Abstract
Chronic myeloid leukemia (CML) is a stem cell–derived leukemia in which neoplastic cells exhibit the Philadelphia chromosome and the related oncoprotein BCR-ABL1. The disease is characterized by an accumulation of myeloid precursor cells in the peripheral blood and bone marrow (BM). A small fraction of neoplastic cells in the CML clone supposedly exhibits self-renewal and thus long-term disease-propagating ability. However, so far, little is known about the phenotype, function, and target expression profiles of these leukemic stem cells (LSCs). Recent data suggest that CML LSCs aberrantly express the interleukin-2 receptor alpha chain CD25. Whereas normal CD34+/CD38− BM stem cells display only low amounts of CD25 or lack CD25 altogether, CD34+/CD38− LSCs express CD25 strongly in more than 90% of all patients with untreated CML. As a result, CD25 can be used to identify and quantify CML LSCs. In addition, it has been shown that CD25 serves as a negative growth regulator of CML LSCs. Here, we review the value of CD25 as a novel marker and potential drug target in CML LSCs.
Chronic myeloid leukemia (CML) is a stem cell–derived neoplasm characterized by the expansion and accumulation of immature and mature myeloid cells in the bone marrow (BM) and peripheral blood (PB), the Philadelphia chromosome (Ph), and the Ph-associated oncoprotein BCR-ABL1 [1–4]. In chronic phase (CP) CML, BCR-ABL1 is considered a major driver of disease evolution and oncogenesis. In line with this notion, the BCR-ABL1 inhibitor imatinib induces complete cytogenetic responses (CCyRs) and major molecular responses (MMRs) in most patients with CML [5–8]. However, some of these patients relapse during treatment with imatinib due to intrinsic and/or acquired resistance of leukemic cells [9–21]. Intrinsic resistance is independent of BCR-ABL1 and is often found in leukemic stem cells (LSCs) [9–16]. Acquired resistance of CML cells against imatinib and other BCR-ABL1-targeting drugs is often mediated by BCR-ABL1 mutations [16–18]. For these patients, novel, more potent BCR-ABL1-targeting drugs have been developed. These drugs include nilotinib, dasatinib, bosutinib, and ponatinib [19–22]. A number of clinical trials have shown that these drugs can induce clinically meaningful cytogenetic and molecular responses in most patients with imatinib-resistant CML [23–27]. However, not all patients with tyrosine kinase inhibitor (TKI)-resistant CML are long-term responders. In fact, these patients may relapse after treatment with one or more second-generation BCR-ABL1-targeting TKIs. One particular problem is primary drug resistance of CML LSCs.
The concept of LSCs was established some time ago with the intention of explaining cellular hierarchies and developing curative drug therapies through the elimination of leukemia-initiating and -propagating cells [28–30]. Based on the LSC concept, the leukemic clone can be divided into two fractions, a bulk population of more mature cells and LSCs. Whereas most cells in the bulk have no long-term proliferative capacity, LSCs exhibit self-renewing and long-term disease-propagating ability [28–30]. This concept has major clinical implications. In particular, the LSC concept predicts that any drug can act in a curative manner only when eliminating most or all LSCs and implies that relapses develop from residual LSCs that escaped therapy [10–13]. Indeed, LSCs are known to exhibit multiple forms of drug resistance [9–15]. In CML, the intrinsic form of resistance is common to most or all LSCs and is considered to be at least in part independent of BCR-ABL1 [9–12]. In contrast, the acquired form of TKI resistance arises during the course of disease in distinct, LSC-derived subclones and is often triggered by BCR-ABL1 mutations.
In patients with CP CML, LSCs are considered to reside in a CD34+/CD38− cell population [9–14,31–33]. Normal hematopoietic stem cells (HSCs) also exhibit this phenotype. Therefore, additional markers need to be used to discriminate between normal (residual) HSCs and CML LSCs. However, little is known about biomarkers and phenotypes of CML LSCs. Recently, a number of more or less specific markers and potential drug targets have been identified in CML LSCs [34–36]. The long-term aim of this research is to develop robust LSC markers for LSC isolation, for diagnostic purposes and prognostication, as well as for the design of LSC-eradicating treatment concepts [35–41]. One novel marker of CML LSCs is the interleukin-2 (IL-2) receptor alpha chain CD25 [34–37,42,43]. Here, we provide an overview of markers and targets displayed by CML LSCs, with special focus on the expression and function of CD25.
Cell surface structures expressed on CML LSCs and their potential clinical value
A number of previous and more recent studies have shown that growth, self-renewal, and distribution of CML LSCs are regulated by a network of cytokines, cytokine receptors, chemokines, and niche-related surface molecules [32,43–47]. In common with normal BM stem cells, CML LSCs express a number of growth factor receptors, including the IL-3 receptor (CD123/CD131), stem cell factor receptor (KIT = CD117), granulocyte colony-stimulating factor receptor (CD114), granulocyte/macrophage colony-stimulating factor receptor (CD116/CD131), and fms like tyrosine kinase 3 (FLT3) (CD135) [43,47–50]. In addition, CML LSCs reportedly express CXCR4 (CD184), a homing-related antigen that serves as a receptor for stromal cell–derived factor-1 (SDF-1) in the stem cell niche [36,51,52]. CML LSCs also display CD33, CD44, CD47, CD52, and CD93 [36,48,50,53]. Table 1 shows a summary of cell surface antigens expressed on CML LSCs and normal BM stem cells. Several of these cell surface antigens, such as CD123, are expressed at higher levels on CML LSCs compared with normal BM HSCs [36,48,50,53]. In addition, it has been shown that CD34+/CD38− LSCs in CP CML display several surface antigens in an aberrant manner. These surface markers include CD25, CD26, and IL-1 receptor-accessory protein (IL-1RAP) [34–36,43,54]. Other surface markers expressed more or less specifically on CML LSCs, but not or only at a low level on normal BM stem cells, include CD56 and CD93 [50,55] (Table 1). The CLL-1 antigen (CD371) that is specifically displayed by CD34+/CD38− LSCs in a subset of patients with acute myeloid leukemia (AML) [56] is only weakly expressed or undetectable on CML LSCs [50] (Table 1).
Table 1.
Expression of cell surface markers and targets on LSCs in CML and AML and comparison with normal BM stem cells
| Expression on CD34+/CD38− cells |
||||
|---|---|---|---|---|
| Target/Marker | CD | CML LSCs | AML LSCs | Normal stem cells |
| T-Span-29 | CD9 | ++ | + | +/− |
| IL-2RA | CD25 | ++ | +/− | −/+ |
| DPPIV | CD26 | ++ | −/+a | – |
| Siglec-3 | CD33 | ++ | ++ | + |
| Pgp-1 | CD44 | ++ | ++ | ++ |
| IAP | CD47 | ++ | ++ | ++ |
| Campath-1 | CD52 | + | +/− | +/− |
| NCAM | CD56 | + | −/+ | −/+ |
| Thy-1 | CD90 | + | – | ++ |
| Lectin-R | CD93 | + | + | +/− |
| Tactile | CD96 | – | −/+ | – |
| Endoglin | CD105 | ++ | ++ | ++ |
| G-CSFR | CD114 | + | +/− | + |
| GM-CSFR | CD116 | +/− | −/+ | −/+ |
| KIT/SCFR | CD117 | ++ | ++ | ++ |
| IL-3RA | CD123 | ++ | ++ | + |
| AC133 | CD133 | ++ | + | ++ |
| FLT3 | CD135 | + | + | +/− |
| CXCR4 | CD184 | + | + | ++ |
| CLL1 | CD371 | +/− | +/− | – |
| IL-1RAP | n.c. | + | +/− | – |
| HLA-DR | n.c. | ++ | +/− | ++ |
AC133 = CD133 (prominin or PROM1); CLL1 = C-type lectin-like molecule-1; CSFR = granulocyte-macrophage colony-stimulating factor receptor; CXCR4 = C-X-C chemokine receptor type 4; DPPIV = dipeptidyl-peptidase IV; FLT3 = fms like tyrosine kinase 3; G-CSFR = granulocyte colony-stimulating factor receptor; GM-CSFR = granulocyte macrophage colony-stimulating factor; HLA-DR = human leukocyte antigen; n.c. = not yet clustered; NCAM = neural cell adhesion molecule; SCFR = stem cell factor receptor; T-Span-29 = Tetraspanin 29.
Expression score: ++, expressed on (L)SCs in >90% of all patients/donors; +, expressed on (L)SCs in 75–90% of all donors; +/−, expressed on (L)SCs in 50–75% of all donors; −/+, expressed on (L)SCs in 15–50% of donors; –, expressed on (L)SCs in <15% of donors.
In a subset of patients with FLT3-mutated AML, LSCs display CD26.
Whereas in untreated patients with CP CML, LSCs display an almost invariable profile of aberrantly expressed surface antigens (CD25+/CD26+/IL-1RAP+/CD56+), this is not the case in patients with accelerated phase (AP) or blast phase (BP) of CML [36,43]. In fact, in AP or BP patients, one or more of the aberrantly expressed LSCs markers may no longer be detectable on CD34+/CD38− cells. Likewise, it has been shown that CD34+/CD38− LSCs express CD25 in more than 90% of patients with untreated CML, but express CD25 in only about one half of patients with AP CML [43]. Even in some CP patients, not all of the aberrantly displayed LSC markers may be detectable in the stem cell fraction, which has practical implications.
Based on the high specificity of the aberrant profile of CML LSCs, diagnostic LSC phenotyping and LSC quantification in CML have been considered recently. The approach of LSC phenotyping has not yet been validated, however, and although CD25 expression on LSCs has been reported [35,38,42], more prospective studies may be required to translate this new diagnostic approach into routine application. For such diagnostic phenotyping of LSCs, we recommend that at least three specific LSC markers (preferably CD25, CD26, and IL-1RAP) are used. With these three markers, along with CD19 and/or CD20, the specificity for CML is more than 90%. Expression of all three markers on LSCs is highly indicative of the presence of BCR-ABL1+ CML. In fact, CD26 and IL-1RAP are not expressed on CD34+/CD38− LSCs in other hematopoietic malignancies, with a few exceptions: patients with BCR-ABL1p210+ acute lymphoblastic leukemia (often referred to as primary lymphoid blast phase of CML), a subset of patients with FLT3 ITD+ AML, and a few patients with primary JAK2 V617F+ myelofibrosis [36]. As mentioned before, aberrantly expressed LSC markers can also be used to quantify CML LSCs at diagnosis and during therapy [36,57]. The number of CD26+ LSCs at diagnosis has been shown to correlate with the patients’ response to BCR-ABL1-targeting TKIs [57]. However, during therapy with imatinib or other TKIs, the numbers of CD26+ LSCs decrease rapidly in the PB and BM and disappear after a few weeks unless the clone is resistant. In all of these patients, monitoring of BCR-ABL1 mRNA by quantitative polymerase chain reaction is by far a more sensitive approach to quantify minimal residual disease (MRD) than LSC phenotyping. Therefore, LSC phenotyping in CML is recommended only at diagnosis and at relapse, not as an MRD parameter. With regard to CD25, another important issue has to be considered: recent data suggest that, in some patients with CML who achieve CCyR/MMR during treatment with imatinib, the CD34+/CD38− stem cells, detectable in the BM and/or PB after debulking of the dominant clone, are negative for BCR-ABL1 and CD26, but still express CD25 (IS and PV, unpublished observation). It is tempting to speculate that these CD25+ stem cells are remaining clonal stem cells derived from an earlier (pre-malignant) phase of LSC evolution [58,59]. However, the robust molecular data needed to confirm this hypothesis are lacking, and whether these cells are indeed residual preleukemic neoplastic stem cells (pre-L-NSCs) or normal HSCs that display CD25 in a “reactive” manner (e.g., inflammation- or cytokine-induced) remains unknown.
CML LSCs display the alpha chain of the IL-2 receptor
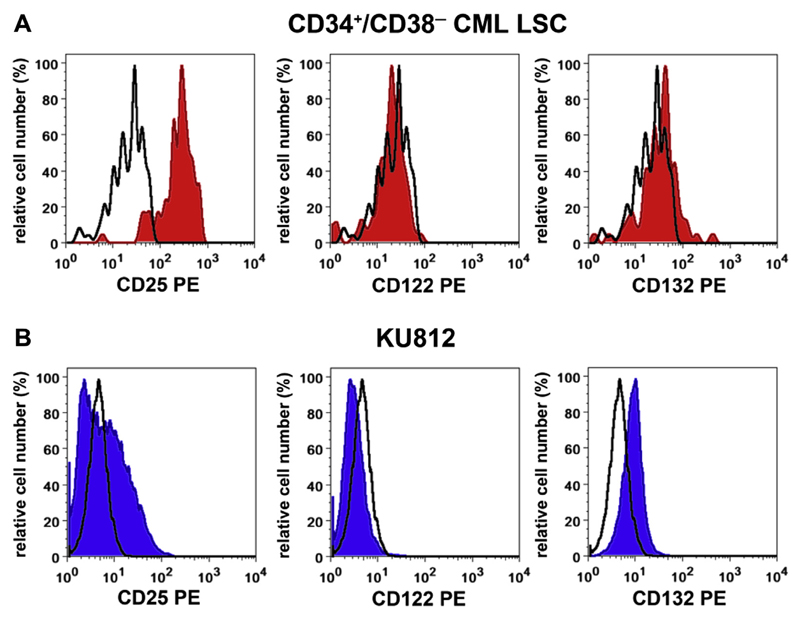
The IL-2 receptor complex on lymphohematopoietic cells consists of an alpha chain (IL-2RA = CD25), a functional beta chain (CD122), and a gamma chain (CD132). CML LSCs display CD25, but do not express substantial amounts of CD122 or CD132 (Fig. 1A) [43,50]. Correspondingly, IL-2 is unable to trigger the growth or survival of CML LSCs, even when relatively high doses of the cytokine are applied [43]. The multipotent BCR-ABL1+ cell line KU812 was also found to express CD25, but did not express substantial amounts of CD122 or CD132 (Fig. 1B) [43]. Moreover, KU812 cells are unresponsive to physiologic concentrations of IL-2 [43].
Figure 1.
Expression of CD25 on CD34+/CD38− LSCs in patients with CML and on KU812 cells. (A) Primary CML cells were stained with fluorochrome-labeled monoclonal antibodies against CD34, CD38, CD45, CD25, CD122, and CD132. LSCs were defined as CD45+/CD34+/CD38− cells. As assessed by multicolor flow cytometry, LSCs were found to co-express CD25 (left panel), but did not express CD122 (middle panel) or CD132 (right panel) (red histograms). The isotype-matched control antibody is also shown (black open histograms). (B) The Ph+ cell line KU812 was stained with antibodies against CD25, CD122, and CD132 (blue histograms). It can be seen that KU812 cells also express CD25 and do not express substantial amounts of CD122 or CD132.
Expression of CD25 on CML LSCs is dependent on signal transducer and activator of transcription 5 (STAT5) activity
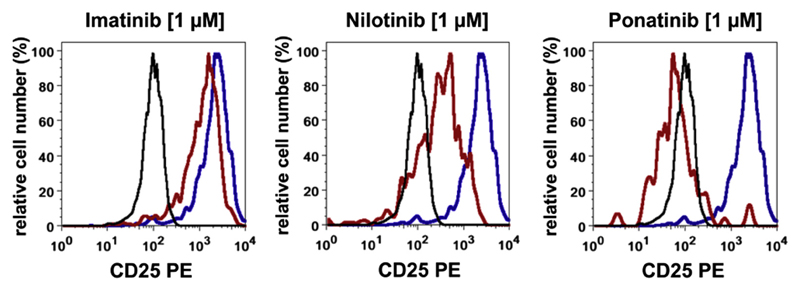
It is generally thought that activated STAT5 is a major trigger of CD25 expression on various physiologic and neoplastic cells. In addition, BCR-ABL1 is known to induce STAT5 activation. Correspondingly, drug-induced or short hairpin RNA (shRNA)-mediated knock-down of STAT5 expression in KU812 cells is followed by reduced expression of CD25 [43]. Consistent with these observations, the STAT5-targeting drug pimozide was found to decrease CD25 expression on CD34+/CD38− CML LSCs [43]. A similar effect was obtained with BCR-ABL1-targeting drugs. In particular, nilotinib and ponatinib were found to suppress expression of CD25 on CML LSCs (Fig. 2) and KU812 cells [43]. In contrast, imatinib did not inhibit the expression of CD25 on CML LSCs (Fig. 2). This discrepancy is best explained by intrinsic LSCs resistance against imatinib, a phenomenon that may be associated with a poor uptake of imatinib into LSCs or rapid efflux of the drug from LSCs after uptake [9,60,61]. STAT5 dependence of CD25 expression in CML LSCs is not restricted to the human system. In fact, transduction of BM stem cells obtained from C57BL/6 mice with STAT5 results in expression of CD25 on Lin−/Sca-1+/Kit+ cells [43]. Because STAT5 activation has been recognized as a major event in disease evolution and the JAK–STAT pathway may be a novel target of therapy, the observation that CD25 expression on CML LSCs is dependent on STAT5 activity is of potential interest. Indeed, STAT5 target genes are thought to play a role in disease evolution in CML.
Figure 2.
Effects of BCR-ABL1-targeting TKIs on CD25 expression on CD34+/CD38− CML LSCs. CML MNCs were incubated in control medium or TKIs (left histogram: imatinib, middle histogram: nilotinib, right histogram: ponatinib; each 1 μmol/L) at 37°C for 24 hours. Thereafter, CD25 expression on CD34+/CD38− LSCs was analyzed by multicolor flow cytometry. Black open histograms represent the matched isotype control; blue histograms represent CD25 expression on CML LSCs kept in control medium; and red histograms represent expression of CD25 on CML LSCs after treatment with TKI. As illustrated, the expression of CD25 on CML LSCs is not altered by imatinib, but decreases on exposure to nilotinib or ponatinib. MNCs = Mononuclear cells.
Aberrant expression of CD25 in CML cells is restricted to LSCs
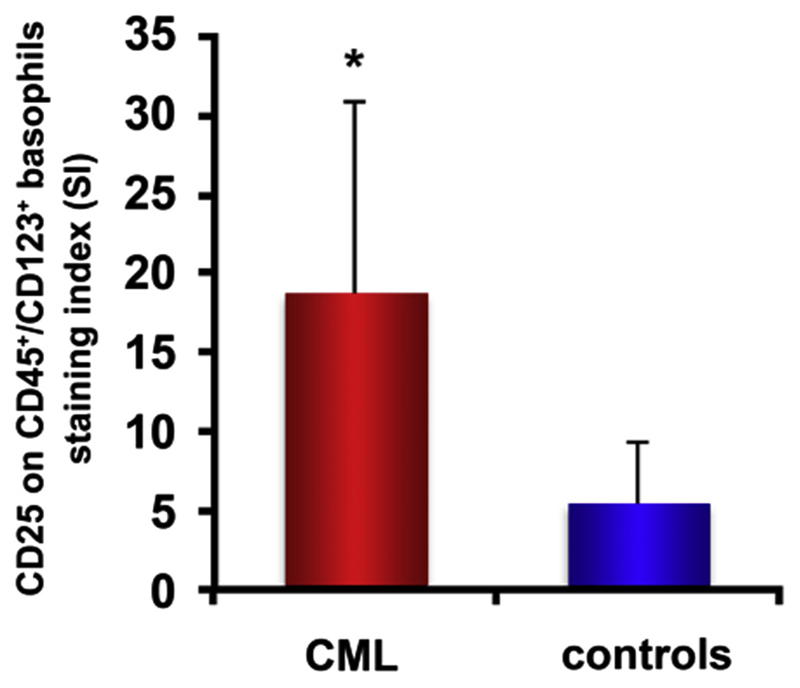
Under physiologic conditions, CD25 is only expressed on a few hematopoietic cell types, including activated T lymphocytes, activated B lymphocytes, and basophils. A remarkable observation has been that, in CML patients, aberrant expression of CD25 is restricted to LSCs. In other words, although BCR-ABL1 and activated STAT5 are expressed in most clonal cells in patients with CML, the STAT5-dependent target gene CD25 is only upregulated and expressed aberrantly on LSCs [43]. More mature (CD34+/CD38+) progenitor cells display a low amount of CD25 or do not express CD25 in CML, similar to normal BM progenitors [43]. Similarly, basophils in CML and in healthy donors express CD25. However, the levels of CD25 on CML basophils are higher than those detectable on normal blood basophils [43,62] (Fig. 3).
Figure 3.
CML basophils express higher levels of CD25 than do basophils obtained from healthy donors. Expression of CD25 on CD123++/CD45+ basophils was analyzed by multicolor flow cytometry using fluorochrome-labeled antibodies against CD45, CD123, and CD25. Cells were obtained from five patients with CP CML (left red bar) and five healthy controls (right blue bar). Expression of CD25 was determined as the MFI and is expressed as the SI according to the following formula: SI = MFI test antibody: MFI isotype-control antibody. *p < 0.05 compared with CD25 expression on normal basophils. MFI = Median fluorescence intensity; SI = staining index.
Functional role of CD25 expression in CML LSCs
The exact functional role of CD25 on CML LSCs remains to be determined. In initial studies, CD25 has been described as a growth-promoting molecule in murine CML LSCs [42]. In addition, it was hypothesized that CD25 could also act as a growth-promoting regulator in human CML LSCs. However, more recently, CD25 was found to act as a growth-inhibitory molecule on human CML LSCs [43]. In particular, a shRNA against CD25 was found to enhance the proliferation of primary human CD34+ CML cells. In addition, a shRNA specific for CD25 was found to promote growth and survival of KU812 cells in vitro (Fig. 4) and in vivo in an NSG xenotransplantation model [43]. Moreover, lentivirus-mediated transduction of CD25 into the CD25-negative cell lines K562, K562-R, and KCL-22 resulted in reduced cell proliferation [43]. Collectively, these data suggest that CD25 serves as a growth-suppressing molecule in CML stem and progenitor cells. So far, it remains unknown whether CD25 can also act as a growth-suppressing molecule of LSCs in patients with other myeloid leukemias such as AML. More recent data suggest that CD25 can act as a tumor suppressor in a murine model of T-cell leukemia [63].
Figure 4.
Evaluation of CD25 as a functional target on CML cells. CD25 shRNA-transduced cells were mixed 1:1 with KU812 cells transduced with a RDM shRNA and cultured at 37°C for 14 days. Expression of CD25+ cells was analyzed three times a week by flow cytometry using a phycoerythrin-conjugated antibody against CD25. Results are expressed as the percentage of CD25+ cells and represent the mean ± standard deviation from three independent experiments. *p < 0.05 compared with the percentage of CD25+ cells measured on day 0. RDM = Random.
Identification of CD25 as a drug-inducible suppressor-type target in CML LSCs
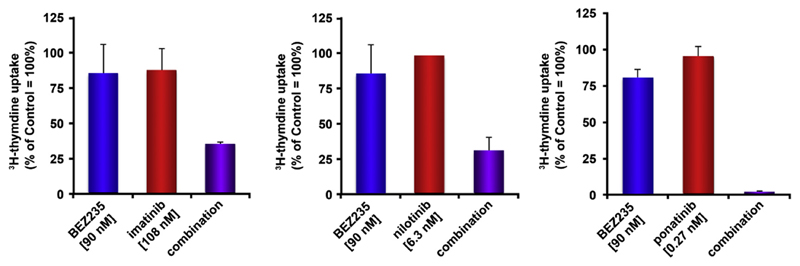
Because BCR-ABL1 TKIs downregulate the expression of CD25 on CML LSCs, a new fascinating hypothesis is that CD25 may also serve as a drug-sensitive suppressor and that resistance against TKIs is facilitated by drug-induced downregulation of CD25 [43]. Based on this notion, drug screens were performed with the aim of identifying drug classes that promote the expression of CD25 on CML LSCs. In these screens, inhibitors of the phosphoinositide 3-kinase (PI3K)–mechanistic target of rapamycin (mTOR) pathway were identified as specific inducers of CD25 expression [43]. In particular, the dual PI3K and mTOR blocker BEZ235 and the mTOR-targeting drug rapamycin not only inhibit growth of CML cells, but also promote expression of CD25 on CML LSCs as well as on KU812 cells [43]. More importantly, KU812 cells transduced with shRNA against CD25 were found to be less sensitive against BEZ235 compared with untransfected cells [43]. To exploit CD25 as a potential drug-inducible LSC inhibitor, BEZ235 was combined with various BCR-ABL1 TKIs to induce synergistic antineoplastic effects. Indeed, when combining these drugs, highly synergistic effects were obtained in KU812 cells [43] (Fig. 5). IL-2R-targeting antibodies and IL-2R-targeting IL-2-toxin conjugates (e.g., denileukin-diftitox) have already been applied in clinical trials. Whether such drugs can be used to attack and kill CML LSCs or LSCs in other hematopoietic neoplasms is being investigated.
Figure 5.
The PI3K/mTOR blocker BEZ235 synergizes with BCR-ABL1-targeting TKIs in inhibiting the proliferation of KU812 cells. KU812 cells were incubated in control medium, medium supplemented with BEZ235 (90 nmol/L), or medium containing BCR-ABL1-targeting drugs (imatinib, 108 nmol/L; nilotinib, 6.3 nmol/L; ponatinib, 0.27 nmol/L) alone or in combination at a fixed ratio of drug concentrations at 37°C for 48 hours (BEZ235 + imatinib, left panel; BEZ235 + nilotinib, middle panel; BEZ235 + ponatinib, right panel). After incubation, 3H-thymidine uptake was measured. Results are expressed as a percentage of control (100%), and represent the mean ± standard deviation from triplicates.
Expression of CD25 on LSCs or pre-L-NSCs in other hematopoietic malignancies
A number of previous and more recent studies suggest that CD25 is expressed not only on LSCs in CML, but also on LSCs in other myeloid neoplasms, including AML [36,43,64]. In addition, CD25 is expressed on CD34+/CD38− stem cells in patients with Ph+ ALL and in a subset of patients with primary myelofibrosis. Finally, as mentioned previously, CD25 may be expressed on pre-L-NSCs in preleukemic conditions. Whether normal CD34+/CD38− (nonmutated) HSCs (e.g., after cytokine exposure) can also express CD25 remains unknown.
Clinical application and translational relevance
So far, the exact value of CD25 as a potential biomarker or target in CML LSCs remains uncertain. From a clinical point of view, CD25 may serve as a diagnostic LSC marker in CML. However, expression of CD25 on LSCs is not specific for CML, but is also seen in AML [64]. In addition, even normal (apparently healthy) BM stem cells may sometimes express CD25. Therefore, the application of CD25 alone is not recommended in CML. Rather, additional diagnostic LSC markers, such as CD26 and IL-1RAP (or CD56 and/or CD93), should be applied. When applied together, the diagnostic accuracy (at diagnosis) in CML is very high (more than 90%). In addition, when combined with CD26 and/or IL-1RAP, CD25 can be used as an LSC marker in the follow-up during treatment with TKIs. However, it should be mentioned that the sensitivity of this approach is limited. In fact, BCR-ABL1 monitoring is much more sensitive compared with LSC phenotyping. Whether CD25 can be used as a target of therapy in CML is not known. Because CD25 may acts as a tumor suppressor in CML LSCs, a functional block of CD25 may not be an advisable approach. However, CD25 may be used as an immunotherapy target using toxin conjugates. The elimination of preleukemic and leukemic stem cells could be an advantage of CD25-targeting therapy. Another approach would be to promote CD25 expression through inhibition of the PI3K–mTOR pathway [43].
Concluding remarks
The alpha chain of the IL-2R, CD25, has been identified recently as a novel biomarker expressed aberrantly on the surface of CML LSCs. Expression of CD25 on CML LSCs depends on STAT5 activity and is seen in virtually all patients with CML. However, in contrast to other LSC markers, CD25 is also detectable on LSCs in patients with AML. Although the functional role of its expression on LSCs remains uncertain, recent data suggest that CD25 serves as a negative regulator of growth of CML LSCs. In addition, recent data suggest that TKI-induced downregulation of CD25 contributes to TKI resistance and that drugs promoting the expression of CD25 in CML cells exert strong synergistic effects when combined with BCR-ABL1 TKIs. In conclusion, CD25 appears to be a novel marker and drug-sensitive suppressor in CML cells. Future studies should define the exact diagnostic and clinical value of CD25 in CML.
Acknowledgments
We thank Karin Bauer, Daniela Berger, Gabriele Stefanzl, Alexandra Keller, Dubravka Smiljkovic, Susanne Herndlhofer, and Georg Greiner for skillful technical assistance. This work was supported by the Austrian Science Fund (Grant Nos. F4701-B20 and F4704-B20).
Footnotes
Conflict of interest disclosure
PV received research funding from Novartis, Celgene, Ariad, Incyte, and Deciphera and honoraria from Novartis, BMS, Celgene, Ariad, Incyte, and Pfizer. GH received research funding from Gilead and honoraria from Novartis, Ariad, and Amgen. WRS received research funding from Amgen and honoraria from Novartis and Amgen. The remaining authors declare no competing financial interests.
References
- 1.Rowley JD. Letter: A new consistent chromosomal abnormality in chronic myelogenous leukaemia identified by quinacrine fluorescence and Giemsa staining. Nature. 1973;243:290–293. doi: 10.1038/243290a0. [DOI] [PubMed] [Google Scholar]
- 2.Faderl S, Talpaz M, Estrov Z, Kantarjian HM. Chronic myelogenous leukemia: biology and therapy. Ann Intern Med. 1999;131:207–219. doi: 10.7326/0003-4819-131-3-199908030-00008. [DOI] [PubMed] [Google Scholar]
- 3.Deininger MW, Goldman JM, Melo JV. The molecular biology of chronic myeloid leukemia. Blood. 2000;96:3343–3356. [PubMed] [Google Scholar]
- 4.Melo JV, Barnes DJ. Chronic myeloid leukaemia as a model of disease evolution in human cancer. Nat Rev Cancer. 2007;7:441–453. doi: 10.1038/nrc2147. [DOI] [PubMed] [Google Scholar]
- 5.Hughes TP, Kaeda J, Branford S, et al. International Randomised Study of Interferon versus STI571 (IRIS) Study Group Frequency of major molecular responses to imatinib or interferon alfa plus cytarabine in newly diagnosed chronic myeloid leukemia. N Engl J Med. 2003;349:1423–1432. doi: 10.1056/NEJMoa030513. [DOI] [PubMed] [Google Scholar]
- 6.O’Brien SG, Guilhot F, Larson RA, et al. Imatinib compared with interferon and low-dose cytarabine for newly diagnosed chronicphase chronic myeloid leukemia. N Engl J Med. 2003;348:994–1004. doi: 10.1056/NEJMoa022457. [DOI] [PubMed] [Google Scholar]
- 7.Druker BJ. Perspectives on the development of a molecularly targeted agent. Cancer Cell. 2002;1:31–36. doi: 10.1016/s1535-6108(02)00025-9. [DOI] [PubMed] [Google Scholar]
- 8.Druker BJ, Guilhot F, O’Brien SG, et al. IRIS Investigators Five-year follow-up of patients receiving imatinib for chronic myeloid leukemia. N Engl J Med. 2006;355:2408–2417. doi: 10.1056/NEJMoa062867. [DOI] [PubMed] [Google Scholar]
- 9.Jiang X, Zhao Y, Smith C, et al. Chronic myeloid leukemia stem cells possess multiple unique features of resistance to BCR-ABL targeted therapies. Leukemia. 2007;21:926–935. doi: 10.1038/sj.leu.2404609. [DOI] [PubMed] [Google Scholar]
- 10.Corbin AS, Agarwal A, Loriaux M, Cortes J, Deininger MW, Druker BJ. Human chronic myeloid leukemia stem cells are insensitive to imatinib despite inhibition of BCR-ABL activity. J Clin Invest. 2011;121:396–409. doi: 10.1172/JCI35721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Valent P. Emerging stem cell concepts for imatinib-resistant chronic myeloid leukaemia: Implications for the biology, management, and therapy of the disease. Br J Haematol. 2008;142:361–378. doi: 10.1111/j.1365-2141.2008.07197.x. [DOI] [PubMed] [Google Scholar]
- 12.Hamilton A, Helgason GV, Schemionek M, et al. Chronic myeloid leukemia stem cells are not dependent on Bcr-Abl kinase activity for their survival. Blood. 2012;119:1501–1510. doi: 10.1182/blood-2010-12-326843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Copland M, Jorgensen HG, Holyoake TL. Evolving molecular therapy for chronic myeloid leukaemia—Are we on target? Hematology. 2005;10:349–359. doi: 10.1080/10245330500234195. [DOI] [PubMed] [Google Scholar]
- 14.Kavalerchik E, Goff D, Jamieson CH. Chronic myeloid leukemia stem cells. J Clin Oncol. 2008;26:2911–2915. doi: 10.1200/JCO.2008.17.5745. [DOI] [PubMed] [Google Scholar]
- 15.Jiang X, Forrest D, Nicolini F, et al. Properties of CD34+ CML stem/progenitor cells that correlate with different clinical responses to imatinib mesylate. Blood. 2010;116:2112–2121. doi: 10.1182/blood-2009-05-222471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.La Rosee P, Deininger MW. Resistance to imatinib: Mutations and beyond. Semin Hematol. 2010;47:335–343. doi: 10.1053/j.seminhematol.2010.06.005. [DOI] [PubMed] [Google Scholar]
- 17.Branford S, Rudzki Z, Walsh S, et al. High frequency of point mutations clustered within the adenosine triphosphate-binding region of BCR/ABL in patients with chronic myeloid leukemia or Ph-positive acute lymphoblastic leukemia who develop imatinib (STI571) resistance. Blood. 2002;99:3472–3475. doi: 10.1182/blood.v99.9.3472. [DOI] [PubMed] [Google Scholar]
- 18.Hochhaus A, Kreil S, Corbin AS, et al. Molecular and chromosomal mechanisms of resistance to imatinib (STI571) therapy. Leukemia. 2002;16:2190–2196. doi: 10.1038/sj.leu.2402741. [DOI] [PubMed] [Google Scholar]
- 19.Shah NP, Tran C, Lee FY, Chen P, Norris D, Sawyers CL. Overriding imatinib resistance with a novel ABL kinase inhibitor. Science. 2004;305:399–401. doi: 10.1126/science.1099480. [DOI] [PubMed] [Google Scholar]
- 20.Weisberg E, Manley PW, Breitenstein W, et al. Characterization of AMN107, a selective inhibitor of native and mutant Bcr-Abl. Cancer Cell. 2005;7:129–141. doi: 10.1016/j.ccr.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 21.Martinelli G, Soverini S, Rosti G, Cilloni D, Baccarani M. New tyrosine kinase inhibitors in chronic myeloid leukemia. Haematologica. 2005;90:534–541. [PubMed] [Google Scholar]
- 22.Rusconi F, Piazza R, Vagge E, Gambacorti-Passerini C. Bosutinib: A review of preclinical and clinical studies in chronic myelogenous leukemia. Expert Opin Pharmacother. 2014;15:701–710. doi: 10.1517/14656566.2014.882898. [DOI] [PubMed] [Google Scholar]
- 23.Kantarjian H, Giles F, Wunderle L, et al. Nilotinib in imatinib-resistant CML and Philadelphia chromosome-positive ALL. N Engl J Med. 2006;354:2542–2551. doi: 10.1056/NEJMoa055104. [DOI] [PubMed] [Google Scholar]
- 24.Talpaz M, Shah NP, Kantarjian H, et al. Dasatinib in imatinib-resistant Philadelphia chromosome-positive leukemias. N Engl J Med. 2006;354:2531–2541. doi: 10.1056/NEJMoa055229. [DOI] [PubMed] [Google Scholar]
- 25.Cortes JE, Kantarjian HM, Brümmendorf TH, et al. Safety and efficacy of bosutinib (SKI-606) in chronic phase Philadelphia chromosome-positive chronic myeloid leukemia patients with resistance or intolerance to imatinib. Blood. 2011;118:4567–4576. doi: 10.1182/blood-2011-05-355594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cortes JE, Kantarjian H, Shah NP, et al. Ponatinib in refractory Philadelphia chromosome-positive leukemias. N Engl J Med. 2012;367:2075–2088. doi: 10.1056/NEJMoa1205127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cortes JE, Kim DW, Pinilla-Ibarz J, et al. A phase 2 trial of ponatinib in Philadelphia chromosome-positive leukemias. N Engl J Med. 2013;369:1783–1796. doi: 10.1056/NEJMoa1306494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lapidot T, Sirard C, Vormoor J, et al. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 1994;367:645–648. doi: 10.1038/367645a0. [DOI] [PubMed] [Google Scholar]
- 29.Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997;3:730–737. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- 30.Dick JE. Stem cell concepts renew cancer research. Blood. 2008;112:4793–4807. doi: 10.1182/blood-2008-08-077941. [DOI] [PubMed] [Google Scholar]
- 31.Petzer AL, Eaves CJ, Lansdorp PM, Ponchio L, Barnett MJ, Eaves AC. Characterization of primitive subpopulations of normal and leukemic cells present in the blood of patients with newly diagnosed as well as established chronic myeloid leukemia. Blood. 1996;88:2162–2171. [PubMed] [Google Scholar]
- 32.Holyoake TL, Jiang X, Jorgensen HG, et al. Primitive quiescent leukemic cells from patients with chronic myeloid leukemia spontaneously initiate factor-independent growth in vitro in association with up-regulation of expression of interleukin-3. Blood. 2001;97:720–728. doi: 10.1182/blood.v97.3.720. [DOI] [PubMed] [Google Scholar]
- 33.Eisterer W, Jiang X, Christ O, et al. Different subsets of primary chronic myeloid leukemia stem cells engraft immunodeficient mice and produce a model of the human disease. Leukemia. 2005;19:435–441. doi: 10.1038/sj.leu.2403649. [DOI] [PubMed] [Google Scholar]
- 34.Bruns I, Czibere A, Fischer JC, et al. The hematopoietic stem cell in chronic phase CML is characterized by a transcriptional profile resembling normal myeloid progenitor cells and reflecting loss of quiescence. Leukemia. 2009;23:892–899. doi: 10.1038/leu.2008.392. [DOI] [PubMed] [Google Scholar]
- 35.Gerber JM, Gucwa JL, Esopi DM, et al. Genome-wide comparison of the transcriptomes of highly enriched normal and chronic myeloid leukemia stem and progenitor cell populations. Oncotarget. 2013;4:715–728. doi: 10.18632/oncotarget.990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Herrmann H, Sadovnik I, Cerny-Reiterer S, et al. Dipeptidylpeptidase IV (CD26) defines leukemic stem cells (LSCs) in chronic myeloid leukemia. Blood. 2014;123:3951–3962. doi: 10.1182/blood-2013-10-536078. [DOI] [PubMed] [Google Scholar]
- 37.Valent P, Sadovnik I, Ráčil Z, et al. DPPIV (CD26) as a novel stem cell marker in Ph+ chronic myeloid leukaemia. Eur J Clin Invest. 2014;44:1239–1245. doi: 10.1111/eci.12368. [DOI] [PubMed] [Google Scholar]
- 38.Warfvinge R, Geironson Ulfsson L, Sommarin MN, et al. Single-cell molecular analysis defines therapy response and immunophenotype of stem cell subpopulations in CML. Blood. 2017;129:2384–2394. doi: 10.1182/blood-2016-07-728873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Misaghian N, Ligresti G, Steelman LS, et al. Targeting the leukemic stem cell: The Holy Grail of leukemia therapy. Leukemia. 2009;23:25–42. doi: 10.1038/leu.2008.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Essers MA, Trumpp A. Targeting leukemic stem cells by breaking their dormancy. Mol Oncol. 2010;4:443–450. doi: 10.1016/j.molonc.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Valent P. Targeting of leukemia-initiating cells to develop curative drug therapies: Straightforward but nontrivial concept. Curr Cancer Drug Targets. 2011;11:56–71. doi: 10.2174/156800911793743655. [DOI] [PubMed] [Google Scholar]
- 42.Kobayashi CI, Takubo K, Kobayashi H, et al. The IL-2/CD25 axis maintains distinct subsets of chronic myeloid leukemia-initiating cells. Blood. 2014;123:2540–2549. doi: 10.1182/blood-2013-07-517847. [DOI] [PubMed] [Google Scholar]
- 43.Sadovnik I, Hoelbl-Kovacic A, Herrmann H, et al. Identification of CD25 as STAT5-dependent growth regulator of leukemic stem cells in Ph+ CML. Clin Cancer Res. 2016;22:2051–2061. doi: 10.1158/1078-0432.CCR-15-0767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jiang X, Lopez A, Holyoake T, Eaves A, Eaves C. Autocrine production and action of IL-3 and granulocyte colony-stimulating factor in chronic myeloid leukemia. Proc Natl Acad Sci U S A. 1999;96:12804–12809. doi: 10.1073/pnas.96.22.12804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bhatia R, Munthe HA, Williams AD, Zhang F, Forman SJ, Slovak ML. Chronic myelogenous leukemia primitive hematopoietic progenitors demonstrate increased sensitivity to growth factor-induced proliferation and maturation. Exp Hematol. 2000;28:1401–1412. doi: 10.1016/s0301-472x(00)00545-2. [DOI] [PubMed] [Google Scholar]
- 46.Holyoake TL, Jiang X, Drummond MW, Eaves AC, Eaves CJ. Elucidating critical mechanisms of deregulated stem cell turnover in the chronic phase of chronic myeloid leukemia. Leukemia. 2002;16:549–558. doi: 10.1038/sj.leu.2402444. [DOI] [PubMed] [Google Scholar]
- 47.Belloc F, Airiau K, Jeanneteau M, et al. The stem cell factor-c-KIT pathway must be inhibited to enable apoptosis induced by BCR-ABL inhibitors in chronic myelogenous leukemia cells. Leukemia. 2009;23:679–685. doi: 10.1038/leu.2008.364. [DOI] [PubMed] [Google Scholar]
- 48.Florian S, Sonneck K, Hauswirth AW, et al. Detection of molecular targets on the surface of CD34+/CD38- stem cells in various myeloid malignancies. Leuk Lymphoma. 2006;47:207–222. doi: 10.1080/10428190500272507. [DOI] [PubMed] [Google Scholar]
- 49.Wisniewski D, Affer M, Willshire J, Clarkson B. Further phenotypic characterization of the primitive lineage- CD34+CD38-CD90+CD45RA- hematopoietic stem cell/progenitor cell sub-population isolated from cord blood, mobilized peripheral blood and patients with chronic myelogenous leukemia. Blood Cancer J. 2011;1:e36. doi: 10.1038/bcj.2011.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sadovnik I, Herrmann H, Blatt K, et al. Evaluation of cell surface markers and targets in leukemic stem cells (LSCs) reveals distinct expression profiles, unique drug effects, and specific checkpoint regulation in AML LSCs and CML LSCs. Blood. 2016;128:4234. [Google Scholar]
- 51.Dürig J, Rosenthal C, Elmaagacli A, et al. Biological effects of stroma-derived factor-1 alpha on normal and CML CD34+ haemopoietic cells. Leukemia. 2000;14:1652–1660. doi: 10.1038/sj.leu.2401875. [DOI] [PubMed] [Google Scholar]
- 52.Peled A, Hardan I, Trakhtenbrot L, et al. Immature leukemic CD34+CXCR4+ cells from CML patients have lower integrin-dependent migration and adhesion in response to the chemokine SDF-1. Stem Cells. 2002;20:259–266. doi: 10.1634/stemcells.20-3-259. [DOI] [PubMed] [Google Scholar]
- 53.Herrmann H, Cerny-Reiterer S, Gleixner KV, et al. CD34(+)/CD38(-) stem cells in chronic myeloid leukemia express Siglec-3 (CD33) and are responsive to the CD33-targeting drug gemtuzumab/ozogamicin. Haematologica. 2012;97:219–226. doi: 10.3324/haematol.2010.035006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Järås M, Johnels P, Hansen N, et al. Isolation and killing of candidate chronic myeloid leukemia stem cells by antibody targeting of IL-1 receptor accessory protein. Proc Natl Acad Sci U S A. 2010;107:16280–16285. doi: 10.1073/pnas.1004408107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kinstrie R, Horne GA, Morrison H, et al. CD93 is a novel biomarker of leukemia stem cells in chronic myeloid leukemia. Blood. 2015;126:49. [Google Scholar]
- 56.van Rhenen A, van Dongen GA, Kelder A, et al. The novel AML stem cell associated antigen CLL-1 aids in discrimination between normal and leukemic stem cells. Blood. 2007;110:2659–2666. doi: 10.1182/blood-2007-03-083048. [DOI] [PubMed] [Google Scholar]
- 57.Culen M, Borsky M, Nemethova V, et al. Quantitative assessment of the CD26+ leukemic stem cell compartment in chronic myeloid leukemia: Patient-subgroups, prognostic impact, and technical aspects. Oncotarget. 2016;7:33016–33024. doi: 10.18632/oncotarget.9108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Valent P, Bonnet D, De Maria R, et al. Cancer stem cell definitions and terminology: The devil is in the details. Nat Rev Cancer. 2012;12:767–775. doi: 10.1038/nrc3368. [DOI] [PubMed] [Google Scholar]
- 59.Valent P, Bonnet D, Wöhrer S, et al. Heterogeneity of neoplastic stem cells: Theoretical, functional, and clinical implications. Cancer Res. 2013;73:1037–1045. doi: 10.1158/0008-5472.CAN-12-3678. [DOI] [PubMed] [Google Scholar]
- 60.Burger H, van Tol H, Boersma AWM, et al. Imatinib mesylate (STI571) is a substrate for the breast cancer resistance protein (BCRP)/ABCG2 drug pump. Blood. 2004;104:2940–2942. doi: 10.1182/blood-2004-04-1398. [DOI] [PubMed] [Google Scholar]
- 61.Brendel C, Scharenberg C, Dohse M, et al. Imatinib mesylate and nilotinib (AMN107) exhibit high-affinity interaction with ABCG2 on primitive hematopoietic stem cells. Leukemia. 2007;21:1267–1275. doi: 10.1038/sj.leu.2404638. [DOI] [PubMed] [Google Scholar]
- 62.Stain C, Stockinger H, Scharf M, et al. Human blood basophils display a unique phenotype including activation linked membrane structures. Blood. 1987;70:1872–1879. [PubMed] [Google Scholar]
- 63.Jena N, Sheng J, Hu JK, et al. CDK6-mediated repression of CD25 is required for induction and maintenance of Notch1-induced T-cell acute lymphoblastic leukemia. Leukemia. 2016;30:1033–1043. doi: 10.1038/leu.2015.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Saito Y, Kitamura H, Hijikata A, et al. Identification of therapeutic targets for quiescent, chemotherapy-resistant human leukemia stem cells. Sci Transl Med. 2010;2:17ra9. doi: 10.1126/scitranslmed.3000349. [DOI] [PMC free article] [PubMed] [Google Scholar]