Abstract
Trophoblast (TB) comprises the outer cell layers of the mammalian placenta that make direct contact with the maternal uterus and, in species with a highly invasive placenta, maternal blood. It has its origin as trophectoderm, a single epithelial layer of extra-embryonic ectoderm that surrounds the embryo proper at the blastocyst stage of development. Here, we briefly compare the features of TB specification and determination in the mouse and the human. We then review research on a model system that has been increasingly employed to study TB emergence, namely the BMP4 (bone morphogenetic protein-4)-directed differentiation of human embryonic stem cells (ESCd), and discuss why outcomes using it have proved so uneven. We also examine the controversial aspects of this model, particularly the issue of whether or not the ESCd represents TB at all. Our focus here has been to explore similarities and potential differences between the phenotypes of ESCd, trophectoderm, placental villous TB, and human TB stem cells. We then explore the role of BMP4 in the differentiation of human pluripotent cells to TB and suggest that it converts the ESC into a totipotent state that is primed for TB differentiation when self-renewal is blocked. Finally we speculate that the TB formed from ESC is homologous to the trophectoderm-derived, invasive TB that envelopes the implanting conceptus during the second week of pregnancy.
Keywords: bone morphogenetic protein-4, cytotrophoblast, implantation, inner cell mass, invasion, placenta, trophectoderm, trophoblast stem cells, syncytiotrophoblast
BMP4-driven differentiation of human embryonic and induced pluripotent stem cells provides a useful model for studying the specification and early development of early placental trophoblast.
Introduction
Trophoblast (TB), a tissue comprised of trophoblasts (from the Greek words trephein, to feed and blastos, germinator), forms the outer epithelial layers of the mammalian placenta and is the essential intermediary tissue linking the maternal system to the fetus. Its primary functions are to anchor the conceptus to the wall of the reproductive tract, extract nutrients from the mother, facilitate the exchange of gases and excretory material, protect the fetus from immune attack, and secrete hormones that act on the maternal system to promote the physiological interests of the conceptus. These are roles common to all types of placentae including those encountered in nonmammalian vertebrates [1] where the placental lineage is not set aside in the very early stages of embryogenesis.
Although placental TB of mammals consists of multiple, specialized cell types, most likely with their own stem cell populations [2–4], all the sublineages are considered to originate from trophectoderm (TE), a single layer of extra-embryonic ectoderm that surrounds the embryo proper at the blastocyst stage of development. As discussed below, TE is specified by a series of events, possibly beginning as early as the 4-cell stage of development in the case of the mouse, and culminating at blastocyst. By this stage, cells of the TE are morphologically and functionally distinct from those of the inner cell mass (ICM) and committed to the TB lineage.
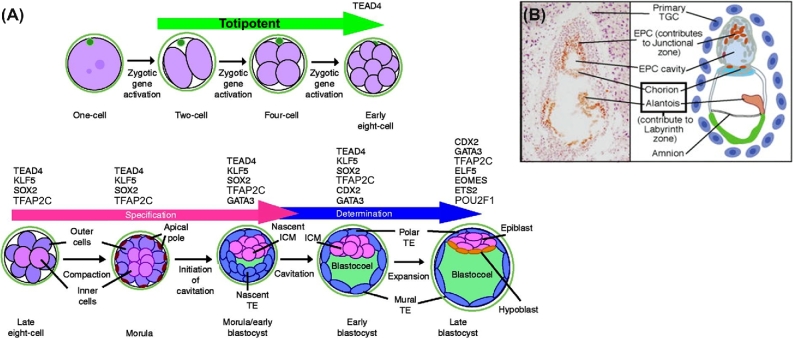
It takes about three and a half days and five rounds of cell division for a mouse zygote to progress to the early blastocyst stage of development, at which point it possesses approximately 32 cells, of which one-third to one-quarter are part of the ICM, while the remainder comprise TE (Figure 1A) [5]. A second cell fate decision is initiated in the ICM soon after TE has emerged, namely the segregation of primitive endoderm (hypoblast) and epiblast [6, 7] but will not be discussed further here. Development of the zygote to the blastocyst, whether in vivo or in vitro, follows a somewhat comparable morphological and temporal patterns across different species, e.g. cow, pig, human, mouse, and rat, [8–12], suggesting that the process is guided by similar genetic mechanisms across eutherian mammals.
Figure 1.
(A) Development of the mouse preimplantation embryo from the zygote to expanded blastocyst. Transcription factors controlling specification and determination of TE are listed above the images by stage of expression. (B) Development of trophoblast progenitors and extraembryonic tissues in a cross section of E7.5 mouse conceptus. TGC: trophoblast giant cells, EPC: ectoplacental cone. Modified from Knott & Paul [5] with permission.
At implantation, however, the mouse and human follow divergent paths of TB development [5, 13]. In the former, attachment to the endometrium is mediated initially by mural TE and, shortly afterwards by polar TE overlying the ICM, which, through proliferation and subsequent expansion, forms the ectoplacental cone (EPC in Figure 1B), with the chorion at its base, embedded in the secondary decidual zone of the maternal endometrium (Figure 1B). The labyrinth, which mediates nutrient exchange with maternal blood sinusoids and is the functional equivalent of human chorionic villi, develops from the chorionic region at about E8.5 coincident with chorioallantoic attachment. The EPC contributes to the junctional zone, a region of the mature placenta that forms between the maternal decidual tissue and the labyrinth layer (Figure 1B). Its ontological relationship to the extravillous TB of the human placenta remains unclear.
In the human, events take a different course (Figure 2). The blastocyst attaches to the uterine epithelium at its polar end, and TB penetrates the endometrium by means of a highly invasive syncytial mass migrating ahead of a zone of proliferating cytotrophoblast (cytoTB) [14–16]. This syncytium comes to rest within the maternal deciduum and creates lacunae filled with maternal blood. However, this syncytial structure seems to be relatively short-lived. The formation of this primitive placenta has been studied in archived human samples [14–16] and in a few non-human primates [17, 18], but events appear to be recapitulated when human blastocysts are cultured in vitro [19, 20], such that by ∼ day 12 post-fertilization, the epiblast is enveloped by TB, with cytoTB to the interior and syncytiotrophoblast (STB) to the exterior (Figure 2). Histological examination of in vivo samples indicates that soon after day 12 post-coitus primary chorionic villi derived from the underlying cytoTB begin to form, penetrate the syncytial zone, and initiate the formation of early villous TB. Extravillous TB derived from those cytoTB columns that contact the endometrium proliferates extensively to form a cytoTB shell that surrounds the entire fetus and anchors it to the endometrium [21]. The shell is also the origin of the extravillous TB that modifies maternal spiral arteries and of other extravillous TB populations that penetrate deeply into the endometrium and myometrium where they have been suggested to be the progenitors of so-called multinucleated giant cells. When these developmental events occur abnormally, later complications of pregnancy, including preeclampsia, may occur [21].
Figure 2.
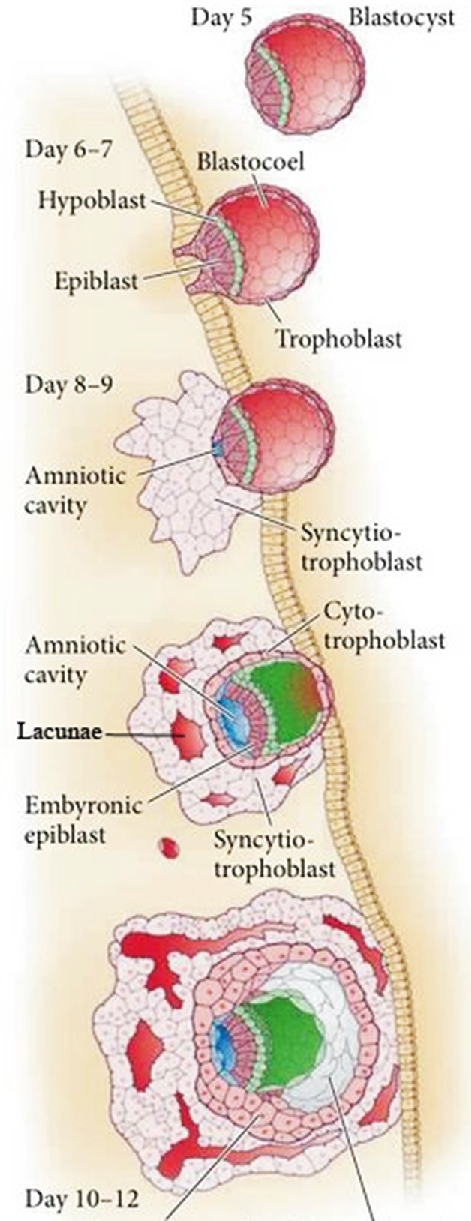
Morphological transition of the human conceptus during implantation. At day 6–7, a sub-population of TE at the polar end of the conceptus initiates attachment to uterine epithelial cells. Day 8–9, invasive STB penetrates the uterine epithelium, tunnels into the decidualizing endometrium, and begins to erode out lacunae (in red). The cytoTB layer (pink) between the embryo proper and the advancing syncytial mass continues to proliferate to replenish the STB. By day 12, the conceptus has penetrated more deeply and is completely enveloped in STB. Subsequently, primary villi originating from the cytoTB breach the STB to initiate formation of the villous placenta (not shown). Modified from Figure 12.17, page 397 in Developmental Biology/Scott F. Gilbert 11th Ed [2016] with permission.
After the sections that follow, we provide a brief review of what is known about the molecular events associated with TE emergence in the mouse and the extent to which this process matches the limited amount of information available for the human. We shall then consider the evidence that TB can be derived directly from human embryonic stem cells (ESC) of the so-called primed or epiblast type after exposing them to the growth factor bone morphogenetic protein-4 (BMP4) a contention that has proved controversial. In particular, critics have protested that the cells are extra-embryonic mesoderm and not TB at all [22]. However, it soon became clear that the proportion of TB relative to mesoendoderm could be much increased by excluding fibroblast growth factor 2 (FGF2) from the differentiation medium and by adding inhibitors targeting the autocrine actions of FGF2 and members of the transforming growth factor beta (TGFB) superfamily, namely ACTIVIN/NODAL/TGFB, that are necessary to maintain pluripotency in ESC [23–27]. A more recent assessment is that the ESC-generated cells represent a population that has not fully differentiated to the kind of TB associated with first trimester placenta [28].
In the sections that follow, we present confirmatory evidence that BMP4 can induce TB differentiation and summarize how alterations in protocol, especially in the culture conditions, can have consequences for the efficiency of the process. Three other questions will then be addressed. First, is there a logical rejoinder to the premise put forward by embryologists that epiblast-derived ESCs have already progressed beyond the first cell fate decision and should no longer have the potential to differentiate to TB? Second, if these ESC-derived cells are, as we believe, authentic TB, what placental cell type do they represent? Finally is there a relationship between early stage, ESC-derived TB, and the TB stem cells recently described by Okae at al. [29]?
Acquisition of trophoblast fate during mouse development
Hundreds of papers and many careers have been spent analyzing the emergence of TE and its segregation from ICM in the mouse, a process that is essentially complete by the 32-cell stage of development. To review all this information here would add little of value to what has been written in numerous reviews on the topic, including the following [5, 30–33]. A general but not necessarily a consensus view is that the process is not initiated by pre-patterning of the oocyte cytoplasm, as occurs in the initial commitment steps of invertebrates and amphibians. Nonetheless, distinctions between still totipotent, individual blastomeres in terms of their gene expression [34–37] and epigenetic status [36, 38] are evident as early as the 4-cell stage and appear to mark blastomeres more likely to assume an embryonic versus extraembryonic fate. Subsequent distinctions between pre-TE and pre-ICM blastomeres are likely reinforced by positon-dependent cell–cell interactions initiating different cell signaling pathways, a gain of cell polarity by outer blastomeres destined to become TE, a loss of expression of genes conferring pluripotency, and an increase in expression of several transcription factors that impose an emerging TE cell phenotype (Figure 1A). Together, these and other observations suggest that early lineage biases may have been established even before the morula stage, although a TE fate may not be sealed until the 32-cell stage when cavitation has begun.
Transcription factors implicated in trophectoderm specification in the mouse
As gene knockout studies in the mouse began to be performed in increasing numbers during the 1990s, it became clear that lack of expression of a range of transcription factors could provide an embryonic-lethal phenotype due to TB failure [5, 39, 40]. In some cases, for example, in caudal type homeobox 2 (Cdx2) [41] and eomesodermin (Eomes) [42] knockouts, early cleavage stages were unaffected, but lethality occurred at or soon after implantation. In other instances, as in the case of distal-less homeobox 3 (Dlx3) [43] and ETS proto-oncogene 2 (Ets2) [44], the demise of the conceptus was delayed until just before the labyrinth placenta had begun to form, reinforcing the notion that placentation is a progressive, but staged, process requiring the input of numerous transcriptional regulators and epigenetic directives.
The isolation of mouse TB stem cells from outgrowths of polar TE of preimplantation embryos (E3.5) [45] and extraembryonic/chorionic ectoderm at the base of the EPC 2 days later [46], creating TB stem cells by reprogramming with ectopic genes under the influence of strong promoters, and defining the culture conditions that allowed such cells to self-renew and differentiate identified other transcription factors linked to the early TB phenotype [39]. More recently, single cell RNAseq analysis performed on mouse blastocysts has provided an increased level of sophistication to TE profiling [47]. None of the transcription factors so far identified in early TE with the possible exception of glial cells missing homolog 1 (GCM1), a transcription factor that plays a role in controlling the formation of STB [48], has a unique TB association, but, as a group, they are co-associated with TE and TSC specification and determination (Figure 1A; morula), and useful markers for recognizing TB [28]. Among these genes are the TEA domain family member Tead4, the caudal-type homeobox factor Cdx2, the T-box gene Eomes, the SRY-box gene Sox2, the estrogen-related receptor Esrrb, the AP-2 family member Tfap2, the Ets family members Ets2 and Elf5, the GATA motif-containing factor Gata3, and Yes-associated protein 1 Yap1 [5, 39, 40, 49]. Exactly how these particular gene products and others act together in concert is far from clear.
There have been attempts to define networks of transcription factors that contribute to the emergence of TB in embryos and to the self-renewal and undifferentiated state of TB stem cells [6]. Some networks are better studied than others. TEAD4, for example, whose knockdown prevents the transition of morulae to blastocysts, controls expression of Cdx2, Gata3, and Eomes in outer blastomeres [50]. ELF5 forms complexes with EOMES and TFAP2C and binds a number of downstream genes, with the complexes acting as molecular switches governing the balance between TSC proliferation and differentiation [49]. CDX2 is a bit of a puzzle. It is expressed as early as the 8-cell stage in surface-located blastomeres [6], but is no longer regarded a master regulator of TE specification, since Cdx2–/– embryos, unlike Tead4–/– embryos, develop as far as the blastocyst stage. One role may be to downregulate expression of key pluripotency genes and provide a final push toward emergence of functional TE.
Differences in trophectoderm specification between mouse and human
There is no reason to believe that the molecular events that lead to the formation of a mouse blastocyst are not re-capitulated to some extent in the human, even though the process takes somewhat longer [51]. On the other hand, in the human, the segregation of TE, epiblast, and primitive endoderm lineages occur most simultaneously, rather than as two sequential cell fate decisions [52]. This three-way lineage split is evident from single cell transcriptome analysis performed just before the morula–blastocyst transition (early day 5). Principal component analysis differentiates three groups of cell corresponding to TB, hypoblast, and epiblast, each expressing a complement of lineage specific genes [52].
There are also some interesting differences between human and mouse in the transcription factors expressed during TE emergence [51–53]. For example, CDX2 is not expressed in the human embryo until the blastocyst has formed. CDX2 also has moderately low expression relative to the genes encoding several other transcription factors linked to TE specification such as GATA2 and GATA3 [52]. These data are more consistent with CDX2 playing a part in the final transition to a functioning epithelium than as a master regulator for TE specification. The genes for several other transcription factors considered pivotal in the mouse, such as ELF5 and EOMES, appear not to be transcribed to any significant extent in human TE [52, 53]. Another anomaly relates to Tead4, which encodes a key regulator of TE specification in the mouse [50]. TEAD4 is expressed weakly in human embryos, although its paralog, TEAD1, is upregulated in TE relative to epiblast and extra-embryonic endoderm, with transcript levels significantly increasing between E5 and E7. Another marker, vestigial like family member 1 (VGLL1), associated with human TB [54] and human TB stem cells [29] but apparently absent from the mouse, has been proposed to be involved in TE specification, possibly substituting for the role played by YAP1 in the mouse [54], although transcript levels for the latter are relatively high in human TB stem cells [29]. Despite some conflicting observations, these data indicate that certain inferences drawn from the mouse regarding TE lineage specification may not necessarily pertain to the human. In addition, single cell RNAseq of human embryos at days 6 and 7 has indications of a subpopulation of TE located at the polar end of the blastocyst where attachment is initiated (Figure 2, day 6–7). These cells show enrichment in transcripts encoding proteins associated with the initiation of syncytialization, for example, GCM1, ovo like transcriptional repressor 1 (OVOL1), endogenous retrovirus group members (ERVW-1, ERVW-2, ERVFRD-1), glycoprotein hormones, alpha polypeptide (CGA,) and placental growth factor (PGF) [52], which is consistent with a role for non-villous STB in the implantation process (Figure 2, day 8–9).
Finally, until this year [29], it has proved difficult to derive well-characterized TB stem cells from either human blastocysts or more mature TB from first trimester placentae. It is now apparent that the past failures were not because such cells did not exist at these stages of development [30, 51]. Rather, the culture conditions employed were inappropriate to sustain long-term self-renewal [29].
BMP4 and trophoblast differentiation from human epiblast stem cells
Human ESC lines, beginning with those described by Thomson et al. [55], have almost invariably been derived from outgrowths of ICM of “spare” E5 or E6 human blastocysts derived from in vitro fertilization programs. Currently, these cells are readily maintained in a self-renewing undifferentiated state on a defined medium supplemented with FGF2 and a TGFB family member, such as ACTIVIN A. These factors provide a substitute for and are possible components of the medium conditioned by mouse embryonic fibroblasts used in the early days of human ESC derivation. The morphologies of the flattened colonies assumed by human ESC (hESC), plus a number of other phenotypic features, are distinct from ESC originating from mouse blastocysts, but they do resemble and have similar growth requirements to mouse ESC generated from mouse epiblast at about E5.5 [56, 57]. For this reason, the hESC have historically been named either epiblast-type or primed and, in terms of their differentiation potential, a step past the leukemia inhibitory factor (LIF)-dependent naïve state of mouse ESC. The general view is that naïve type ESC hold higher developmental potential than the primed or epiblast type. However, it is now recognized that the two states, primed versus naïve, are interconvertible for both species by adding or removing specific inhibitors and switching growth factors in the culture medium [58–60].
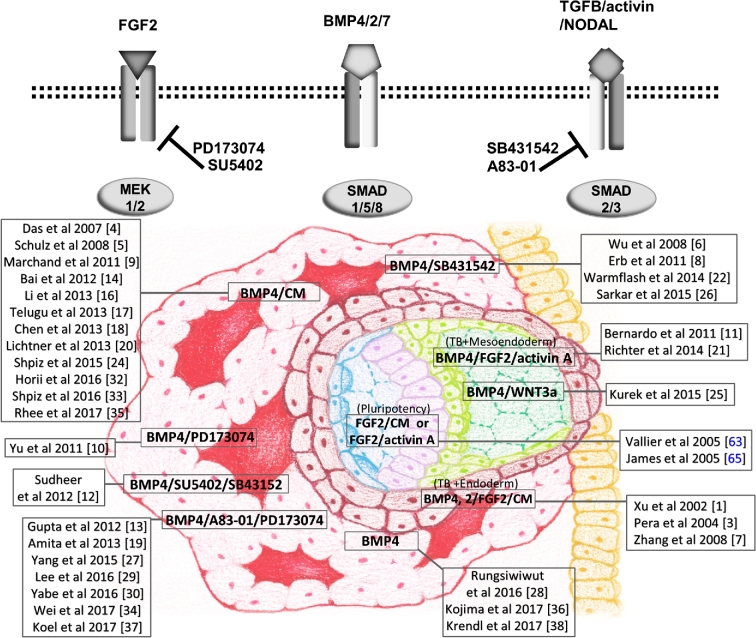
In 2002, Xu et al. [61] showed that when four different human ESC lines with what we now know to be primed/epiblast type features were exposed to members of the BMP (bone morphogenetic protein) growth factor family, they changed their morphologies, with a few cells becoming multinuclear and positive for human chorionic gonadotropin (hCG). The colonies also began to express a range of markers characteristic of TB and secreted hCG, and progesterone into the medium. Differentiation state progressed with day of culture and with the concentration of added growth factor, of which BMP4 was the most potent [61, 62]. What is not clear from this paper is whether or not the medium was free of FGF2 when the BMP-driven TB differentiation was initiated, because markers of mesoderm and endoderm were also detectable in microarray analyses of differentiating colonies [61]. The presence or absence of FGF2 in the culture medium becomes an important consideration when reviewing subsequent papers based on this initial report (Supplementary Table S1), because the presence of FGF2, ACTIVIN-A, and BMP4 together can drive lineage directionality of hESC selectively toward TB, mesoderm, or endoderm depending upon the relative concentration of each factor in the medium (Figure 3). For example, hESCs are converted predominantly toward a mesoderm fate by BMP4 in the presence of FGF2 and inhibitors that minimize SMAD2/3 signaling and toward endoderm when such signaling is unimpeded [22, 63]. Similarly, if all three growth factors (BMP4, FGF2, ACTIVIN-A) are present, there is a bias toward endoderm. Our contention is that when BMP4 is added and FGF2 and ACTIVIN-A/NODAL/TGFB signaling suppressed differentiation is entirely towards TB. Another complication is that the differentiating cultures themselves either continue to or begin to express genes encoding FGF2 and BMP family members, e.g. BMP4 and BMP7, as they differentiate [64]. In other words, simply omitting these factors from the culture medium may not be sufficient to suppress their autocrine effects and inhibit differentiation along the mesoderm and endoderm lineages [64, 65]. It was this rationale that prompted the addition of FGF2 and ACTIVIN-A/NODAL/TGFB signaling inhibitors to the differentiation cocktail in order to provide complete directionality toward TB [66].
Figure 3.
Cartoon illustrating the interacting roles of FGF2, ACTIVIN A, and BMP4 in lineage specification, especially in directing TB differentiation from epiblast type ESC and iPSC. The upper part of the figure shows how inhibitors can be used to block FGF2 signaling (PD173074, SU5402) and ACTIVIN A/TGFB/NODAL signaling (A83–01, SB431542) in conjunction with BMP4-directed differentiation to TB. Blocking signaling via both the SMAD2/3 and MEK1/2 pathways in the presence of BMP4 appears to be optimal for unidirectional differentiation toward TB. The lower part of the cartoon illustrates the various combinations of reagents, including medium conditioned by mouse embryonic fibroblasts (CM) that have been employed to drive hESC to TB, mesoendoderm, and mixtures of TB and mesoendoderm. Names (adjacent to a drawing of an implanting day 9–10 human conceptus) are relevant references with citation numbers listed in Supplementary Table S1, except Vallier et al. [63] and James et al. [65] that are only cited in the main text. In the conceptus, note the pluripotent, bilaminar epiblast (blue and mauve), extra-embryonic mesoderm (green), progenitor cytoTB (maroon), and the advancing mass of STB (pink) enclosing lacunae filled with maternally derived fluid acting as a nutrient support external to the embryo proper. At this stage of pregnancy, the conceptus has mainly passed through the uterine epithelium (yellow) and lodged in the uterine wall.
From our literature search, we estimate that at least 38 papers have employed the BMP system to model differentiation of human TB (Supplementary Table S1). In general, it has been assumed that the TB that emerged was homologous to early villous TB because the colonies contained varying amounts of STB and mononucleated cells and produced large amounts of placental hormones. True villi were not expected, because the cultures were two dimensional and did not contain an extra-embryonic mesoderm component to provide connective tissue and blood vessels. Even so, outcomes have been inconsistent with regard to the efficiency and extent of TB formation, most likely because protocols varied, if not in principal, in detail. For example, in protocols where BMP4 alone was used to drive differentiation, it is not always clear whether or not FGF2 was present in the medium at the time that BMP4 was added [22, 67–70]. To add to this confusion it is often uncertain whether or not conditioned medium had been employed during the differentiation stages of the protocols [71–74]. In other cases, FGF2 was certainly present, [23, 61, 75, 76], occasionally at high concentrations [26]. Only a few reports mention whether FGF2 was specifically excluded at the time of BMP4 addition [24, 77–79]. Under mixed FGF2/BMP4 conditions, particularly if the cultures had been held under ambient O2 conditions, there would almost certainly be a hefty mesoderm component to the colonies (Figure 3) [27]. In other cases, the rate of differentiation of the ESC, for example in terms of assuming a keratin 7 (KRT7)+ phenotype, timing of hCG production, and amounts of placental hormones released have been relatively anemic [74, 80, 81], raising the question as to the quality and potency of the BMP4 used to drive differentiation (Supplementary Table S1). Our own experience with one commercial supplier (supplier 3 in Supplementary Table S1) is that the BMP4 product available had only a fraction of the potency at inducing differentiation as the one sold by a competitor (supplier 1), although it was somewhat cheaper. As far as we can judge, only two of the citations in Supplementary Table S1 used supplier 3. One suggested that BMP4 plus FGF2 led to mesoderm formation [82]. The second [80] reported a much slower rate of differentiation and lower hCG production than observed by others who used comparable BMP4 concentrations. Unpublished experiments from our laboratory have also indicated that the efficiency of TB differentiation and hCG production can be influenced by the starting sizes of colonies and initial colony density at the time of BMP4 addition. These variables are rarely described in publications.
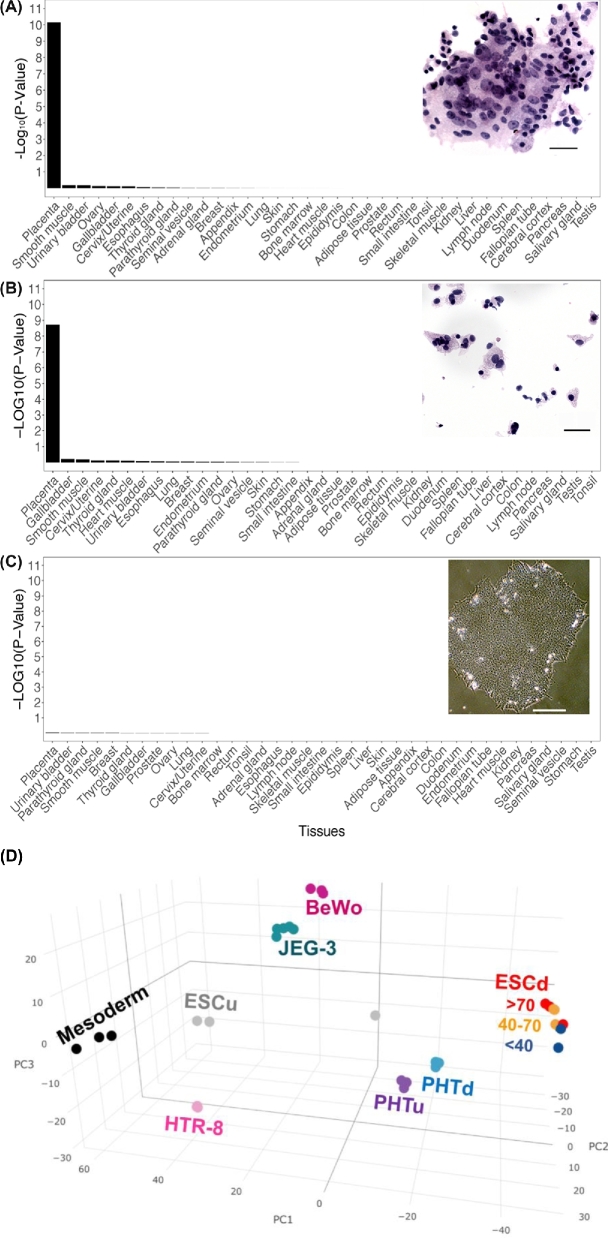
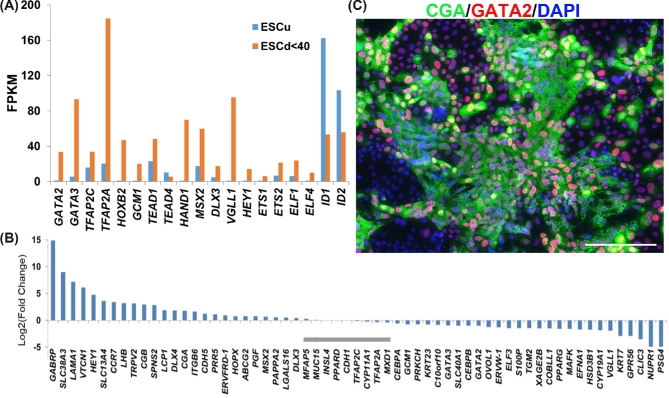
The progress of differentiation of a single colony from the time of BAP addition, when the larger colonies are about 250 μm in diameter until day 6 when they often can reach 3 mm, is shown in Supplementary Figure S1A. Areas of emerging STB can be visualized by staining with crystal violet (Supplementary Figure S1A, day 6) and comprise about 10% areas of the colonies at day 6. They also become evident by immunostaining for the alpha subunit of hCG, CGA, which appears to be localized mainly in cytoplasmic granules (Figure 5C). Most cells shown in Figure 5C are weakly positive for GATA2 and those that fluoresce most strongly tend to be located around the periphery of the CGA+ regions rather than deep within it. It will require single cell RNAseq to define the heterogeneity of the various cell populations better.
Figure 5.
The distinctive trophoblastic signature of the ESC-derived TB. Data were derived from RNAseq results of Yabe et al. [80] on cell populations fractionated by size from colonies of ESC. Enrichment of placenta-specific genes in the stem cell derived STB (ESCd > 70) (A), cytoTB (ESCd < 40) (B), and undifferentiated ESC (ESCu) (C). The respective images are clumped sheets of isolated STB, cytoTB (ESCd < 40), and ESC colonies, respectively. In A and B, cells were isolated from ESCd after 8 days of differentiation [80] and stained by hematoxylin and eosin. The enrichments are presented as –Log10(P-value). The bars for the images are 20, 20, and 100 μm, respectively. (D) PCA plot based on the 1000 most highly expressed genes in STB (>70 μm; red), in ESCd cells of intermediate size (40–70 μm; yellow), in the predominant mononuclear cells (<40 μm; blue), and in undifferentiated ESC (gray). The ESCd cell fractions are compared with the transformed cell lines (BeWo, HTR-8, and JEG-3), mononucleated cytoTB and STB (PHTu and PHTd) from term placentas, and mesoderm cells also generated from ESC. More detailed procedures and original figures are provided by Jain et al. [81].
As suggested above, the starting size of the colonies influences differentiation patterns. Small clumps yield, as expected, smaller colonies at day 6, but also ones whose shape is less compact and whose internal features appear different from those of the larger colonies (Supplementary Figure S1B). It may be noteworthy that where passage had been initiated through dispersion of the colonies to single cells and small clumps by using Accutase, an enzyme mixture with both trypsin and collagenase activities, and a RHO-kinase inhibitor, hCG production was quite low [77]. Clearly, unless protocols become consistent, different outcomes in terms of differentiation end points should be anticipated.
Phenotypic assessment of embryonic stem cell-derived trophoblast
Supplementary Table S1 also summarizes some of the methods that have been employed to establish a TB phenotype in the BMP-induced model of TB differentiation. These diagnostic procedures have included immunolocalization of numerous traditional TB/TE markers, RT-PCR for expression of specific genes, enzyme linked immunoassays of hormone production, western blotting, and flow cytometry. Each one of these procedures based on only a few markers, could, of course, provide a wrong diagnosis [83]. However, many combined markers provide strong evidence that the ESC can be differentiated into TB, in some cases with high efficiency.
RNAseq and microarray analyses have provided the most clear-cut evidence that BMP-directed differentiation of ESC gives rise to a predominantly TB phenotype. Moreover, when inhibitors of FGF2 and ACTIVIN-A signaling are provided simultaneously, differentiation is more or less exclusively toward TB [66, 84], and the minor diversion to mesoderm and endoderm noted with BMP4 alone [81] is completely avoided. Not only does the process occur quickly and relatively efficiently but areas within the colonies progressively advance to display markers for different TB sublineages, e.g. STB and extravillous TB. Not unexpectedly, therefore, an analysis of the top 1000 genes expressed in ESC-derived TB relative to control ESC at day 8 of differentiation shows a significant enrichment of terms linked to placental development [85]. Additionally, this same list of 1000 genes contains a highly significant enrichment of placenta-specific genes, meaning that, according to the Human Gene Atlas, the expression of these genes is at least fivefold higher in the placenta relative to their expression in any other tissues [86] (Figure 4). Perhaps most intriguingly, the ESC-derived TB display a high level of expression of genes implicated in migration and invasion compared to commonly used immortalized TB cell lines, such as JEG3, BeWo, and HTR8 cells, and to primary cells from term placenta [85]. In addition, the transcriptome is highly enriched for mRNA encoding extracellular matrix materials, including many collagens, laminins, and proteoglycans not found associated with villous TB [84]. Finally, principal component analysis of the RNAseq data clearly demonstrates that the ESCd constitutes a distinct form of TB relative to that associated with placental villi at term and to bear little resemblance to JEG3, BeWo, and HTR8 cells [85] (Figure 4D).
Figure 4.
(A) Comparative expression of transcription factors that play roles in TE emergence in undifferentiated human ESC (ESCu; blue bars) and differentiated ESC (ESCd; red bars). Transcript levels are shown in FPKM (Fragments Per Kilobase of transcript per Million mapped reads). RNAseq data are from Yabe et al. [80]. (B) Fold-change differences in relative expressions of known trophoblast gene markers in STB from ESCd (>70 μm size fraction from BAP-differentiated ESC) and from STB generated from placenta at term (PHTd). Data are from Yabe et al. [80]. Gray horizontal bars indicate where differences in gene expression are not significant (q > 0.05). All other values differ between the two types of cell. (C) Immunofluorescent detection of CGA (green) and GATA2 (red) in a human iPSC cell line cultured in the presence of BAP for 6 days. The image represents cells in a part of the iPSC colony that include STB areas (see live images in Supplementary Figure S1, day 6) [27]. Nuclear materials are stained with DAPI (blue). The scale bar represents 200 μm.
Microarray analyses have also been designed to follow the time course of differentiation in response to BMP4 treatment of ESC and to make inferences about the transcriptional networks involved in specifying TB [25, 77, 81, 87, 88]. What is apparent is that several transcription factors associated with mouse [5] and human TE [52], including GATA3 and transcription factor AP-2 gamma (TFAP2C), are upregulated early and continue to be expressed, while transcription factors associated with pluripotency become quickly downregulated. Krendl et al. [81] sorted putative TB cells on the basis of expression of the surface antigen ENEP (glutamyl aminopeptidase). By combining transcriptome and chromatin occupancy analyses during the early stages of differentiation, they inferred that initial commitment to the TB lineage probably required the coordinated input of at least four key transcription factors, GATA2, GATA3, TFAP2A, and TFAP2C, which appear to play an instructional role in early specification steps for TB and also direct the switch from pluripotency by downregulating key pluripotency factors. Liu et al. [89] compared ESC and BMP-directed ESC (ESCd) with regard to nucleotide sequences within DNase I-sensitive sites and found that in the ESCd the open chromatin sites were enriched in binding sites for transcription factors linked to TE emergence, including GATA, ETS, and TFAP family members. Our laboratory has also noted that GATA2, GATA3, TFAP2A, and TFAP2C are significantly upregulated as ESC differentiate to TB (Figure 5A). Together these data suggest that that the specification of TB occurring in ESC when they are exposed to a BMP-directed differentiation regimen is quite similar to what occurs when TE is specified during the morula to blastocyst transition in human embryos.
The role of BMP4 in the differentiation of human pluripotent cells to trophoblast
The observation that BMP4 can initiate TB differentiation from hESC and iPSC has been perplexing. In naïve-type mouse ESC, BMP4 enhances pluripotency, rather than destabilizes it [90, 91]. On the other hand, there is evidence that BMP produced by inside cells of the mouse morula might be necessary to prime outer cells for their transition to TE [92]. Perhaps in a similar manner, the addition of BMP4 to human pluripotent cell cultures may be essential only to initiate differentiation, as it is required for no more than the first 24 h or so [66]. Recently, our laboratory demonstrated that a similar short-term treatment of human ESC and iPSC cell lines allowed novel ESC lines to be isolated [93]. These cells were selected by trypsin dispersion and an ability to be cultured on a gelatin substratum, properties not seen in primed-type hESC and iPSC. They also responded rapidly to PD173074 (P) in the absence of both FGF2 and BMP4 by conversion to TB and especially STB, while an A83–01/PD173074 (AP) combination favored increased expression of major histocompatibility complex HLA-G, a marker of extravillous TB (Figure 6B and D). When these cells were used to create teratomas in immunocompetent mice, small areas of TB were detectable in the tumors by immunohistochemistry, and hCG became elevated in the blood of the host animals. We speculate that BMP4 can prime hESC to a self-renewing, totipotent state that has enhanced potential for both embryonic and extraembryonic lineage development. A somewhat similar switch to totipotency was noted when human and mouse ESC cells were treated with a cocktail of low molecular weight pharmaceuticals [60], and earlier studies with mouse naïve-type ESC had indicated that LIF could support expansion of cells with totipotent potential that contributed to TB in chimeras [94]. Additionally, when adult cells were reprogrammed within live mice, circulating iPSC could contribute to TB [95]. These data indicate that both naïve-type and primed/epiblast-type ESC, whether of mouse or human origin, have the potential to differentiate to TB, given the right prompting.
Figure 6.
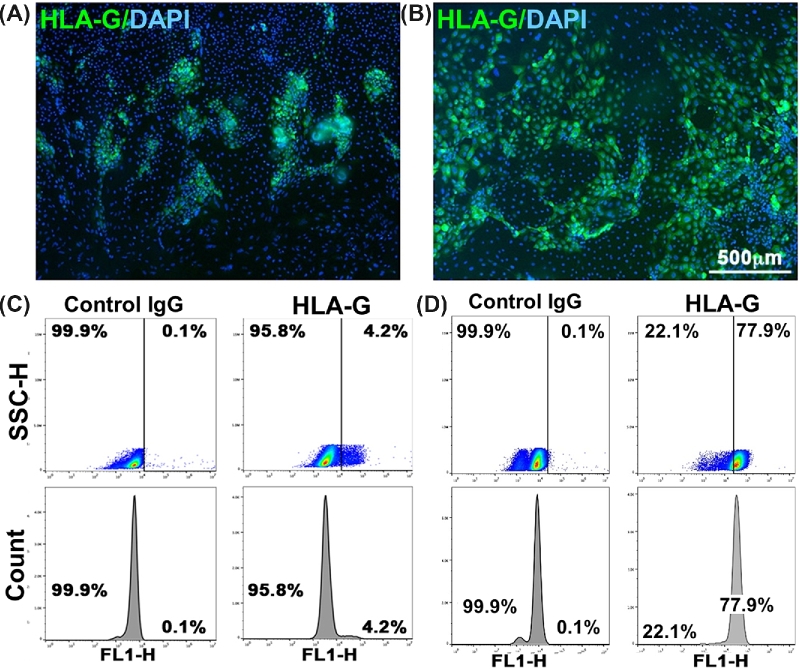
Expression of HLA-G in ESCd. (A) Immunofluorescent image from a colony of H1 ESC differentiated for 6 days in the presence of BMP4, A83–01, and PD173074 (BAP conditions). (B) Immunofluorescent image from a colony of H1 ESCBP (stable cell line from a BMP4-primed H1 cell [93] that had been differentiated to TB in the presence of A83–01, and PD173074 (AP conditions for 6 days). Cells had been fixed and then immunostained with anti-HLA-G (mouse monoclonal antibody 4H84, Santa Cruz Biotech. sc-21799; green immunofluorescence). Nuclear material was stained with DAPI (blue). The scale bar is 0.5 mm. (C) Flow cytometry histograms for HLA-G expression in H1 cells after 6 days of BAP treatment. (D) Flow cytometry histograms for HLA-G expression in H1 ESCBP after 6 days of AP treatment. The negative controls in each case employed the same cells and differentiation conditions, but during immunostaining the cells were exposed to a control IgG rather than anti-HLA-G antibody (4H84). Flow cytometry was performed as described by Amita et al. [66] and Yang et al. [93]. SSC-H: side scatter height, FL1-H: relative intensity of GFP florescence.
What is the nature of the trophoblast that forms from embryonic stem cell?
The evidence that human ESC and iPSC can be converted to some form of TB now seems to be unequivocal based on a vast range of markers, the ability to produce placental hormones, and lack of evidence for significant contribution from ectoderm, endoderm, and mesoderm, but what stage of TB development do these cells represent? As shown in Figure 4D, principal component analysis of RNAseq data demonstrates unequivocally that the ESCd are distinct, not only from term villous TB but also from various transformed TB cell lines [85]. Moreover, the STB that forms in the colonies fails to show a close ontological relationship with the STB associated with a mature placenta. On the other hand, Lee et al. [28] have claimed that “BMP-treated human ESC have not fully differentiated to TB” on the basis of four criteria: a largely hypermethylated ELF5 promoter, lack of expression of the chromosome 19-encoded miRNA (C19MC), a particular profile of HLA-class I molecules (and specifically lack of expression of HLA-G), and a lack of certain “positive trophoblast markers.” If the authors mean that ESCd are not in vitro homologs of first trimester TB, we would agree. Certainly, it is hardly surprising that the ELF5 promoter is not hypo-methylated in view of the fact the ELF5 gene is barely expressed in ESCd [84], but neither is ELF5 expressed in human blastocyst TE [52, 53]. We also agree that the C19MC RNAs are only weakly expressed in ESCd [96]. The third criterion, a lack of expression of HLA-G in ESCd, cited by both Bernardo et al. [22] and Lee et al. [28], is simply wrong. HLA-G mRNA is conspicuously present as judged by RNAseq analyses [84] and quantitative RT-PCR [66]. Additionally, the protein is readily detected with the 4H84 monoclonal antibody by immunofluorescence imaging (Figure 6A and B), flow cytometry (Figure 6C and D) [66, 93], and western blotting [66, 93]. Unlike Lee et al. [28], two other groups [74, 88] have found that flow cytometry after tagging cells with MEMG-9 provides a useful means of identifying populations of HLA-G+ cells in ESC cells differentiated to TB. Together, these experiments minimize any concern that the 4H84 reagent is less specific than MEMG-9 [92]. Others have also identified HLA-G in ESCd by a variety of approaches [70, 74, 88, 97]. Finally, HLA-G+ cells can be purified from ESCd colonies by collection on immunobeads coated with MEMG-9 [97]. The last of the four criteria of Lee et al., [28] lack of other “positive trophoblast markers,” is puzzling in light of what has been discussed earlier and data such as those shown in Figure 5B, which compares relative expression of a combination of 61 marker genes in ESCd [84]. Clearly most, but not all, of these genes are expressed in both ESCd and villous TB from term placentae, although not in equivalent proportions.
Given that the ESCd embodies a distinct form of TB, what is the in vivo counterpart of these cells? We have hypothesized that BMP-treated human ESC correspond to a stage during very early placental development, possibly representing the TB that surrounds the embryo proper as it implants [84, 98]. The following observations are consistent with this hypothesis:
Other than in placental villi, STB is found at two other sites during placental development: in the TB layer encompassing the conceptus as it implants and in placental giant cells embedded in the myometrium [21, 99]. The rapid emergence of STB in the BMP-treated ESC favors, but does not prove, the first possibility.
Expression of CGA and CGB family members is at least a magnitude higher in STB from ESC than from villous STB, perhaps reflecting the need for massive hCG production by a diminutive conceptus during the critical peri-implantation phase when the corpus luteum, wavering on the cusp of regression, must be rescued if the pregnancy is to survive [84].
In vivo, placental lactogen (CSH1 and CSH2) produced by villous STB [100, 101] becomes detectable in maternal serum after 4–5 weeks’ gestation [102]. By contrast, ESCd do not express either gene for placental lactogen, suggesting that they represent early cells.
The transcription factor heart and neural crest derivatives expressed 1 (HAND1) is highly expressed in ESCd and human blastocyst TE but not in placental villous TB. In rodents, HAND1 has a key role in the differentiation of the invasive TB giant cells [103].
ESCd have a transcriptome consistent with invasiveness, migration [85], and matrix remodeling [84] that is far more exaggerated than that observed in placental villous TB, choriocarcinoma cell lines (BeWo and JEG3), and a transformed extravillous TB cell line (HTR8/SVneo). Consistent with these observations, BMP4-differentiated ESC develop a capacity to invade through Matrigel [97] and type I collagen matrices (R. Karvas, R.M. Roberts & L. C. Schulz, unpublished data).
Is there a link between the differentiation of human embryonic stem cells and trophoblast stem cells?
Between the time that this review was submitted and received back for revision, a new approach toward studying human TB specification and development was described with a publication describing the generation of human TB stem cells. Okae et al. [29] used two sources of tissue as starting material: cytoTB cells isolated from first trimester (weeks 6–9 of gestation) and outgrowths of frozen-thawed blastocysts (5–6 days post fertilization). There were a number of surprises. First, the medium employed did not rely on FGF4 as a growth factor, which was critical for producing mouse TB stem cells [45]. Instead, Okae et al. [29] crafted a medium supplemented with a WNT activator (CHIR), EGF, and inhibitors of ACTIVIN-A/TGFB, HDAC, and RHO-associated protein kinase (ROCK) signaling [29]. These cell lines were able to differentiate along the three known TB lineages (cytoTB, extravillous TB, and STB), were self-renewing, and could be grafted under the skin of NOD-SCID mice to form hCG-secreting lesions. They also could be coaxed into forming TB organoids. The second surprise was that, with the exception of just a few genes, the cell lines derived from TE and villous TB origin had almost identical transcriptome profiles. This was unexpected because TE does not transition directly into villous structures, but instead gives rise to the invasive primitive placenta described earlier in this review, which we propose to be the in vivo homolog of the ESCd.
Are there then features of these TB stem cells shared with ESCd, acknowledging the fact that the former are essentially undifferentiated and self-renewing whereas the latter are progressing along a pathway of differentiation? We compared the transcriptomes of both types of cell using the Supplemental Data Sheet “Expression levels of Refseq genes shown as log2(FPKM + 1)” of Okae et al. [29] with our own data for ESCd [84]. Most interestingly, the undifferentiated TB stem cells express a number of marker genes that distinguish the ESCd from term villous TB, including V-set domain containing T cell activation inhibitor 1 (VTCN1), lumican (LUM), WAP four-disulfide core domain 2 (WFDC2), actin, alpha, cardiac muscle 1 (ACTC1), cadherin 3CDH3), receptor activity modifying protein 1 (RAMP1), collagen type I alpha (COL1A1, and A2). In addition, both cell types express genes encoding TB-associated transcription factors, including TP63, GATA2 and -3, TFAP2A and -C, TEAD1 and -4, GCM1, msh homeobox 2 (MSX2), DLX3, VGLL1, ELF1 and -4, and most notably HAND1, which is silent in villous TB. Interestingly, ELF5 and CDX2 are barely expressed in either cell type. Transcripts for some major ESCd markers, such as gamma-aminobutyric acid type A receptor pi subunit (GABRP), are more or less absent from the TB stem cells, but this was not unexpected, as the latter represent the undifferentiated progenitor state of human TB. Consistent with the observations of Li et al. [70], these data suggest that along their path to TB the ESCd transiently pass through a stage where a stem cell phenotype materializes, but the medium to support the cells does not permit that phenotype to stabilize and self-renew.
Closing comments
We conclude that the TB derived from ESC (ESCd) probably represents an unusual, possibly short-lived, but functional form of TB that emerges as the embryo begins to implant. It seems likely that it precedes the subsequent villous and extravillous forms of TB that dominate the more advanced villous placenta of the mid first trimester. The transcriptome signature of the ESCd is likely, therefore, to be somewhat intermediate between that of TE and that of villous TB. The early stage of placentation that we suggest is represented by the ESCd exists at a precarious time for the pregnancy to proceed, when wastage is high and the conceptus particularly vulnerable to environmental influences. The ESCd system may prove to be a valuable model for studying implantation and events associated with TB specification. It may also provide a means for screening environmental chemicals [104] and pathogens suspected as threats to the conceptus [98].
Supplementary data
Supplementary data are available at BIOLRE online.
Supplementary Figure S1. Phase images of live cells from one individual colony of H1 hESC highlight the morphological changes as these cells are differentiated by BAP treatment. (A) Images of a single hESC colony were captured prior to BAP treatment (day 0) and every 24 h after initiation of BAP treatment (days 1–6). During the early phase of colony expansion (days 1–4), the cell population is relatively homogenous in appearance, but this phase is followed by differentiation into a more heterogeneous population, including emergence of STB (visualized as bright areas, days 5 and 6). At day 6, cells were fixed and stained with crystal violet to define areas of emerging STB as dark purple. (B) Differences in the seeding size of hESC colonies influence the differentiation patterns. Left: an average size colony (∼100 cells) at day 0 gives rise to a well-differentiated heterogeneous TB population by day 6. Right: a smaller sized colony generates a smaller, less compact colony at day 6, which also appeared to be more homogeneous than the colony on the Left. The scale bar is 200 μm in phase contrast images and 1 mm in in the stained image.
Supplementary Table S1. Summary of studies in which one or more bone morphogenetic proteins were used to drive human pluripotent stem cells along the extra-embryonic lineage, including trophoblast and mesoderm.
Acknowledgments
The research from this laboratory was supported by NIH grants HD-067759 and HD 077108. We thank Yuchen Tian for drawing Fig. 3 and our colleagues Laura C. Schulz, Danny J. Schust, and Rowan Karvas for helpful discussion.
Footnotes
Grant Support: This study was sponsored by National Institutes of Health (NIH) Grants HD-067759 and HD 077108
Competing interests: The authors declare no conflicts of interest.
References
- 1. Roberts RM, Green JA, Schulz LC. The evolution of the placenta. Reproduction 2016; 152 (5):R179–R189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chang CW, Parast MM. Human trophoblast stem cells: real or not real? Placenta 2017; 60 (Suppl 1):S57–S60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Simmons DG, Fortier AL, Cross JC. Diverse subtypes and developmental origins of trophoblast giant cells in the mouse placenta. Dev Biol 2007; 304(2):567–578. [DOI] [PubMed] [Google Scholar]
- 4. Simmons DG, Cross JC. Determinants of trophoblast lineage and cell subtype specification in the mouse placenta. Dev Biol 2005; 284(1):12–24. [DOI] [PubMed] [Google Scholar]
- 5. Knott JG, Paul S. Transcriptional regulators of the trophoblast lineage in mammals with hemochorial placentation. Reproduction 2014; 148(6):R121–R136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zernicka-Goetz M, Morris SA, Bruce AW. Making a firm decision: multifaceted regulation of cell fate in the early mouse embryo. Nat Rev Genet 2009; 10(7):467–477. [DOI] [PubMed] [Google Scholar]
- 7. Rossant J. Stem cells and lineage development in the mammalian blastocyst. Reprod Fertil Dev 2007; 19(1):111–118. [DOI] [PubMed] [Google Scholar]
- 8. Niakan KK, Han J, Pedersen RA, Simon C, Pera RA. Human pre-implantation embryo development. Development 2012; 139(5):829–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cao S, Han J, Wu J, Li Q, Liu S, Zhang W, Pei Y, Ruan X, Liu Z, Wang X, Lim B, Li N. Specific gene-regulation networks during the pre-implantation development of the pig embryo as revealed by deep sequencing. BMC Genomics 2014; 15(1):4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sahakyan A, Plath K. Transcriptome encyclopedia of early human development. Cell 2016; 165(4):777–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hosseini SM, Dufort I, Caballero J, Moulavi F, Ghanaei HR, Sirard MA. Transcriptome profiling of bovine inner cell mass and trophectoderm derived from in vivo generated blastocysts. BMC Dev Biol 2015; 15(1):49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhao XM, Cui LS, Hao HS, Wang HY, Zhao SJ, Du WH, Wang D, Liu Y, Zhu HB. Transcriptome analyses of inner cell mass and trophectoderm cells isolated by magnetic-activated cell sorting from bovine blastocysts using single cell RNA-seq. Reprod Dom Anim 2016; 51(5):726–735. [DOI] [PubMed] [Google Scholar]
- 13. Cross JC, Werb Z, Fisher SJ. Implantation and the placenta: key pieces of the development puzzle. Science 1994; 266(5190):1508–1518. [DOI] [PubMed] [Google Scholar]
- 14. Boyd JD, Hamilton WJ.. The Human Placenta. Cambridge: Heffer & Sons; 1970. [Google Scholar]
- 15. Benirschke K, Kaufmann P, Baergen RN. Pathology of the Human Placenta. New York: Springer-verlag; 2006. [Google Scholar]
- 16. Hertig AT, Rock J, Adams EC. A description of 34 human ova within the first 17 days of development. Am J Anat 1956; 98(3):435–493. [DOI] [PubMed] [Google Scholar]
- 17. Enders AC, King BF. Early stages of trophoblastic invasion of the maternal vascular system during implantation in the macaque and baboon. Am J Anat 1991; 192(4):329–346. [DOI] [PubMed] [Google Scholar]
- 18. Enders AC, Lantz KC, Peterson PE, Hendrickx AG. Symposium: reproduction in baboons. From blastocyst to placenta: the morphology of implantation in the baboon. Hum Reprod Update 1997; 3(6):561–573. [DOI] [PubMed] [Google Scholar]
- 19. Deglincerti A, Croft GF, Pietila LN, Zernicka-Goetz M, Siggia ED, Brivanlou AH. Self-organization of the in vitro attached human embryo. Nature 2016; 533(7602):251–254. [DOI] [PubMed] [Google Scholar]
- 20. Shahbazi MN, Jedrusik A, Vuoristo S, Recher G, Hupalowska A, Bolton V, Fogarty NM, Campbell A, Devito LG, Ilic D, Khalaf Y, Niakan KK et al. Self-organization of the human embryo in the absence of maternal tissues. Nat Cell Biol 2016; 18(6):700–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Burton GJ, Jauniaux E. The cytotrophoblastic shell and complications of pregnancy. Placenta 2017; 60:134–139. [DOI] [PubMed] [Google Scholar]
- 22. Bernardo AS, Faial T, Gardner L, Niakan KK, Ortmann D, Senner CE, Callery EM, Trotter MW, Hemberger M, Smith JC, Bardwell L, Moffett A et al. BRACHYURY and CDX2 mediate BMP-induced differentiation of human and mouse pluripotent stem cells into embryonic and extraembryonic lineages. Cell Stem Cell 2011; 9(2):144–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhang P, Li J, Tan Z, Wang C, Liu T, Chen L, Yong J, Jiang W, Sun X, Du L, Ding M, Deng H. Short-term BMP-4 treatment initiates mesoderm induction in human embryonic stem cells. Blood 2008; 111(4):1933–1941. [DOI] [PubMed] [Google Scholar]
- 24. Erb TM, Schneider C, Mucko SE, Sanfilippo JS, Lowry NC, Desai MN, Mangoubi RS, Leuba SH, Sammak PJ. Paracrine and epigenetic control of trophectoderm differentiation from human embryonic stem cells: the role of bone morphogenic protein 4 and histone deacetylases. Stem Cells Dev 2011; 20(9):1601–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sudheer S, Bhushan R, Fauler B, Lehrach H, Adjaye J. FGF inhibition directs BMP4-mediated differentiation of human embryonic stem cells to syncytiotrophoblast. Stem Cells Dev 2012; 21(16):2987–3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yu P, Pan G, Yu J, Thomson JA. FGF2 sustains NANOG and switches the outcome of BMP4-induced human embryonic stem cell differentiation. Cell Stem Cell 2011; 8(3):326–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Das P, Ezashi T, Schulz LC, Westfall SD, Livingston KA, Roberts RM. Effects of fgf2 and oxygen in the bmp4-driven differentiation of trophoblast from human embryonic stem cells. Stem Cell Res 2007; 1(1):61–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lee CQ, Gardner L, Turco M, Zhao N, Murray MJ, Coleman N, Rossant J, Hemberger M, Moffett A. What is trophoblast? a combination of criteria define human first-trimester trophoblast. Stem Cell Rep 2016; 6(2):257–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Okae H, Toh H, Sato T, Hiura H, Takahashi S, Shirane K, Kabayama Y, Suyama M, Sasaki H, Arima T. Derivation of human trophoblast stem cells. Cell Stem Cell 2018; 22(1):50–63.e6. [DOI] [PubMed] [Google Scholar]
- 30. Soncin F, Natale D, Parast MM. Signaling pathways in mouse and human trophoblast differentiation: a comparative review. Cell Mol Life Sci 2015; 72(7):1291–1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Latos PA, Hemberger M. From the stem of the placental tree: trophoblast stem cells and their progeny. Development 2016; 143(20):3650–3660. [DOI] [PubMed] [Google Scholar]
- 32. Leung CY, Zernicka-Goetz M. Mapping the journey from totipotency to lineage specification in the mouse embryo. Curr Opin Genet Dev 2015; 34:71–76. [DOI] [PubMed] [Google Scholar]
- 33. Wennekamp S, Mesecke S, Nedelec F, Hiiragi T. A self-organization framework for symmetry breaking in the mammalian embryo. Nat Rev Mol Cell Biol 2013; 14(7):452–459. [DOI] [PubMed] [Google Scholar]
- 34. Tabansky I, Lenarcic A, Draft RW, Loulier K, Keskin DB, Rosains J, Rivera-Feliciano J, Lichtman JW, Livet J, Stern JN, Sanes JR, Eggan K. Developmental bias in cleavage-stage mouse blastomeres. Curr Biol 2013; 23(1):21–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Plachta N, Bollenbach T, Pease S, Fraser SE, Pantazis P. Oct4 kinetics predict cell lineage patterning in the early mammalian embryo. Nat Cell Biol 2011; 13(2):117–123. [DOI] [PubMed] [Google Scholar]
- 36. White MD, Angiolini JF, Alvarez YD, Kaur G, Zhao ZW, Mocskos E, Bruno L, Bissiere S, Levi V, Plachta N. Long-lived binding of Sox2 to DNA predicts cell fate in the four-cell mouse embryo. Cell 2016; 165(1):75–87. [DOI] [PubMed] [Google Scholar]
- 37. Goolam M, Scialdone A, Graham SJL, Macaulay IC, Jedrusik A, Hupalowska A, Voet T, Marioni JC, Zernicka-Goetz M. Heterogeneity in Oct4 and Sox2 targets biases cell fate in 4-cell mouse embryos. Cell 2016; 165(1):61–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Torres-Padilla ME, Parfitt DE, Kouzarides T, Zernicka-Goetz M. Histone arginine methylation regulates pluripotency in the early mouse embryo. Nature 2007; 445(7124):214–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Roberts RM, Fisher SJ. Trophoblast stem cells. Biol Reprod 2011; 84(3):412–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cockburn K, Rossant J. Making the blastocyst: lessons from the mouse. J Clin Invest 2010; 120(4):995–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Strumpf D, Mao CA, Yamanaka Y, Ralston A, Chawengsaksophak K, Beck F, Rossant J. Cdx2 is required for correct cell fate specification and differentiation of trophectoderm in the mouse blastocyst. Development 2005; 132(9):2093–2102. [DOI] [PubMed] [Google Scholar]
- 42. Russ AP, Wattler S, Colledge WH, Aparicio SA, Carlton MB, Pearce JJ, Barton SC, Surani MA, Ryan K, Nehls MC, Wilson V, Evans MJ. Eomesodermin is required for mouse trophoblast development and mesoderm formation. Nature 2000; 404(6773):95–99. [DOI] [PubMed] [Google Scholar]
- 43. Morasso MI, Grinberg A, Robinson G, Sargent TD, Mahon KA. Placental failure in mice lacking the homeobox gene Dlx3. Proc Natl Acad Sci USA 1999; 96(1):162–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yamamoto H, Flannery ML, Kupriyanov S, Pearce J, McKercher SR, Henkel GW, Maki RA, Werb Z, Oshima RG. Defective trophoblast function in mice with a targeted mutation of Ets2. Genes Dev 1998; 12(9):1315–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tanaka S, Kunath T, Hadjantonakis AK, Nagy A, Rossant J. Promotion of trophoblast stem cell proliferation by FGF4. Science 1998; 282(5396):2072–2075. [DOI] [PubMed] [Google Scholar]
- 46. Uy GD, Downs KM, Gardner RL. Inhibition of trophoblast stem cell potential in chorionic ectoderm coincides with occlusion of the ectoplacental cavity in the mouse. Development 2002; 129:3913–3924. [DOI] [PubMed] [Google Scholar]
- 47. Dey KK, Hsiao CJ, Stephens M. Visualizing the structure of RNA-seq expression data using grade of membership models. PLoS Genet 2017; 13(3):e1006599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Cross JC, Baczyk D, Dobric N, Hemberger M, Hughes M, Simmons DG, Yamamoto H, Kingdom JC. Genes, development and evolution of the placenta. Placenta 2003; 24(2-3):123–130. [DOI] [PubMed] [Google Scholar]
- 49. Latos PA, Sienerth AR, Murray A, Senner CE, Muto M, Ikawa M, Oxley D, Burge S, Cox BJ, Hemberger M. Elf5-centered transcription factor hub controls trophoblast stem cell self-renewal and differentiation through stoichiometry-sensitive shifts in target gene networks. Genes Dev 2015; 29(23):2435–2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yagi R, Kohn MJ, Karavanova I, Kaneko KJ, Vullhorst D, DePamphilis ML, Buonanno A. Transcription factor TEAD4 specifies the trophectoderm lineage at the beginning of mammalian development. Development 2007; 134 (21):3827–3836. [DOI] [PubMed] [Google Scholar]
- 51. Niakan KK, Eggan K. Analysis of human embryos from zygote to blastocyst reveals distinct gene expression patterns relative to the mouse. Dev Biol 2013; 375(1):54–64. [DOI] [PubMed] [Google Scholar]
- 52. Petropoulos S, Edsgard D, Reinius B, Deng Q, Panula SP, Codeluppi S, Reyes AP, Linnarsson S, Sandberg R, Lanner F. Single-cell RNA-Seq reveals lineage and X chromosome dynamics in human preimplantation embryos. Cell 2016; 167(1):285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Blakeley P, Fogarty NM, Del Valle I, Wamaitha SE, Hu TX, Elder K, Snell P, Christie L, Robson P, Niakan KK. Defining the three cell lineages of the human blastocyst by single-cell RNA-seq. Development 2015; 142(20):3613–3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Soncin F, Khater M, To C, Pizzo D, Farah O, Wakeland A, Arul Nambi Rajan K, Nelson KK, Chang C-W, Moretto-Zita M, Natale DR, Laurent LC et al. Comparative analysis of mouse and human placentae across gestation reveals species-specific regulators of placental development. Development 2018; 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science 1998; 282(5391):1145–1147. [DOI] [PubMed] [Google Scholar]
- 56. Brons IG, Smithers LE, Trotter MW, Rugg-Gunn P, Sun B, Chuva de Sousa Lopes SM, Howlett SK, Clarkson A, Ahrlund-Richter L, Pedersen RA, Vallier L. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature 2007; 448(7150):191–195. [DOI] [PubMed] [Google Scholar]
- 57. Tesar PJ, Chenoweth JG, Brook FA, Davies TJ, Evans EP, Mack DL, Gardner RL, McKay RD. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature 2007; 448(7150):196–199. [DOI] [PubMed] [Google Scholar]
- 58. Gafni O, Weinberger L, Mansour AA, Manor YS, Chomsky E, Ben-Yosef D, Kalma Y, Viukov S, Maza I, Zviran A, Rais Y, Shipony Z et al. Derivation of novel human ground state naive pluripotent stem cells. Nature 2013; 504(7479):282–286. [DOI] [PubMed] [Google Scholar]
- 59. Davidson KC, Mason EA, Pera MF. The pluripotent state in mouse and human. Development 2015; 142(18):3090–3099. [DOI] [PubMed] [Google Scholar]
- 60. Yang Y, Liu B, Xu J, Wang J, Wu J, Shi C, Xu Y, Dong J, Wang C, Lai W, Zhu J, Xiong L et al. Derivation of pluripotent stem cells with in vivo embryonic and extraembryonic potency. Cell 2017; 169(2):243–257.e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Xu RH, Chen X, Li DS, Li R, Addicks GC, Glennon C, Zwaka TP, Thomson JA. BMP4 initiates human embryonic stem cell differentiation to trophoblast. Nat Biotechnol 2002; 20(12):1261–1264. [DOI] [PubMed] [Google Scholar]
- 62. Lichtner B, Knaus P, Lehrach H, Adjaye J. BMP10 as a potent inducer of trophoblast differentiation in human embryonic and induced pluripotent stem cells. Biomaterials 2013; 34(38):9789–9802. [DOI] [PubMed] [Google Scholar]
- 63. Vallier L, Touboul T, Chng Z, Brimpari M, Hannan N, Millan E, Smithers LE, Trotter M, Rugg-Gunn P, Weber A, Pedersen RA. Early cell fate decisions of human embryonic stem cells and mouse epiblast stem cells are controlled by the same signalling pathways. PLoS One 2009; 4(6):e6082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ezashi T, Telugu BP, Roberts RM. Model systems for studying trophoblast differentiation from human pluripotent stem cells. Cell Tissue Res 2012; 349(3):809–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. James D, Levine AJ, Besser D, Hemmati-Brivanlou A. TGF /activin/nodal signaling is necessary for the maintenance of pluripotency in human embryonic stem cells. Development 2005; 132(6):1273–1282. [DOI] [PubMed] [Google Scholar]
- 66. Amita M, Adachi K, Alexenko AP, Sinha S, Schust DJ, Schulz LC, Roberts RM, Ezashi T. Complete and unidirectional conversion of human embryonic stem cells to trophoblast by BMP4. Proc Natl Acad Sci USA 2013; 110(13):E1212–E1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Chen Y, Wang K, Chandramouli GV, Knott JG, Leach R. Trophoblast lineage cells derived from human induced pluripotent stem cells. Biochem Biophys Res Commun 2013; 436(4):677–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Warmflash A, Sorre B, Etoc F, Siggia ED, Brivanlou AH. A method to recapitulate early embryonic spatial patterning in human embryonic stem cells. Nat Methods 2014; 11(8):847–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Liu LX, Zeng H, Liu EY, Chen FP. Tissue factor expression and methylation regulation in differentiation of embryonic stem cells into trophoblast. Asian Pacific J Trop Med 2014; 7(7):557–561. [DOI] [PubMed] [Google Scholar]
- 70. Li Y, Moretto-Zita M, Soncin F, Wakeland A, Wolfe L, Leon-Garcia S, Pandian R, Pizzo D, Cui L, Nazor K, Loring JF, Crum CP et al. BMP4-directed trophoblast differentiation of human embryonic stem cells is mediated through a Δ Np63+ cytotrophoblast stem cell state. Development 2013; 140(19):3965–3976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Wu Z, Zhang W, Chen G, Cheng L, Liao J, Jia N, Gao Y, Dai H, Yuan J, Cheng L, Xiao L. Combinatorial signals of activin/nodal and bone morphogenic protein regulate the early lineage segregation of human embryonic stem cells. J Biol Chem 2008; 283(36):24991–25002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Shpiz A, Kalma Y, Frumkin T, Telias M, Carmon A, Amit A, Ben-Yosef D. Human embryonic stem cells carrying an unbalanced translocation demonstrate impaired differentiation into trophoblasts: an in vitro model of human implantation failure. Mol Hum Reprod 2015; 21(3):271–280. [DOI] [PubMed] [Google Scholar]
- 73. Shpiz A, Ben-Yosef D, Kalma Y. Impaired function of trophoblast cells derived from translocated hESCs may explain pregnancy loss in women with balanced translocation (11;22). J Assist Reprod Genet 2016; 33(11):1493–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Kojima J, Fukuda A, Taira H, Kawasaki T, Ito H, Kuji N, Isaka K, Umezawa A, Akutsu H. Efficient production of trophoblast lineage cells from human induced pluripotent stem cells. Lab Invest 2017; 97(10):1188–1200. [DOI] [PubMed] [Google Scholar]
- 75. Wu Z, Zhang W, Chen G, Cheng L, Liao J, Jia N, Gao Y, Dai H, Yuan J, Xiao L. Combinatorial signals of activin/nodal and bone morphogenic protein regulate the early lineage segregation of human embryonic stem cells. J Biol Chem 2008; 283(36):24991–25002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Sarkar P, Randall SM, Collier TS, Nero A, Russell TA, Muddiman DC, Rao BM. Activin/nodal signaling switches the terminal fate of human embryonic stem cell-derived trophoblasts. J Biol Chem 2015; 290(14):8834–8848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Marchand M, Horcajadas JA, Esteban FJ, McElroy SL, Fisher SJ, Giudice LC. Transcriptomic signature of trophoblast differentiation in a human embryonic stem cell model. Biol Reprod 2011; 84(6):1258–1271. [DOI] [PubMed] [Google Scholar]
- 78. Rungsiwiwut R, Numchaisrika P, Ahnonkitpanit V, Virutamasen P, Pruksananonda K. Triploid human embryonic stem cells derived from tripronuclear zygotes displayed pluripotency and trophoblast differentiation ability similar to the diploid human embryonic stem cells. J Reprod Dev 2016; 62(2):167–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Wei Y, Zhou X, Huang W, Long P, Xiao L, Zhang T, Zhong M, Pan G, Ma Y, Yu Y. Generation of trophoblast-like cells from the amnion in vitro: a novel cellular model for trophoblast development. Placenta 2017; 51:28–37. [DOI] [PubMed] [Google Scholar]
- 80. Koel M, Vosa U, Krjutskov K, Einarsdottir E, Kere J, Tapanainen J, Katayama S, Ingerpuu S, Jaks V, Stenman UH, Lundin K, Tuuri T et al. Optimizing bone morphogenic protein 4-mediated human embryonic stem cell differentiation into trophoblast-like cells using fibroblast growth factor 2 and transforming growth factor-β/activin/nodal signalling inhibition. Reprod Biomed Online 2017; 35(3):253–263. [DOI] [PubMed] [Google Scholar]
- 81. Krendl C, Shaposhnikov D, Rishko V, Ori C, Ziegenhain C, Sass S, Simon L, Muller NS, Straub T, Brooks KE, Chavez SL, Enard W et al. GATA2/3-TFAP2A/C transcription factor network couples human pluripotent stem cell differentiation to trophectoderm with repression of pluripotency. Proc Natl Acad Sci USA 2017; 114(45):E9579–E9588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Richter A VL, Hrafnkelsdottir HE, Runarsson JF, Omarsdottir AR, Ward-van Oostwaard D, Mummery C, Valdimarsdottir G. BMP4 promotes EMT and mesodermal commitment in human embryonic stem cells via SLUG and MSX2. Stem Cells 201432(3):636–648. [DOI] [PubMed] [Google Scholar]
- 83. Roberts RM, Loh KM, Amita M, Bernardo AS, Adachi K, Alexenko AP, Schust DJ, Schulz LC, Telugu BP, Ezashi T, Pedersen RA. Differentiation of trophoblast cells from human embryonic stem cells: to be or not to be? Reproduction 2014; 147(5):D1–D12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Yabe S, Alexenko AP, Amita M, Yang Y, Schust DJ, Sadovsky Y, Ezashi T, Roberts RM. Comparison of syncytiotrophoblast generated from human embryonic stem cells and from term placentas. Proc Natl Acad Sci USA 2016; 113(19):E2598–E2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Jain A, Ezashi T, Roberts RM, Tuteja G. Deciphering transcriptional regulation in human embryonic stem cells specified towards a trophoblast fate. Sci Rep 2017; 7(1):17257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson A, Kampf C, Sjostedt E, Asplund A, Olsson I, Edlund K et al. Tissue-based map of the human proteome. Science 2015; 347(6220):1260419–1260419. [DOI] [PubMed] [Google Scholar]
- 87. Schulz LC, Ezashi T, Das P, Westfall SD, Livingston KA, Roberts RM. Human embryonic stem cells as models for trophoblast differentiation. Placenta 2008; 29(Suppl A):10–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Horii M, Li Y, Wakeland AK, Pizzo DP, Nelson KK, Sabatini K, Laurent LC, Liu Y, Parast MM. Human pluripotent stem cells as a model of trophoblast differentiation in both normal development and disease. Proc Natl Acad Sci USA 2016; 113:E3882–E3891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Liu Y, Ding D, Liu H, Sun X. The accessible chromatin landscape during conversion of human embryonic stem cells to trophoblast by bone morphogenetic protein 4. Biol Reprod 2017; 96:1267–1278. [DOI] [PubMed] [Google Scholar]
- 90. Ying QL, Nichols J, Chambers I, Smith A. BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell 2003; 115:281–292. [DOI] [PubMed] [Google Scholar]
- 91. Hassani SN, Totonchi M, Sharifi-Zarchi A, Mollamohammadi S, Pakzad M, Moradi S, Samadian A, Masoudi N, Mirshahvaladi S, Farrokhi A, Greber B, Arauzo-Bravo MJ et al. Inhibition of TGFβ signaling promotes ground state pluripotency. Stem Cell Rev 2014; 10:16–30. [DOI] [PubMed] [Google Scholar]
- 92. Graham SJ, Wicher KB, Jedrusik A, Guo G, Herath W, Robson P, Zernicka-Goetz M. BMP signalling regulates the pre-implantation development of extra-embryonic cell lineages in the mouse embryo. Nat Commun 2014; 5:5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Yang Y, Adachi K, Sheridan MA, Alexenko AP, Schust DJ, Schulz LC, Ezashi T, Roberts RM. Heightened potency of human pluripotent stem cell lines created by transient BMP4 exposure. Proc Natl Acad Sci USA 2015; 112:E2337–E2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Morgani SM, Canham MA, Nichols J, Sharov AA, Migueles RP, Ko MS, Brickman JM. Totipotent embryonic stem cells arise in ground-state culture conditions. Cell Rep 2013; 3:1945–1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Abad M, Mosteiro L, Pantoja C, Canamero M, Rayon T, Ors I, Grana O, Megias D, Dominguez O, Martinez D, Manzanares M, Ortega S et al. Reprogramming in vivo produces teratomas and iPS cells with totipotency features. Nature 2013; 502:340–345. [DOI] [PubMed] [Google Scholar]
- 96. Bayer A, Lennemann NJ, Ouyang Y, Sadovsky E, Sheridan MA, Roberts RM, Coyne CB, Sadovsky Y. Chromosome 19 microRNAs exert antiviral activity independent from type III interferon signaling. Placenta 2018; 61:33–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Telugu BP, Adachi K, Schlitt JM, Ezashi T, Schust DJ, Roberts RM, Schulz LC. Comparison of extravillous trophoblast cells derived from human embryonic stem cells and from first trimester human placentas. Placenta 2013; 34:536–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Sheridan MA, Yunusov D, Balaraman V, Alexenko AP, Yabe S, Verjovski-Almeida S, Schust DJ, Franz AW, Sadovsky Y, Ezashi T, Roberts RM. Vulnerability of primitive human placental trophoblast to Zika virus. Proc Natl Acad Sci USA 2017; 114:E1587–E1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. James JL, Carter AM, Chamley LW. Human placentation from nidation to 5 weeks of gestation. Part I: What do we know about formative placental development following implantation? Placenta 2012; 33:327–334. [DOI] [PubMed] [Google Scholar]
- 100. Ozawa M, Sakatani M, Hankowski KE, Terada N, Dobbs KB, Hansen PJ. Importance of culture conditions during the morula-to-blastocyst period on capacity of inner cell-mass cells of bovine blastocysts for establishment of self-renewing pluripotent cells. Theriogenology 2012; 78:1243–1251.e2. [DOI] [PubMed] [Google Scholar]
- 101. Boyd PA, Brown RA, Coghill GR, Slidders W, Stewart WJ. Measurement of the mass of syncytiotrophoblast in a range of human placentae using an image analysing computer. Placenta 1983; 4:255–262. [DOI] [PubMed] [Google Scholar]
- 102. Boime I, Boothby M, Hoshina M, Daniels-McQueen S, Darnell R. Expression and structure of human placental hormone genes as a function of placental development. Biol Reprod 1982; 26:73–91. [DOI] [PubMed] [Google Scholar]
- 103. Alvarez M, Guzman MG, Pupo M, Morier L, Bravo J, Rodriguez R. Study of biologic attributes of Cuban dengue 2 virus after serial passage in primary dog kidney cells. Int J Infect Dis 2001; 5:35–39. [DOI] [PubMed] [Google Scholar]
- 104. Roberts RM, Yabe S, Yang Y, Ezashi T. A human stem cell model for creating placental syncytiotrophoblast, the major cellular barrier that limits fetal exposure to xenobiotics. In: Sahu SC. (ed.) Stem Cells in Toxicology and Medicine 1st ed: Wiley & Sons; 2016: 179–195. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.