Abstract
The collective somatic mutations observed in a cancer are the outcome of multiple mutagenic processes that have been operative over the lifetime of a patient. Each process leaves a characteristic imprint — a mutational signature — on the cancer genome, which is defined by the type of DNA damage and DNA repair processes that result in base substitutions, insertions and deletions or structural variations. With the advent of whole-genome sequencing, researchers are identifying an increasing array of these signatures. Mutational signatures can be used as a physiological readout of the biological history of a cancer and also have potential use for discerning ongoing mutational processes from historical ones, thus possibly revealing new targets for anticancer therapies.
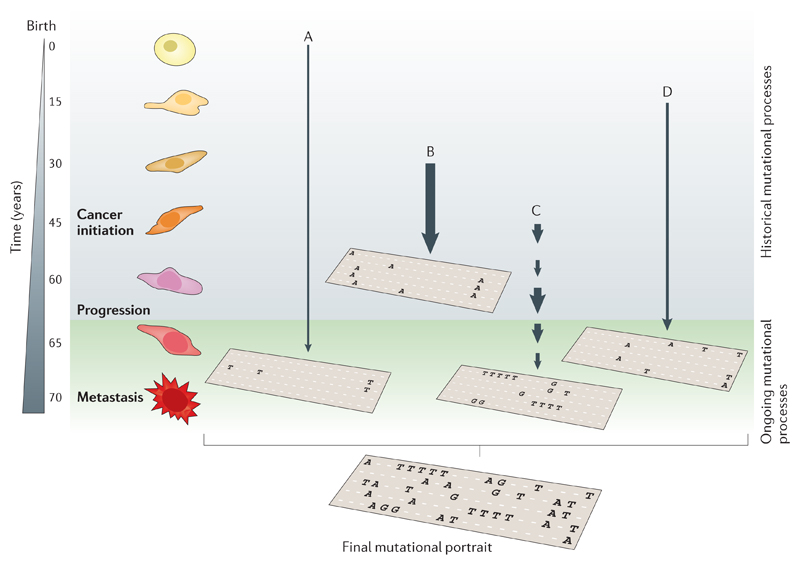
Until recently, cancer research was focused on the discovery of driver mutations (that is, key somatic mutations that are causally implicated in oncogenesis and that confer selective advantages during the evolution of a cancer)1. However, a cancer contains more than a mere handful of driver mutations. Each cancer bears many thousands of passenger mutations that may not be causative of cancer development but that are a rich source of historical information1–3. Although they are not the focus of positive selection, these bystander mutations are the product of, and therefore bear the ‘scars’ of, the biological perturbations (that is, the mutational processes) that have occurred throughout the development of a cancer1–3. Each mutational process leaves a characteristic pattern — a mutational signature — on the cancer genome, which is defined by the type of DNA damage that has occurred as a result of a plethora of exogenous and endogenous DNA damaging agents, as well as by the DNA repair or replicative mechanisms that were successively activated. Irrespective of the nature of these mutagenic or repair mechanisms, the final catalogue of mutations is also determined by the strength and duration of exposure to each mutational process2 (FIG. 1). Additionally, cancers are likely to comprise different cell populations (that is, subclonal populations), which can be variably exposed to each mutational process; this promotes the complexity of the final landscape of somatic mutations in a cancer genome3. The final mutational portrait, which is obtained after a cancer has been removed by surgery and then sequenced, is therefore a composite of multiple mutational signatures (FIG. 1).
Figure 1. Active mutational processes over the course of cancer development.
Each mutational process leaves a characteristic imprint — a mutational signature — in the cancer genome and comprises both a DNA damage component and a DNA repair component. In this hypothetical cancer genome, arrows indicate the duration and intensity of exposure to a mutational process. The final mutational portrait is the sum of all of the different mutational processes (A–D) that have been active in the entire lifetime. Ongoing mutational processes reflect active biological processes in the cancer that could be exploited either as biomarkers to monitor treatment response or as therapeutic anticancer targets. By contrast, historical mutational processes are no longer active. Signature A represents deamination of methylated cytosines, which is ongoing through life. Signature B can be matched up with the signatures of tobacco smoking, Signature C can represent bursts of APOBEC (apolipoprotein B mRNA editing enzyme, catalytic polypeptide)-induced deamination, and Signature D represents a DNA repair pathway that is awry.
The advent of next-generation sequencing technology4 has led to an extraordinary surge in the speed and scale of sequencing5,6. Large-scale sequencing of all protein-coding exons (using whole-exome sequencing) or even whole cancer genomes (using whole-genome sequencing) is achievable in a single experiment7,8. These sequencing efforts yield many thousands of mutations per cancer and thus provide sufficient power to detect mutational signatures. Mathematical algorithms can then be applied to these big, complex and multidimensional data sets to extract individual mutational signatures9,10 and to quantify these in the cancer of each patient2,9,10 (BOX 1). The number of mutations that contribute to each signature is a proxy for the amount of exposure to each mutational process, which can vary considerably from one cancer to another. Mutational signatures therefore provide an account of not only the mechanism that has gone awry in the cancer cell but also the degree to which it has been affected by this perturbation. Nevertheless, in-depth knowledge of the underlying individual mutational processes is still lacking. A better understanding of how particular mutational signatures arise is important in order to distinguish ongoing mutational processes from historical ones (FIG. 1). Historical mutational processes are informative of past exposures, and mutational signatures that underlie these processes therefore have an important message regarding cancer prevention and public health. However, they have limited value as biomarkers or therapeutic targets, as they are no longer actively promoting cancer development. By contrast, ongoing mutational processes could be used as prognostic indicators, as predictors of therapeutic sensitivity or as targets of disease control.
Box 1. Extracting mutational signatures from complex data sets.
Exploring the sequence context of somatic substitutions in cancer
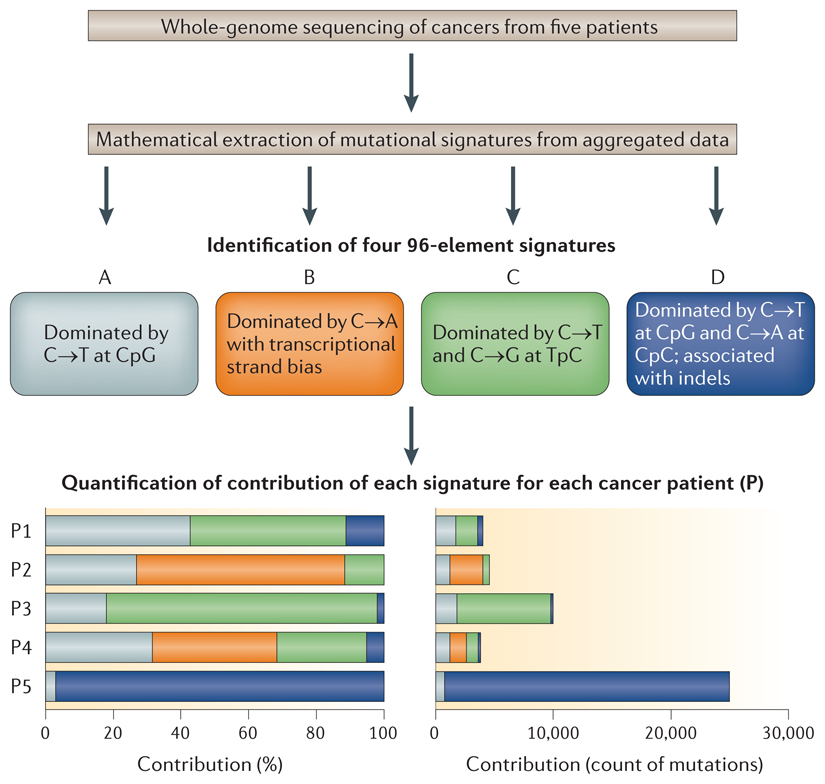
Mathematical approaches can be used to identify mutational patterns in a data set of base substitutions pooled from several cancer samples obtained from multiple patients (see the figure). The process of extracting multiple patterns from a complex multidimensional pool of data is described as a blind source separation problem. In this case, the multidimensional data set comprises the 96 possible combinations of base pair substitution mutations (that is, when the immediate flanking sequence context is taken into account). Adding an additional flanking base to the one immediately adjacent to the mutation would give 1,536 possible mutations, and the number of mutations per cancer genome will then become the statistically limiting factor. Several different mathematical methods can be applied to solve this problem.
A mathematical approach for extracting mutational signatures
Non-negative matrix factorization (NMF) and model selection is simply one of many approaches that have previously been developed to factorize or reduce complex multidimensional data sets in order to identify common, defining underlying patterns that make up a pooled data set113. Consider that each 96-element data set is akin to a ‘face’ of a cancer. Each of these cancer faces is similar to a human face and is a complex assembly of features; nevertheless, it is recognizable as an individual face. The application of NMF to a pool of images of faces yields interpretable underlying ‘features’ that are shared across the group of faces, such as the eyes, nose and mouth. The aggregate of somatic substitutions of each cancer is essentially the face of a cancer, and each extracted feature is equivalent to an individual mutational signature. In this case, aggregated data are parsed through NMF in order to obtain the signatures that underlie the data sets, and 96-element signatures are extracted (see the figure, Signatures A–D).
Quantifying the amount of each signature in each cancer
For each mutational signature, NMF allows estimation of the relative contribution of each signature to the final mutational catalogue of individual cancers (see the figure). The amount of each signature can be quantified in each cancer either as a proportional contribution or as absolute numbers. NMF can therefore both highlight cancers that are driven predominantly by a single mutational signature and identify cancers that have a combination of many different signatures (see the figure). NMF can identify even the lowest levels of signatures that are ubiquitously present9.
In this Review, we present examples of mutational signatures according to different classes of mutations, including base substitutions, insertions and deletions (indels), and structural variations (also known as genomic rearrangements). We emphasize how different DNA damaging agents and DNA repair and replication pathways contribute mechanistically in the generation of each signature type, and our main purpose is to show the wealth of biology that could be discovered in the totality of somatic mutations.
Mutational signatures of base substitutions
Historically, simple analyses of somatic base substitutions as six-bar mutational spectra (C∙G→A∙T, C∙G→G∙C, C∙G→T∙A, T∙A→A∙T, T∙A→C∙G and T∙A→G∙C) have been useful in highlighting typical but crude mutation patterns that show how mutational spectra can be specific to tumour type and related to exogenous carcinogens. For example, mutations associated with smoking-related damage in lung cancers are mainly G∙C→T∙A transversions11, whereas mutations associated with ultraviolet (UV) radiation exposure in skin cancers comprise predominantly C∙G→T∙A transitions8. However, the flanking sequence context of a mutation (that is, the neighbouring bases immediately 5′ and 3′ to the mutated base) is known to affect mutation rates in the genome12 and should therefore be taken into consideration when defining a mutational signature. As there are 6 classes of base substitutions and 16 possible sequence contexts for each mutated base (A, C, G or T at the 5′ base and A, C, G or T at the 3′ base), 96 different mutated trinucleotides are possible2,9,10. The following convention has been adopted to describe mutations; for example, a cytosine mutation flanked by a 5′ thymine and a 3′ guanine is represented as TpCpG, and the mutated base is underlined.
In a recent mathematical analysis, 21 different mutational signatures were identified in 96-trinucleotide format from the somatic mutations of >7,000 sequenced primary human cancers of 30 different cancer types9. Although some of these signatures were known (for example, an excess of C∙G→T∙A transitions particularly at dipyrimidines (Signature 7) has previously been shown to be associated with UV radiation and is found in cutaneous malignancies13), many were novel. Importantly, each base substitution signature represents a pattern that consists of 96 elements, which vary in their relative amounts. A particular element — such as C∙G→T∙A transitions at TpCpN (where N denotes any base) — may be the overriding feature within a mutational signature, but the element is not considered to be a signature per se.
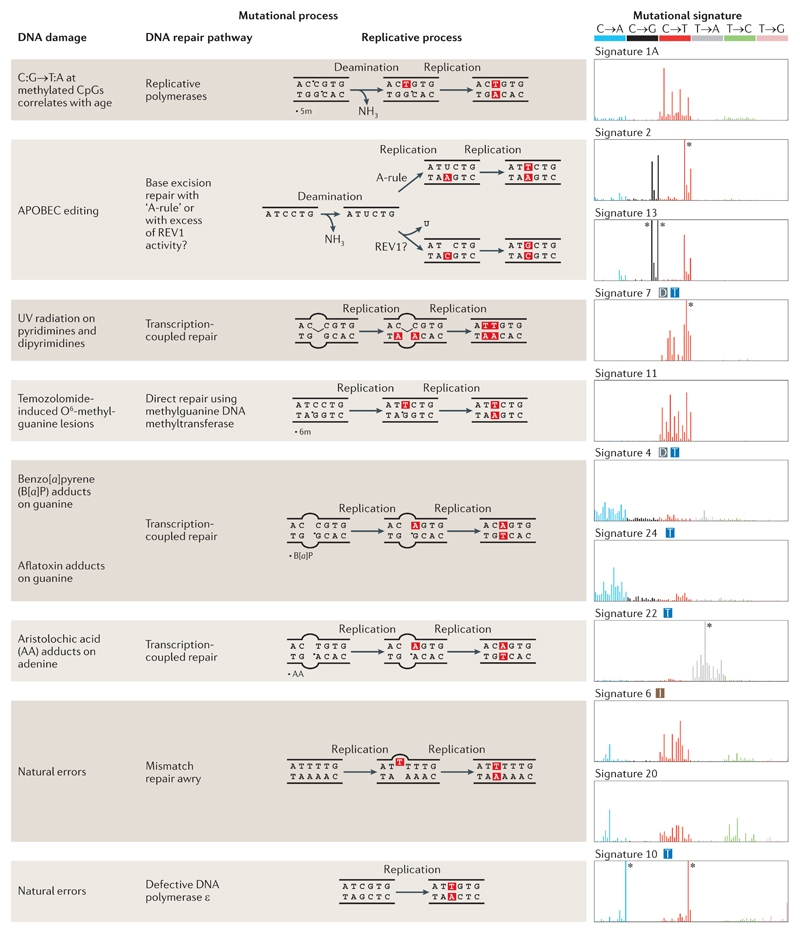
Below, we consider some examples of base substitution signatures on the basis of the different categories of mutational processes that underlie each signature. Mechanistically, each mutational process comprises both a DNA damage component and a DNA repair or replicative component (FIG. 2). Each type of DNA damage has its own predilection for specific nucleotides, which can produce recognizable patterns of mutagenesis. The most prominent base substitution signatures are illustrated (FIG. 2) to show the 96-element pattern of each signature, as well as the DNA damage and repair or replication components that constitute the determinant mutational process.
Figure 2. Summary of known mutational signatures, and the components of DNA damage and repair that constitute the mutational processes.
There are marked differences among the 96-element mutational signatures, which are dominated by specific elements, including enrichment of various base substitutions (shown in the graphs on the right), transcriptional strand bias (T), excess of dinucleotide mutations (D), and association with insertions and deletions (I). The asterisks mark instances at which the limits of the y axes, which represent the likelihood of specific mutations being present in a signature, are exceeded. 5m, 5′ methyl group; 6m, O6 methyl group; APOBEC, apolipoprotein B mRNA editing enzyme, catalytic polypeptide; REV1, DNA repair protein REV1; UV, ultraviolet.
Mechanisms underlying substitution signatures
Endogenous DNA damage
Several different 96-element mutational signatures have been linked to mutagenic processes that are attributed to deamination, which occurs spontaneously in all DNA bases that contain primary amines albeit at markedly different rates. Common deamination reactions include 5-methylcytosine→thymine, cytosine→uracil and adenine→hypoxanthine reactions.
The hydrolytic deamination of 5-methylcytosines at CpG dinucleotides14 has occurred so frequently throughout evolution that it is thought to be the reason for the depletion of the number of methylated CpGs observed in the human genome14. Despite the reduction in absolute numbers of these sites, it remains one of the most mutagenic sequence motifs, and a net effect of C∙G→T∙A transitions is observed at methylated CpG dinucleotides. Consistent with this phenomenon, C∙G→T∙A substitutions at NpCpG are characteristic of two of the most frequent mutational signatures — Signatures 1A and 1B — which have collectively been documented in at least 25 different cancer types9. These signatures possibly represent one biological process but tend to be separated mathematically because of limitations regarding the number of samples in the data sets examined so far and the algorithm used9,15. Intriguingly, a correlation between the burden of mutations associated with these signatures and the patient age at the time of cancer diagnosis has been reported for several cancer types, including adult cancers (for example, acute myeloid leukaemia, breast cancer, glioma, head and neck cancers, kidney clear cell cancer, malignant melanoma and ovarian cancer) and paediatric cancers (for example, acute lymphoblastic leukaemia and neuroblastoma) in both males and females9. This suggests that this mutational process is occurring in cells prior to malignant transformation.
The deamination process of cytosine to uracil is thought to be catalysed by members of the cytidine deaminase family (which include activation-induced cytidine deaminase (AICDA) and the APOBEC (apolipoprotein B mRNA editing enzyme, catalytic polypeptide) enzymes). AICDA is the most well characterized of this family of DNA editing enzymes; it has a role in antibody diversification and shows a strong preference for deaminating cytosine residues that are flanked by a 5′ purine16. By contrast, the APOBECs — which have variable roles, including restriction of retroviruses and mobile retroelements — show various sequence specificities, for example, APOBEC1, APOBEC3A, APOBEC3B and APOBEC3C show a preference for a TpC sequence context in experimental systems such as yeast and human cell lines17–19. First characterized in breast cancers2,5,20, signatures with a thymine preceding a mutated cytosine (TpCpN; Signatures 2 and 13) have been observed in 16 other cancer types9. Particular members of the cytidine deaminase family (APOBEC3A, APOBEC3B and APOBEC1) have been speculated to underlie this phenomenon given the similarity between sequence specificity observed in cancers and that observed in vitro2,19. Aggregated expression-based analyses have shown correlations with the burden on mutated cytosines at a TpCpN context, which led the authors to suggest that APOBECs are a mutagenic source of these signatures21,22. There is additional support for a role of APOBECs in the generation of the DNA damage component of these signatures. Intriguingly, mutations associated with Signatures 2 and 13 show a high degree of strand coordination: they arise on the same parental allele and are on the same DNA strand; that is, successive mutations can be C→T then C→G followed by C→T, or G→A then G→C followed by G→A, but not C→T, G→A followed by C→T)2,9,23. This strand-coordinated nature of TpCpN mutations argues in favour of APOBEC-related activity, as APOBECs preferentially cause deamination of stretches of single-stranded DNA (ssDNA)23–25. Furthermore, a germline copy-number polymorphism involving the neighbouring APOBEC3A and APOBEC3B genes that essentially deletes all of the genomic region encompassing APOBEC3B apart from its 3′ untranslated region has been shown to act as a modest susceptibility allele in breast cancer23. Carriers of at least 1 copy of the deletion polymorphism have a 2.37-fold increased relative risk of harbouring cancers that comprise Signatures 2 and 13. Interestingly, although these two signatures are likely to arise through the same DNA damage mechanism of APOBECs, Signature 13 is dominated by C∙G→G∙C transversions. In other words, the sequence context of mutated cytosine bases is shared with Signature 2 (TpCpN) because the DNA damaging enzyme is possibly identical; however, the excess of transversions in Signature 13 relative to transitions in Signature 2 suggests subtly different involvement of repair or replicative polymerases (see below) between the two signatures (FIG. 2).
Adenine can deaminate to hypoxanthine at a rate of 10% of the cytosine deamination rate26. The product pairs preferentially with cytosine during replication and can give rise to A∙T→G∙C transitions27. Several signatures characterized by A∙T→G∙C transitions (Signatures 5, 12, 16 and 21) have been found in primary human cancers, although none has been specifically attributed to this mutational process so far.
Free radical species such as reactive oxygen species or nitrogen oxide species are generated endogenously as by-products of normal cellular metabolism, including apoptosis and the inflammatory response, as well as by exposure to exogenous agents such as ionizing radiation28. Their interaction with DNA can lead to >25 different oxidative DNA base lesions29. One of the best studied oxidative DNA lesions of reactive oxygen species is 8-oxo-2′-deoxyguanosine. It has been shown to favour hydrogen bonding with adenine, which gives rise to G∙C→T∙A transversions with evidence for GpGpG sequence specificity in vitro30,31. A mutational signature derived from primary human cancers has not been attributed to this oxidative DNA lesion, although two novel signatures are noted to mainly comprise G∙C→T∙A mutations (Signatures 8 and 18)9.
Exogenous DNA damage
Environmental sources of DNA damage can be physical or chemical (BOX 2). Non-ionizing UV radiation is an example of a physical agent with enough energy to excite molecular bonds that cause covalent modifications between neighbouring pyrimidine nucleotides. These modifications result in pyrimidine dimers: (6–4) pyrimidine photoproducts ((6–4)PPs) and cyclobutane pyrimidine dimers (CPDs)13,32. Consistent with this finding, a preponderance of C∙G→T∙A mutations at dipyrimidines (that is, two adjacent pyrimidines) and an excess of CC∙GG→TT∙AA double substitutions (Signature 7) (FIG. 2) are characteristic features of cutaneous cancers that are associated with UV exposure, such as squamous cell skin carcinomas and malignant melanomas9,33. Indeed, the effect is so pronounced that CC∙GG→TT∙AA double substitutions can constitute up to 25% of the total (and often very large) mutation burden in those cancers9 and can be used as a clear indicator of UV-related DNA damage. Mechanistically, Signature 7 is caused by deamination of cytosines to uracil within (6–4)PPs or CPDs at sites of stalled transcription complexes33, which triggers the activity of transcription-coupled repair (TCR; see below). This process explains why the signature shows a transcriptional strand bias9 (that is, a lower prevalence of mutations on the transcribed strand than on the non-transcribed strand).
Box 2. An overview of types of DNA damage and causal agents.
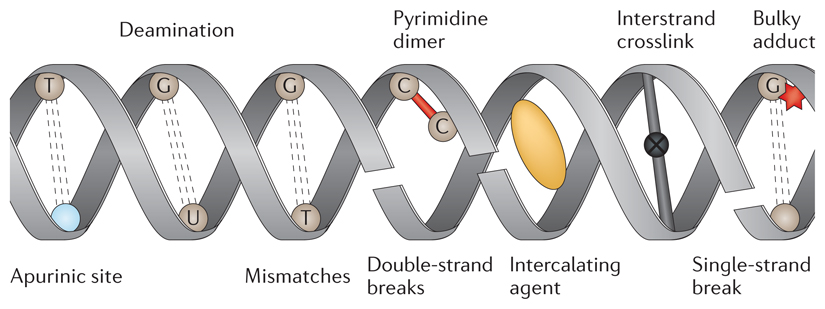
DNA is under a constant stream of attack from various exogenous and endogenous sources. Each mutagen can cause damage either directly or indirectly to the nucleotides in the genome. Moreover, each mutagenic agent shows a predilection for damaging specific nucleotides, which can produce recognizable patterns of mutagenesis.
Sources of DNA damage include endogenous factors such as spontaneous or enzymatic conversions. The N-glycosidic bond that links a nucleobase and a pentose sugar to form a nucleoside is labile. This fact underlies the common occurrence of spontaneous base loss in DNA (~104 bases per cell per day)27, which results in the formation of apurinic or apyrimidinic sites (see the figure). Depurination occurs more readily than depyrimidination, which makes apurinic sites more common than apyrimidinic sites, and A∙T→T∙A or G∙C→T∙A transversions arise depending on the purine that is lost.
Other types of endogenous DNA damage include deamination, replication errors and free radical species. Free radical species are generated either as a by-product of metabolism or through exposure to exogenous physical agents, such as ionizing radiation, which can induce the formation of double-strand breaks. By contrast, non-ionizing ultraviolet radiation is responsible for biochemical modifications, such as the formation of pyrimidine dimers, which can be mutagenic when left unrepaired. Other external agents that are known to cause DNA damage include chemical compounds, for example, platinum-based compounds such as cisplatin, which can cause bulky adducts or interstrand and intrastrand crosslinks; intercalating agents such as benzo[a]pyrenes, daunorubicin and actinomycin-D; DNA alkylating agents such as nitrogen mustards, methyl methanesulphonate (MMS), N-nitroso-N-methylurea (NMU) and N-ethyl-N-nitrosourea (ENU); and psoralens.
Chemical compounds intercalate or covalently bind to DNA in various ways and can produce particular mutational signatures. For example, chemotherapeutic alkylating agents such as cyclophosphamide and temozolomide result in C∙G→T∙A transitions5 (Signature 11)9, whereas benzo[a]pyrene (B[a]P) diol epoxides — a carcinogenic by-product of tobacco smoking34,35 — cause G∙C→T∙A transversions and have a predilection for methylated CpG dinucleotides11 (Signature 4)9 (FIG. 2). Psoralens, which are a type of phototherapeutic agent used for inflammatory conditions such as psoriasis, lead to pyrimidine mutations at a TpA sequence context36,37; aristolochic acid, which is a plant extract linked to nephropathy and urothelial tumours, is associated with a T∙A→A∙T signature38 (Signature 22) (FIG. 2). These examples highlight the variability in mutational signatures that can be produced through a myriad of exposure to chemicals. The ability to pinpoint chemical mutagens to specific signatures means that a patient’s history of past exposure to specific chemicals could be revealed by analysing their tumours. Many other chemical compounds are known to cause DNA damage (BOX 2), although specific signatures remain to be discovered and assigned to these agents.
DNA repair processes
It is impossible to exhaustively describe all repair pathways here; hence, we give brief descriptions that focus on how each repair pathway leaves its molecular mark on a genome and how its disruption can result in specific mutational signatures (FIG. 3).
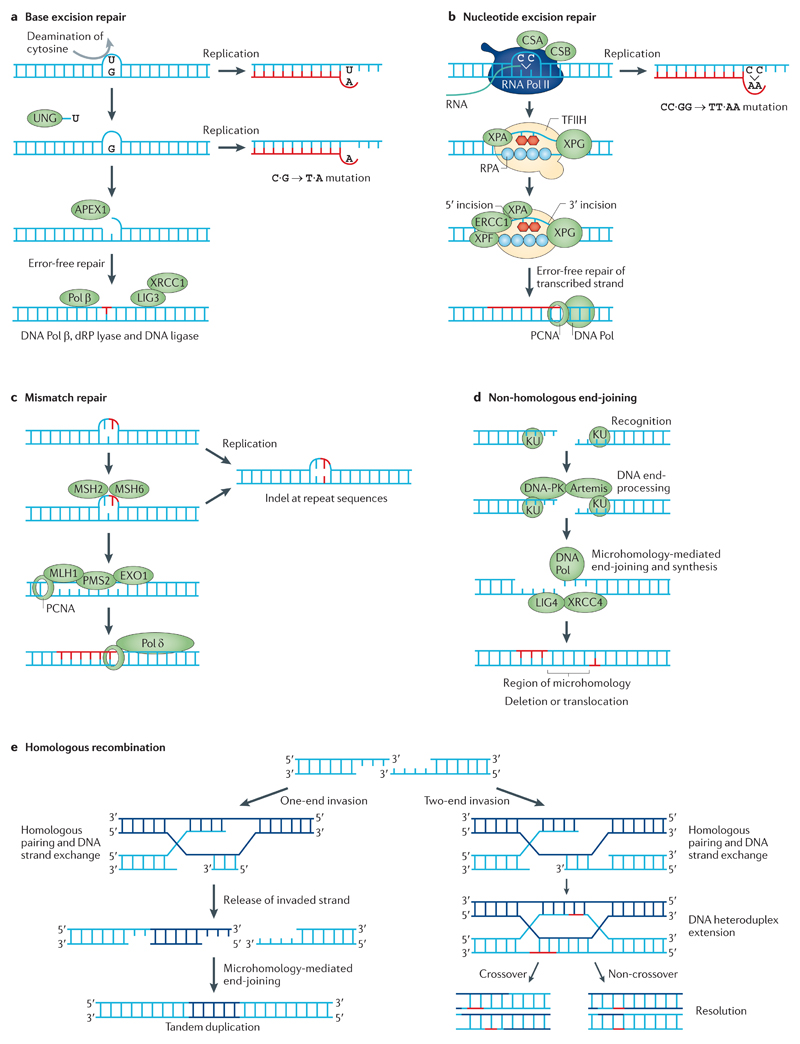
Figure 3. DNA repair pathways and mutational consequences.
a | Base excision repair (BER) typically mediates the removal and replacement of a single base residue. Substrates include uracil residues in DNA (which are created by deamination of cytosines) and damaged bases caused by reactive oxygen species, hydrolytic reactions and methylation. A damaged base is removed by a specific DNA glycosylase; here, the uracil is removed by uracil-DNA glycosylase (UNG). The resulting apurinic or apyrimidinic site is incised by DNA-(apurinic or apyrimidinic site) lyase APEX1. The 5′-deoxyribose-phosphate (dRP) residue is removed by a dRP lyase, which leaves a one-nucleotide gap that is filled in by DNA polymerase β (Pol β). Replication before completion of repair leads to base misinsertion and potentially C∙G→T∙A mutations. b | Nucleotide excision repair (NER) can remove various helix-distorting adducts, including those caused by ultraviolet radiation and cisplatin. The distorted region is recognized either during global genome repair by XPC (DNA repair protein complementing XP-C cells)–RAD23B (not shown) or during transcription, and two incisions are made on either side of the adduct to excise the damaged DNA. The resulting 27–29-nucleotide gap is filled by Pol δ or Pol ε and, under some circumstances, Pol κ. Replication before repair may result in mutations. c | Mismatch repair (MMR) is an excision repair process that removes mismatched bases or misinserted bases in DNA. It is initiated by the DNA mismatch recognition proteins MSH2 and MSH6; a segment of DNA is excised between the mismatch and a nearby nick by the MMR endonuclease PMS2 and exonuclease 1 (EXO1). The gap that is left in the DNA is filled by Pol δ. Failed MMR results in a high mutation load in microsatellite repeat sequences. d | DNA double-strand breaks (DSBs) can be repaired by non-homologous end-joining (NHEJ), which is often mediated by microhomology at ends. DSBs caused by ionizing radiation or by enzymes that cleave DNA usually do not yield DNA ends that can be ligated directly. End-trimming and resynthesis of bases are therefore required to join breaks, which may give rise to mutations. e | An alternative strategy for DSB repair is homologous recombination (HR). HR only operates when a double-stranded copy of the sequence is available, for example, as a sister chromatid in late S or G2 phase of the cell cycle, which may give rise to tandem duplication. CSA and CSB are also known as DNA excision repair protein ERCC8 and ERCC6, respectively; DNA-PK, DNA-dependent protein kinase; indel, insertion and deletion; LIG3, DNA ligase 3; PCNA, proliferating cell nuclear antigen. Figure from REF. 114, Nature Publishing Group.
In base excision repair (BER), a base lesion is identified by a DNA glycosylase that recognizes, hydrolytically cleaves and removes the altered base, which gives rise to an apurinic or apyrimidinic site39 (FIG. 3a). Unrepaired apurinic or apyrimidinic sites are particularly mutagenic, as incorrect bases are easily introduced during replication. Subsequently, DNA-(apurinic or apyrimidinic site) lyase APEX1 incises the DNA strand 5′ to the apurinic or apyrimidinic site. The replicative DNA polymerase β (Pol β) catalyses the elimination of the 5′-deoxyriboso-phosphate residue and then fills the one-nucleotide gap. Finally, the nick is sealed by the DNA ligase III–XRCC1 complex40,41 (FIG. 3a). Multiple mutation patterns have been associated with engineered defects of certain DNA glycosylases in mouse embryonic fibroblasts. For example, defects in single-strand selective monofunctional uracil DNA glycosylase (SMUG1) have been linked to C∙G→T∙A transitions42, whereas disruption of the DNA glycosylase OGG1 has been associated with G∙C→T∙A transversions43. However, 96-element signatures extracted from human cancers have not been attributed to defects in specific components of the BER pathway so far.
Nucleotide excision repair (NER) is a nonspecific repair process that is activated upon sensing of bulky DNA distortions, for example, bulky adducts caused by B[a]Ps and by aromatic amines such as aflatoxin, as well as modifications due to platinum-based compounds, psoralens and UV-induced lesions (that is, CPDs and (6–4)PPs) (reviewed in REF. 44) (FIG. 3b). A particular class of NER that is coupled to transcription is TCR44. A consequence of TCR is that DNA damage on the transcribed strand is repaired more efficiently than that on the non-transcribed strand. The activity of TCR is appreciable in several mutational signatures. For example, C∙G→T∙A transitions that constitute the UV-associated Signature 7 show transcriptional strand bias; that is, fewer mutations are found on the transcribed strand than on the non-transcribed strand45. This bias is also seen in other mutational signatures, including those caused by B[a]Ps34 (Signature 4) and aristolochic acid46 (Signature 22). Several novel signatures that show transcriptional strand bias (Signatures 5, 8, 12 and 16) have additionally been identified9, which suggests that these could be caused by DNA damaging agents that are repaired by TCR. However, BER was recently shown to also display transcriptional strand bias47, which suggests that there are alternative mechanisms that can generate strand bias in these signatures.
The post-replicative mismatch repair (MMR) system recognizes and repairs misincorporated bases, as well as erroneous indels that arise during DNA replication and DNA recombination repair activity (extensively reviewed in REFS 48,49) (FIG. 3c). MMR reduces the rate of replication-associated errors by 100-fold to 1 in 10−9 (reviewed in REF. 48). Hence, defects in the MMR pathway increase the spontaneous mutation rate50. Mutations in MMR-related proteins affect genomic stability and result in microsatellite instability51. MMR-related base substitution signatures have not been previously shown in experimental systems. Nevertheless, a base substitution signature extracted from primary human cancers (Signature 6) — which is characterized by C∙G→T∙A transitions at an NpCpG sequence context and C∙G→A∙T transversions at CpCpC — has been associated with MMR deficiency (biallelic somatic mutations in MMR genes and particularly those affecting MLH1 methylation)9. Furthermore, cancers that contained a high proportion of this signature also showed thousands of small 1-bp indels, which is a feature associated with microsatellite instability9. More recently, additional signatures (Signature 20 and a new pattern, Signature 26) have been additionally associated with MMR deficiency (S.N.-Z., unpublished observations). These may relate to specific MMR defects, although the data required to confirm this are not currently available.
DNA replication errors
Given the size of the human genome (~3 × 109 nucleotides), even the smallest error rate during DNA synthesis can result in many mutations, which underscores the replication machinery as a source of mutagenesis. DNA polymerases use a template DNA strand to select nucleotides for incorporation into the nascent strand during both DNA replication and synthesis associated with DNA repair; however, replication mismatches can be generated on the nascent strand (reviewed in REF. 52). The high-fidelity B family DNA polymerases Pol δ and Pol ε have an error rate of 1 in 10−7 for every nucleotide synthesized owing to intrinsic proofreading properties53. Somatic and germline mutations in Pol ε have been associated with Signature 10 in colorectal and endometrial carcinomas9,54,55, and they result in a striking pattern of C∙G→A∙T and C∙G→T∙A mutations at TpCpG (FIG. 2). It has been suggested that the increased rate of mutagenesis associated with mutations in Pol ε exceeds that expected after loss of proofreading capacity, which indicates a distinct defect of replication fidelity or an active mutagenic process56.
An additional factor that affects the likelihood of nucleotide misincorporation by replicative DNA polymerases such as Pol δ and Pol ε is the balance of the cellular deoxynucleoside triphosphate (dNTP) pool. Loss of the usual constraints on cell cycle regulation during cancer development causes an increased demand on a potentially reduced dNTP pool. Perturbations of the dNTP pool can lead to insertion–deletion loops and erroneous base incorporation; they can also affect proofreading efficiency57 and be another source of replication-related mutagenesis58–60.
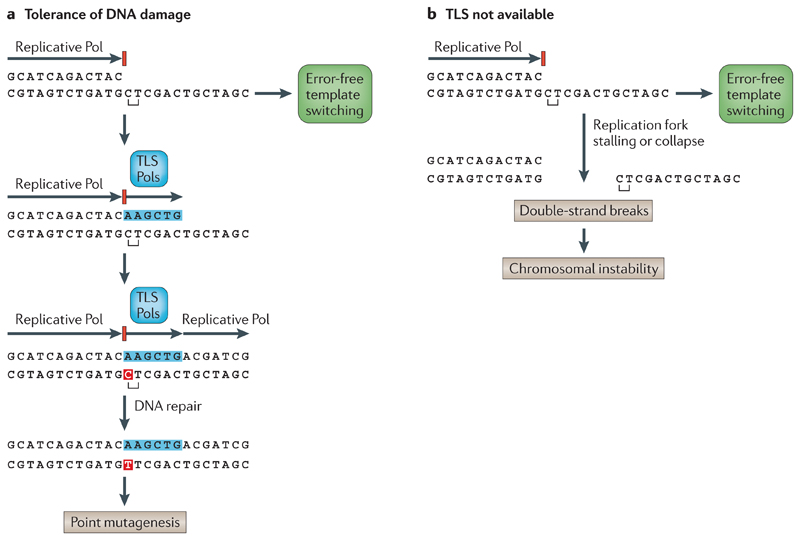
A collection of low-fidelity error-prone polymerases — such as Pol η, Pol ι, Pol κ and DNA repair protein REV1 — can replicate damaged DNA or non-informative DNA templates. These translesion polymerases have a higher error rate (which ranges between 1 in 10−4 and 1 in 10−1) than nuclear DNA replication polymerases because they lack proofreading capacity and are poor discriminators of mismatched, non-fitting nucleotides (reviewed in REF. 61). This phenomenon known as DNA damage tolerance (reviewed in REFS 61–63) is crucial to allow completion of replication at the cost of introducing errors — which may be fixed later by excision repair pathways — and to avoid replication fork collapse (FIG. 4). However, by providing this escape route, translesion polymerases can produce a myriad of potential mutational spectra. For example, the preference for insertion of an adenine opposite an apurinic or apyrimidinic site (that is, the ‘A-rule’)64 results in different signatures depending on the original base at the site: adenine loss would lead to A∙T→T∙A, whereas guanine loss would lead to C∙G→A∙T. A signature characterized by T∙A→G∙C transversions at ApTpN and TpTpN trinucleotides (Signature 9), which is seen in haematological malignancies that have undergone somatic hypermutation at the immunoglobulin (IG) loci, has been attributed to the activity of Pol η65, although the precise mechanism remains unclear. Moreover, the error-prone polymerase REV1 generates a C∙G→G∙C signature61,66 (FIG. 2).
Figure 4. Bypass of replication forks blocked by lesions.
a | In the presence of a translesion DNA synthesis (TLS) polymerase (Pol), a lesion can be bypassed by TLS, which can result in point mutagenesis. An error-free alternative to bypass a stalled replication fork is template switching. Point mutations are marked in red. b | In the absence of a TLS Pol, a translesion bypass is not possible (although some template switching still occurs), and the stalled replication fork collapses. This leads to double-strand breaks and chromosomal instability. Figure from REF. 114, Nature Publishing Group.
Taken together, these studies indicate that irrespective of the damaged base, the resulting mutation — regardless of whether it is a transition or transversion — is likely to be determined by the replicative process.
Mutational signatures of indels
Indel signatures
Modern mathematical methods for extracting base substitution signatures can be integrated with other mutation classes (such as indels) to provide important insights. Although indels can be of any size, we focus on small indels (<100 bp) here. Currently, calling somatic indels from next-generation short-read sequencing data still yields a high rate of false positives owing to technical difficulties associated with mapping of short-read sequencing data and to limitations of mutation-calling algorithms. Additionally, as fewer indels are generally identified in human cancers than base substitutions9, the power to detect patterns of indel generation is relatively limited. Nevertheless, some early patterns can be derived from analyses based on the size of indels and on the characteristics of the deletion junctions.
Small 1–3-bp indels within repetitive sequences correlate with a base substitution signature that is characterized by an excess of C∙G→T∙A mutations at NpCpG (Signature 6)9. Individual cancers can be overloaded both by mutations associated with Signature 6 and by small indels, as has been reported in colorectal, uterine, kidney, liver, prostate, oesophageal and pancreatic cancers. By contrast, larger indels (between 4 bp and ~50 bp) that show a degree of sequence similarity between the indel motif and the immediate junction sequence (that is, microhomology) have been associated with a base substitution signature that is characterized by a fairly uniform distribution of mutations across all 96 possible base substitution types (Signature 3)9. This signature has been reported in breast, ovarian and pancreatic cancers9.
Mechanisms of indel signature formation
The two contrasting indel signatures described above are thought to arise as a result of defects in the DNA repair machinery. For example, loss of MMR in humans leads to microsatellite instability — an indel phenomenon that can be recognized owing to variation in repeat length at mononucleotide or dinucleotide repetitive sequences, which is frequently observed in colorectal carcinomas67,68. The mechanistic importance of post-replicative MMR as a constraint on the generation of indels during replication is emphasized by studies showing that spontaneous indel error rates in repetitive sequences increase by many orders of magnitude when MMR is inactivated, and this is shown to be an overwhelming feature of cancers of individuals with inherited germline mutations in MMR genes69,70. Consistent with these reports, the excess of Signature 6 and the associated abundance of small indels (1–3 bp) at polynucleotide tracts are concomitant with inactivation of the MMR genes in affected cancers9. In addition, a signature of indels on a background of MMR deficiency is highly reproducible in experimental systems71.
Overlapping microhomology is often considered to be a signature of non-homologous end-joining (NHEJ) repair of DNA double-strand breaks (DSBs), in which short segments of homology are aligned to mediate the joining of the two DNA fragments72,73 (FIG. 3d). Signature 3 is associated with inactivating mutations of BRCA1 (breast cancer 1, early onset) and BRCA2 (REF. 9). The protein products of BRCA1 and BRCA2 are involved in error-free homologous recombination-based DSB repair74,75, in which BRCA1 controls resection of DNA ends76 and BRCA2 is required for loading of RAD51 onto ssDNA77. Thus, the increased frequency of microhomology-mediated indels in BRCA1- or BRCA2-null cancers might reflect the requirement for alternative methods of DSB repair in these cancers. However, it remains unclear how defects in these two distinct components of the homologous recombination pathway can result in a final characteristic readout of a somatic base substitution that correlates with a signature of larger, microhomology-mediated indels (Signature 3). This signature may either reflect the supplementary roles of BRCA1 or BRCA2 in the response to DNA damage or be a result of the increased recruitment of error-prone polymerases to compensate for the inability to use homologous recombination to bypass a lesion.
Mutational signatures of structural variations
The landscape of somatically acquired rearrangements is extremely diverse and ranges from very few mutations to tens or hundreds of mutations per cancer78. Some cancer-associated rearrangements are functional driver events and are under strong selection, including amplification of oncogenic regions, whole-exon or whole-gene deletions, losses of whole chromosomal arms that involve tumour suppressor genes and translocations that produce oncogenic fusion genes79. However, most rearrangements are passenger events78. The ability to call somatic rearrangements from next-generation sequencing data is still fraught with suboptimal sensitivity and specificity owing to the limitations of current rearrangement-calling algorithms. Hence, cancer genome data sets are not as comprehensively characterized for structural variations as they are for base substitution mutations. Nevertheless, the patterns of somatic rearrangements, their spatial distribution throughout the genome and the junctional features at breakpoints of available rearrangement data sets reveal some mechanisms of damage and repair that are involved in the generation of somatic structural variations.
Structural variations arise from DSBs through either direct or indirect mechanisms, which can determine the resulting molecular signature. Primary DSBs are due to direct lesions that cause breaks in the sugar-phosphate backbone (for example, by ionizing radiation), whereas secondary DSBs are the result of complex DNA lesions which, when encountered by a replication fork, induce replication collapse80,81. Each type of DSB repair mechanism will leave its own characteristic imprint of activity in the genome.
Microhomology-mediated end-joining (MMEJ) is a subtype of NHEJ (FIG. 3d), in which the ligation is facilitated by microhomologies between ssDNA exposed at the DNA ends as a result of limited end-processing activities. MMEJ is commonly involved in somatic structural variation from primary cancers and cell lines78, as well as from experimental DSB repair models72, particularly in systems in which homologous recombination is defective. In mammalian somatic cells, NHEJ and MMEJ activity on double-ended DSBs occurs throughout the cell cycle82, whereas homologous recombination acts on replication-associated or G2-induced double-ended DSBs at which a homologous sister chromatid is available83,84. The near-constant action of NHEJ throughout the cell cycle makes its contribution almost ubiquitous in all forms of structural variations. The mark of NHEJ is essentially the absence of sequence homology or, more commonly, the presence of MMEJ at breakpoints in a distribution that is different to that expected if microhomology had occurred randomly78,85. Unsurprisingly, MMEJ of all forms of structural variations has been reported86–88.
Tandem duplications
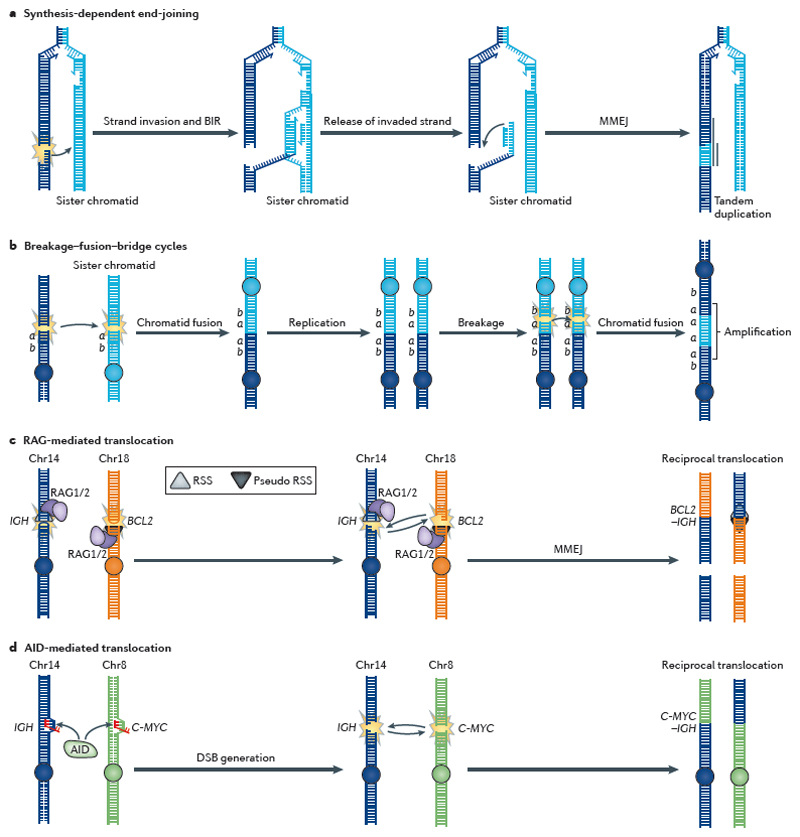
Rearrangements of tandem duplications (that is, identical sequences duplicated in head-to-tail formation) with microhomology junctions have been reported in breast2,78 and ovarian cancers89. Some of these cancers have shown biallelic loss of BRCA1 (REFS 2,78). Interestingly, a specific homologous recombination subpathway that is distinct from RAD51-mediated homologous recombination has been implicated in the generation of tandem duplications90. In this pathway, DNA ends at DSBs that occur at replication forks invade the sister chromatid to restart replication in a process known as break-induced replication (BIR)91 (FIG. 5a). The invaded strand can be released by branch migration and the new extended double-stranded DNA end repaired by MMEJ, which leaves a tandem duplication90. This combination — termed synthesis-dependent end-joining (SDEJ) — has previously been described to provide an explanation for tandem duplications in mammalian genomes92. Specifically, SDEJ is initiated in a similar manner to all homologous recombination events by resection of the DNA end, followed by strand invasion of the sister chromatid and DNA extension on the D-loop93 (FIG. 3e; FIG. 5a). However, unlike the synthesisdependent strand annealing model for DSB repair94, synthesis on the lagging strand is also initiated on the sister chromatid, and the released DNA molecule will be partly double-stranded. When this is ligated onto the opposite DNA end (using MMEJ), a tandem duplication is produced as replication extends beyond the original breakpoint (FIG. 5a). Upsetting the balance of error-free homologous recombination-based DSB repair could result in upregulation of other components of DSB repair, such as SDEJ, and result in the tandem duplication signature. This possibility reflects the complex nature of homologous recombination, which involves several pathways with specific enzymatic requirements.
Figure 5. Gene rearrangements in cancer.
Gene rearrangements in cancer arise primarily from DNA double-strand breaks (DSBs). a | Synthesis-dependent end-joining (SDEJ), which is involved in repairing replication-associated DSBs, results in tandem duplications. Break-induced replication (BIR) initiates synthesis on the sister chromatid after strand invasion. Reversed branch migration of the Holliday junction formed following strand invasion can release the invaded strand, which contains extra DNA material from the sister chromatid and is fused to the original end by microhomology-mediated end-joining (MMEJ), resulting in a tandem duplication90. b | Sister chromatid fusion causes gene amplification by breakage–fusion–bridge cycles96,97. In this process, two adjacent DSBs on sister chromatids are substrates for non-homologous end-joining (NHEJ), which rejoins the sister chromatids. After replication, these are again broken to form anther fusion chromosome carrying four gene a copies. c | The V(D)J recombination-activating (RAG) proteins recognize either the correct recombination signal sequences (RSSs) or almost identical (that is, pseudo) RSSs at which they initiate DSBs; they then mediate interchromosomal translocation rather than regular recombination within the V(D)J segments. This can create an immunoglobulin H (IGH)–BCL2 (B-cell CLL/lymphoma 2) fusion gene that drives cancer. d | Activation-induced cytidine deaminase (AID) is involved in class switch recombination and deaminates cytosines to uracils in transcribed regions, which are then processed by DNA repair enzymes into a DSB. If DSBs coexist in the IGH and C-MYC genes, then they can recombine by interchromosomal translocation to produce an IGH–C-MYC fusion gene. Chr, chromosome.
Clustered structural variations
Somatic structural variations — for example, oncogenic amplifications such as HER2 (also known as ERBB2) in breast cancer — are regional or topographically clustered95. These somatic events show high levels of copy number (>5) and many types of microscopic rearrangements within a macroscopic region. They are also recurrent (as a result of positive selection) and show concomitant elevated levels of expression of the relevant oncogene95. The exact mechanisms that cause gene amplification in cancer remain unclear. The model originally proposed by Barbara McClintock in 1938 suggests that intrachromosomal cycles of breakage–fusion–bridge initiated by a DSB can promote progressive acquisition of additional genomic alterations that result in localized amplification96,97 (FIG. 5b). If this hypothesis is true, then DNA replication is likely to be interspersed with the accumulation of structural variations throughout the development of a cancer, even though the structural variations may have accumulated over a fairly short period. This hypothesis is distinct from a phenomenon called chromothripsis, which comprises the formation of tens to hundreds of locally clustered structural variations that show a characteristic pattern of copy-number ‘oscillations’ (~2–3 copy-number states) with scattered losses of DNA fragments85. This type of structural variation is also locoregional but distinct from gene amplification, as it was arisen purportedly in a single cataclysmic moment in the history of a cancer. Both intrachromosomal and interchromosomal rearrangements arise from chromothripsis, which can lead to the formation of small circular marker chromosomes (double-minutes) that may subsequently amplify (that is, increase in copy number), particularly if they harbour an oncogene85. Recently, the term chromoplexy was given to the appearance of complex rearrangements that involve multiple chromosomes linked in a chain of rearrangements98. No specific pathophysiological mechanism has been implicated in this descriptive term.
Profound mechanistic insights can be gained from the detailed study of rearrangements that show marked colocalization with base substitution hypermutations — a phenomenon termed kataegis. Although all types of rearrangements have been described to harbour this unusual signature, which so far seems to be stochastic, it is the highly clustered base substitutions that show distinctive features: they comprise C∙G→T∙A transitions2 and C∙G→G∙C transversions9 with a marked predilection for a TpC or GpA sequence context and a striking strand coordination. Although the precise mechanism that underlies the kataegis signature is uncertain, an excess of these base substitutions at this specific sequence context has been found around induced DNA DSBs19,99, which has prompted speculation that these clustered mutations occur at end-resected DSBs that expose ssDNA — the particular substrate of the APOBEC family of cytidine deaminases.
Chromosomal instability
Cancers are often characterized by chromosomal instability, which includes numerical or structural chromosomal aberrations. Historically, chromosomal instability is a feature that is defined at a macroscopic or chromosomal scale using techniques such as spectral karyotyping. Biologically, chromosomal instability has been attributed to replication stress in studies involving colorectal cancer cells100 that arises from the activation of oncogenes such as HRAS, CCNE1 (which encodes cyclin E), MOS and cell division cycle 6 (CDC6)101–103. These activated oncogenes induce the deregulation of cyclin-dependent kinase 2 (CDK2), which is involved in replication origin firing104,105. Interestingly, oncogene-induced replication stress has been shown to result in genetic instability and DSB formation specifically at fragile sites106,107, which are hot spots for gene rearrangements107. Currently, it remains unclear how chromosomal instability translates to a genomic signature at the base-pair level.
Structural variation and immune loci
The generation of double-ended DSBs can be physiological. It is a necessary part of maturation at the IG locus of cells of the immune system. This deliberate activity may be achieved by V(D)J recombination-activating protein 1 (RAG1) and RAG2 (REF. 108), as well as by activation-induced cytidine deaminase (AID)-mediated class switch recombination109 or somatic hypermutation79. Intriguingly, the role of these proteins can be appreciated as signatures in various haematological malignancies. For example, the RAG proteins, which show sequence specificity for a recombination signal sequence, underpin rearrangements between the IGH locus and the B-cell CLL/lymphoma 2 (BCL2) gene that drive follicular lymphoma110 (FIG. 5c), whereas the AID protein is required for C-MYC–IGH chromosomal translocations that drive Burkitt’s lymphoma111,112 (FIG. 5d). In these malignancies, detailed analyses of the distribution of translocations in lymphocytes using genome-wide approaches have provided insights into the nonrandom nature of AID-mediated rearrangements86–88. These studies are further supported by observations in genome-sequenced haematological malignancies such as B-cell leukaemias and lymphomas9. Similar to kataegis, foci of substitutions are found to be coupled to rearrangements but, unlike kataegis, they are not stochastic; that is, they show recurrence at the IGH and C-MYC loci. They also show a preference for a purine preceding a mutated cytosine9; this sequence specificity differs from that of kataegis but is consistent with that of AID-mediated translocation. Breakpoint analyses have shown that MMEJ is involved in the ligation of the broken ends86–88.
Conclusions
Each complex and multidimensional cancer genome may carry one or more mutational signatures (that is, the imprints of all of the mutational processes that have occurred throughout cancer development). The enduring mutational signatures in cancer genomes are the final physiological readout of the biology that has gone wrong throughout the development of the cancer, the readout of mutagenic damage from environmental or endogenous sources, as well as that of the repair and replicative processes that have been operative. The studies presented here show how technological advances in sequencing the human genome have led to a deeper appreciation of somatic mutational signatures in human cancers. By studying these enormous data sets in great detail, mechanistic insights can be gained. However, it must be highlighted that many recently discovered signatures are novel and remain to be understood. There is demand for experimental evidence even for signatures with clear candidate processes.
Several important observations should be highlighted. First, the overarching 96-element pattern of each signature is essentially identical between cancer samples of different patients, even of disparate cancer types, which suggests that similar processes are operating in different individuals. However, the individual somatic mutations that make up each signature in patient-specific cancers are highly variable between patients9. Second, some signatures seem to be the final readout of a deregulated pathway regardless of the precise somatic or germline mutation that underlies the perturbation (for example, the biallelic somatic and germline mutations of BRCA1 and BRCA2 in Signature 3). In these cases, knowledge of signatures could inform clinical decision making, for example, regarding potential sensitivity to therapeutics in the absence of precise genotypic information. The relationship between mutational signatures and clinical response to therapeutics requires investigation. Coupled to systematic characterization by experimental manipulation of model systems and detailed annotation of the resulting signatures, this will take us a step closer to more tailored treatments.
Given that mutational signatures are revealing the consequence of abrogated pathways, knowledge of the presence of a particular signature may enable targeting of the underlying mutational processes and thus provide a more successful path for cancer disease control. To this end, it is important for future work to determine the mutational processes that are still ongoing, either through serial biopsies from patients or through cell-line-based experiments. In addition, therapeutic strategies that selectively target processes responsible for specific signatures could complement current genotype-specific strategies. In the future, molecular genomic profiling should incorporate all mutations regardless of whether they are causative or consequential.
Acknowledgements
The authors thank the Knut and Alice Wallenberg Foundation, the Swedish Research Council, Swedish Cancer Society, the Swedish Pain Relief Foundation and the Torsten and Ragnar Söderberg Foundation (all to T.H.). S.N-Z. is personally funded through a Wellcome Trust Intermediate Fellowship (WT100183MA) and is a Wellcome-Beit Prize Fellow.
Footnotes
Competing interests statement
The authors declare no competing interests.
Driver mutations Genetic changes that give selective advantages to clones during cancer development.
Somatic mutations Mutations that are acquired as opposed to inherited.
Passenger mutations Genetic changes that do not confer any selective advantage in cancer development.
Mutational processes Biological activities that generate mutations; each of these processes comprises both a DNA damage component and a DNA repair component. These processes can be ongoing or historical depending on whether the biological processes that cause the acquisition of mutations in a cancer are active or inactive, respectively.
Mutational signature The pattern of mutations produced by a mutational process.
Mutational portrait The total genetic changes observed in a cancer genome; that is, the sum of all mutational signatures occurring in a lifetime.
Base substitutions A type of mutation in which one base is replaced by another in DNA.
Insertions and deletions (Indels). A type of mutation that arises from the insertion or deletion of one or more nucleotides within a DNA sequence.
Structural variations Large-scale genomic changes (typically >1 kb) such as deletions, tandem duplications, amplifications, inversions and translocations.
Transversions Mutations that involve different classes of nucleotides; that is, purine-to-pyrimidine or pyrimidine-to-purine mutations.
Transitions Mutations that involve the same class of nucleotides; that is, purine-to-purine or pyrimidine-to-pyrimidine mutations.
Deamination A biochemical reaction that removes an amine group from a molecule.
Transcriptional strand bias Bias in mutation load between the transcribed strand and the non-transcribed strand.
Microsatellite instability Variability in the length of base pair repeated sequences (<5 bp) that is caused by replication slippage and that is normally kept stable by mismatch repair.
Replication fork collapse A condition at a replication fork in which the integrity of a DNA molecule is impaired and can result in a DNA double-strand break.
Somatic hypermutation Regional hypermutation at the immunoglobulin locus that generates antibody diversity.
Synthesis-dependent end-joining (SDEJ). A process in which a DNA end at a double-strand break is extended using the intact sister chromatid as template. The DNA end is released from the sister chromatid and rejoined by end-joining.
Chromothripsis An event with tens or hundreds of locally clustered rearrangements that result in distinct oscillations of copy-number states.
Chromoplexy A rearrangement event that involves multiple chromosomes.
Kataegis A base substitution hypermutation that comprises C∙G→T∙A transitions and C∙G→G∙C transversions with a predilection for a thymine preceding the mutated cytosine (that is, a TpC context); it usually macroscopically colocalizes with structural variation.
Chromosomal instability A process that results in failure to maintain euploidy after mitosis and that is caused by either numerical or structural chromosomal aberrations.
Replication stress A condition in which progression of a replication fork is hindered.
References
- 1.Stratton MR, Campbell PJ, Futreal PA. The cancer genome. Nature. 2009;458:719–724. doi: 10.1038/nature07943. [This is an overview of cancer genomes with a description of various types of somatic mutations acquired during the multistep process of cancer development.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nik-Zainal S, et al. Mutational processes molding the genomes of 21 breast cancers. Cell. 2012;149:979–993. doi: 10.1016/j.cell.2012.04.024. [This study presents catalogues of somatic mutations from 21 breast cancers, the respective mutational signatures of which were extracted by mathematical methods.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nik-Zainal S, et al. The life history of 21 breast cancers. Cell. 2012;149:994–1007. doi: 10.1016/j.cell.2012.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bentley DR, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Greenman C, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–158. doi: 10.1038/nature05610. [This study shows the prevalence of somatic mutations in human cancer genomes, which indicates that most of the mutations do not drive oncogenesis. Nevertheless, it provides evidence for driver mutations that are actively involved in tumour development.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wood LD, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 7.Pleasance ED, et al. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature. 2010;463:191–196. doi: 10.1038/nature08658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pleasance ED, et al. A small-cell lung cancer genome with complex signatures of tobacco exposure. Nature. 2010;463:184–190. doi: 10.1038/nature08629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alexandrov LB, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [In this study, >20 distinct mutational signatures have been extracted from several cancer types, which shows the presence of the APOBEC-mediated signature in various cancers.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alexandrov LB, Nik-Zainal S, Wedge DC, Campbell PJ, Stratton MR. Deciphering signatures of mutational processes operative in human cancer. Cell Rep. 2013;3:246–259. doi: 10.1016/j.celrep.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pfeifer GP, et al. Tobacco smoke carcinogens, DNA damage and p53 mutations in smoking-associated cancers. Oncogene. 2002;21:7435–7451. doi: 10.1038/sj.onc.1205803. [DOI] [PubMed] [Google Scholar]
- 12.Ellegren H, Smith NG, Webster MT. Mutation rate variation in the mammalian genome. Curr Opin Genet Dev. 2003;13:562–568. doi: 10.1016/j.gde.2003.10.008. [DOI] [PubMed] [Google Scholar]
- 13.Pfeifer GP, You YH, Besaratinia A. Mutations induced by ultraviolet light. Mutat Res. 2005;571:19–31. doi: 10.1016/j.mrfmmm.2004.06.057. [DOI] [PubMed] [Google Scholar]
- 14.Lutsenko E, Bhagwat AS. Principal causes of hot spots for cytosine to thymine mutations at sites of cytosine methylation in growing cells. A model, its experimental support and implications. Mutat Res. 1999;437:11–20. doi: 10.1016/s1383-5742(99)00065-4. [DOI] [PubMed] [Google Scholar]
- 15.Nikolaev SI, et al. A single-nucleotide substitution mutator phenotype revealed by exome sequencing of human colon adenomas. Cancer Res. 2012;72:6279–6289. doi: 10.1158/0008-5472.CAN-12-3869. [DOI] [PubMed] [Google Scholar]
- 16.Pham P, Bransteitter R, Petruska J, Goodman MF. Processive AID-catalysed cytosine deamination on single-stranded DNA simulates somatic hypermutation. Nature. 2003;424:103–107. doi: 10.1038/nature01760. [DOI] [PubMed] [Google Scholar]
- 17.Landry S, Narvaiza I, Linfesty DC, Weitzman MD. APOBEC3A can activate the DNA damage response and cause cell-cycle arrest. EMBO Rep. 2011;12:444–450. doi: 10.1038/embor.2011.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suspene R, et al. Somatic hypermutation of human mitochondrial and nuclear DNA by APOBEC3 cytidine deaminases, a pathway for DNA catabolism. Proc Natl Acad Sci USA. 2011;108:4858–4863. doi: 10.1073/pnas.1009687108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Taylor BJ, et al. DNA deaminases induce break-associated mutation showers with implication of APOBEC3B and 3A in breast cancer kataegis. Elife. 2013;2:e00534. doi: 10.7554/eLife.00534. [This paper shows that kataegis observed in the breast cancer genome can stem from AID- or APOBEC-mediated cytidine deamination in the proximity of DNA breaks.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stephens P, et al. A screen of the complete protein kinase gene family identifies diverse patterns of somatic mutations in human breast cancer. Nature Genet. 2005;37:590–592. doi: 10.1038/ng1571. [DOI] [PubMed] [Google Scholar]
- 21.Burns MB, et al. APOBEC3B is an enzymatic source of mutation in breast cancer. Nature. 2013;494:366–370. doi: 10.1038/nature11881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roberts SA, et al. An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nature Genet. 2013;45:970–976. doi: 10.1038/ng.2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nik-Zainal S, et al. Association of a germline copy number polymorphism of APOBEC3A and APOBEC3B with burden of putative APOBEC-dependent mutations in breast cancer. Nature Genet. 2014;46:487–491. doi: 10.1038/ng.2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Byeon IJ, et al. NMR structure of human restriction factor APOBEC3A reveals substrate binding and enzyme specificity. Nature Commun. 2013;4:1890. doi: 10.1038/ncomms2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holtz CM, Sadler HA, Mansky LM. APOBEC3G cytosine deamination hotspots are defined by both sequence context and single-stranded DNA secondary structure. Nucleic Acids Res. 2013;41:6139–6148. doi: 10.1093/nar/gkt246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karran P, Lindahl T. Hypoxanthine in deoxyribonucleic acid: generation by heat-induced hydrolysis of adenine residues and release in free form by a deoxyribonucleic acid glycosylase from calf thymus. Biochemistry. 1980;19:6005–6011. doi: 10.1021/bi00567a010. [DOI] [PubMed] [Google Scholar]
- 27.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 28.Hussain SP, Hofseth LJ, Harris CC. Radical causes of cancer. Nature Rev Cancer. 2003;3:276–285. doi: 10.1038/nrc1046. [DOI] [PubMed] [Google Scholar]
- 29.Evans MD, Dizdaroglu M, Cooke MS. Oxidative DNA damage and disease: induction, repair and significance. Mutat Res. 2004;567:1–61. doi: 10.1016/j.mrrev.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 30.Oikawa S, Kawanishi S. Site-specific DNA damage at GGG sequence by oxidative stress may accelerate telomere shortening. FEBS Lett. 1999;453:365–368. doi: 10.1016/s0014-5793(99)00748-6. [DOI] [PubMed] [Google Scholar]
- 31.Oikawa S, Tada-Oikawa S, Kawanishi S. Site-specific DNA damage at the GGG sequence by UVA involves acceleration of telomere shortening. Biochemistry. 2001;40:4763–4768. doi: 10.1021/bi002721g. [DOI] [PubMed] [Google Scholar]
- 32.Cadet J, Sage E, Douki T. Ultraviolet radiation-mediated damage to cellular DNA. Mutat Res. 2005;571:3–17. doi: 10.1016/j.mrfmmm.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 33.Hendriks G, et al. Transcription-dependent cytosine deamination is a novel mechanism in ultraviolet light-induced mutagenesis. Curr Biol. 2010;20:170–175. doi: 10.1016/j.cub.2009.11.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schiltz M, et al. Characterization of the mutational profile of (+)-7R,8S-dihydroxy-9S,10R-epoxy-7,8,9,10-tetrahydrobenzo[a]pyrene at the hypoxanthine (guanine) phosphoribosyltransferase gene in repair-deficient Chinese hamster V-H1 cells. Carcinogenesis. 1999;20:2279–2285. doi: 10.1093/carcin/20.12.2279. [DOI] [PubMed] [Google Scholar]
- 35.Wiseman RW, Miller EC, Miller JA, Liem A. Structure–activity studies of the hepatocarcinogenicities of alkenylbenzene derivatives related to estragole and safrole on administration to preweanling male C57BL/6J x C3H/HeJ F1 mice. Cancer Res. 1987;47:2275–2283. [PubMed] [Google Scholar]
- 36.Papadopoulo D, Laquerbe A, Guillouf C, Moustacchi E. Molecular spectrum of mutations induced at the HPRT locus by a cross-linking agent in human cell lines with different repair capacities. Mutat Res. 1993;294:167–177. doi: 10.1016/0921-8777(93)90025-c. [DOI] [PubMed] [Google Scholar]
- 37.Yang SC, Lin JG, Chiou CC, Chen LY, Yang JL. Mutation specificity of 8-methoxypsoralen plus two doses of UVA irradiation in the hprt gene in diploid human fibroblasts. Carcinogenesis. 1994;15:201–207. doi: 10.1093/carcin/15.2.201. [DOI] [PubMed] [Google Scholar]
- 38.Feldmeyer N, et al. Further studies with a cell immortalization assay to investigate the mutation signature of aristolochic acid in human p53 sequences. Mutat Res. 2006;608:163–168. doi: 10.1016/j.mrgentox.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 39.Krokan HE, Standal R, Slupphaug G. DNA glycosylases in the base excision repair of DNA. Biochem J. 1997;325:1–16. doi: 10.1042/bj3250001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Caldecott KW. Single-strand break repair and genetic disease. Nature Rev Genet. 2008;9:619–631. doi: 10.1038/nrg2380. [DOI] [PubMed] [Google Scholar]
- 41.Robertson AB, Klungland A, Rognes T, Leiros I. DNA repair in mammalian cells: base excision repair: the long and short of it. Cell Mol Life Sci. 2009;66:981–993. doi: 10.1007/s00018-009-8736-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.An Q, Robins P, Lindahl T, Barnes DE. C→T mutagenesis and γ-radiation sensitivity due to deficiency in the Smug1 and Ung DNA glycosylases. EMBO J. 2005;24:2205–2213. doi: 10.1038/sj.emboj.7600689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smart DJ, Chipman JK, Hodges NJ. Activity of OGG1 variants in the repair of pro-oxidant-induced 8-oxo-2′-deoxyguanosine. DNA Repair (Amst.) 2006;5:1337–1345. doi: 10.1016/j.dnarep.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 44.Nouspikel T. DNA repair in mammalian cells: nucleotide excision repair: variations on versatility. Cell Mol Life Sci. 2009;66:994–1009. doi: 10.1007/s00018-009-8737-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bohr VA, Smith CA, Okumoto DS, Hanawalt PC. DNA repair in an active gene: removal of pyrimidine dimers from the DHFR gene of CHO cells is much more efficient than in the genome overall. Cell. 1985;40:359–369. doi: 10.1016/0092-8674(85)90150-3. [DOI] [PubMed] [Google Scholar]
- 46.Poon SL, et al. Genome-wide mutational signatures of aristolochic acid and its application as a screening tool. Sci Transl Med. 2013;5:197ra101. doi: 10.1126/scitranslmed.3006086. [DOI] [PubMed] [Google Scholar]
- 47.Guo J, Hanawalt PC, Spivak G. Comet-FISH with strand-specific probes reveals transcription-coupled repair of 8-oxoguanine in human cells. Nucleic Acids Res. 2013;41:7700–7712. doi: 10.1093/nar/gkt524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pena-Diaz J, Jiricny J. Mammalian mismatch repair: error-free or error-prone? Trends Biochem Sci. 2012;37:206–214. doi: 10.1016/j.tibs.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 49.Jiricny J. The multifaceted mismatch-repair system. Nature Rev Mol Cell Biol. 2006;7:335–346. doi: 10.1038/nrm1907. [DOI] [PubMed] [Google Scholar]
- 50.Tiraby G, Fox MS, Bernheimer H. Marker discrimination in deoxyribonucleic acid-mediated transformation of various Pneumococcus strains. J Bacteriol. 1975;121:608–618. doi: 10.1128/jb.121.2.608-618.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shibata D, Peinado MA, Ionov Y, Malkhosyan S, Perucho M. Genomic instability in repeated sequences is an early somatic event in colorectal tumorigenesis that persists after transformation. Nature Genet. 1994;6:273–281. doi: 10.1038/ng0394-273. [DOI] [PubMed] [Google Scholar]
- 52.McCulloch SD, Kunkel TA. The fidelity of DNA synthesis by eukaryotic replicative and translesion synthesis polymerases. Cell Res. 2008;18:148–161. doi: 10.1038/cr.2008.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shevelev IV, Hübscher U. The 3′–5′ exonucleases. Nature Rev Mol Cell Biol. 2002;3:364–376. doi: 10.1038/nrm804. [DOI] [PubMed] [Google Scholar]
- 54.Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cancer Genome Atlas Research Network et al. Integrated genomic characterization of endometrial carcinoma. Nature. 2013;497:67–73. doi: 10.1038/nature12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kane DP, Shcherbakova PV. A common cancer-associated DNA polymerase ε mutation causes an exceptionally strong mutator phenotype, indicating fidelity defects distinct from loss of proofreading. Cancer Res. 2014;74:1895–1901. doi: 10.1158/0008-5472.CAN-13-2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Roberts JD, Kunkel TA. Fidelity of a human cell DNA replication complex. Proc Natl Acad Sci USA. 1988;85:7064–7068. doi: 10.1073/pnas.85.19.7064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bester AC, et al. Nucleotide deficiency promotes genomic instability in early stages of cancer development. Cell. 2011;145:435–446. doi: 10.1016/j.cell.2011.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jones RM, et al. Increased replication initiation and conflicts with transcription underlie cyclin E-induced replication stress. Oncogene. 2013;32:3744–3753. doi: 10.1038/onc.2012.387. [DOI] [PubMed] [Google Scholar]
- 60.Petermann E, Woodcock M, Helleday T. Chk1 promotes replication fork progression by controlling replication initiation. Proc Natl Acad Sci USA. 2010;107:16090–16095. doi: 10.1073/pnas.1005031107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sale JE, Lehmann AR, Woodgate R. Y-family DNA polymerases and their role in tolerance of cellular DNA damage. Nature Rev Mol Cell Biol. 2012;13:141–152. doi: 10.1038/nrm3289. [This is a review on our current understanding of translesion synthesis and the associated Y-family DNA polymerases.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Knobel PA, Marti TM. Translesion DNA synthesis in the context of cancer research. Cancer Cell Int. 2011;11:39. doi: 10.1186/1475-2867-11-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Klarer AC, McGregor W. Replication of damaged genomes. Crit Rev Eukaryot Gene Expr. 2011;21:323–336. doi: 10.1615/critreveukargeneexpr.v21.i4.30. [DOI] [PubMed] [Google Scholar]
- 64.Strauss BS. The “A” rule revisited: polymerases as determinants of mutational specificity. DNA Repair (Amst.) 2002;1:125–135. doi: 10.1016/s1568-7864(01)00014-3. [DOI] [PubMed] [Google Scholar]
- 65.Puente XS. Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature. 2011;475:101–105. doi: 10.1038/nature10113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kunz BA, Straffon AF, Vonarx EJ. DNA damage-induced mutation: tolerance via translesion synthesis. Mutat Res. 2000;451:169–185. doi: 10.1016/s0027-5107(00)00048-8. [DOI] [PubMed] [Google Scholar]
- 67.Thibodeau SN, Bren G, Schaid D. Microsatellite instability in cancer of the proximal colon. Science. 1993;260:816–819. doi: 10.1126/science.8484122. [DOI] [PubMed] [Google Scholar]
- 68.Ionov Y, Peinado MA, Malkhosyan S, Shibata D, Perucho M. Ubiquitous somatic mutations in simple repeated sequences reveal a new mechanism for colonic carcinogenesis. Nature. 1993;363:558–561. doi: 10.1038/363558a0. [DOI] [PubMed] [Google Scholar]
- 69.Bhattacharyya NP, Skandalis A, Ganesh A, Groden J, Meuth M. Mutator phenotypes in human colorectal carcinoma cell lines. Proc Natl Acad Sci USA. 1994;91:6319–6323. doi: 10.1073/pnas.91.14.6319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Karran P. Microsatellite instability and DNA mismatch repair in human cancer. Semin Cancer Biol. 1996;7:15–24. doi: 10.1006/scbi.1996.0003. [DOI] [PubMed] [Google Scholar]
- 71.Kuraguchi M, et al. Tumor-associated Apc mutations in Mlh1−/− Apc1638N mice reveal a mutational signature of Mlh1 deficiency. Oncogene. 2000;19:5755–5763. doi: 10.1038/sj.onc.1203962. [DOI] [PubMed] [Google Scholar]
- 72.Weinstock DM, Brunet E, Jasin M. Formation of NHEJ-derived reciprocal chromosomal translocations does not require Ku70. Nature Cell Biol. 2007;9:978–981. doi: 10.1038/ncb1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yun MH, Hiom K. CtIP–BRCA1 modulates the choice of DNA double-strand-break repair pathway throughout the cell cycle. Nature. 2009;459:460–463. doi: 10.1038/nature07955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Moynahan ME, Chiu JW, Koller BH, Jasin M. Brca1 controls homology-directed DNA repair. Mol Cell. 1999;4:511–518. doi: 10.1016/s1097-2765(00)80202-6. [DOI] [PubMed] [Google Scholar]
- 75.Moynahan ME, Pierce AJ, Jasin M. BRCA2 is required for homology-directed repair of chromosomal breaks. Mol Cell. 2001;7:263–272. doi: 10.1016/s1097-2765(01)00174-5. [DOI] [PubMed] [Google Scholar]
- 76.Bunting SF, et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell. 2010;141:243–254. doi: 10.1016/j.cell.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Davies AA, et al. Role of BRCA2 in control of the RAD51 recombination and DNA repair protein. Mol Cell. 2001;7:273–282. doi: 10.1016/s1097-2765(01)00175-7. [DOI] [PubMed] [Google Scholar]
- 78.Stephens PJ, et al. Complex landscapes of somatic rearrangement in human breast cancer genomes. Nature. 2009;462:1005–1010. doi: 10.1038/nature08645. [This study analyses somatic rearrangements in the breast cancer genome using paired-end sequencing strategy, which reveals that these rearrangements are mostly intrachromosomal.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nussenzweig A, Nussenzweig MC. Origin of chromosomal translocations in lymphoid cancer. Cell. 2010;141:27–38. doi: 10.1016/j.cell.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Groth P, et al. Homologous recombination repairs secondary replication induced DNA double-strand breaks after ionizing radiation. Nucleic Acids Res. 2012;40:6585–6594. doi: 10.1093/nar/gks315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Arnaudeau C, Lundin C, Helleday T. DNA double-strand breaks associated with replication forks are predominantly repaired by homologous recombination involving an exchange mechanism in mammalian cells. J Mol Biol. 2001;307:1235–1245. doi: 10.1006/jmbi.2001.4564. [DOI] [PubMed] [Google Scholar]
- 82.Riballo E, et al. A pathway of double-strand break rejoining dependent upon ATM, Artemis, and proteins locating to γ-H2AX foci. Mol Cell. 2004;16:715–724. doi: 10.1016/j.molcel.2004.10.029. [DOI] [PubMed] [Google Scholar]
- 83.Rothkamm K, Kruger I, Thompson LH, Lobrich M. Pathways of DNA double-strand break repair during the mammalian cell cycle. Mol Cell Biol. 2003;23:5706–5715. doi: 10.1128/MCB.23.16.5706-5715.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Saleh-Gohari N, Helleday T. Conservative homologous recombination preferentially repairs DNA double-strand breaks in the S phase of the cell cycle in human cells. Nucleic Acids Res. 2004;32:3683–3688. doi: 10.1093/nar/gkh703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Stephens PJ, et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 2011;144:27–40. doi: 10.1016/j.cell.2010.11.055. [This is the first study to characterize chromothripsis in a human cancer genome.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Klein IA, et al. Translocation-capture sequencing reveals the extent and nature of chromosomal rearrangements in B lymphocytes. Cell. 2011;147:95–106. doi: 10.1016/j.cell.2011.07.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chiarle R, et al. Genome-wide translocation sequencing reveals mechanisms of chromosome breaks and rearrangements in B cells. Cell. 2011;147:107–119. doi: 10.1016/j.cell.2011.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hakim O, et al. DNA damage defines sites of recurrent chromosomal translocations in B lymphocytes. Nature. 2012;484:69–74. doi: 10.1038/nature10909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ng CK, et al. The role of tandem duplicator phenotype in tumour evolution in high-grade serous ovarian cancer. J Pathol. 2012;226:703–712. doi: 10.1002/path.3980. [DOI] [PubMed] [Google Scholar]
- 90.Costantino L, et al. Break-induced replication repair of damaged forks induces genomic duplications in human cells. Science. 2014;343:88–91. doi: 10.1126/science.1243211. [This paper reports a role of DNA Pol δ in BIR repair.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Haber JE. Lucky breaks: analysis of recombination in Saccharomyces. Mutat Res. 2000;451:53–69. doi: 10.1016/s0027-5107(00)00040-3. [DOI] [PubMed] [Google Scholar]
- 92.Helleday T. Pathways for mitotic homologous recombination in mammalian cells. Mutat Res. 2003;532:103–115. doi: 10.1016/j.mrfmmm.2003.08.013. [DOI] [PubMed] [Google Scholar]
- 93.West SC. Molecular views of recombination proteins and their control. Nature Rev Mol Cell Biol. 2003;4:435–445. doi: 10.1038/nrm1127. [DOI] [PubMed] [Google Scholar]
- 94.Szostak JW, Orr-Weaver TL, Rothstein RJ, Stahl FW. The double-strand-break repair model for recombination. Cell. 1983;33:25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- 95.Baehner FL, et al. Human epidermal growth factor receptor 2 assessment in a case-control study: comparison of fluorescence in situ hybridization and quantitative reverse transcription polymerase chain reaction performed by central laboratories. J Clin Oncol. 2010;28:4300–4306. doi: 10.1200/JCO.2009.24.8211. [DOI] [PubMed] [Google Scholar]
- 96.McClintock B. The production of homozygous deficient tissues with mutant characteristics by means of the aberrant mitotic behavior of ring-shaped chromosomes. Genetics. 1938;23:315–376. doi: 10.1093/genetics/23.4.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ma C, Martin S, Trask B, Hamlin JL. Sister chromatid fusion initiates amplification of the dihydrofolate reductase gene in Chinese hamster cells. Genes Dev. 1993;7:605–620. doi: 10.1101/gad.7.4.605. [DOI] [PubMed] [Google Scholar]
- 98.Baca SC, et al. Punctuated evolution of prostate cancer genomes. Cell. 2013;153:666–677. doi: 10.1016/j.cell.2013.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Roberts SA, et al. Clustered mutations in yeast and in human cancers can arise from damaged long single-strand DNA regions. Mol Cell. 2012;46:424–435. doi: 10.1016/j.molcel.2012.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Burrell RA, et al. Replication stress links structural and numerical cancer chromosomal instability. Nature. 2013;494:492–496. doi: 10.1038/nature11935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Di Micco R, et al. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature. 2006;444:638–642. doi: 10.1038/nature05327. [DOI] [PubMed] [Google Scholar]
- 102.Bartkova J, et al. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature. 2006;444:633–637. doi: 10.1038/nature05268. [DOI] [PubMed] [Google Scholar]
- 103.Spruck CH, Won KA, Reed SI. Deregulated cyclin E induces chromosome instability. Nature. 1999;401:297–300. doi: 10.1038/45836. [DOI] [PubMed] [Google Scholar]
- 104.Zimmerman KM, Jones RM, Petermann E, Jeggo PA. Diminished origin-licensing capacity specifically sensitizes tumor cells to replication stress. Mol Cancer Res. 2013;11:370–380. doi: 10.1158/1541-7786.MCR-12-0491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Takeda DY, Dutta A. DNA replication and progression through S phase. Oncogene. 2005;24:2827–2843. doi: 10.1038/sj.onc.1208616. [DOI] [PubMed] [Google Scholar]
- 106.Tsantoulis PK, et al. Oncogene-induced replication stress preferentially targets common fragile sites in preneoplastic lesions. A genome-wide study. Oncogene. 2008;27:3256–3264. doi: 10.1038/sj.onc.1210989. [DOI] [PubMed] [Google Scholar]
- 107.Barlow J, et al. A novel class of early replicating fragile sites that contribute to genome instability in B cell lymphomas. Cell. 2013;152:620–632. doi: 10.1016/j.cell.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gellert M, et al. V(D)J recombination: links to transposition and double-strand break repair. Cold Spring Harb Symp Quant Biol. 1999;64:161–167. doi: 10.1101/sqb.1999.64.161. [DOI] [PubMed] [Google Scholar]
- 109.Neuberger MS, Harris RS, Di Noia J, Petersen-Mahrt SK. Immunity through DNA deamination. Trends Biochem Sci. 2003;28:305–312. doi: 10.1016/S0968-0004(03)00111-7. [DOI] [PubMed] [Google Scholar]
- 110.Vaandrager JW, Schuuring E, Philippo K, Kluin PM. V(D)J recombinase-mediated transposition of the BCL2 gene to the IGH locus in follicular lymphoma. Blood. 2000;96:1947–1952. [PubMed] [Google Scholar]
- 111.Robbiani DF, et al. AID is required for the chromosomal breaks in c-myc that lead to c-myc/IgH translocations. Cell. 2008;135:1028–1038. doi: 10.1016/j.cell.2008.09.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ramiro AR, et al. AID is required for c-myc/IgH chromosome translocations in vivo. Cell. 2004;118:431–438. doi: 10.1016/j.cell.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 113.Berry MW, Browne M, Langville AN, Pauca VP, Plemmons RJ. Algorithms and applications for approximate nonnegative matrix factorization. Comput Statist Data Analysis. 2007;52:155–173. [Google Scholar]
- 114.Lange SS, Takata K, Wood RD. DNA polymerases and cancer. Nature Rev Cancer. 2011;11:96–110. doi: 10.1038/nrc2998. [DOI] [PMC free article] [PubMed] [Google Scholar]