Abstract
Background
Next-generation sequencing (NGS) is a useful tool for detecting genomic alterations in circulating tumor DNA (ctDNA). To date, most ctDNA tests have been performed on patients with widely metastatic disease. Patients with peritoneal carcinomatosis (metastases) present unique prognostic and therapeutic challenges. We therefore explored preoperative ctDNA in patients with peritoneal metastases undergoing surgery.
Methods
Patients referred for surgical resection of peritoneal metastases underwent preoperative blood-derived ctDNA analysis (clinical-grade NGS (68 to 73 genes)). ctDNA was quantified as the percentage of altered circulating cell-free DNA (% cfDNA).
Results
Eighty patients had ctDNA testing: 46 (57.5%) women; median age, 55.5 years. The following diagnoses were included: 59 patients (73.8%), appendix cancer; 11 (13.8%), colorectal; five (6.3%), peritoneal mesothelioma; two (2.5%), small bowel; one (1.3%), each of cholangiocarcinoma, ovarian, and testicular cancer. Thirty-one patients (38.8%) had detectable preoperative ctDNA alterations, most frequently in the following genes: TP53 (25.8% of all alterations detected) and KRAS (11.3%). Among 15 patients with tissue DNA NGS, 33.3% also had ctDNA alterations (overall concordance = 96.7%). Patients with high ctDNA quantities (≥ 0.25% cfDNA, n=25) had a shorter progression-free survival (PFS) than those with lower ctDNA quantities (n=55; 7.8 vs. 15.0 months; hazard ratio (95% confidence interval), 3.23 (1.43 to 7.28), P = 0.005, univariate; (p = 0.044, multivariate)).
Conclusions
A significant proportion of patients with peritoneal metastases referred for surgical intervention have detectable ctDNA alterations preoperatively. Patients with high levels of ctDNA have a worse prognosis independent of histologic grade.
INTRODUCTION
Detection and investigation of molecular alterations in cancer has led to advances in understanding of tumor biology, use of targeted cancer therapies, and provides potential assessment of response to therapy.1–3 Molecular tests from tumor tissue require biopsies or resection, are often performed on archival tissue, and do not represent the heterogeneous genomic constitution of many malignancies.4,5 Cell-free circulating tumor DNA (ctDNA) is measurable in the plasma using next-generation sequencing (NGS) techniques, and is less invasive than obtaining tumor-derived genetic material.6 The half-life of cell-free DNA (cfDNA) in the bloodstream is between 16 minutes and 2.5 hours, such that it may serve as a ‘real-time’ snapshot of the of the genomic status of a patient’s cancer. cfDNA is rapidly lysed in the circulation via nucleases, followed by primarily renal excretion as well as uptake by the liver and spleen, where it is ultimately degradaded by macrophages.7 This reproducible, minimally invasive, and spatially unbiased technology has been used to identify targeted therapies, measure residual disease, and assess response to therapy.8–10
The peritoneum is a common site of tumor metastasis, with approximately 55,000 new cases of peritoneal metastasis occurring annually in the US.11 Patients with peritoneal carcinomatosis (metastases) have an uncertain, but often poor prognosis, which is influenced by tumor histology and extent of disease.12 Treatment options for peritoneal metastases are limited as many peritoneal tumors are poorly vascularized and can be surrounded by a viscous layer of mucin, thus limiting drug delivery.13 Surgical approaches with complete cytoreductive surgery (CRS) and intraperitoneal chemotherapy in select patients and histologies (including appendiceal, colorectal, and ovarian cancers, and peritoneal mesothelioma) with disease confined to the peritoneal cavity have demonstrated improved outcomes versus treatment with systemic chemotherapy, or in some cases cytoreduction, alone.14–19 Imaging of peritoneal metastases for operative candidacy and recurrence is imprecise and often underestimates the actual disease burden.20,21 Given the difficulty in treatment, imaging, and estimation of prognosis in patients with peritoneal metastases, use of a reliable, non-invasive, molecular diagnostic test has potentially high utility in this disease; although it is not known whether DNA shed from peritoneal metastases reach the systemic circulation in sufficient quantity to serve as a potential predictive or prognostic biomarker.22
Herein, we sought to determine the rate of detection and type of ctDNA alterations in patients with peritoneal metastases undergoing surgical resection.
MATERIALS AND METHODS
Patients
This is a prospective study of ctDNA analysis in patients referred for surgical management of peritoneal metastases. All patients had potentially resectable peritoneal metastases, and were referred to our institution’s peritoneal malignancy program for consideration for cytoreductive surgery (CRS) and hyperthermic intraperitoneal chemotherapy (HIPEC) from May 20th, 2015, to January 1st, 2017. This study was approved by the University of California San Diego (UCSD) institutional review board (IRB) and patients gave consent prior to study enrollment in accordance with IRB guidelines.
Procedures and Follow-Up
Patients were taken to the operating room with the intent of removing peritoneal metastases. If all or nearly all visible disease could be removed, then patients typically underwent cytoreductive surgery and intraperitoneal chemotherapy, per standardized techniques at our institution;23 which include resection of tumor nodules and/or involved viscera followed by 90 minute hyperthermic peritoneal chemoperfusion with mitomycin C (for colorectal or appendiceal primary tumors) or doxorubicin/cisplatin (for ovarian cancer and peritoneal mesothelioma). If complete cytoreduction was not possible, patients underwent palliative debulking procedures, with the goal of minimizing peritoneal metastasis-related symptoms and morbidity. Complete resection was defined as removal of all visible (gross) peritoneal metastases.
Histology of the resected specimen(s) was categorized by grade, with low-grade histology including patients with acellular mucin and low-grade mucinous carcinoma peritonei; and high-grade histology including patients with any grade of adenocarcinoma or high-grade mucinous carcinoma peritonei. Mesothelioma cases were not included in the grade analyses.
Postoperative treatment and surveillance were at the discretion of the referring oncologist; and due to referral patterns, complete information of all subsequent care and observation was not available for every patient. However, patients generally underwent surveillance imaging every three to six months after surgery. Postoperative systemic treatment was typically administered in those patients with high-grade malignancies with incomplete resections or without sufficient preoperative systemic treatment.
Next Generation Sequencing
Next generation digital sequencing of cell-free ctDNA extracted from plasma was performed by Guardant Health (Guardant360, www.guardanthealth.com/guardant360/), a Clinical Laboratory Improvement Amendments (CLIA)-certified, College of American Pathologists (CAP)-accredited, and New York State Department of Health-approved clinical laboratory (Guardant Health, Inc., Redwood City, CA). At the time of this study, this test identified potential tumor-related genomic alterations via exon sequencing of 68 to 73 cancer-related genes, 18 gene amplifications, 6 gene fusions, and 23 gene insertion/deletions. This assay has high analytic sensitivity (detects single molecules of somatic tumor DNA in 10 mL blood samples), high clinical sensitivity (detects 85%+ of the single nucleotide variants detected in tissue in advanced cancer patients), and high analytic specificity (>99.9999%).24 All cell-free DNA (cfDNA) was sequenced, including the germline cell-free DNA that is derived from leukocyte lysis and the somatic cfDNA. Single nucleotide variants were quantitated as the mutant allele fraction which is the number of ctDNA fragments divided by the number of wild type DNA fragments that overlap the same mutated nucleotide base position. Gene amplifications were reported as absolute gene copy number in plasma. In each sequencing run, a normal control sample was included. Each ctDNA alteration was quantified as a percentage, or allele frequency, of circulating cell-free DNA (% cfDNA). Variants of uncertain significance (VUS) were included in the ctDNA analysis in order to capture the most comprehensive DNA alteration profile. These alterations have uncertain functional consequences and clinical significance; but they may be reflective of tumor growth, turn-over, size, heterogeneity, vascularization, disease progression, or treatment. All samples were drawn prior to operative management of peritoneal metastases, typically 1–2 weeks before the operation.
Some patients also underwent tissue genomic analysis from resected peritoneal tumors, performed at the discretion of their treating clinician, which were compared with the ctDNA alterations. Tissue DNA analysis was performed from formalin-fixed, paraffin-embedded (FFPE) tumor samples in a CLIA-certified clinical laboratory for genomic profiling (Foundation Medicine, Cambridge, MA). At the time of this study, this test analyzes the entire coding sequence of 315 cancer-related genes and select introns from 28 genes.
Statistics
Continuous variables were reported as median and ranges, and dichotomous and categorical variables were reported as proportions in each group. Progression-free survival (PFS) was calculated from the time of operation to progression (as determined by RECIST on subsequent imaging, endoscopy, or operation) or death, and was analyzed by the Kaplan-Meier method. Univariate and multivariate Cox proportional hazards model were performed to identify predictors of PFS. A P-value of < 0.05 was considered statistically significant. Concordance of ctDNA and tissue DNA alterations (by gene altered) was calculated as follows, considering variants reported on both assays: overall concordance was the sum of present and absent alterations found in tissue and blood among all possible concordant alterations; positive concordance was the number of present alterations in tissue and blood among the number of any present alterations in tissue or blood; and negative concordance was the number of absent alterations in tissue and blood among any absent alteration in tissue or blood. All statistical analyses were performed using SPSS Version 24 (IBM Corp., Armonk, NY) and R Version 3.1.2 (R Foundation for Statistical Computing, Vienna, Austria, http://www.R-project.org/).
RESULTS
Patient and ctDNA Characteristics
This analysis included 80 consecutive patients who had ctDNA analyzed and underwent surgery for peritoneal metastases. Baseline and ctDNA characteristics are listed in Table 1. ctDNA assessments were made a median of 4.0 days prior to surgery (range 0–108 days).
Table 1.
Baseline patient characteristics and ctDNA
| Variable | n (%) |
|---|---|
| Total Patients | 80 (100) |
| Age (yrs) (median, range) | 55.5 (26–73) |
| Number of patients with alterations (includes variants of unknown significance) (%) | 31 (38.8%) |
| Gender | |
| Men | 34 (42.5) |
| Women | 46 (57.5) |
| Procedure | |
| Colectomy | 1 (1.3) |
| CRS/HIPEC | 53 (66.3) |
| Palliative Debulking | 26 (32.5) |
| Complete Resection1 | |
| Yes | 33 (41.3) |
| No | 47 (58.8) |
| Detectable ctDNA Alteration2 | |
| All Patients (n=80) | 31 (38.8)3 |
| Appendiceal Cancer (n=59) | 21 (35.6) |
| Low-Grade Carcinomatosis (n=34) | 9 (26.5) |
| High-Grade Carcinomatosis (n=25) | 12 (48.0) |
| Colon Adenocarcinoma (n=11) | 7 (63.6) |
| Malignant Mesothelioma (n=5) | 2 (40.0) |
| Small Bowel Adenocarcinoma (n=2) | 0 (0) |
| Cholangiocarcinoma (n=1) | 0 (0) |
| Ovarian Cancer (n=1) | 0 (0) |
| Testicular Cancer (n=1) | 1 (100) |
Resection to no residual gross disease (R0/R1)
ctDNA alterations include characterized alterations and variants of unknown significance
% of patients with designated cancer with ctDNA alteration
Mutational Landscape
Altered genes by ctDNA among all cancers, and among the two most common primary sites are listed in Table 2. The quantity of ctDNA for each gene alteration is listed as the median percentage of altered cell-free DNA (% cfDNA). Taking into account the highest % cfDNA of any alteration in each patient (peak % cfDNA), the median value among all patients was 0% (61.2% had no ctDNA alteration), and the mean peak % cfDNA was 0.5%. Among patients with ctDNA detected (n = 31), the median peak % cfDNA was 0.30% (range 0.1 – 10.1). Dividing the entire cohort into high and low % cfDNA groups by comparing patients harboring the highest one-third of peak % cfDNA levels with others resulted in groups containing < or ≥ 0.25% cfDNA. There were 24 distinct genes altered in the entire cohort, including 59 distinct genomic alterations, with a median number of two alterations per patient. The two most common genes altered were TP53 (n=16, 25.8% of all alterations) and KRAS (n=7, 11.3%).
Table 2.
Altered Genes and Proportion of Cell-Free DNA (n=31 patients with detectable alterations)
| Gene | Alterations, n (%1) | Median % cfDNA (range) | ||
|---|---|---|---|---|
| All Cancers (n=31) | Appendiceal Cancer (n=21) | Colon Cancer (n=7) | ||
| TP53 | 16 (25.8) | 12 (32.4) | 3 (15.0) | 0.40 (0.10–4.00) |
| KRAS | 7 (11.3) | 3 (8.1) | 4 (20.0) | 0.52 (0.20–1.65) |
| EGFR | 4 (6.5) | 3 (8.1) | 1 (5.0) | 0.13 (0.10–0.30) |
| NF1 | 4 (6.5) | 4 (10.8) | 0.65 (0.40–4.10) | |
| TERT | 4 (6.5) | 3 (8.1) | 1 (5.0) | 4.46 (0.30–10.10) |
| APC | 3 (4.8) | 1 (2.7) | 1 (5.0) | 0.10 (0.10–0.40) |
| GNAS | 3 (4.8) | 1 (2.7) | 1 (5.0) | 0.20 (0.20–3.60) |
| ARID1A | 2 (3.2) | 1 (2.7) | 1 (5.0) | 0.80 (0.20–1.40) |
| ATM | 2 (3.2) | 1 (2.7) | 1 (5.0) | 0.80 (0.30–1.30) |
| MET | 2 (3.2) | 1 (5.0) | 0.40 (0.20–0.60) | |
| PIK3CA | 2 (3.2) | 1 (2.7) | 1 (5.0) | 0.75 (0.10–1.40) |
| ARAF | 1 (1.6) | 1 (2.7) | 0.3 | |
| CCND1 | 1 (1.6) | 1 (2.7) | Amplification | |
| CCND2 | 1 (1.6) | 1 (5.0) | Amplification | |
| ERBB2 | 1 (1.6) | 1 (2.7) | 0.2 | |
| FGFR2 | 1 (1.6) | 1 (2.7) | 0.2 | |
| HNF1A | 1 (1.6) | 0.3 | ||
| JAK2 | 1 (1.6) | 1 (2.7) | 1.8 | |
| MYC | 1 (1.6) | 1 (5.0) | Amplification | |
| NOTCH | 1 (1.6) | 1 (5.0) | 1.42 | |
| PDGFRA | 1 (1.6) | 1 (2.7) | 0.4 | |
| RAF1 | 1 (1.6) | 1 (2.7) | 0.1 | |
| RIT1 | 1 (1.6) | 1 (5.0) | 0.1 | |
| SMAD4 | 1 (1.6) | 1 (5.0) | 0.2 | |
| Total Alterations | 62 | 37 | 20 | 0.30 (0.10–10.10) |
| Median Number Alterations/Patient (range) | 1 (1–9) | 1 (1–9) | 2 (1–6) | |
| Total Number Distinct Alterations | 59 | 36 | 19 | |
| Total Number Distinct Genes Altered | 24 | 17 | 15 | |
% of total alterations in designated cancer types; alterations include characterized alterations and VUS
Patients were stratified into high-grade (n = 39) versus low-grade (n = 36) tumors. The median number (range) of alterations per patient in high-grade versus low-grade tumors was 1 (0 to 9) versus 0 (0 to 3) (P = 0.010, student’s t test). The median (range) peak % cfDNA was 0.20 (0 to 10.01) versus 0 (0 to 4.10) (P = 0.276, student’s t test).
The extent of peritoneal disease (as measured by the peritoneal cancer index25), mucinous tumors, the presence of signet ring cells, and neoadjuvant chemotherapy within three months of surgery did not correlate with the level (< or ≥ 0.25% cfDNA) of ctDNA (by univariate logistic regression).
Concordance of Tissue and ctDNA Genetic Alterations
Fifteen patients underwent tissue DNA NGS from tumor removed during the index operation (procedure following ctDNA measurement). Results of tissue DNA and ctDNA alterations are shown in Table 3 by the gene(s) altered. Among the nine appendiceal primary patients that underwent tissue genomic profiling, three had low-grade disease (patients 4, 9, and 13 in Table 3). The median time between tissue acquisition and blood draw was 3.0 days (range, 1 to 108 days). Three patients were tested with a 68 gene ctDNA panel, one patient was tested with a 70 gene ctDNA panel, and one with a 73 gene ctDNA panel. Five patients with tissue alterations also had alterations in the blood (33.3% of 15 patients). All five patients with ctDNA alterations also had tissue alterations. There were a total of 17 variants identified that were covered by both the ctDNA panel and the tissue panel. An additional 15 variants were identified by tissue NGS, but these genes/regions were not covered in the ctDNA panel, and an additional one variant identified in ctDNA was not covered by the tissue NGS panel. Of the 17 alterations covered by both test types, 6 of 17 (35.3%) were concordant in both sample types, 7 of 17 (41.2%) were detected only in tissue, and 4 of 17 (23.5%) were detected only in ctDNA. Among patients with detectable ctDNA alterations (n = 5), the overall, positive, and negative concordance was 96.7%, 35.3%, and 96.6%, respectively, with tissue DNA alterations.
Table 3.
Tumor Tissue Genomic Profiles (n=15 patients)*
| Patient | Primary Cancer | Tissue DNA Alterations1 | ctDNA Alterations1 | |
|---|---|---|---|---|
| Not in ctDNA Panel | In ctDNA Panel | |||
| 1 | Colon |
KRAS, APC, PIK3CA |
KRAS, NOTCH, TP53 |
|
| 2 | Appendix | GNAS | GNAS | |
| 3 | Colon | PIK3R1 |
TP53 APC, KRAS |
TP53 RIT1 (not in tissue panel) |
| 4 | Appendix | GNAS | ||
| 5 | Appendix | RBM10 |
KRAS,
SMAD4, TP53 |
|
| 6 | Small Bowel | CHEK2 | KRAS | |
| 7 | Appendix | SMAD2 | KRAS | |
| 8 | Appendix |
MAP2K4,
SPTA1, TGFBR2 |
GNAS,
KRAS |
|
| 9 | Appendix | TGFBR2 |
GNAS,
KRAS, PIK3CA |
|
| 10 | Appendix |
KRAS,
SMAD4, TP53 |
||
| 11 | Appendix | TGFBR2 |
GNAS,
KRAS |
|
| 12 | Colon | AKT3, BARD1, FGF23, FGF6, KDM5A, KDM6A, |
CCND2,
KRAS, MYC |
CCND2, KRAS, MET, MYC |
| 13 | Appendix | TGFBR2 |
GNAS,
KRAS |
|
| 14 | Colon | BTG1, CARD11, CREBBP, FAM123B, PIK3R2, PMS2, SOX9, TGFBR2 |
APC, KRAS, TP53 |
APC, KRAS |
| 15 | Bile Duct |
CDKN2A,
KRAS, MTOR, TP53 |
||
Tissue NGS was performed at Foundation Medicine based on tissue obtained at the time of index surgery. ctDNA was generally obtained within two weeks prior to surgery.
Include characterized alterations and VUS
Correlation of ctDNA and Progression-Free Survival
Median follow-up for the entire cohort was 6.8 months (range 0.1–27.9 months). The median progression-free survival (PFS) in the entire cohort was 14.3 months (95% CI, 10.5 to 18.1 months). Median PFS was 15.0 months for those with low (< 0.25% cfDNA) ctDNA levels versus 7.8 months for those with high (≥ 0.25% cfDNA) ctDNA levels (HR 3.23, CI 95% 1.43 to 7.28, P = 0.005; Figure 1). PFS for high versus low ctDNA levels among subgroups (incomplete/incomplete resections, low/high-grade histology) is also shown in Figure 1.
Figure 1. Progression-Free Survival (n=80 patients).
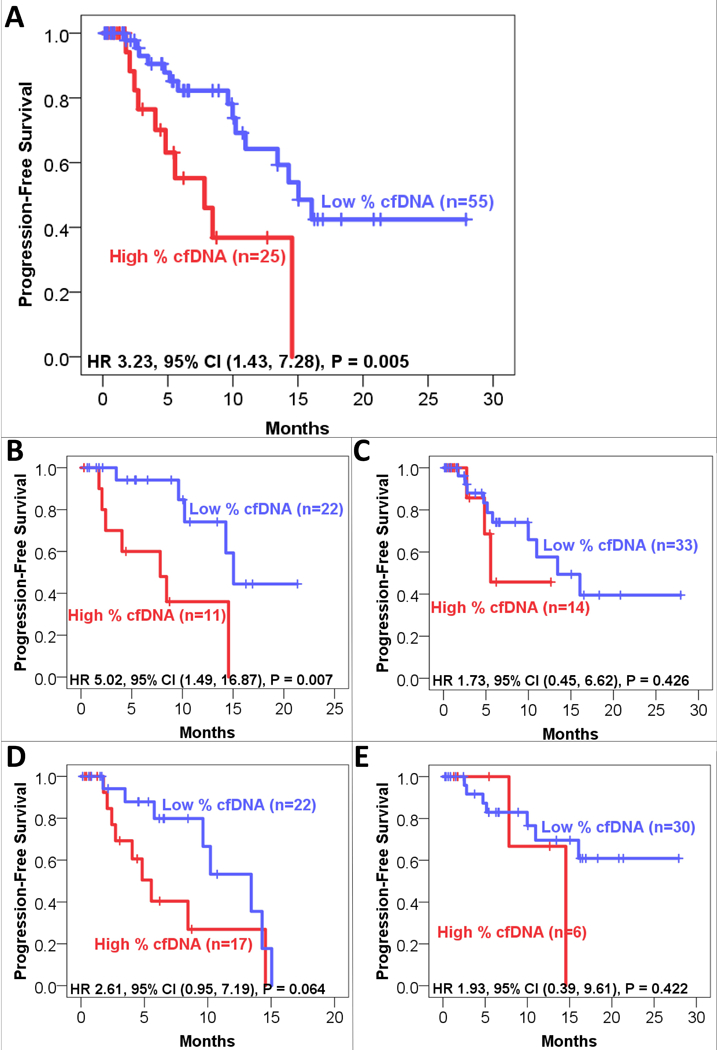
Kaplan-Meier analysis of progression-free survival (PFS) in patients with high (≥ 0.25%) or low (<0.25%) ctDNA level, measured as the alteration with highest percentage of altered cfDNA (peak % cfDNA). Patients who had not progressed at the time of last follow-up were censored (tick marks). A. All patients. B. Patients with complete resection. C. Patients in whom complete resection was not possible. D. Patients with high-grade histology. E. Patients with low-grade histology.
Univariate and multivariate analyses by Cox proportional hazards model were performed to identify predictors of PFS (Table 4). On univariate analysis, high ctDNA (> 0.25% cfDNA) and high-grade histology were predictors of worse PFS (HR 3.23 for high vs. low levels ctDNA, P = 0.005; HR 1.99 for high- vs. low-grade, P = 0.002). These two factors remained independent predictors of PFS on multivariate analysis (HR 2.36 for high vs. low levels ctDNA, P = 0.044; HR 3.30 for high- vs. low-grade, P = 0.009). Excluding the 15 patients who had ctDNA drawn greater than two weeks prior to surgery, high ctDNA levels still correlated with worse PFS by univariate Cox proportional hazards model (HR 4.18, p=0.006). Analyzing the entire cohort by the presence/absence of ctDNA (n=31/49), we found the presence of ctDNA correlated with worse PFS (HR 2.32, p=0.035) by univariate analysis.
Table 4.
Univariate and Multivariate Predictors of PFS (by Cox Proportional Hazards Model)*.
| Variable | Univariate Analysis | Multivariate Analysis | |||
|---|---|---|---|---|---|
| Median PFS (months) (95% CI) | HR (95% CI) | P-value | HR (95% CI) | P-value | |
| Preoperative ctDNA Alterations % cfDNA1 | |||||
| < 0.25% (n=55) | 15.0 (11.8, 18.3) | 3.23 (1.43, 7.28) | 0.005 | 2.36 (1.02, 5.45) | 0.044 |
| ≥ 0.25% (n=25) | 7.8 (3.6, 12.0) | ||||
| Complete Resection | |||||
| Yes (n=33) | 14.5 (10.3, 18.8) | 0.96 (0.65, 1.41) | 0.837 | ||
| No (n=47) | 13.4 (6.5, 20.4) | ||||
| Grade2 | |||||
| Low (n=36) | Not reached | 1.99 (1.29, 3.09) | 0.002 | 3.30 (1.34, 8.11) | 0.009 |
| High (n=39) | 9.6 (7.1, 12.1) | ||||
Multivariate model was developed using variables that were significant (P < 0.05) in univariate analysis. All HRs are given for ≥ 0.25% vs. < 0.25 peak % cfDNA, No vs. Yes Complete Resection, and High vs. Low Grade, respectively
ctDNA as highest % of altered circulating cell-free DNA (peak % cfDNA)
Excluding patients with mesothelioma
Abbreviations: cfDNA = cell free DNA; CI = confidence interval; HR = hazard ratio; PFS = progression-free survival
DISCUSSION
Repeatable, minimally invasive, cancer-specific biomarkers such as ctDNA allow potential opportunities for diagnosis, treatment, and surveillance of malignancy. In this study of patients with peritoneal metastases from a variety of primary tumors referred for surgical management, we found detectable ctDNA alterations in 38.8%. This is the first study evaluating ctDNA in patients with peritoneal metastases treated with surgical resection. The rate of detectable alterations and the quantity of ctDNA varied by histology, consistent with other studies.26 Detectable ctDNA alterations also vary by stage of disease, and patients with peritoneal metastases are thought to have higher levels than those with localized disease.27 The two most common alterations - in TP53 and KRAS - are typical of gastrointestinal malignancies and represent the most commonly mutated genes in human cancer.28 Furthermore, inactivation of TP53 is typically a truncal driver event, and shed from all tumor cells.29 Thus, it would be expected to be at highest % cfDNA and most likely to be detectable. Overall, the levels of ctDNA in our patients were significantly lower than reported previously, possibly because prior studies focused on patients with widely metastatic disease and high tumor burden.30,31
We found high concordance rates with tissue DNA alterations (96.7% overall concordance), among those patients in whom tissue NGS was obtained, which has also been shown in other studies.32 However, the positive concordance rates were overall much lower (35.3%) than the negative concordance rate (96.6%). Differences between tissue and circulating tumor-derived DNA alterations may be due, in part, to tumor heterogeneity, tumor cellularity, and the extent of tumor cell DNA shedding. Indeed, ctDNA analysis may not suffer from the sampling bias of tissue-based genomic analyses, since tumor DNA shed into the blood derives from multiple sites while tissue NGS reflects only the small tissue sample assayed.5 Differences in the sensitivity of the assays may also explain these differences.
We found that high levels of ctDNA (≥ 0.25% cfDNA) correlated with worse PFS. This was true despite controlling for the grade of tumor, as high-grade tumors have higher rates and levels of ctDNA alterations. This finding may allow use of ctDNA to better predict prognosis, particularly in malignancies such as appendiceal cancer, which has a 3-fold variance in risk of recurrence among various subtypes.33 The correlation between ctDNA and PFS may also have implications for surveillance of patients using ctDNA (regardless of the genomic alteration), and investigation in this area is ongoing at our institution.
The primary limitations of ctDNA include the need for tumor-encoded mutations to be detected in the circulation. DNA in the circulation may also come from sources other than the tumor in question, such as a second primary or myeloid pre-malignant condition known as clonal hematopoiesis of indeterminate origin.34 Other limitations of ctDNA, as it becomes more sensitive, may theoretically include the ability to discern genomic mutations derived from benign tissue.35 However, the methods used herein are approaching a practical limit with sensitivity as low as 1–2 DNA molecular fragments in 10 mL of blood without evidence of ctDNA from benign lesions. Additional limitations of this project include modest sample size, single-institution setting, and incomplete follow-up due to referral patterns. Also, due to cost and complexity, not all patients had tissue molecular profiling completed from the index operation for comparison with the ctDNA results.
In summary, ctDNA alterations are detectable in many patients undergoing surgical treatment of peritoneal metastases. Patients with high preoperative levels of ctDNA have a worse prognosis. Liquid biopsy for ctDNA analysis in this population is a promising new method for prognostication that merits additional studies. Further investigations of longitudinal ctDNA analysis are ongoing, which may yield additional information regarding the role of ctDNA interrogation in diagnosis, treatment, and surveillance.
Synopsis.
This paper examines preoperative circulating tumor DNA (ctDNA) in patients undergoing surgery for peritoneal metastasis at a single high-volume center; describing the ctDNA mutational landscape, concordance with tissue DNA alterations, and correlation with progression-free survival.
ACKNOWLEDGEMENTS
Funded in part by the Joan and Irwin Jacobs Fund philanthropic fund and by National Cancer Institute grant P30 CA016672.
Disclosures
Dr. Kurzrock has research funding from Incyte, Genentech, Merck Serono, Pfizer, Sequenom, Foundation Medicine, and Guardant Health, as well as consultant fees from Sequenom, Loxo and Actuate Therapeutics, speaker fees from Roche, and an ownership interest in Curematch, Inc.
Ms. Raymond and Dr. Lanman are employees at Guardant Health, Inc.
REFERENCES
- 1.Schwaederle M, Zhao M, Lee JJ, et al. Impact of Precision Medicine in Diverse Cancers: A Meta-Analysis of Phase II Clinical Trials. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. November 10 2015;33(32):3817–3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sinicrope FA, Okamoto K, Kasi PM, Kawakami H. Molecular Biomarkers in the Personalized Treatment of Colorectal Cancer. Clinical gastroenterology and hepatology : the official clinical practice journal of the American Gastroenterological Association. May 2016;14(5):651–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ghadimi BM, Jo P. Cancer Gene Profiling for Response Prediction. Methods in molecular biology (Clifton, N.J.). 2016;1381:163–179. [DOI] [PubMed] [Google Scholar]
- 4.Tsimberidou AM, Iskander NG, Hong DS, et al. Personalized medicine in a phase I clinical trials program: the MD Anderson Cancer Center initiative. Clinical cancer research : an official journal of the American Association for Cancer Research. November 15 2012;18(22):6373–6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gerlinger M, Rowan AJ, Horswell S, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. The New England journal of medicine. March 8 2012;366(10):883–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Diehl F, Schmidt K, Choti MA, et al. Circulating mutant DNA to assess tumor dynamics. Nature medicine. September 2008;14(9):985–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wan JCM, Massie C, Garcia-Corbacho J, et al. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nature reviews. Cancer. April 2017;17(4):223–238. [DOI] [PubMed] [Google Scholar]
- 8.Janku F, Angenendt P, Tsimberidou AM, et al. Actionable mutations in plasma cell-free DNA in patients with advanced cancers referred for experimental targeted therapies. Oncotarget. May 20 2015;6(14):12809–12821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Siravegna G, Bardelli A. Genotyping cell-free tumor DNA in the blood to detect residual disease and drug resistance. Genome biology. 2014;15(8):449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Husain H, Melnikova VO, Kosco K, et al. Monitoring Daily Dynamics of Early Tumor Response to Targeted Therapy by Detecting Circulating Tumor DNA in Urine. Clinical cancer research : an official journal of the American Association for Cancer Research. April 18 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Surveillance, Epidemiology, and End Results (SEER) Program. SEER*Stat Database: Incidence - SEER 9 Regs Research Data, Nov 2013 Sub (1973–2011) ed. National Cancer Institute, DCCPS, Surveillance Research Program, Surveillance Systems Branch2013.
- 12.Franko J, Shi Q, Goldman CD, et al. Treatment of colorectal peritoneal carcinomatosis with systemic chemotherapy: a pooled analysis of north central cancer treatment group phase III trials N9741 and N9841. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. January 20 2012;30(3):263–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hasovits C, Clarke S. Pharmacokinetics and pharmacodynamics of intraperitoneal cancer chemotherapeutics. Clinical pharmacokinetics. April 1 2012;51(4):203–224. [DOI] [PubMed] [Google Scholar]
- 14.Chua TC, Moran BJ, Sugarbaker PH, et al. Early- and long-term outcome data of patients with pseudomyxoma peritonei from appendiceal origin treated by a strategy of cytoreductive surgery and hyperthermic intraperitoneal chemotherapy. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. July 10 2012;30(20):2449–2456. [DOI] [PubMed] [Google Scholar]
- 15.Franko J, Ibrahim Z, Gusani NJ, Holtzman MP, Bartlett DL, Zeh HJ, Cytoreductive surgery and hyperthermic intraperitoneal chemoperfusion versus systemic chemotherapy alone for colorectal peritoneal carcinomatosis. Cancer. August 15 2010;116(16):3756–3762. [DOI] [PubMed] [Google Scholar]
- 16.Yan TD, Deraco M, Baratti D, et al. Cytoreductive surgery and hyperthermic intraperitoneal chemotherapy for malignant peritoneal mesothelioma: multi-institutional experience. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. December 20 2009;27(36):6237–6242. [DOI] [PubMed] [Google Scholar]
- 17.Cashin PH, Mahteme H, Spang N, et al. Cytoreductive surgery and intraperitoneal chemotherapy versus systemic chemotherapy for colorectal peritoneal metastases: A randomised trial. European journal of cancer (Oxford, England : 1990). January 2016;53:155–162. [DOI] [PubMed] [Google Scholar]
- 18.van Driel WJ, Koole SN, Sikorska K, et al. Hyperthermic Intraperitoneal Chemotherapy in Ovarian Cancer. The New England journal of medicine. January 18 2018;378(3):230–240. [DOI] [PubMed] [Google Scholar]
- 19.Verwaal VJ, Bruin S, Boot H, van Slooten G, van Tinteren H. 8-year follow-up of randomized trial: cytoreduction and hyperthermic intraperitoneal chemotherapy versus systemic chemotherapy in patients with peritoneal carcinomatosis of colorectal cancer. Annals of surgical oncology. September 2008;15(9):2426–2432. [DOI] [PubMed] [Google Scholar]
- 20.Pasqual EM, Bertozzi S, Bacchetti S, et al. Preoperative assessment of peritoneal carcinomatosis in patients undergoing hyperthermic intraperitoneal chemotherapy following cytoreductive surgery. Anticancer research. May 2014;34(5):2363–2368. [PubMed] [Google Scholar]
- 21.de Bree E, Koops W, Kroger R, van Ruth S, Witkamp AJ, Zoetmulder FA. Peritoneal carcinomatosis from colorectal or appendiceal origin: correlation of preoperative CT with intraoperative findings and evaluation of interobserver agreement. Journal of surgical oncology. May 1 2004;86(2):64–73. [DOI] [PubMed] [Google Scholar]
- 22.McMullen JRW, Selleck M, Wall NR, Senthil M. Peritoneal carcinomatosis: limits of diagnosis and the case for liquid biopsy. Oncotarget. March 22 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baumgartner JM, Tobin L, Heavey SF, Kelly KJ, Roeland EJ, Lowy AM. Predictors of progression in high-grade appendiceal or colorectal peritoneal carcinomatosis after cytoreductive surgery and hyperthermic intraperitoneal chemotherapy. Annals of surgical oncology. May 2015;22(5):1716–1721. [DOI] [PubMed] [Google Scholar]
- 24.Lanman RB, Mortimer SA, Zill OA, et al. Analytical and Clinical Validation of a Digital Sequencing Panel for Quantitative, Highly Accurate Evaluation of Cell-Free Circulating Tumor DNA. PloS one. 2015;10(10):e0140712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Portilla AG, Shigeki K, Dario B, Marcello D. The intraoperative staging systems in the management of peritoneal surface malignancy. Journal of surgical oncology. September 15 2008;98(4):228–231. [DOI] [PubMed] [Google Scholar]
- 26.Bettegowda C, Sausen M, Leary RJ, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. February 19 2014;6(224):224ra224.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fang WL, Lan YT, Huang KH, et al. Clinical significance of circulating plasma DNA in gastric cancer. International journal of cancer. June 15 2016;138(12):2974–2983. [DOI] [PubMed] [Google Scholar]
- 28.Schwaederle M, Chattopadhyay R, Kato S, et al. Genomic alterations in circulating tumor DNA from diverse cancer patients identified by next-generation sequencing. Cancer research. August 14 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rivlin N, Brosh R, Oren M, Rotter V. Mutations in the p53 Tumor Suppressor Gene: Important Milestones at the Various Steps of Tumorigenesis. Genes & cancer. April 2011;2(4):466–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schwaederle M, Husain H, Fanta PT, et al. Detection rate of actionable mutations in diverse cancers using a biopsy-free (blood) circulating tumor cell DNA assay. Oncotarget. March 01 2016;7(9):9707–9717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schwaederle MC, Patel SP, Husain H, et al. Utility of Genomic Assessment of Blood-Derived Circulating Tumor DNA (ctDNA) in Patients with Advanced Lung Adenocarcinoma. Clinical cancer research : an official journal of the American Association for Cancer Research. May 24 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schwaederle M, Husain H, Fanta PT, et al. Use of Liquid Biopsies in Clinical Oncology: Pilot Experience in 168 Patients. Clinical cancer research : an official journal of the American Association for Cancer Research. November 15 2016;22(22):5497–5505. [DOI] [PubMed] [Google Scholar]
- 33.Austin F, Mavanur A, Sathaiah M, et al. Aggressive management of peritoneal carcinomatosis from mucinous appendiceal neoplasms. Annals of surgical oncology. May 2012;19(5):1386–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Genovese G, Kahler AK, Handsaker RE, et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. The New England journal of medicine. December 25 2014;371(26):2477–2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kato S, Lippman SM, Flaherty KT, Kurzrock R. The Conundrum of Genetic “Drivers” in Benign Conditions. Journal of the National Cancer Institute. August 2016;108(8). [DOI] [PMC free article] [PubMed] [Google Scholar]


