Abstract
In nature, specific antibodies can be generated as a result of an adaptive selection and expansion of lymphocytes with suitable protein binding properties. We attempted to mimic the antibody-antigen recognition by displaying multiple chemical diversity elements on a defined macrocyclic scaffold. Encoding of the displayed combinations was achieved using distinctive DNA tags, resulting in a library size of 35,393,112. Specific binders could be isolated against a variety of proteins, including carbonic anhydrase IX, horseradish peroxidase, tankyrase 1, human serum albumin, alpha-1 acid glycoprotein, calmodulin, prostate specific antigen and tumor necrosis factor. Similar to antibodies, the encoded display of multiple chemical elements on a constant scaffold enabled practical applications, such as fluorescence microscopy procedures or the selective in vivo delivery of payloads to tumors. Furthermore, the versatile structure of the scaffold facilitated the generation of protein specific chemical probes, as illustrated by photo-crosslinking.
Versatile and specific recognition of target proteins is a fundamentally important feature of the humoral arm of the adaptive immune system. Indeed, antibodies can be raised against virtually any antigen, by suitable immunization procedures1. This natural process of clonal selection and amplification can be mimicked in vitro by the construction of large combinatorial antibody libraries and the implementation of efficient selection systems (e.g., phage display technology2), which allow the generation of “man-made” antibodies3–5. The selected antibodies from phage display libraries can also be used for pharmaceutical applications; for example, the TNF-inhibitor Humira™ (one of the best-selling drugs in the world) was generated using this technology.
There is a great scientific and industrial interest to isolate molecules which are substantially smaller than antibodies, but which retain the ability to recognize various target proteins with high binding affinity and specificity. Such molecules would be less immunogenic, should penetrate tissues more efficiently and therefore could be advantageous for certain pharmaceutical applications6. Large combinatorial libraries of polypeptides, developed from display methods, (e.g. phage display7, mRNA display8,9, yeast display10 and ribosome display11) have demonstrated the ability to yield specific binders against different proteins. However, those polypeptides are still mainly composed of proteinogenic amino acids and are developed using biosynthetic methodologies.
Serving as a chemical solution for exponentially generating molecular diversity, DNA-encoded chemical libraries (DECLs) are increasingly being employed for the isolation of small-molecule binders against target proteins of interest12,13. The technology couples the power of genetics with chemical synthesis, allowing the generation of large sets of synthetic molecules, each linked to a distinctive DNA fragment serving as amplifiable identification barcode14,15. Compared with conventional screening methodologies such as high-throughput screening and non-encoded combinatorial libraries, DECL technology allows the rapid and rather inexpensive construction of very large chemical libraries of typically millions to billions of compounds, which can be quickly interrogated for target binding by affinity-based selection procedures followed by decoding with high-throughput DNA sequencing (HTDS)16.
Most of the DECLs reported so far by both academia and industry were constructed by split-and-pool synthetic procedures16–18, aiming at drug-like molecules complying with Lipinski’s rule of five (RO5)19. Further approaches for the construction of DECLs have been proposed featuring, e.g., DNA-based routing20, DNA-templated synthesis21–23, or fragment-based strategies24–27. While RO5-type DECLs may preferentially yield binders for targets with defined pockets, such as proteases28, kinases17 or phosphatases29, the recognition of large surfaces of target proteins remains a challenge. The intrinsically larger size and complexity of macrocycles suggests their usefulness for the recognition of larger target surfaces, however, the pharmaceutical properties of macrocyclic binders may be difficult to optimize, since modification on the cyclic backbone may lead to unexpected conformational changes7,30.
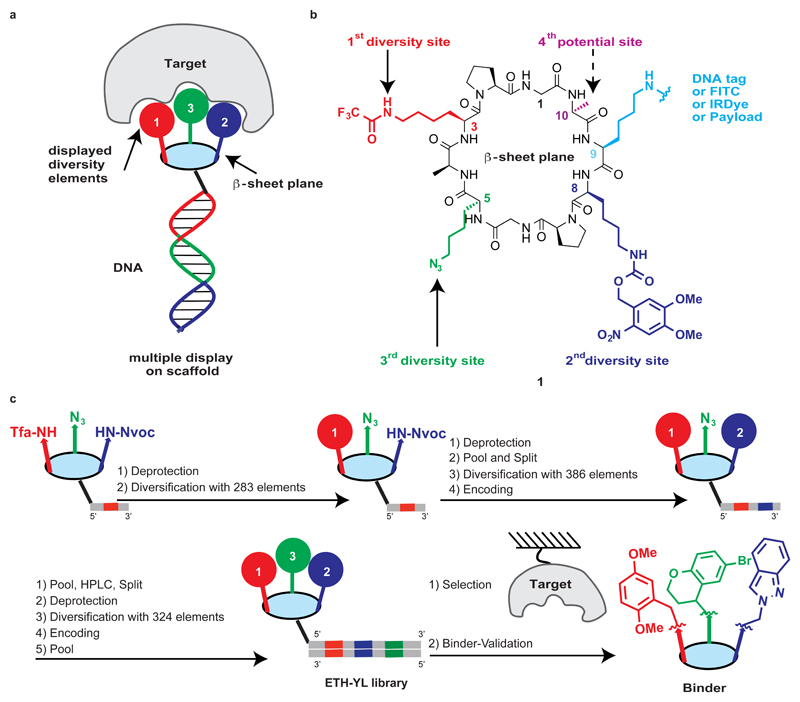
In this article, we explored a strategy featuring the encoded combinatorial display of multiple chemical diversity elements (DEs) on a structurally-defined macrocyclic scaffold, in order to achieve a versatile and specific recognition of different target proteins. To that aim, we sought out a fixed macrocyclic scaffold with antiparallel β-sheets, previously described by Manfred Mutter and Pascal Dumy, which serves as a defined platform for the presentation of multiple chemical diversity elements into one side of the β-sheet plane (Fig. 1a)31–33. The constant macrocyclic scaffold also contains a further chemically addressable site, which facilitates the encoding of individual synthetic combinations with distinctive DNA tags, serving as amplifiable identification barcodes for further selection procedures. Alternatively, the same site can be modified with various chemical entities, thus allowing binder validation experiments or chemical biology applications34. Our strategy of directed display of multiple diversity elements on a constant macrocyclic scaffold yielded specific binders against various target proteins and the resulting binders exhibited antibody-like properties, enabling biochemical and biological applications.
Figure 1. Design, synthesis, encoding and selection of the multiple display DNA-encoded chemical library.
a, Protein recognition by encoded multiple display of chemical elements on a constant macrocyclic scaffold. b, scaffold 1 containing three diversity sites, one potential diversity site and one site for DNA tagging. c, Scheme of library construction by a split-and-pool strategy employing three rounds of coupling and encoding with DNA tags. The performance of selection procedures on immobilized target proteins of interest allows the isolation of binders, whose binding affinities can be confirmed after resynthesis. A schematic representation of a possible macrocycle is illustrated as an example.
Results
Library design and synthesis
We first synthesized the cyclic peptide scaffold 1 (Fig. 1b) by using a solid-phase peptide synthesis approach, followed by cyclisation of the pre-organized linear precursor. Three lysine side-chains (highlighted in red at position 3, green at position 5 and dark blue at position 8, pointing to one side of the β-sheet plane; Fig. 1b) were masked with mutually orthogonal protecting groups and subsequently coupled with sets of chemical diversity elements, under DNA-compatible conditions (Supplementary Fig. 3). The lysine at position 9 (highlighted in light blue; Fig. 1b), pointing to the other side of the β-sheet plane, was used for the stepwise incorporation of DNA tags or for later stage functionalization. Position 10 (highlighted in magenta; Fig. 1b) was reserved as a fourth potential diversity site, which could be used for the conjugation of additional diversity elements or as a chemical handle for specific needs (e.g., photo-crosslinking).
Library synthesis was accomplished with three rounds of chemical coupling and encoding (Fig. 1c). Scaffold 1 was conjugated to 283 individual oligonucleotides, which encoded the first diversity site (Tfa site). The resulting conjugates were further coupled with 281 carboxylic acids acting as diversity elements (DE-1 representing the first diversity element, in all 3D cubes of Fig. 2), while the two residual oligonucleotides were used as controls (“Tfa-on” and “Tfa-off”). After Nvoc deprotection by UV irradiation, the conjugates were pooled to a mixture, followed by splitting into 386 vials for the subsequent modification with a second set of different carboxylic acids (DE-2 representing the second diversity element in Fig. 2). The resulting conjugates were encoded by an enzymatic ligation procedure. After a further pool-and-split step, the third site diversification (DE-3 representing the third diversity element in Fig. 2) was performed either through a copper-catalyzed cycloaddition with a set of terminal alkynes or by Staudinger reduction, followed by the acylation with a set of carboxylic acids. The last encoding step, featuring Klenow polymerization with partially complementary oligonucleotides, led to the formation of double-stranded DNA, corresponding to an encoded library comprising 35,393,112 displayed combinations on a constant macrocyclic scaffold (ETH-YL library).
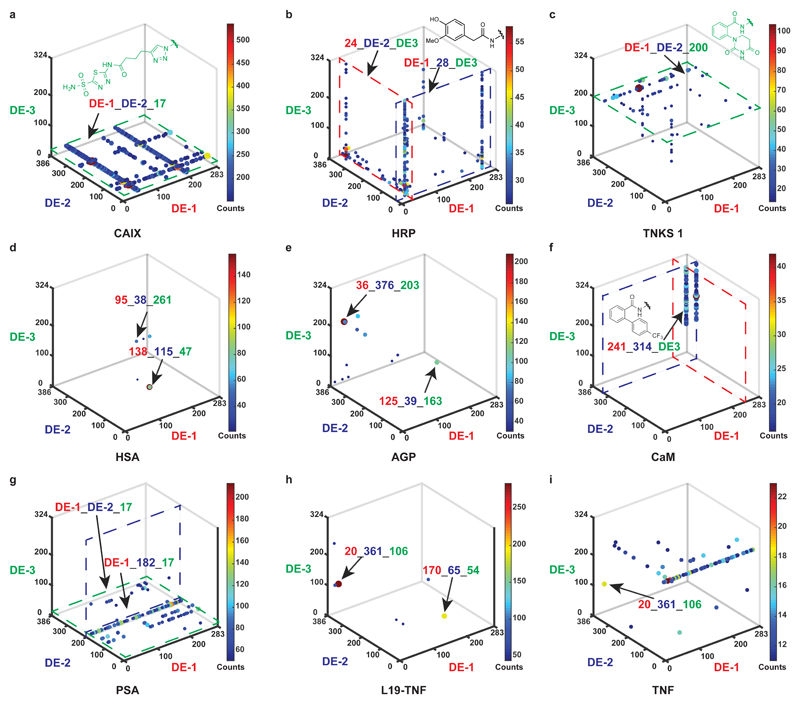
Figure 2. Selection fingerprints from the multiple display DNA-encoded chemical library.
a, High-throughput DNA sequencing (HTDS) plot of a selection against CAIX. The x-, y- and z-axes represent the first diversity element (DE-1, red), second diversity element (DE-2, dark blue), and third diversity element (DE-3, green) of the library, respectively. Jet colour scale and dot size represent the HTDS sequence counts. Total sequence counts (TSC) = 2,357,282, cut-off level = 150. b, HTDS plot of a selection against HRP. TSC = 4,269,414, cut-off level = 25. c, HTDS plot of a selection against TNKS 1. TSC = 2,525,256, cut-off level = 10. d, HTDS plot of a selection against HSA. TSC = 3,344,294, cut-off level = 20. e, HTDS plot of a selection against AGP. TSC = 1,929,066, cut-off level = 30. f, HTDS plot of a selection against CaM. TSC = 1,931,086, cut-off level = 15. g, HTDS plot of a selection against PSA. TSC = 2,244,812, cut-off level = 50. h, HTDS plot of a selection against L19-TNF. TSC = 2,353,006, cut-off level = 40. i, HTDS plot of a selection against TNF. TSC = 2,357,282, cut-off level = 10.
The coupling efficiency of all employed chemical diversity elements to DNA was evaluated in model reactions before library construction35,36. Shifts of the average mass of conjugate mixtures after the second and third coupling step were monitored by LC-MS17, and only diversity elements with conversion yields of over 80 % were included in the final library (Supplementary Figs. 10 and 12). The efficiency of the DNA-encoding procedure was also checked by gel electrophoresis and LC-MS (Supplementary Figs. 14 and 15). HTDS data of the library pool prior to selection exhibited a uniform distribution of sequence counts, providing additional support for the homogeneity of the library construction (Supplementary Fig. 16).
Selection experiments and binder validation
Affinity-based selections of our library with biotinylated proteins were performed according to a previously published procedure and decoded by HTDS37. Fig. 2 presents the results of affinity selections against nine different target proteins as “fingerprints”: The frequency of the sequenced DNA codes (counts) for each DE-1, DE-2, DE-3 combination can be plotted as individual dots, using both a Jet colour code (from blue to red) and the dot size (from small to large) to rank the counts of the displayed combinations, in a 3D cube with the axes DE-1, DE-2, and DE-3.
Carbonic anhydrase IX (CAIX), horseradish peroxidase (HRP) and tankyrase 1 (TNKS 1) served as positive controls, as the library contained diversity elements that were known to bind to these proteins. As expected, selections against CAIX revealed one plane of enriched combinations (DE-1_DE-2_17, highlighted in green), which corresponds to acetazolamide, a nanomolar binder to this protein (Fig. 2a)38. Selections against HRP (Fig. 2b) revealed an enriched phenolic structure (24_DE-2_DE-3 and DE-1_28_DE-3, respectively)39. Fig. 2c shows a plane of enriched compounds, featuring the presence of a chemical moiety (DE-1_DE-2_200, highlighted in green) that had previously been reported as a TNKS 1 binder40.
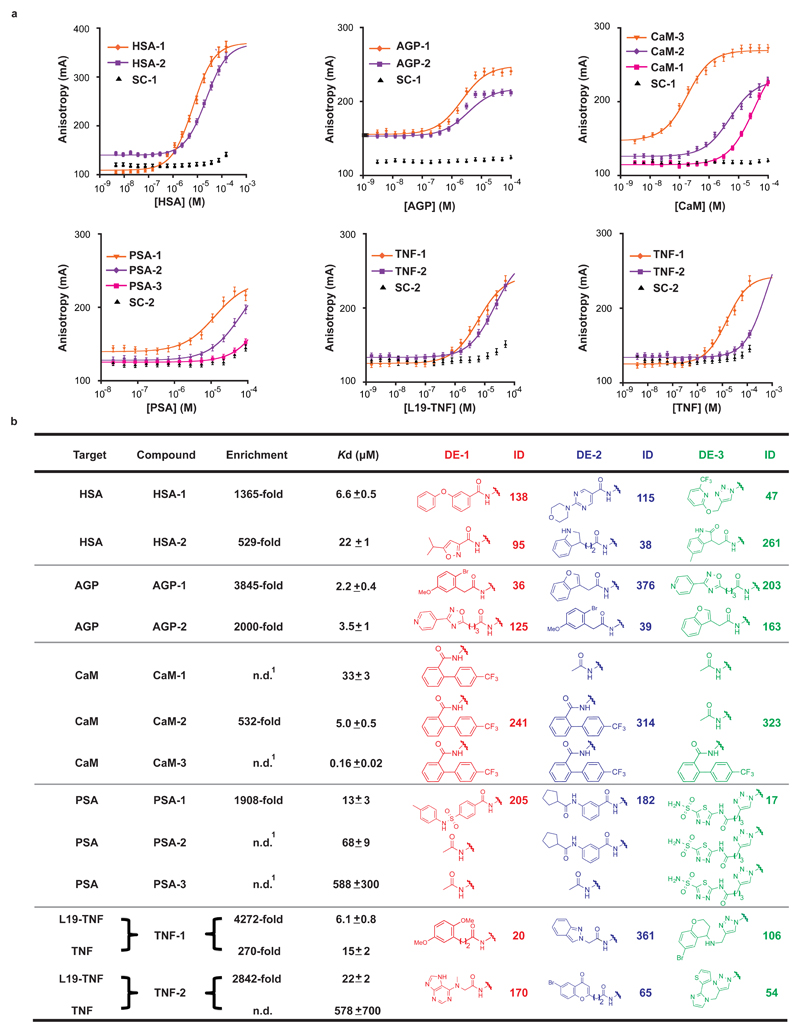
The fingerprint of the human serum albumin (HSA) selection (Fig. 2d) showed a scattered pattern of displayed combinations. We synthesized two enriched combinations (138_115_47 and 95_38_261) as fluorescein isothiocyanate (FITC)-labelled derivatives in the absence of DNA and used them in fluorescence polarization (FP) measurements, revealing dissociation constants in the single-digit micromolar range (Kd = 6.6 μM and 22 μM, respectively, Fig. 3).
Figure 3. Binder validation of selected combinations against HSA, AGP, CaM, PSA, L19-TNF and TNF.
a, fluorescence polarization measurements of selected fluorescently-labeled compounds against HSA, AGP, CaM, PSA, L19-TNF and TNF. A shift of the sigmoidal curves to the left for selected compounds indicates a contribution of the displayed chemical diversity elements to the binding affinity, compared to the unmodified scaffold controls (SC-1 and SC-2). Error bars indicate the standard deviation of three measurements. b, Enrichments and dissociation constants of synthesized compounds, chemical structures and corresponding identification numbers (ID) of the three diversity elements. For calculation of enrichment see Supplementary Table 7. SC-1 and SC-2 are two respective scaffold controls capped at the diversity sites; see Supplementary Figs. 21 and 44 for detailed structures. n.d. = not determined.1 = not included in the library.
The selection against alpha-1 acid glycoprotein (AGP) revealed a preferential enrichment of two identical sets of three chemical diversity elements, which were located at different positions on the scaffold (Fig. 3b). This corresponds to a “pseudo-rotation” of the three displayed chemical moieties along the defined scaffold. Synthetic FITC-labelled derivatives of these combinations (AGP-1 and AGP-2) showed a similar AGP binding affinity (2.2 μM and 3.5 μM, respectively, Fig. 3) in FP measurements. AGP-linear, a linear analogue of AGP-1, was synthesized and exhibited a 7-fold decrease in binding affinity (Kd = 15 μM), indicating a contribution of the cyclic nature of the scaffold to protein recognition (Supplementary Fig. 29).
A further selection against calmodulin (CaM) evidenced a preferential enrichment of an identical biphenyl moiety at the first and the second diversity site, respectively (Fig. 2f). The corresponding bivalent binder CaM-2 had a Kd = 5 μM (Fig. 3), while the monovalent binder CaM-1 bound to CaM with a Kd = 33 μM. Inclusion of a third binding moiety led to the trivalent derivative CaM-3 with a submicromolar dissociation constant (Kd = 0.16 μM).
The fingerprint of a selection against human prostate specific antigen (PSA), a tumor marker, revealed lines of enriched combinations in a plane, corresponding to DE-3 = 17 (highlighted in green; Fig. 2g). This finding implies that, while DE-3 = 17 plays an essential role, additional diversity elements contribute to protein recognition. The enriched combination 205_182_17 (PSA-1) showed a Kd = 13 μM, while the progressive removal of individual diversity elements led to a continuous loss of binding affinity (PSA-2 and PSA-3; Fig. 3).
We also performed selections against human tumor necrosis factor, used both as recombinant protein (TNF) and as TNF-antibody fusion (L19-TNF). We identified two diversity element combinations (20_361_106 and 170_65_54) as individual spots in the L19-TNF selection fingerprint (Fig. 2h), with high enrichment (Fig. 3b). The corresponding synthetic derivatives TNF-1 and TNF-2 exhibited Kd = 6.1 μM and 22 μM, respectively. The combination 20_361_106 was enriched also in the TNF selection fingerprint (Fig. 2i), indicating that binding was directed against the TNF moiety. The Kd of TNF-1 to TNF was found to be 15 μM, while TNF-2 did not exhibit considerable TNF binding (Fig. 3).
The utility of a constant macrocyclic scaffold lends itself to a further investigation of the impact of the flexibility of the scaffold and the length of the lysine sidechains, which serve as linkers between the diversity elements and the scaffold itself, on the binding affinity of the selected binders. A previous study reported that a substitution of two L-proline residues with D-proline in the macrocyclic backbone leads to an even more restricted β-hairpin conformation33. For the TNF binder D-TNF-1 and the PSA binder D-PSA-1, this substitution could be implemented, resulting in neither a significant loss nor gain of binding affinity (Supplementary Figs. 54 and 67). Shortening the linker sidechains by 1, 2 or 3 methylene groups, however, lead to a loss in binding affinity for the PSA binder (Supplementary Fig. 54). Interestingly, in the case of the TNF binder, the substitution of the diversity elements-displaying lysine with ornithine residues lead to a 6-fold increase in binding affinity of TNF-1, while further shortening of the linker sidechains had a detrimental effect on Kd values (Supplementary Fig. 67).
In vitro and in vivo applications of selected binders
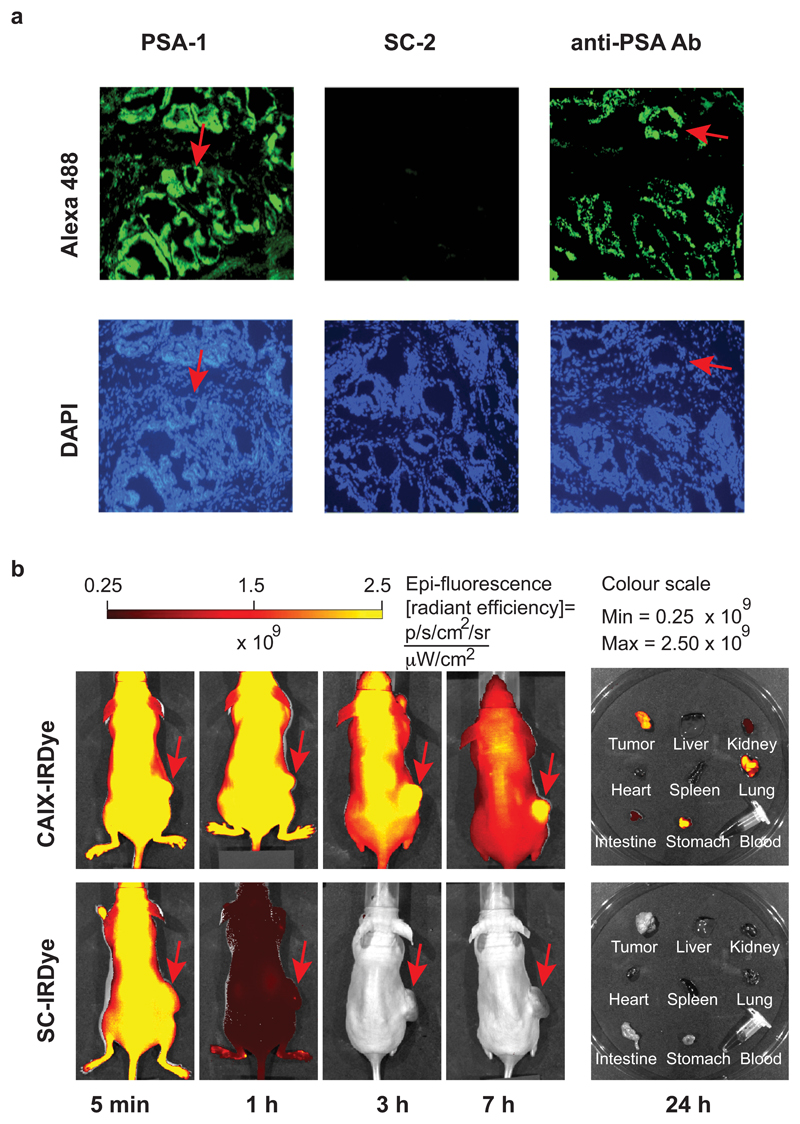
In order to test whether the selected synthetic binders could be used like antibodies in certain applications, we first investigated the performance of the binder PSA-1 in a fluorescence microscopy analysis of human prostate tissue expressing PSA. The synthetic FITC-labelled derivative was incubated with the tissue sections and detected, using an anti-FITC antibody as secondary reagent. This procedure stained the glandular structures, which are known to express PSA. The same structures were also stained by an anti-PSA antibody, which served as a positive control (Fig. 4a). As negative control, the FITC-labelled capped scaffold SC-2 was used. In a second application, we injected mice (bearing a subcutaneously-grafted tumor, expressing CAIX) with CAIX-IRDye, a derivative conjugated with acetazolamide as diversity elements and carrying a near-infrared dye for detection. Imaging at different time points revealed a preferential accumulation of the CAIX binder in the tumor, while no preferential tumor uptake could be observed when the capped scaffold SC-IRDye was used as a negative control (Fig. 4b).
Figure 4. Immunofluoresence performance of selected PSA binder and in vivo performance of CAIX binder.
a, Upper panels: micrographs illustrating PSA localization by binding to PSA-1, SC-2, and to a mouse anti-PSA antibody. The sections were incubated with PSA-1 or SC-2 (0.5 μM each), treated with rabbit anti-FITC antibody, and detected by goat anti-rabbit antibody-Alexa 488. For comparison, the third section was incubated with mouse anti-PSA antibody and detected with goat anti-mouse antibody-Alexa 488. Lower panels: DAPI staining of the same sections. Some glandular structures are indicated with red arrows. DAPI: 4′, 6-Diamidine-2′-phenylindole. b, Evaluation of the targeting performance of IRDye-680 conjugates CAIX-IRDye and of the control SC-IRDye, using near-infrared fluorescence imaging of BALB/c nu/nu mice bearing SKRC-52 xenografts, after intravenous administration of 3 nmol of conjugates. Images were acquired with identical parameters at 5 min, 1 h, 3 h and 7 h after injection. Right: imaging of excised organs and blood of injected mice 24 h after injection. See Supplementary Figs. 68 and 70 for detailed structures.
Development of chemical probes
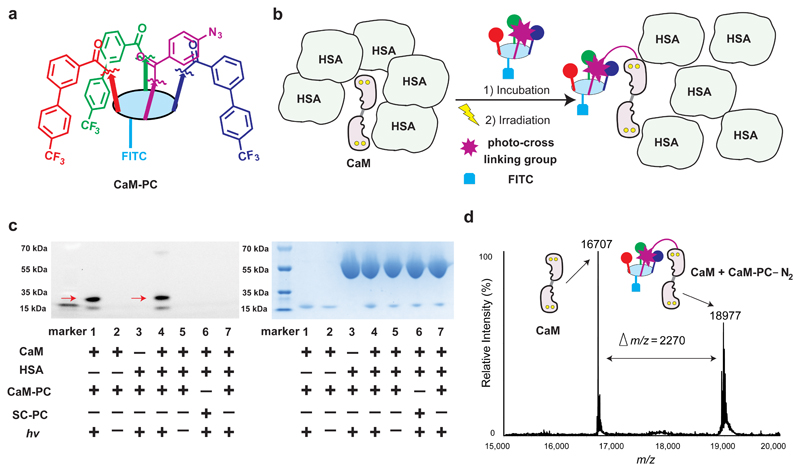
We installed a photo-crosslinking moiety on the fourth diversity site of the scaffold, in order to convert binders into chemical probes for specific protein labelling applications. As an illustrative example, we fused a photo-crosslinking moiety (phenyl azide) to the trivalent CaM binder (CaM-3), generating a probe (CaM-PC; Fig. 5a). Incubation of this probe with CaM in the presence of a 5-fold excess of HSA, followed by UV irradiation (Fig. 5b), led to the preferential photo-crosslinking of the probe to CaM rather than to HSA, as evidenced by gel electrophoretic analysis (lane 4 in Fig. 5c). A control probe (SC-PC), consisting of the scaffold equipped with the photo-crosslinking moiety but devoid of the CaM recognition elements, did not label CaM, as expected (lane 6, Fig. 5c). The formation of a covalent conjugate with CaM under irradiation was further confirmed by mass spectrometric analysis (Fig. 5d).
Figure 5. Performance of a chemical probe developed from selected CaM binder.
a, Structure of the chemical probe CaM-PC developed from CaM-3. b, specific labelling of CaM by CaM-PC in presence of excess HSA. c, SDS-PAGE analysis reveals the formation of a covalent adduct (indicated with red arrows) between the CaM-PC probe and CaM; left panel: fluorescence imaging of FITC, a smaller proteolytic fragment of calmodulin is also visible; right: Coomassie Brilliant Blue staining; left lane, marker; lane 1, CaM (3 μM)/CaM-PC (10 μM), hv; lane 2, same as lane 1, no hv; lane 3, HSA (15 μM)/CaM-PC (10 μM), hv; lane 4, CaM (3 μM)/HSA (15 μM)/CaM-PC (10 μM), hv; lane 5: same as lane 4, no hv; lane 6, same as lane 4 with scaffold control SC-PC instead of CaM-PC; lane 7, same as lane 4, with addition of soluble competitor CaM-3-amino (100 μM). See Supplementary Figs. 76 and 77 for full gel images. d, ESI-TOF-MS spectrum of the reaction mixture containing CaM-PC and CaM following UV irradiation (hv), revealing the formation of a covalent adduct. CaM (10 μM)/CaM-PC (10 μM), hv. See Supplementary Figs. 72 and 74 for detailed structures.
Discussion
We have described the synthesis of a DNA-encoded chemical library based on the display of multiple diversity elements on a constant macrocyclic scaffold, containing over 35 million combinations. We used optimized DNA-compatible synthetic procedures41, in order to ensure high library quality, which is a prerequisite for good performance in selections and for the identification of structure-activity relationships40,42,43.
The encoded library yielded binders against various, structurally diverse target proteins. This feature is reminiscent of the performance of large combinatorial libraries of antibodies (e.g., phage display libraries), which produce useful binding specificities against the majority of target proteins. The typical dissociation constants for binders isolated from our library were in the single-digit micromolar range, while phage display libraries containing billions of clones typically yield antibody fragments with Kd values in the 10 nM to 1 μM range44–46. Interestingly, it has previously been reported that the use of a smaller portion (containing only 10 million library members) of a 65 billion-membered phage-display library in selection experiments yielded binders with Kd values ranging between 0.8 μM and 12 μM44. These findings suggest that binders of higher affinity may be isolated from larger libraries based on the design strategy reported in this article, featuring the incorporation of more chemical diversity elements on the same scaffold. On the other hand, the structure-activity relationships obtained from selection experiment of our library as fingerprints for a given target allow the construction of a next-generation library by using structural analogues of the enriched chemical diversity elements on the scaffold, in analogy to the stepwise process often used for the affinity maturation of antibodies47–49. Alternatively, we have shown that fine-tuning of certain structural features of the constant scaffold may also have an impact on dissociation constants (Supplementary Fig. 67).
An antibody recognizes its antigen as a result of numerous interactions between its multiple residues in the hypervariable loops and the contact residues of the antigen. Interestingly, specific antibodies could also be isolated from “designer” combinatorial libraries, in which the multiple antigen-contacting sites of the hypervariable loops contained a restricted set of diversity elements50. To the other extreme, the fixed macrocyclic scaffold described in this article displays only three spatially oriented sets of chemical diversity elements. This chemical strategy was sufficient to generate specific binders against different target proteins, providing further evidence that certain antibody properties for recognition may be “minimized” (i.e., converted into smaller chemical structures, capable of similar performance)51. The diversity elements that could be employed in our strategy, are not limited to proteinogenic aminoacids as required by antibodies, and can be chosen from a broader chemical space.
There is a growing interest in the field of macrocycles for drug discovery52. However, it might be difficult to improve their pharmaceutical properties, as modifications of the backbone can lead to conformational changes30. The nature of the constant macrocyclic scaffold for this study allows the display of diversity elements in a pre-defined spatial orientation (as shown both by NMR studies32 and by X-ray crystallography33), and facilitates the incorporation of additional chemical moieties, such as photo-crosslinkers, fluorophores or other payloads, for further applications.
In summary, we have shown that an encoded library, containing over 35 million displayed combinations on a constant macrocyclic scaffold, yielded specific binders against multiple target proteins, including “difficult” targets (e.g., TNF). The binders discovered from our library enabled biochemical applications, (e.g., immunofluorescence tissue staining procedures and in vivo delivery of payloads to tumors) with antibody-like performance and the development of a chemical probe. Our strategy may facilitate chemical biology research and the development of novel classes of pharmaceutical products.
Methods
Detailed methods, synthetic procedures and characterization of the presented ETH-YL library and compounds are described in the Supplementary Information.
Library synthesis
The scaffold 1 was conjugated to amino-modified oligonucleotides (code 1: 5’-amino-C6-GGAGCTTCTGAATTCTGTGTG CTGNNNNNNCGAGTCCCATGGCGC-3’-OH, N representing variable bases serving as code) using a reported EDC/HOAt method36. The conjugates were precipitated with ethanol before purification by HPLC. Tfa-deprotection was performed with ammonium hydroxide (25 % aq), followed by immobilization on diethylaminoethyl (DEAE) sepharose. The first diversification elements were coupled to oligonucleotides with EDC-HCl/HOAt on pseudo-solid phase35 and resulting DNA-conjugates were then isolated by ethanol precipitation. The individual conjugates with the first diversity elements were subjected to irradiation at 365 nm at 0 °C for 60 min, followed by equimolar pooling of the 283 conjugates. The first pool was further divided into 386 aliquots and coupled with the second diversity elements using the same EDC-HCl/HOAt conditions. Individual conjugates with second diversity elements were employed in an enzymatic ligation step. To the conjugates, 5’-phosphorylated-oligonucleotides (code 2: 5’-phosphate-CGGATCGACGNNNNNNNGCGTCAGGCAGC-3’-OH) and a “splint” oligonucleotide (5’-OH-CGTCGATCCGGCGCCATGGG-3’-OH) were added, followed by addition of ligase buffer and T4 DNA ligase. The ligation was allowed for 6 h at 22 °C. Equimolar amounts of the 386 conjugates were combined to generate the second pool, followed by ethanol precipitation and HPLC purification. A portion of the second pool was divided into 136 aliquots and was immobilized on DEAE speharose. The resin was washed with aq. AcOH (3 × 0.5 mL, 10 mM), H2O (3 × 0.5 mL) and DMSO: H2O: tBuOH = 4 : 3 : 1 (3 × 0.5 mL). To the resin-immobilized conjugates, a solution of the corresponding alkyne diversity element (20 mM), TBTA (10 mM), CuSO4 (2.5 mM) and ascorbate (10 mM) in DMSO: H2O: tBuOH = 4 : 3 : 1 (0.2 mL) was added. The slurry was agitated for 4 hours at 25 ºC. The coupling solution was removed and the resin washed with DMSO: H2O: tBuOH = 4 : 3 : 1 (6 × 0.5 mL), aq. EDTA (3 × 0.5 mL, 50 mM) and aq. AcOH (3 × 0.5 mL, 10 mM). The conjugates were eluted from the resin by incubation with aq. AcOH (3 × 0.2 mL, 3 M) for 5 min. A further portion of the second pool was dissolved in Tris-HCl (500 mM, pH 8.0) and treated with TCEP-HCl (100 mM). The Staudinger reduction lasted for 12 hours at 25 °C and the reduction products were isolated by ethanol precipitation. The reduction products were further split into 185 aliquots and conjugated with 185 carboxylic acids using the above-mentioned EDC-HCl/HOAt method. Individual conjugates with the third diversity elements were further subjected to polymerase extension using oligonucleotides of following sequence for encoding (code 3: 5’-OH-GCTCTGCACGGTCGCNNNNNNNGCTGCCTGACGC-3’-OH). Equimolar amounts of the 324 conjugates were combined to generate the final library of 35 million combinations. For further details, see Supplementary Information.
Fluorescence polarization measurement with FITC-labelled binders
Freshly dissolved FITC labelled binders (7.5 μL, final concentration 50 nM, 1 % DMSO) were incubated at 22 °C for 10 min in a black 384-well plate (Greiner, non-binding) in PBS with increasing concentrations of protein to a final volume of 15 μL. The fluorescence anisotropy was measured on a Spectra Max Paradigm multimode plate reader (Molecular Devices). Experiments were performed in triplicate and the mean anisotropy values were fitted to equation 3, see Supplementary Information.
Mass spectrometry analysis of CaM-probe covalent complex
In a 96-well plate, CaM (10 μM) in Dulbecco’s Phosphate Buffered Saline (10 % DMSO) was incubated with the probe (CaM-PC, 10 μM) for 10 min at 0 °C with gentle shaking. The samples were irradiated at 365 nm for 15 min at 0 °C. Protein samples were purified on Vivaspin 500 centrifugal concentrators and dissolved in 100 μL PBS. A Xevo G2-XS Q-TOF with electrospray ionization source was used for detection.
Supplementary Material
Supplementary information is available in the online version of the paper.
Acknowledgements
This work was supported by ETH Zürich, the Swiss National Science Foundation (310030B_163479/1 Grant and CRSII2_160699/1 Sinergia Grant), the ERC Advanced Grant “Zauberkugel”, Philochem AG and the Start-up Funding for “Hundred Young-Talent Scheme” Professorship provided by the Chongqing University in China (0247001104406). The authors thank G. Annuziato for help with synthesis, C. Aquino and L. Optiz for help with high-throughput DNA sequencing, M. Wichert, R. Franzini, G. Zimmerman, A. Dal Corso, W. Decurtins, A. Schmid, M. Xi and J. Zhang for helpful discussions. Instant JChem (ChemAxon) was used for structure and data management. (http://www.chemaxon.com)
Footnotes
Data availability. The authors declare that the main data supporting the findings of this study are available within the Article and its Supplementary Information. Extra data and materials are available from the corresponding authors upon reasonable request. A Life Sciences Reporting Summary for this paper is available.
Author contributions
Y. L., J. S. and D. N. designed the project. Y. L. constructed the library. R. L. provided target proteins. Y. L. designed and performed the selections. Y. L. and J. S. analyzed high-throughput DNA screening data. Y. L. performed synthesis and hit validation experiments and performed the photo-crosslinking experiments. Y. L. and F. P. performed the immunofluorescence experiments. Y. L. and S. C. performed in vivo experiments. D. B. performed the biotinylation of target proteins. Y. L., J. S. and D. N. wrote the manuscript.
Competing financial interests
D. N. is a co-founder and shareholder of Philochem AG (Otelfingen, Switerland), J. S. is a board member of Philochem AG.
References
- 1.Murphy K, Janeway CA, Mowat A. Janeway's immunobiology. 8th edn. Garland Science; 2012. [Google Scholar]
- 2.Smith GP. Filamentous fusion phage: novel expression vectors that display cloned antigens on the virion surface. Science. 1985;228:1315–1317. doi: 10.1126/science.4001944. [DOI] [PubMed] [Google Scholar]
- 3.Clackson T, Hoogenboom HR, Griffiths AD, Winter G. Making antibody fragments using phage display libraries. Nature. 1991;352:624–628. doi: 10.1038/352624a0. [DOI] [PubMed] [Google Scholar]
- 4.Lerner RA, Kang AS, Bain JD, Burton DR, Barbas CF., 3rd Antibodies without immunization. Science. 1992;258:1313–1314. doi: 10.1126/science.1455226. [DOI] [PubMed] [Google Scholar]
- 5.Winter G, Griffiths AD, Hawkins RE, Hoogenboom HR. Making antibodies by phage display technology. Annu Rev Immunol. 1994;12:433–455. doi: 10.1146/annurev.iy.12.040194.002245. [DOI] [PubMed] [Google Scholar]
- 6.Thurber GM, et al. Single-cell and subcellular pharmacokinetic imaging allows insight into drug action in vivo. Nat Commun. 2013;4:1504. doi: 10.1038/ncomms2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heinis C, Rutherford T, Freund S, Winter G. Phage-encoded combinatorial chemical libraries based on bicyclic peptides. Nat Chem Biol. 2009;5:502–507. doi: 10.1038/nchembio.184. [DOI] [PubMed] [Google Scholar]
- 8.Wilson DS, Keefe AD, Szostak JW. The use of mRNA display to select high-affinity protein-binding peptides. Proc Natl Acad Sci U S A. 2001;98:3750–3755. doi: 10.1073/pnas.061028198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kawakami T, et al. Diverse backbone-cyclized peptides via codon reprogramming. Nat Chem Biol. 2009;5:888–890. doi: 10.1038/nchembio.259. [DOI] [PubMed] [Google Scholar]
- 10.Boder ET, Wittrup KD. Yeast surface display for screening combinatorial polypeptide libraries. Nat Biotechnol. 1997;15:553–557. doi: 10.1038/nbt0697-553. [DOI] [PubMed] [Google Scholar]
- 11.Hanes J, Pluckthun A. In vitro selection and evolution of functional proteins by using ribosome display. Proc Natl Acad Sci U S A. 1997;94:4937–4942. doi: 10.1073/pnas.94.10.4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goodnow RA, Jr, Dumelin CE, Keefe AD. DNA-encoded chemistry: enabling the deeper sampling of chemical space. Nat Rev Drug Discov. 2017;16:131–147. doi: 10.1038/nrd.2016.213. [DOI] [PubMed] [Google Scholar]
- 13.Lerner RA, Brenner S. DNA-Encoded Compound Libraries as Open Source: A Powerful Pathway to New Drugs. Angew Chem Int Ed. 2017;56:1164–1165. doi: 10.1002/anie.201612143. [DOI] [PubMed] [Google Scholar]
- 14.Brenner S, Lerner RA. Encoded combinatorial chemistry. Proc Natl Acad Sci U S A. 1992;89:5381–5383. doi: 10.1073/pnas.89.12.5381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Needels MC, et al. Generation and screening of an oligonucleotide-encoded synthetic peptide library. Proc Natl Acad Sci U S A. 1993;90:10700–10704. doi: 10.1073/pnas.90.22.10700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mannocci L, et al. High-throughput sequencing allows the identification of binding molecules isolated from DNA-encoded chemical libraries. Proc Natl Acad Sci U S A. 2008;105:17670–17675. doi: 10.1073/pnas.0805130105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clark MA, et al. Design, synthesis and selection of DNA-encoded small-molecule libraries. Nat Chem Biol. 2009;5:647–654. doi: 10.1038/nchembio.211. [DOI] [PubMed] [Google Scholar]
- 18.Litovchick A, et al. Encoded Library Synthesis Using Chemical Ligation and the Discovery of sEH Inhibitors from a 334-Million Member Library. Sci Rep. 2015;5:10916. doi: 10.1038/srep10916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Franzini RM, Randolph C. Chemical Space of DNA-Encoded Libraries. J Med Chem. 2016;59:6629–6644. doi: 10.1021/acs.jmedchem.5b01874. [DOI] [PubMed] [Google Scholar]
- 20.Halpin DR, Harbury PB. DNA Display I. Sequence-Encoded Routing of DNA Populations. PLOS Biol. 2004;2:e173. doi: 10.1371/journal.pbio.0020173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gartner ZJ, et al. DNA-templated organic synthesis and selection of a library of macrocycles. Science. 2004;305:1601–1605. doi: 10.1126/science.1102629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hansen MH, et al. A yoctoliter-scale DNA reactor for small-molecule evolution. J Am Chem Soc. 2009;131:1322–1327. doi: 10.1021/ja808558a. [DOI] [PubMed] [Google Scholar]
- 23.Li Y, Zhao P, Zhang M, Zhao X, Li X. Multistep DNA-templated synthesis using a universal template. J Am Chem Soc. 2013;135:17727–17730. doi: 10.1021/ja409936r. [DOI] [PubMed] [Google Scholar]
- 24.Wichert M, et al. Dual-display of small molecules enables the discovery of ligand pairs and facilitates affinity maturation. Nat Chem. 2015;7:241–249. doi: 10.1038/nchem.2158. [DOI] [PubMed] [Google Scholar]
- 25.Reddavide FV, Lin W, Lehnert S, Zhang Y. DNA-Encoded Dynamic Combinatorial Chemical Libraries. Angew Chem Int Ed. 2015;54:7924–7928. doi: 10.1002/anie.201501775. [DOI] [PubMed] [Google Scholar]
- 26.Li G, et al. Design, preparation, and selection of DNA-encoded dynamic libraries. Chem Sci. 2015;6:7097–7104. doi: 10.1039/c5sc02467f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Daguer JP, et al. DNA display of fragment pairs as a tool for the discovery of novel biologically active small molecules. Chem Sci. 2015;6:739–744. doi: 10.1039/c4sc01654h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deng H, et al. Discovery of Highly Potent and Selective Small Molecule ADAMTS-5 Inhibitors That Inhibit Human Cartilage Degradation via Encoded Library Technology (ELT) J Med Chem. 2012;55:7061–7079. doi: 10.1021/jm300449x. [DOI] [PubMed] [Google Scholar]
- 29.Barluenga S, et al. Novel PTP1B inhibitors identified by DNA display of fragment pairs. Bioorganic & Med Chem Lett. 2016;26:1080–1085. doi: 10.1016/j.bmcl.2015.11.102. [DOI] [PubMed] [Google Scholar]
- 30.Maianti JP, et al. Anti-diabetic activity of insulin-degrading enzyme inhibitors mediated by multiple hormones. Nature. 2014;511:94–98. doi: 10.1038/nature13297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mutter M, Hersperger R, Gubernator K, Muller K. The construction of new proteins: V. A template-assembled synthetic protein (TASP) containing both a 4-helix bundle and beta-barrel-like structure. Proteins. 1989;5:13–21. doi: 10.1002/prot.340050104. [DOI] [PubMed] [Google Scholar]
- 32.Dumy P, Eggleston IM, Esposito G, Nicula S, Mutter M. Solution structure of regioselectively addressable functionalized templates: An NMR and restrained molecular dynamics investigation. Biopolymers. 1996;39:297–308. doi: 10.1002/(sici)1097-0282(199609)39:3<297::aid-bip3>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 33.Peluso S, et al. Crystal Structure of a Synthetic Cyclodecapeptide for Template-Assembled Synthetic Protein Design. ChemBioChem. 2001;2:432–437. doi: 10.1002/1439-7633(20010601)2:6<432::AID-CBIC432>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 34.Salamon H, Klika Škopić M, Jung K, Bugain O, Brunschweiger A. Chemical Biology Probes from Advanced DNA-encoded Libraries. ACS Chem Biol. 2016;11:296–307. doi: 10.1021/acschembio.5b00981. [DOI] [PubMed] [Google Scholar]
- 35.Franzini RM, et al. Systematic evaluation and optimization of modification reactions of oligonucleotides with amines and carboxylic acids for the synthesis of DNA-encoded chemical libraries. Bioconjug Chem. 2014;25:1453–1461. doi: 10.1021/bc500212n. [DOI] [PubMed] [Google Scholar]
- 36.Li Y, et al. Optimized Reaction Conditions for Amide Bond Formation in DNA-Encoded Combinatorial Libraries. ACS Comb Sci. 2016;18:438–443. doi: 10.1021/acscombsci.6b00058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Decurtins W, et al. Automated screening for small organic ligands using DNA-encoded chemical libraries. Nat Protoc. 2016;11:764–780. doi: 10.1038/nprot.2016.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krall N, et al. A Small-Molecule Drug Conjugate for the Treatment of Carbonic Anhydrase IX Expressing Tumors. Angew Chem Int Ed. 2014;53:4231–4235. doi: 10.1002/anie.201310709. [DOI] [PubMed] [Google Scholar]
- 39.Gilabert MaA, et al. Differential substrate behaviour of phenol and aniline derivatives during oxidation by horseradish peroxidase: kinetic evidence for a two-step mechanism. Biochimica et Biophysica Acta (BBA) - Proteins and Proteomics. 2004;1699:235–243. doi: 10.1016/j.bbapap.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 40.Franzini RM, et al. Identification of structure-activity relationships from screening a structurally compact DNA-encoded chemical library. Angew Chem Int Ed. 2015;54:3927–3931. doi: 10.1002/anie.201410736. [DOI] [PubMed] [Google Scholar]
- 41.Malone ML, Paegel BM. What is a “DNA-Compatible” Reaction? ACS Comb Sci. 2016;18:182–187. doi: 10.1021/acscombsci.5b00198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Deng H, et al. Discovery, SAR, and X-ray Binding Mode Study of BCATm Inhibitors from a Novel DNA-Encoded Library. ACS Med Chem Lett. 2015;6:919–924. doi: 10.1021/acsmedchemlett.5b00179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Satz AL. Simulated Screens of DNA Encoded Libraries: The Potential Influence of Chemical Synthesis Fidelity on Interpretation of Structure-Activity Relationships. ACS Comb Sci. 2016;18:415–424. doi: 10.1021/acscombsci.6b00001. [DOI] [PubMed] [Google Scholar]
- 44.Griffiths AD, et al. Isolation of high affinity human antibodies directly from large synthetic repertoires. EMBO J. 1994;13:3245–3260. doi: 10.1002/j.1460-2075.1994.tb06626.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nissim A, et al. Antibody fragments from a 'single pot' phage display library as immunochemical reagents. EMBO J. 1994;13:692–698. doi: 10.1002/j.1460-2075.1994.tb06308.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vaughan TJ, et al. Human Antibodies with Sub-nanomolar Affinities Isolated from a Large Non-immunized Phage Display Library. Nat Biotech. 1996;14:309–314. doi: 10.1038/nbt0396-309. [DOI] [PubMed] [Google Scholar]
- 47.Gram H, et al. In vitro selection and affinity maturation of antibodies from a naive combinatorial immunoglobulin library. Proc Natl Acad Sci U S A. 1992;89:3576–3580. doi: 10.1073/pnas.89.8.3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jespers LS, Roberts A, Mahler SM, Winter G, Hoogenboom HR. Guiding the selection of human antibodies from phage display repertoires to a single epitope of an antigen. Biotechnology (NY) 1994;12:899–903. doi: 10.1038/nbt0994-899. [DOI] [PubMed] [Google Scholar]
- 49.Pini A, et al. Design and use of a phage display library. Human antibodies with subnanomolar affinity against a marker of angiogenesis eluted from a two-dimensional gel. J Biol Chem. 1998;273:21769–21776. doi: 10.1074/jbc.273.34.21769. [DOI] [PubMed] [Google Scholar]
- 50.Fellouse FA, Wiesmann C, Sidhu SS. Synthetic antibodies from a four-amino-acid code: A dominant role for tyrosine in antigen recognition. Proc Natl Acad Sci U S A. 2004;101:12467–12472. doi: 10.1073/pnas.0401786101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li B, et al. Minimization of a Polypeptide Hormone. Science. 1995;270:1657–1660. doi: 10.1126/science.270.5242.1657. [DOI] [PubMed] [Google Scholar]
- 52.Driggers EM, Hale SP, Lee J, Terrett NK. The exploration of macrocycles for drug discovery - an underexploited structural class. Nat Rev Drug Discov. 2008;7:608–624. doi: 10.1038/nrd2590. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.