenhuyzen et al. 2016). Research conducted on our insects led to pheromone-based monitoring programs (Evenden and Silk 2016) and population dynamic models (Régnière 1987). Researchers at the GLFC have also used our insect stocks to develop insect cell lines (Sohi 1995), further broadening the research potential of our insects. We sell our stocks to researchers throughout the world (http://www.nrcan.gc.ca/forests/research-centres/glfc/13467), thereby facilitating research beyond the core mandate of our organization.
We believe that formally naming and describing our insect stocks meets a variety of needs for the scientific community, especially given the breadth and impact of our insect stocks have on pest management. First, it ensures accurate and precise communication of research results and experimental procedures within scientific publications. The research community is able to clearly document what organisms they are working with, as well as compare results across studies. Second, this process provides an opportunity to describe the detailed history of each insect stock, in particular their geographic provenance. Knowledge of colony history is quickly lost over time, particularly as scientists and technicians retire. Third, the process of formally naming our colonies in a peer-reviewed publication provides us with the ability to track their usage through the scientific literature. We are able to validate current and future investment in our facility when we can easily document the impact our insect colonies have on the research community.
Formalized naming of laboratory stocks is common practice among narrow segments of the research community, but it is not universal. We feel that this practice should extend to most, if not all, insect colonies and biofactories producing insects. These underlying histories provide important information that can influence experimental designs and their results. Without access to this information, the precise identity of colonies can be challenging, if not impossible to obtain. We present this article as a guide for other institutions with long-term laboratory colonies that wish to assign a formal nomenclature to their insect stocks. Clear documentation of insect colonies and their history will improve our understanding of these species, our ability to communicate their use, and our overall scientific rigor.
Acknowledgments
Insect rearing is a truly human endeavor. Many technicians, field crews, researchers, and summer students contributed to the establishment, development, and maintenance of our insect colonies. We wish to acknowledge several staff that were essential to this article. Tim Ladd, Tripti Sharma, and Gene Jones provided detailed information on the history and origin of the EAB stocks. Dr. Daniel Doucet and two anonymous reviewers provided insightful comments on earlier versions of this article. Shelley Hanninen and Sandy Houston provided essential assistance in tracking down unpublished archives on the IPQL unit. We are forever grateful to Kees van Frankenhuyzen for saving scraps of meeting notes that proved invaluable to our research on stock origins.
References Cited
- Ankeny R. A. 2001. The natural history of Caenorhabditis elegans research. Nat. Rev. Genet. 2: 474–479. [DOI] [PubMed] [Google Scholar]
- Bell R., Owens D., Shapiro M., and Tardif J.. 1981. Development of mass rearing technology, pp. 599–633. In C. Doane and M. McManus (eds.), The gypsy moth: research toward integrated pest management Technical Bulletin 1584. United States Department of Agriculture, Washington, DC. [Google Scholar]
- Duan J. J., and Oppel C.. 2012. Critical rearing parameters of Tetrastichus planipennisi (Hymenoptera: Eulophidae) as affected by host plant substrate and host-parasitoid group structure. J. Econ. Entomol. 105: 792–801. [DOI] [PubMed] [Google Scholar]
- Duan J. J., Oppel C. B., Ulyshen M. D., Bauer L. S., and LeLito J.. 2011. Biology and life history of Tetrastichus planipennisi (Hymenoptera: Eulophidae), a larval endoparasitoid of the emerald ash borer (Coleoptera: Buprestidae). Florida Entomol. 94: 933–940. [Google Scholar]
- Duan J. J., Bauer L. S., Abell K. J., Lelito J. P., and Van Driesche R.. 2013. Establishment and abundance of Tetrastichus planipennisi (Hymenoptera: Eulophidae) in Michigan: potential for success in classical biocontrol of the invasive emerald ash borer (Coleoptera: Buprestidae). J. Econ. Entomol. 106: 1145–1154. [DOI] [PubMed] [Google Scholar]
- Ebling P. 2013. The insect production and quarentine laboratories. Frontline Express Bulletin N: 2. [Google Scholar]
- Evenden M. L., and Silk P. J.. 2016. The influence of Canadian research on semiochemical-based management of forest insect pests in Canada. Can. Entomol. 148: S170–S209. [Google Scholar]
- Festing M. F. W. 1993. International index of laboratory animals, 6th ed Michael F.W. Festing, Leicester, UK. [Google Scholar]
- van Frankenhuyzen K., Ebling P., McCron B., Ladd T., Gauthier D., and Vossbrinck C.. 2004. Occurrence of Cystosporogenes sp. (Protozoa, Microsporidia) in a multi-species insect production facility and its elimination from a colony of the eastern spruce budworm, Choristoneura fumiferana (Clem.) (Lepidoptera: Tortricidae). J. Invertebr. Pathol. 87: 16–28. [DOI] [PubMed] [Google Scholar]
- van Frankenhuyzen K., Lucarotti C., and Lavallée R.. 2016. Canadian contributions to forest insect pathology and to the use of pathogens in forest pest management. Can. Entomol. 148: S210–S238. [Google Scholar]
- Grisdale D. 1970. An improved method of rearing large numbers of spruce budworm, Choristoneura fumiferana (Lepidoptera: Tortricidae). Can. Entomol. 102: 1111–1117. [Google Scholar]
- Grisdale D. 1984. A laboratory method for mass rearing the eastern spruce budworm, pp. 223–231. In E. G. King and N. C. Lepella (eds.), Advances and challenges in insect rearing. United States Department of Agriculture, Agricultural Research Service, Washington, DC. [Google Scholar]
- Howe K., Clark M. D., Torroja C. F., Torrance J., Berthelot C., Muffato M., Collins J. E., Humphray S., McLaren K., Matthews L., et al. . 2013. The zebrafish reference genome sequence and its relationship to the human genome. Nature. 496: 498–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings B. H. 2011. Drosophila-a versatile model in biology & medicine. Mater. Today 14: 190–195. [Google Scholar]
- Keena M. A. 2005. Pourable artificial diet for rearing Anoplophora glabripennis (Coleoptera : Cerambycidae) and methods to optimize larval survival and synchronize development. Ann. Entomol. Soc. Am. 98: 536–547. [Google Scholar]
- Kuno G. 2010. Early history of laboratory breeding of Aedes aegypti (Diptera: Culicidae) focusing on the origins and use of selected strains. J. Med. Entomol. 47: 957–971. [DOI] [PubMed] [Google Scholar]
- Macoris M., Andreghetti M., Nelson K., Gerbeleto V., and Junior A.. 2005. Standardization of bioassays for monitoring resistance to insecticides in Aedes aegypti. Dengue Bull. 29: 176–182. [Google Scholar]
- McGuire M., Galan-Wong L., and Tamez-Guerra P.. 1997. Bacteria: bioassay of Bacillus thrungiensis against lepidoptera larvae, pp. 91–100. In L., Lacey (ed.), Manual of Insect Pathology. Academic Press Inc, San Diego, CA. [Google Scholar]
- McMorran A. 1965. A synthetic diet for the spruce budworm, Choristoneura fumiferana (Clem.) (Lepidoptera: Tortricidae). Can. Entomol. 97: 58–62. [Google Scholar]
- Phifer-Rixey M., and Nachman M. W.. 2015. Insights into mammalian biology from the wild house mouse Mus musculus. eLife. 2015: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Régnière J. 1987. Temperature-dependent development of eggs and larvae of Choristonera fumiferana (Clem.) (Lepidoptera: Tortricidae) and simulation of its seasonal history. Can. Entomol. 119: 717–728. [Google Scholar]
- Rubin G. M., and Lewis E. B.. 2000. A brief history of Drosophila’s contributions to genome research. Science. 287: 2216–2218. [DOI] [PubMed] [Google Scholar]
- Snell G. 1941. Biology of the laboratory mouse, 1st ed Dover Publications, Inc, New York, NY. [Google Scholar]
- Sohi S. 1995. Development of lepidopteran cell lines, pp. 397–412. In C., Richardson (ed.), Baculovirus expression protocols. Methods in molecular biology, Vol. 39 Humana Press, New York, NY. [DOI] [PubMed] [Google Scholar]
- Wang L., Wang S., Li Y., Paradesi M. S. R., and Brown S. J.. 2007. BeetleBase: the model organism database for Tribolium castaneum. Nucleic Acids Res. 35: 476–479. [DOI] [PMC free article] [PubMed] [Google Scholar]