Abstract
7T MRI scanner provides MR images with higher resolution and better contrast than 3T MR scanners. This helps many medical analysis tasks, including tissue segmentation. However, currently there is a very limited number of 7T MRI scanners worldwide. This motivates us to propose a novel image post-processing framework that can jointly generate high-resolution 7T-like images and their corresponding high-quality 7T-like tissue segmentation maps, solely from the routine 3T MR images. Our proposed framework comprises two parallel components, namely (1) reconstruction and (2) segmentation. The reconstruction component includes the multi-step cascaded convolutional neural networks (CNNs) that map the input 3T MR image to a 7T-like MR image, in terms of both resolution and contrast. Similarly, the segmentation component involves another paralleled cascaded CNNs, with a different architecture, to generate high-quality segmentation maps. These cascaded feedbacks between the two designed paralleled CNNs allow both tasks to mutually benefit from each another when learning the respective reconstruction and segmentation mappings. For evaluation, we have tested our framework on 15 subjects (with paired 3T and 7T images) using a leave-one-out cross-validation. The experimental results show that our estimated 7T-like images have richer anatomical details and better segmentation results, compared to the 3T MRI. Furthermore, our method also achieved better results in both reconstruction and segmentation tasks, compared to the state-of-the-art methods.
1 Introduction
In the past years, many efforts have been put into strengthening of the magnetic field of MRI, leading to ultra-high-field (7T) MRI scanner. This highly advanced imaging technology provides MR images with much higher resolution and contrast in comparison to the routine 3T MRI. Figure 1 shows sharper white matter (WM), gray matter (GM), and cerebrospinal fluid (CSF) tissue boundaries provided by 7T MRI. However, 7T MRI scanners are much more expensive and less available in research and clinical sites, compared with 3T MRI scanners [1,2].
Fig. 1.
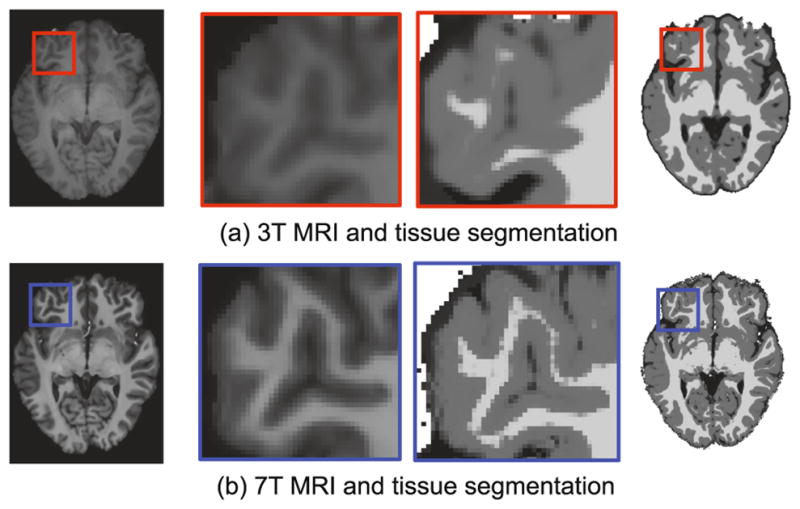
(a) 3T brain MRI and its corresponding tissue segmentation map. (b) 7T brain MRI and its corresponding tissue segmentation map. Clearly, the 7T brain MRI shows better tissue segmentation results compared to those of 3T brain MRI.
To somehow compensate for the lack of availability of 7T MRI scanners in health care, several reconstruction and synthesis methods have been recently proposed to improve the resolution and quality of 3T MRI. Subsequently, better tissue segmentation maps can be also improved by applying the existing segmentation tools such as FAST [3] on the reconstructed high-resolution MR images, for segmenting the MRI brain tissue into WM, GM, and CSF. However, such high-quality segmentation strategy is limited by the decoupling of the segmentation and the resolution-enhancement learning tasks. Alternatively, the image segmentation task can be jointly integrated with the image reconstruction task in a unified framework, where the reconstruction task helps generate more accurate image segmentation map and in turn the segmentation task helps generate a better high-resolution image reconstruction. This motivates us to propose a deep learning framework for joint reconstruction-segmentation of 3T MR images.
Several 7T-like reconstruction and segmentation methods have been recently proposed for enhancing the resolution and the segmentation of 3T MRI. For instance, in [4], a sparse representation-based method has been proposed to improve the resolution of 3T MRI by using the estimated sparse coefficients via representing the input low-resolution MRI by the training low-resolution images. In another method, Manjon et al. [5] incorporated information from coplanar high-resolution images to enhance the super-resolution of MR images. Roy et al. [6] proposed an image synthesis method, called MR image example-based contrast synthesis (MIMECS), by improving both the resolution and contrast of the images. In another work, Bahrami et al. [7] proposed a sparse representation in multi-level canonical correlation analysis (CCA) to increase the correlation between 3T MRI and 7T MRI for 7T-like MR image reconstruction.
While the existing methods mostly were proposed for improving the resolution of the medical images, in this paper we propose a method for improving both the resolution and tissue segmentation of 3T MR images by reconstructing 7T-like MR images from 3T MR images. To do so, we propose a novel mutual deep-learning based framework, composed of cascaded CNNs, to estimate 7T-like MR images and their corresponding segmentation maps from 3T MR images. Our proposed method has the following key contributions: First, we propose a mutual deep learning framework that both parallels and cascades a series of CNNs to non-linearly map the 3T MR image to 7T MR image, and also segments 3T MR image with a 7T-like MRI segmentation quality. Second, the proposed CNN architecture is used for 3D patch reconstruction, which enforces a better spatial consistency between anatomical structures of the neighboring patches, compared to the previous 2D methods. Third, our proposed architecture leverages on a mutual shared learning between the reconstruction and segmentation tasks, so the jointly learned reconstruction and segmentation mappings can benefit from each another in each cascaded step.
2 Proposed Method
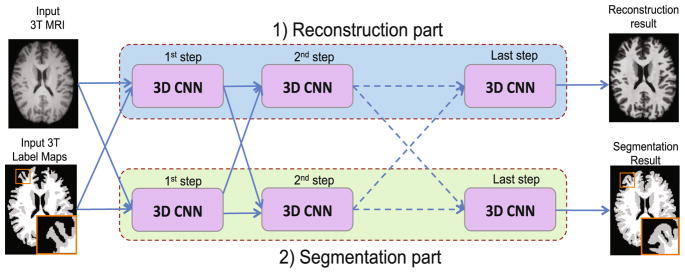
To improve the resolution and segmentation of 3T MR images, we propose a novel multi-step deep architecture based on 3D CNN for joint reconstruction and segmentation, as shown in Fig. 2.
Fig. 2.
Proposed joint reconstruction and segmentation based on cascaded 3D CNNs.
2.1 Proposed 7-Layer 3D Convolutional Neural Network (CNN)
Using a 3T MR image as input, our goal is to train a CNN function f (.) that can non-linearly map a 3T MR image to a 7T MR image. Such mapping is learned using the paired 3T and 7T MR images of training subjects, and then can be applied to a new 3T MR image X for generating its corresponding 7T-like MR image Y = f (X), with image quality similar to the ground-truth 7T MR image. Correspondingly, a second CNN is paralleled within the same architecture to map the segmentation of the 3T MR image to the segmentation of 7T MR image (Fig. 2). Notably, the learned parameters would be distinct for both CNNs in the reconstruction and segmentation tasks. Although both CNNs are trained independently for reconstruction and segmentation tasks, we enforce the sharing of learned features between both tasks in the proposed architecture (Fig. 2). For each CNN, we propose a 7-layer architecture with convolutional and ReLU operations as described below.
In the first layer, the feature map of the input 3T MR image is produced by convolving the input 3T MR image with a convolutional kernel and applying an activation function. Let x denote a patch of size m ×m ×m extracted from the input 3T MR image X. The first layer of our network includes N1 convolution filters of size w × w × w, followed by a ReLU activation function. The input to this layer is the intensity of the patch x, while its output y1 includes N1 feature maps. This layer is formulated as y1 = f1(x) = Ψ (F1 * x + B1), where F1 includes N1 convolutional filters of size w × w × w and B1 includes N1 bias values, each associated with a filter. The symbol * denotes the 3D convolution operation, which convolves each of the N1 filters with the input image to generate N1 feature maps.
The output of the convolutional filters are then thresholded using the ReLU activation function, denoted as Ψ (.). Next, by applying the N1 filters of the first layer to the input 3T MRI patch, we generate the response y1, along with new N1 feature maps. Except for the last layer, all other layers (the 2nd–6th layers) are similar in structure to the first layer, each followed by ReLU, such that yl = fl(yl–1) = Ψ(Fl * yl–1 + Bl), where Fl corresponds to the Nl filters with size of Nl–1 × w × w × w. Bl includes Nl bias values, each associated with a filter.
The last layer uses one convolution to generate one voxel value from the feature maps of the 6th layer. Eventually, the 7th layer convolves the N6 feature maps of the 6th layer with one filter of size N6 × w × w × w, followed by ReLU operation, to output one voxel value, as defined as y = f7(y6) = Ψ (F7 *y6 +B7).
The proposed CNN architecture generates a non-linear mapping from an input 3T MRI patch with size of m ×m × m to the voxel intensity at the center of the corresponding 7T MRI patch. The proposed CNN architecture is also used to regress the segmentation map of the 3T MRI patch to the segmentation map of the 7T MRI patch.
2.2 Proposed Cascaded CNN Architecture for Joint Reconstruction and Segmentation
We incorporate the proposed paralleled 7-layer CNNs in a cascaded deep learning framework for jointly reconstructing and segmenting 7T-like MR images from 3T MR images, thereby generating better reconstructed and segmented images as detailed below.
Reconstruction Part
The reconstruction part includes a chain of 3D CNNs, which aim to improve the resolution of 3T MR images using a multi-step deep-learning architecture. In each step, each CNN inputs both intensity and segmentation maps produced by the paralleled CNN of the previous step. To enforce the spatial consistency between neighboring patches within the same tissue for 7T-like MRI reconstruction, we train the designed CNNs using both the 7T-like MRI reconstruction result (intensity features) and the segmentation map (label features) of the previous step. This operation is performed in multiple progressive steps until the final 7T-like MRI has a finer brain tissue appearance. In the first step, we input the original 3T MRI and the tissue segmentation of the input 3T MR image to the first CNN. The output of the first step is a 7T-like MR image, which will be used to provide intensity features for the next step. The subsequent steps use both the outputted appearance and segmentation maps from the previous step to produce a better 7T-like MR image, which can gradually capture finer anatomical details and also clearer tissue boundaries, compared with the previous step.
Segmentation Part
The segmentation part in the proposed framework has a similar parallel architecture as the reconstruction part (Fig. 2). It involves a chain of 3D CNNs, which progressively improve the segmentation of the 3T MR image using a cascade of CNNs. In such architecture, each CNN inputs both the reconstructed intensity image and the produced segmentation map of the previous step. Specifically, in the first step, we input the original 3T MRI patch and its tissue segmentation to the first CNN. In the following steps, our designed segmentation CNN uses the 7T-like MRI reconstruction result of the previous step in the reconstruction part as the intensity features, together with the produced segmentation map of the previous step. This operation is iterated until the final segmentation map of the 7T-like MR image is generated with sharper anatomical details. In a gradual and deep manner, the proposed architecture improves the segmentation results by iteratively providing more refined context information from the reconstruction part to train the next segmentation CNN in the cascade.
3 Experimental Results
We used 15 pairs of 3T and 7T MR images collected from 15 healthy volunteers. The 3T MR and 7T MR images have resolution of 1 × 1 × 1 mm3 and 0.65 × 0.65 ×0.65 mm3, respectively. All MR images were aligned by registering the 3 T and 7T MR images using FLIRT in FSL package with a 9-DOF transformation.
For brain tissue segmentation, we use FAST in FSL package [3] to (1) generate 3T MRI segmentation maps for training, and (2) also segment the reconstructed 7T-like images into WM, GM and CSF for evaluation. To generate the mapping from 3T MRI to 7T MRI, we use a patch size of m = 15. For CNN, we use a 7-layer network with a filter size of w = 3 from the first layer to the last layer, and N = 64 filters for all layers. We extract overlapping patches with 1-voxel step size. For the proposed architecture, we cascade 3 CNNs. To evaluate the proposed framework, we use leave-one-out cross-validation. Training took 16 h on an iMAC with Intel Quad core i7 (3 GHz) and 16 GB of RAM. Testing took 10 min for each image.
Evaluation for Reconstruction of 7T-like MR Images
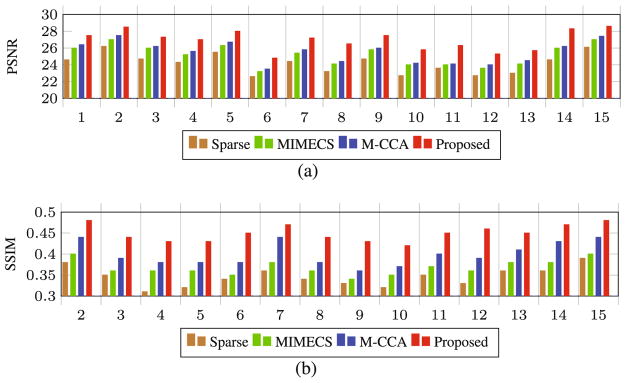
We evaluate our proposed joint reconstruction and segmentation method in improving both the resolution and segmentation of 3T MR images. For reconstruction of 7T-like MR images from 3T MR image, we compare our method with different methods, such as Sparse [4], MIMECS [6], and multi-level canonical correspondence analysis (M-CCA) [7]. Furthermore, we compare our method with the segmentation results based on the 3T MR images and also the reconstructed 7T-like MR images by different methods. For numerical evaluation, we use peak-signal-to-noise ratio (PSNR) and structural similarity (SSIM) measurements. In Fig. 3, PSNR and SSIM measurements have been compared for all 15 subjects by different methods. Our method has both higher PSNR and SSIM than the comparison methods, indicating closer appearance to the ground-truth image by our method. Also, using two-sample t-test, we examine the significance of improvement by our results compared to the results by the comparison methods. We found that our method significantly outperformed the comparison methods (p < 0.01 for each t-test).
Fig. 3.
Comparison of our proposed method with the state-of-the-art methods, in terms of (a) PSNR and (b) SSIM for all 15 subjects using leave-one-out cross-validation.
Evaluation for Brain Tissue Segmentation
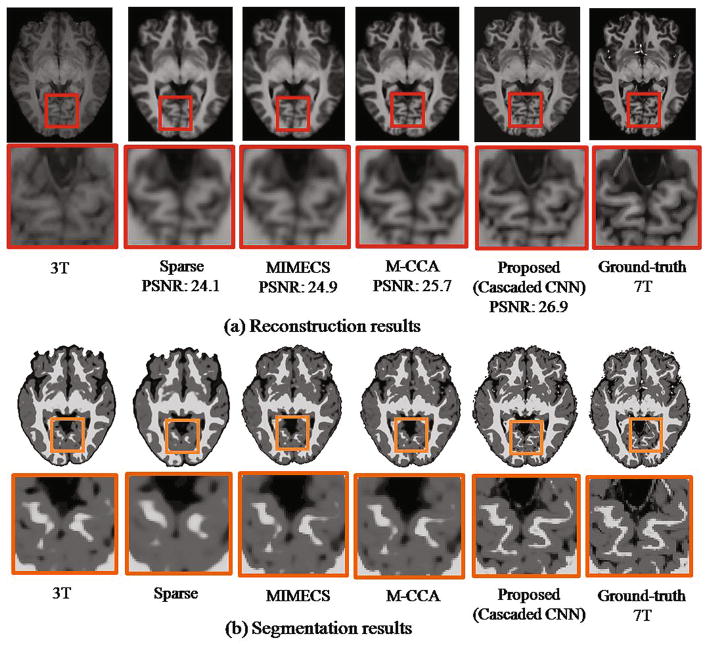
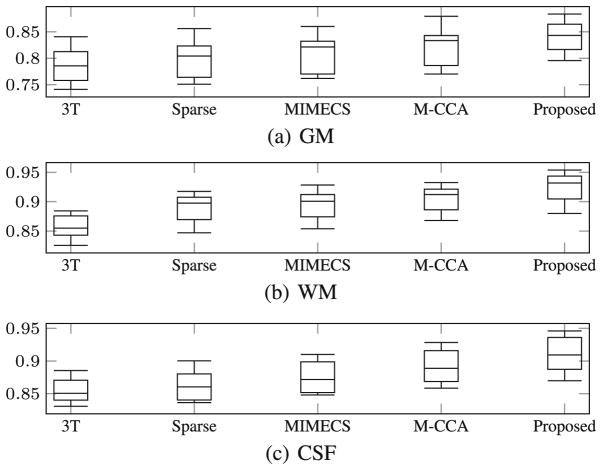
To evaluate our proposed method for brain tissue segmentation of 3T MR images, we generate brain tissue segmentation maps using the proposed method. Figure 4(b) shows the segmentation results for 3T MR image, the reconstructed 7T-like MR images by different methods, and the ground-truth 7T MR image, respectively. For comparison methods, we segmented the reconstructed 7T-like MR images using the same FAST method. The close-up views for the corresponding regions clearly indicate that the segmentation result based on our reconstructed 7T-like MR image captures more anatomical details. Compared to the segmentation result from the original 3T MR image, our segmentation result (based on our reconstructed 7T-like MR image) is much closer to the segmentation result based on the ground-truth 7T MR image, with superior WM, GM, and CSF brain tissue segmentation accuracies, compared to the baseline methods. To further show the superiority of our method, we quantitatively evaluate the segmentation results using different methods. In Fig. 5, we show the distribution of Dice overlap between the segmentation result by each reconstruction method and the segmentation result by 7T MR image in each leave-one-out cross-validation. We provided quantitative evaluations on WM, GM, and CSF separately to clearly show the improvement of our result compared to other methods. Obviously, our method significantly outperforms other reconstruction methods (with p < 0.01 by two-sample t-test). Also, the segmentation of our reconstructed 7T-like MR image is much better than direct segmentation of original 3T MR image.
Fig. 4.
Comparison of different methods in (a) reconstructing 7T-like MRI and (b) contributing for tissue segmentation by the FAST method. Left to right of (a) show the axial views and the close-up views of 3T MRI, the four reconstructed 7T-like MRIs (by Sparse [4], MIMECS [6], M-CCA [7], and our method), and the ground-truth 7T MRI. The segmentation results in (b) are arranged in the same way as in (a).
Fig. 5.
Box plot of the Dice ratio for segmentation of (a) GM, (b) WM, (c) CSF from the reconstructed 7T-like MR image by four different methods using FAST. From left to right of each subfigure, the distribution of Dice ratios on 3T MRI and the 7T-like MR images (by Sparse [6], MIMECS [6], M-CCA [7], and our proposed method) are shown, respectively.
4 Conclusion
In this paper, we have presented a novel paralleled deep-learning-based architecture for joint reconstruction and segmentation of 3T MR images. For the reconstruction results, both visual inspection and quantitative measures show that our proposed method produces better 7T-like MR images compared to other state-of-the-art methods. For evaluating the segmentation results, we applied a segmentation method (FAST) to 7T-like images generated by different reconstruction methods (e.g., Sparse, MIMECs, and M-CCA), and then compared their results with the output segmentation maps of our framework. It is shown that our method has significantly higher accuracy in segmenting WM, GM, and CSF maps, compared to direct segmentation of the original 3T MR images. Also, it is worth to note that this framework is general enough to be applied to MRI of other organs.
References
- 1.Kolka AG, Hendriksea J, Zwanenburg JJM, Vissera F, Luijtena PR. Clinical applications of 7T MRI in the brain. Euro J Radiol. 2013;82:708–718. doi: 10.1016/j.ejrad.2011.07.007. [DOI] [PubMed] [Google Scholar]
- 2.Beisteiner R, Robinson S, Wurnig M, Hilbert M, Merksa K, Rath J, Hillinger I, Klinger N, Marosi C, Trattnig S, Geiler A. Clinical fMRI: evidence for a 7T benefit over 3T. NeuroImage. 2011;57:1015–1021. doi: 10.1016/j.neuroimage.2011.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang Y, Brady M, Smith S. Segmentation of brain MR images through Markov random field model and the expectation-maximization algorithm. IEEE Trans Med Imaging. 2001;20(1):45–57. doi: 10.1109/42.906424. [DOI] [PubMed] [Google Scholar]
- 4.Rueda A, Malpica N, Romero E. Single-image super-resolution of brain MR images using overcomplete dictionaries. Med Image Anal. 2013;17:113–132. doi: 10.1016/j.media.2012.09.003. [DOI] [PubMed] [Google Scholar]
- 5.Manjon JV, Coupe P, Buades A, Collins DL, Robles M. MRI superresolution using self-similarity and image priors. Int J Biomed Imaging. 2010;2010:1–12. doi: 10.1155/2010/425891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roy S, Carass A, Prince JL. Magnetic resonance image example based contrast synthesis. IEEE Trans Med Imaging. 2013;32(12):2348–2363. doi: 10.1109/TMI.2013.2282126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bahrami K, Shi F, Zong X, Shin HW, An H, Shen D. Reconstruction of 7T-like Images from 3T MRI. IEEE Trans Med Imaging. 2016;35(9):2085–2097. doi: 10.1109/TMI.2016.2549918. [DOI] [PMC free article] [PubMed] [Google Scholar]