Abstract
Many poly-ubiquitinated proteins are extracted from membranes or complexes by a conserved ATPase, called Cdc48 in yeast and p97/VCP in mammals, before proteasomal degradation1. Each Cdc48 hexamer contains two stacked ATPase rings (D1 and D2) and six N-terminal (N) domains2. Cdc48 binds various cofactors, including a heterodimer of Ufd1 and Npl43. Here, we report structures of the Cdc48-Ufd1-Npl4 ATPase complex from Chaetomium thermophilum. Npl4 interacts through its UBX-like domain with a Cdc48 N domain, and uses two Zn2+-finger domains to anchor an enzymatically inactive Mpr1/Pad1 N-terminal (MPN) domain, homologous to domains found in several isopeptidases, to the top of the D1 ATPase ring. The MPN domain of Npl4 is located above Cdc48’s central pore, similarly to the MPN of the de-ubiquitinase Rpn11 in the proteasome4. Our results indicate that Npl4 is unique among Cdc48 cofactors, and suggest a mechanism for how poly-ubiquitinated substrates bind to and translocate into the ATPase.
Introduction
Certain poly-ubiquitinated substrates cannot be directly degraded by the proteasome because they are well folded or located in membranes, chromatin, or multimeric complexes. These proteins are generally extracted from such assemblies and unfolded by a conserved ATPase, called Cdc48 in yeast and p97 or VCP in mammals, before being transferred to the proteasome1. Cdc48/p97 belongs to the AAA family of ATPases. The Cdc48/p97 hexamer contains two ATPase rings (D1 and D2) and N-terminal (N) domains that can be captured in different conformations2. Upon ATP binding by the D1 ATPases, the N domains transition from a “down-conformation” coplanar with the D1 ring to an “up-conformation” above the D1 plane5,6. Structural studies also indicate that the D1 and D2 ATPase rings undergo relative rotations upon ATP binding by D26.
Cdc48/p97 binds various cofactors, which determine substrate specificity, target the ATPase to different cellular locations, or modify the ubiquitin chain attached to the substrate7. The exact functions of these cofactors are poorly understood. All known cofactors bind to either the N domain of Cdc48/p97 or its unstructured C-terminal tail7. One of the most important cofactors is the Ufd1/Npl4 heterodimer (UN), which participates in many Cdc48-dependent processes, including ER-associated protein degradation (ERAD), a process in which misfolded proteins are extracted from the ER membrane and degraded by the proteasome8. Like Cdc48, both Ufd1 and Npl4 are evolutionarily conserved and essential for cell viability. Npl4 is a target of the potential cancer drug disulfiram9.
Npl4 contains an N-terminal UBX-like domain that binds to the N domain of Cdc48 and is predicted to have a Zn2+-finger (zf-Npl4) followed by an MPN domain (Fig. 1a)10. MPN domains are found in several Zn2+-dependent isopeptidases, including AMSH/AMSH-LP, the COP9 signalosome subunit CSN5, and the proteasomal de-ubiquitinase (DUB) Rpn1111–13. Apart from the zf-Npl4 domain, mammalian Npl4 contains a C-terminal Zn2+-finger domain that binds ubiquitin, but this domain is absent in yeast orthologs 14. Ufd1 has two short SHP motifs that bind to Cdc48, and a ubiquitin-binding UT3 domain with homology to the N domain of Cdc4815. Ufd1 interacts with Npl4 through its UT6 domain, a segment predicted to be unstructured (Fig. 1a)16. Cdc48 and Npl4 can also interact with Vms1, instead of Ufd1, which then recruits the ATPase complex to mitochondria17.
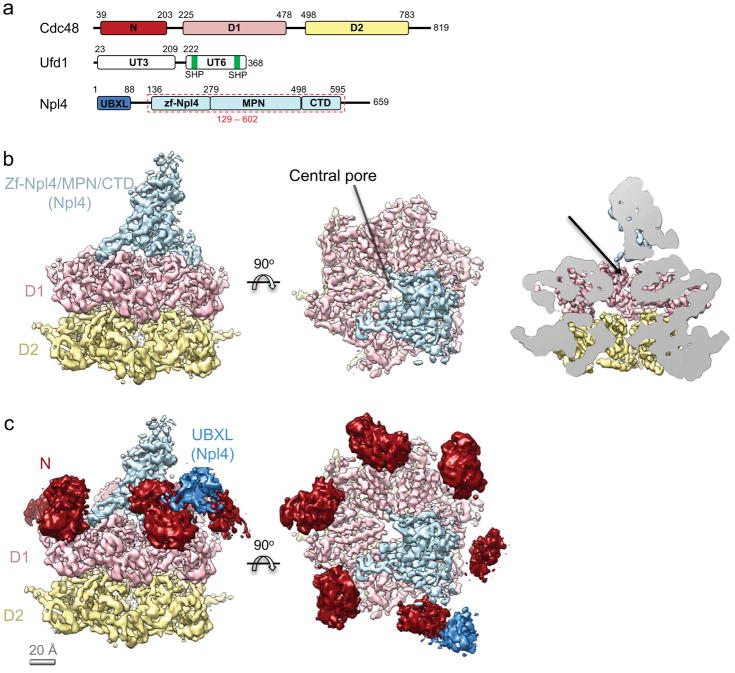
Figure 1. Structure of the Cdc48/UN complex with ATPγS.
a, Domain organization of Chaetomium thermophilum Cdc48 and its co-factors, Ufd1 and Npl4. The Npl4 region indicated with a red dashed box was crystallized. b, Cryo-EM density map of the Cdc48/UN complex, colored as in a. The N domains were masked out in the final refinement step. The rightmost panel shows a side cutaway view. The arrow indicates the putative path of the substrate into the pore. c, The N domains of Cdc48 (dark red) from a map refined without a mask are shown relative to the map obtained with masking. The contiguous extra density next to one of the N domains (dark blue) is assigned to the UBX-like domain of Npl4.
Recent in vitro experiments with purified Cdc48, UN cofactor, and a poly-ubiquitinated model substrate have resulted in some mechanistic insight18. After interaction of the poly-ubiquitin chain with UN, Cdc48 uses ATP hydrolysis in the D2 domain to move the polypeptide through its central pore, thereby unfolding the substrate. ATP hydrolysis in the D1 domain is involved in substrate release from the Cdc48 complex, a process that requires the cooperation of the ATPase with a DUB. The DUB trims the poly-ubiquitin chain, and the remaining oligo-ubiquitin chain is then also translocated through the pore. These experiments indicated that at least two strands of the translocating polypeptide chain can be present in the central pore, as also found for other hexameric AAA ATPases19,20.
The mechanism by which translocation of a polypeptide chain through Cdc48 is initiated is unclear. One unresolved issue is how the poly-ubiquitin chain is recognized by the UN complex. The only well-characterized ubiquitin-binding site is in the UT3 domain of Ufd1 (ref. 21). How a polypeptide chain is moved into the central pore of Cdc48 is even less understood. A substrate segment needs to move through the D1 ring before the D2 ATPases can use their loop residues to grab the polypeptide and pull it through the pore18,22. This is particularly puzzling, because Cdc48 can act on a large variety of folded substrates. By contrast, initiation of translocation by the ATPase ring of the 19S subunit of the proteasome is much easier to understand. Here, the substrate needs a flexible polypeptide segment that inserts into the pore of the single ATPase ring and serves as the initiation site23.
An understanding of the mechanism of Cdc48 requires structural information. So far, several structures of the ATPase itself are available5,6, but there is only limited information on the UN cofactor and its interaction with Cdc48. Previous electron microscopy (EM) structures showed density for the cofactor near the N domains of the ATPase, but the resolution of the reconstructions was insufficient to derive molecular models24,25. Here, we report single-particle cryo-EM and crystal structures that clarify the interaction of the UN cofactor with the Cdc48 ATPase.
Results
Cryo-EM structures of the Cdc48 ATPase complex
We decided to use Cdc48 and UN cofactor from the thermophilic fungus Chaetomium thermophilum, reasoning that the flexibility of protein segments might be reduced compared to orthologs from mesophilic organisms. We first determined cryo-EM structures of Cdc48 alone. C. thermophilum Cdc48 was expressed in E. coli and purified as a hexamer (Supplementary Figs. 1a,b). Structures of Cdc48 were determined in the presence of ADP or ATPγS, and, after 3D classification and refinement, reached overall resolutions of 7.2 Å and 8.2 Å, respectively (Table 1; Supplementary Figs. 2, 3, 4a, 5). As reported for mammalian p97 (ref. 6), both structures showed stacked D1 and D2 ATPase rings, and the best-refining classes had the N domains in the down-conformation in the ADP-bound state and the up-conformation in the ATPγS-bound state (Supplementary Figs. 2, 3, 5a). Some classes in ATPγS were in the down-conformation, perhaps due to slow nucleotide hydrolysis. The conformational switch of the N domains is likely triggered by the change in nucleotide state of the D1 ring5. The thermophilic Cdc48 protein thus recapitulates essential features of the mammalian p97 ATPase.
Table 1.
Data collection and refinement statistics (molecular replacement)
| zf-Npl4/MPN/CTD | |
|---|---|
| Data collection | |
| Space group | P 1 21 1 |
| Cell dimensions | |
| a, b, c (Å) | 58.858, 72.221, 193.543 |
| α, β, γ (°) | 90, 96.714, 90 |
| Resolution (Å) | 96.11 - 2.582 (2.675 - 2.582) |
| Rmerge | 0.06434 (2.031) |
| I/σI | 9.82 (0.49) |
| Completeness (%) | 95.15 (77.79) |
| Redundancy | 2.9 (2.7) |
| Refinement | |
| Resolution (Å) | 2.582 |
| No. reflections | 94503 (7751) |
| Rwork/Rfree | 0.1918/0.2287 |
| No. atoms | 7515 |
| Protein | 7283 |
| Ligand/ion | 4 |
| Water | 228 |
| B-factors | 96.95 |
| Protein | 97.44 |
| Ligand/ion | 120.87 |
| Water | 80.87 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.002 |
| Bond angles (°) | 0.53 |
Data were collected from a single crystal. Highest resolution shell is in parentheses.
Next, we purified a complex of Cdc48 and UN. The UN complex from C. thermophilum was again expressed in E. coli and had the expected 1:1 stoichiometry after purification (Supplementary Fig. 1c). A complex of hexameric Cdc48 and UN (Supplementary Fig. 1d) was subjected to single-particle cryo-EM analysis in the presence of ADP or ATPγS (Table 1; Supplementary Figs. 4–7). The refined structures had overall resolutions of 6.7 Å and 4.3 Å, in ADP and ATPγS, respectively. The presence of the cofactor had only a small effect on the structure of the ATPase rings (Supplementary Figs. 5–7). However, even in the ADP-bound state, a sizable population of the Cdc48 molecules had their N domain in the up-conformation, although the percentage was lower than in ATPγS (~60% versus ~95%; Supplementary Figs. 6, 7). Thus, ATP and cofactor binding act in concert to move the N domains into the up-conformation, a state likely required for initiation of substrate processing18.
The most obvious density contributed by the cofactor complex is a central tower that lies above the D1 ATPase ring (Fig. 1b). In addition, some 3D classes show density close to one of the N domains, which can be attributed to the UBX-like domain of Npl4. This domain is known to bind to a hydrophobic cleft on the N domain of Cdc48, and a previously determined structure of Npl4UBXL-p97N fits well into this region of our cryo-EM density map10 (Fig. 1c; Supplementary Fig. 5c). Deletion of the UBX-like domain abolishes binding of Npl4 to Cdc4826, indicating that the cofactor domains constituting the tower density interact only weakly with the ATPase despite having several contact points. The binding of the UBX-like domain to the Cdc48 N domain is probably a prerequisite for the binding of the other cofactor domains to the ATPase. Although the tower occupies a large fraction of the space above the D1 ring, the central pore remains unobstructed, thus allowing substrate to move into it (Fig. 1b).
Identification of Npl4 domains in the density map
Although the density map of the Cdc48/cofactor complex permitted the visualization of helices, the resolution was insufficient to build a molecular model for the cofactor. We therefore first identified cofactor regions that are in close proximity to the D1 ATPase ring. The Cdc48/UN complex was treated with bis[sulfosuccinimidyl] suberate (BS3), a bifunctional amine-reactive crosslinker. The sample was then digested with trypsin, and crosslinked peptides were identified by mass spectrometry. The data showed that the N terminus of Ufd1 interacts promiscuously with multiple locations in the ATPase and Npl4 (Supplementary Fig. 8a), far outside the calculated dynamic range of the BS3 spacer (5.2–9.2 Å), suggesting that the succeeding UT3 domain is flexible. On the other hand, several lysines in the zf-Npl4 and MPN domains of Npl4 crosslinked specifically to residues on the surface of the D1 ATPase ring (Supplementary Figs. 8a,b). Thus, these domains were the best candidates to form the base of the central tower. Consistent with the location of the cofactor density, no crosslinks were discovered between the Cdc48 D2 domain and either Ufd1 or Npl4.
Using limited proteolysis, we found that the zf-Npl4 and MPN domains form a stable fragment (Supplementary Fig. 8c). A crystal structure of this construct was determined using the central tower density of the cryo-EM map as a molecular replacement model (data collection and refinement statistics in Table 2). The initial low-resolution phases derived from the EM map were extended to high resolution using the X-ray data. Model bias was excluded by calculating a simulated annealing composite omit map (Supplementary Fig. 9). The resulting crystal structure indeed fits well into the cryo-EM map (Figs. 2a,b), indicating that the Npl4 domains undergo only small changes upon Cdc48 binding. The bottom of the tower is formed by the zf-Npl4 domain, the central portion by the MPN domain, and the top portion by a C-terminal domain (CTD) of five α-helices (Figs. 2a,b). A small unassigned region of the central tower density likely corresponds to a segment of UT6 in Ufd1 (Fig. 2a). Indeed, hydrogen/deuterium exchange mass spectrometry experiments showed that several Npl4 peptides in this region were protected when Ufd1 was present (Supplementary Figs. 8d,e). The Npl4-interacting region of UT6 is likely located between the two SHP motifs that anchor Ufd1 to the N domains of Cdc4826. The UT3 domain of Ufd1 is not visible in the density map, further indicating that it is flexible. Interestingly, a domain with the same fold is also flexible in the Pex1/Pex6 ATPase, another AAA ATPase27. These domains might only be fixed when they bind to ubiquitinated substrate.
Table 2.
Cryo-EM data collection, refinement and validation statistics
| Cdc48 (ADP) (EMDB-7477) |
Cdc48 (ATPγS) (EMDB-7478) |
Cdc48-Ufd1-Npl4 (ADP) (EMDB-7479) |
Cdc48-Ufd1-Npl4 (ATPγS) (EMDB-7476) (PDB 6CHS) |
|
|---|---|---|---|---|
| Data collection and processing | ||||
| Magnification | 31,000 | 31,000 | 22,500 | 22,500 |
| Voltage (kV) | 300 | 300 | 300 | 300 |
| Electron exposure (e−/Å2) | 32 | 32 | 89 | 89 |
| Defocus range (μm) | 1.6 – 3.0 | 1.6 – 3.0 | 1.2 – 2.8 | 1.2 – 2.8 |
| Pixel size (Å) | 1.23 | 1.23 | 1.3 | 1.3 |
| Symmetry imposed | C6 | C6 | C1 | C1 |
| Image stacks (no.) | 1440 | 354 | 2551 | 5844 |
| Initial particle images (no.) | 52,348 | 29,313 | 145,947 | 808,059 |
| Final particle images (no.) | 9,751 | 3021 | 52,178 | 91,883 |
| Map resolution (Å) | 7.2 | 8.2 | 6.7 | 4.3 |
| FSC threshold | 0.143 | 0.143 | 0.143 | 0.143 |
| Map sharpening B-factor (Å2) | −694 | −874 | −249 | −162 |
| Refinement | ||||
| Number of protein residues | 3774 | |||
| Number of atoms | 29508 | |||
| R.m.s. deviations | ||||
| Bond lengths (Å) | 0.008 | |||
| Bond angles (°) | 1.16 | |||
| PDB validation | ||||
| Clash score | 12 | |||
| Poor rotamers (%) | 0.29 | |||
| Ramachandran plot | ||||
| Favored (%) | 94.86 | |||
| Allowed (%) | 5.01 | |||
| Disallowed (%) | 0.13 | |||
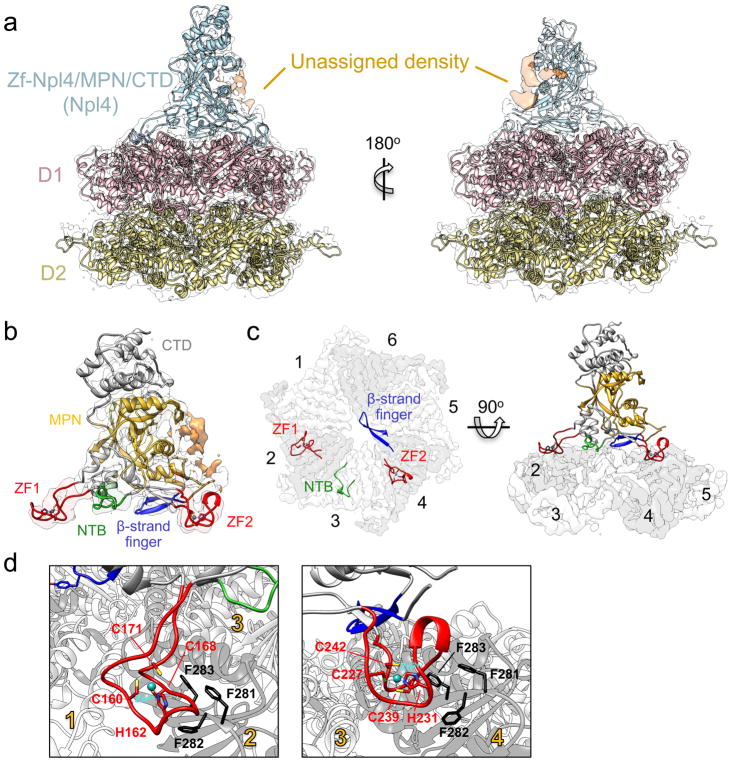
Figure 2. Interactions between Cdc48 and its cofactor.
a, A crystal structure of Npl4 (light blue) and a homology model of Cdc48 (D1 in pink; D2 in yellow) were docked into the cryo-EM map. Unassigned density (orange) likely belongs to parts of the UT6 domain of Ufd1. b, Close-up view of the Npl4 crystal structure docked into the cryo-EM map. The MPN domain is shown in yellow and D1-interacting regions extending from it are highlighted: the two Zn2+ fingers (ZF; red), an N-terminal bundle (NTB; light green), and the ‘β-strand finger’ (blue). c, Top view of D1-interacting regions, colored as in b, with the D1 ring shown as a white/gray surface. For clarity, D2 was omitted. ATPase subunits of the Cdc48 hexamer are numbered. d, Close-up views of the Zn2+ fingers, with Zn2+-coordinating residues in stick representation. The interacting tri-phenylalanine (FFF) sequence in Cdc48 is highlighted.
The MPN domain of Npl4 is anchored to the top of the D1 ATPase rings of Cdc48 via the preceding Zn2+-finger domains, which are both of the CHCC type, i.e., use one His residue and three Cys residues for coordination of the Zn2+ ion (Figs. 2b–d). The Zn2+ fingers form two “stalks” that project into grooves between adjacent subunits of the Cdc48 D1 ring (Figs. 2b,c). When numbered from the position of the N-terminal Zn2+ finger, the interacting grooves are between ATPase subunits 1 and 2 and between subunits 3 and 4 (Fig. 2c). A third stalk is formed by segments preceding the first Zn2+ finger as well as residues located between the two Zn2+ fingers. This N-terminal bundle (NTB) makes only a few contacts with the surface of ATPase subunit 3 and is less conserved than the Zn2+ fingers. Finally, a fourth stalk is formed by two β-strands with a loop at their tip. This loop projects over the axial pore and faces ATPase subunit 6 (Fig. 2c), but it makes no clear contact with the D1 ring. The use of several contact sites precludes the binding of a second cofactor molecule (Supplementary Fig. 9), explaining why one Cdc48 hexamer binds only one UN heterodimer.
Functional tests of Npl4 segments
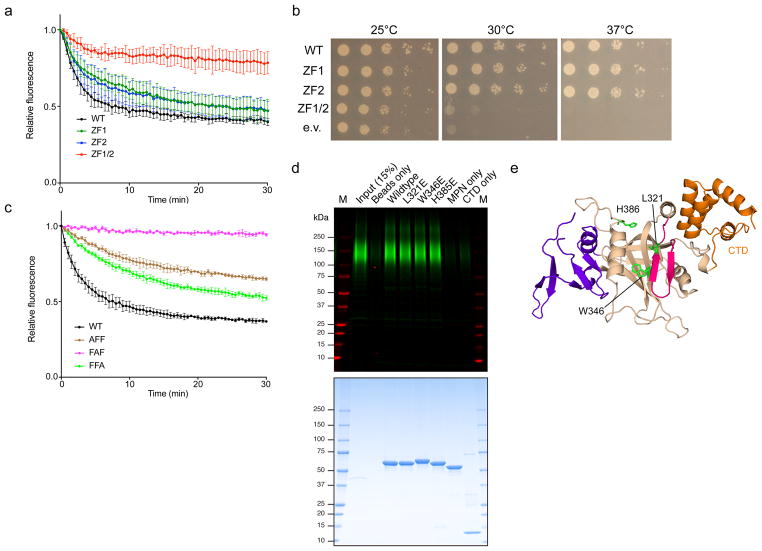
To test the functional role of the Zn2+ fingers, we used S. cerevisiae Cdc48, Npl4, and Ufd1 in an in vitro unfolding assay. A fusion between a short degron and the fluorescent protein mEos3.2 was poly-ubiquitinated and incubated with the ATPase complex; the loss of fluorescence is an indication of Eos unfolding18. The results show that mutation of the central His and Cys residues in either of the individual Zn2+-finger domains had little effect on unfolding, but a defect was seen when both domains were mutated together (Fig. 3a). Similarly, mutants in individual Zn2+ fingers could rescue the temperature-sensitive growth phenotype of an npl4-1 yeast strain, but a mutant in both Zn2+ fingers could not (Fig. 3b). We also tested mutations in the Cdc48 ATPase in the unfolding assay. Both Zn2+-finger domains are in close proximity to a conserved tri-phenylalanine (FFF) sequence in the D1 domain. Indeed, mutation of the first or third Phe reduced the unfolding activity of Cdc48 without affecting hexamer formation (Fig. 3c; Supplementary Fig. 10a). Mutation of the central Phe in the FFF motif abolished unfolding completely, but also reduced hexamerization, consistent with the fact that it faces a hydrophobic pocket in the ATPase domain. The importance of the Zn2+ fingers of Npl4 is supported by a recent report in which these domains were identified as a target of the drug disulfiram9. The role of the “β-strand finger” of Npl4 remains unclear. Although there is a highly conserved tyrosine residue at the tip of the β-strand finger, its mutation or deletion did not alter unfoldase activity in vitro or affect the ability to complement the npl4-1 mutant (Supplementary Fig. 10b). It remains possible that a subclass of Cdc48 substrates is dependent on the β-strand finger.
Figure 3. Functional analysis of Zn2+-finger and MPN domain mutations.
a, Unfolding of poly-ubiquitinated Eos by wild-type S. cerevisiae Cdc48 and the indicated UN variants. ZF1: H139A/C145A. ZF2: H208A/C216A. ZF1/2: H139A/C145A/H208A/C216A. Data are shown as mean ± SD of n=3 technical replicates. See Suppl Data Set 1. b, A npl4-1 temperature-sensitive S. cerevisiae strain was transformed with a plasmid encoding wild-type Npl4 or the indicated Zn2+-finger mutants, spotted in serial dilution, and incubated at the indicated temperatures for two days (30 and 37°C) or three days (25°C). c, As in a, but with mutants in the FFF motif (residues 275-277) of Cdc48. Data are shown as mean ± SD of n=3 technical replicates. See Suppl Data Set 1. d, Binding of poly-ubiquitinated substrate to SBP-tagged C. thermophilum Npl4 (Zn2+-finger/MPN/CTD domains, residues 129-602) or the indicated variants (MPN only: residues 129-519, CTD only: residues 519-602). The bait proteins were bound to streptavidin beads and incubated with dye-labeled, poly-ubiquitinated superfolder GFP. Bound material was analyzed by SDS-PAGE followed by fluorescence scanning (top) and Coomassie blue staining (bottom). M, molecular weight markers. e, The locations of MPN cleft mutants tested in d are shown in stick representation. The MPN, insert-1, insert-2, and CTD are shown in tan, magenta, purple, and orange, respectively.
Comparison with other MPN domains
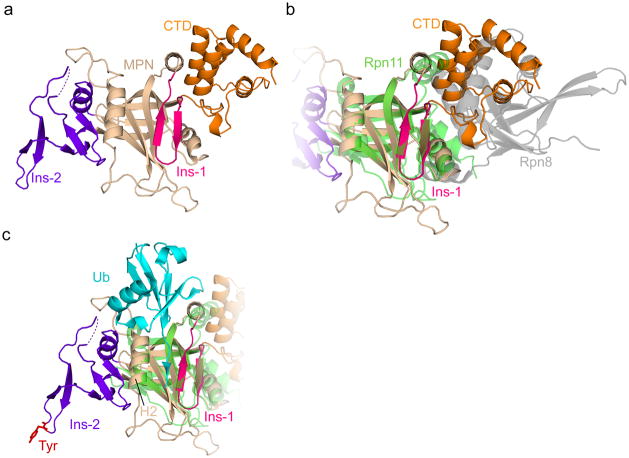
Like other members of the MPN family, the one in Npl4 consists of a core MPN fold with two inserts (insert-1 and -2) (Fig. 4a). Npl4 is enzymatically inactive, as it lacks the Zn2+-binding motif in the core, which is essential for the hydrolytic activity of other MPN domains28. The position of the Npl4 MPN domain above the D1 ATPase ring (Figs. 2a–c) is similar to that of the Rpn11 MPN domain in the 19S regulatory particle of the proteasome. In the case of Rpn11, an enzymatically active MPN domain is located over a ring of six homologous ATPase subunits4,29. The Rpn11 MPN domain dimerizes with the enzymatically inactive MPN domain of Rpn8. The Npl4 MPN domain, however, is a monomer, with its C-terminal helical CTD domain occupying the site that mediates dimerization in Rpn11 (Fig. 4b).
Figure 4. MPN domain of Npl4.
a, Crystal structure of the MPN/CTD domains of Npl4. The core MPN region is in tan, the insert-1 (Ins-1) region in magenta, the insert-2 (Ins-2) region in purple, and the CTD in orange. The dashed line indicates a 9-residue acidic loop unresolved in the crystal structure. b, As in a, but with the Rpn11/Rpn8 structure overlaid (PDB: 5U4P). Rpn11 is in green and Rpn8 in gray. c, As in a, with the Rpn11 MPN (green) and its associated ubiquitin (cyan) overlaid (PDB: 5U4P). The conserved Tyr at the tip of Ins-2 is shown in stick representation in red. H2: MPN helix 2.
The MPN domains of Rpn11 and AMSH-LP accommodate the C-terminal tail of ubiquitin in a cleft between insert-1 and helix 2, positioning the C terminus of ubiquitin next to the active site (shown for Rpn11 in Fig. 4c)30,31. Insert-1 of Npl4 closely resembles the structure assumed by the corresponding region of Rpn11 in the presence of ubiquitin (Fig. 4c), but it adopts this conformation even in the absence of ubiquitin, as observed for AMSH and several other MPN family members32. Given the similarity with Rpn11 and AMSH, it is likely that the cleft in Npl4’s MPN domain also accommodates the C-terminal tail of a ubiquitin molecule, but in our structure the groove is covered by a segment of UT6 of Ufd1, suggesting that the UT6/MPN interaction may be broken to allow ubiquitin binding.
Ubiquitin binding by Npl4
To test whether the cleft of the MPN domain has a role in ubiquitin binding, we incubated a poly-ubiquitinated substrate with a streptavidin-binding peptide (SBP)-tagged version of the Npl4 construct used for crystallization. In agreement with previous results33, this construct indeed bound poly-ubiquitin, as shown after pull-down with streptavidin beads (Fig. 3d,e). However, mutagenesis showed that the cleft of the MPN domain does not contribute much to the interaction. Although this result is at variance with a previous report33, perhaps because the bait in the previous experiments was destabilized by its fusion with glutathione S-transferase, it is consistent with the fact that only a few residues in the MPN cleft of Rpn11 contact ubiquitin34. Rather than by the cleft, most of the affinity for ubiquitin may be provided by the interface between MPN and CTD, as neither domain alone was active (Fig. 3d). Alternatively, both domains might have weak affinity, and the combination is required for avid substrate recognition. Although the elucidation of the exact binding mode of poly-ubiquitin to the UN complex requires a structure of the Cdc48 complex with a ubiquitin chain of defined length, it is clear that the UN cofactor has ubiquitin-binding sites in both Ufd1 and Npl4, similarly to the multiple receptors present in the proteasome35.
Discussion
Our data lead to a model for the interaction of the Cdc48 ATPase with the UN cofactor (Fig. 5a). Npl4 interacts through its UBX-like domain with one of the N domains of Cdc48 hexamer, and uses its two Zn2+-finger domains to anchor its MPN domain to the top of the D1 ATPase ring. Ufd1 interacts through a short and poorly conserved segment of its UT6 domain with Npl4, and through the flanking SHP motifs with other N domains of Cdc48. The UT3 domain of Ufd1 does not seem to interact with either Npl4 or Cdc48. The interface between Npl4 and Ufd1 is surprisingly small and might even be broken when substrate binds to Npl4, but Ufd1 would likely still remain bound to Cdc48 through its SHP motifs.
Figure 5. Model for Cdc48/UN function.
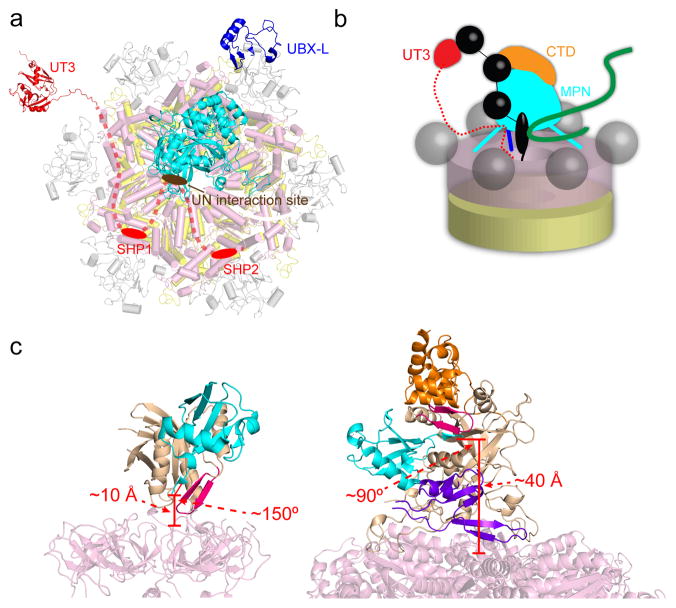
a, Arrangement of the various domains in the Cdc48/UN complex. The UBX-like (PDB: 2PJH) and Zn2+-finger/MPN domains of Npl4 are shown in dark blue and cyan, respectively. The UT3 domain of Ufd1 (in red; PDB: 1ZC1) is flexibly attached to the complex. The Npl4-interacting region of UT6 is shown as a brown oval, and the SHP boxes anchoring Ufd1 to the N domains of Cdc48 as red ovals. The intervening segments are shown as dashed lines. b, Model for the path of a poly-ubiquitinated substrate (black: ubiquitin; green: substrate protein) into the central pore of Cdc48. Four ubiquitin molecules are shown: one associated with UT3, one bound at the interface between CTD and MPN, one located with its C terminus in the MPN cleft, and one in the gap between MPN and the ATPase, which might serve as initiation site for translocation into the pore. c, Comparison between the location of the ubiquitin-bound MPN domain in the proteasome (left) with that predicted in the Cdc48 complex (right). For Rpn11, the core MPN region is in tan and the insert-1 (Ins-1) region in magenta. Ubiquitin is in cyan. For Npl4, the core MPN region is in tan, the insert-1 (Ins-1) region in magenta, the insert-2 (Ins-2) region in purple, and the CTD in orange. The approximate distance from the ubiquitin C terminus to the central pore opening and the approximate angle between the MPN cleft and the central pore axis are marked in red in each case. The proteasome/ubiquitin model was generated from PDBs 5T0H and 5U4P.
Our results in conjunction with previous studies suggest that ubiquitin molecules attached to a substrate can bind to both the UT3 domain of Ufd1 and to the CTD/MPN domains of Npl4 (Fig. 5b)21,33. These two independent interactions can explain, at least in part, why a chain length of at least five ubiquitin molecules is required to obtain maximum binding to the UN complex18,33. An additional ubiquitin molecule would be located in the cleft of the MPN domain (Fig. 5b). The cleft does not provide much affinity, but it would allow the accommodation of the C terminus of this ubiquitin molecule in a similar manner as seen in Rpn11 of the proteasome30. However, in the Cdc48 complex, the putative ubiquitin-binding groove and central ATPase pore are at an approximately right angle, whereas they are more closely aligned in the proteasome (Fig. 5c)36. Furthermore, the catalytic site of Rpn11 is located immediately above the pore in the substrate-engaged proteasome37, whereas in the Cdc48/UN complex, there is a ~40 Å gap between the putative C terminus of ubiquitin and the pore (Fig. 5c). The presence of this gap raises the question of how substrates might initially be inserted into the ATPase. One possibility is that the gap is bridged by an additional ubiquitin moiety proximal to the one bound to the MPN cleft. In this case, Cdc48 might begin translocation not on a segment of the substrate, but instead on a segment of this proximal ubiquitin molecule. This model is attractive in that ubiquitin could serve as a universal initiating signal for translocation into the pore, regardless of the substrate to which it is attached. This would eliminate the need for a flexible region to mediate pore entry and explain why Cdc48, unlike the proteasome, has no requirement for a pre-unfolded segment. However, it clearly remains possible that the substrate itself is the first inserted polypeptide segment. Although the details of pore insertion need to be clarified, ideally with a structure of a substrate-associated Cdc48 complex, our data show that Npl4 is unique among the known Cdc48 cofactors, as it binds directly to the ATPase ring and likely serves as a universal gatekeeper for all Cdc48-dependent reactions that require translocation through the central pore.
Online Methods
Protein expression and purification
Full-length Chaetomium thermophilum and Saccharomyces cerevisiae Cdc48, Ufd1, and Npl4, as well as the Npl4 Zn2+-finger/MPN/CTD fragment (residues 129-602) and its tagged versions and mutants, were overexpressed in E. coli BL21 DE3 RIPL cells as follows. Cdc48, Ufd1, and the Zn2+-finger/MPN/CTD fragment were expressed with N-terminal His14-SUMO tags from the K27SUMO vector38. Full-length Npl4 was expressed untagged from the pET21b vector. Buffers used for Cdc48 purifications contained 5 mM MgCl2 throughout. Cells were grown to an OD600 of 0.6 to 0.8 in Terrific Broth and induced with 0.25 mM isopropyl β-thiogalactopyranoside (IPTG) for 16 hrs at 18°C. Cells were pelleted at 4000xg and resuspended in lysis buffer (50 mM Tris pH 8, 300 mM NaCl, 30 mM imidazole, 5 mM MgCl2). For the UN complex, Ufd1 and Npl4 pellets were mixed at this step. The suspension was supplemented with protease inhibitors and DNAse I, and cells were lysed by sonication. Lysates were cleared by centrifugation in a Ti45 rotor (Beckman) for 30 min at 40,000 rpm and applied to Ni-NTA agarose for 1 hr at 4°C. Beads were washed with lysis buffer, and His-tagged proteins were eluted in 10–20 mL elution buffer (50 mM Tris pH 8, 100 mM NaCl, 400 mM imidazole). The eluates were supplemented with 0.5 mM Tris(2-carboxyethyl)phosphine (TCEP) and 100–200 nM SUMO protease (Ulp1p), then incubated at 4°C for 1 hr to remove His14-SUMO tags. Protein solutions were diluted to <200 mM imidazole with 50 mM Tris pH 8 and loaded onto a MonoQ 10/100 GL column (GE Healthcare) equilibrated in 50 mM Tris pH 8, then eluted by a linear gradient to 500 mM NaCl over 8 column volumes. Peak fractions were pooled and concentrated. UN variants used for binding experiments and unfolding assays were stored at this point. Other proteins were further purified by size-exclusion chromatography as follows. For Cdc48 and Cdc48/UN complexes, proteins were incubated with 1 mM nucleotide (ADP or ATPγS) for 45 min prior to gel filtration, and the gel-filtration buffer (50 mM HEPES pH 7.5, 150 mM NaCl, 5 mM MgCl2, 0.5 mM TCEP) additionally contained 100 μM nucleotide. For Cdc48/UN complexes, UN was added at a 3- to 5-fold molar excess of heterodimer to hexamer prior to gel filtration. For the Npl4 Zn2+-finger/MPN/CTD crystallization construct, the buffer was 25 mM HEPES pH 7.5, 100 mM NaCl. All proteins were separated on a S200 size-exclusion column (GE Healthcare). Samples for cryo-EM analysis were used immediately after gel filtration, while crystallization samples were supplemented with 5% vol/vol 1,2-propanediol and flash frozen in liquid nitrogen.
EM specimen preparation and data collection
Sample homogeneity was first examined by negative-stain EM with 0.7% (wt/vol) uranyl formate, as previously described39. Images were recorded using a 1K × 1K CCD camera (Gatan) on a Philips CM10 electron microscope (FEI) operated at an acceleration voltage of 100 kV and a nominal magnification of 52,000x.
Prior to preparing grids for cryo-EM, all samples were concentrated to 2–3 mg/mL and centrifuged at 13,000 × g for 10 min to remove protein aggregates. NP-40 was added to the samples to a final concentration of 0.05% immediately before vitrification to lower the propensity of the particles to adopt preferred orientations in the ice layer.
The specimens for cryo-EM were frozen using a Cryoplunge 3 (Gatan). A 3.5-μL aliquot of the sample was applied to a glow-discharged Quantifoil Cu 1.2/1.3 grid (Quantifoil). The grid was blotted for 2.5–3.5 s and then plunge frozen in liquid ethane, which was maintained at a temperature of −172°C.
For analysis of Cdc48 alone in the presence of ADP or ATPγS, cryo-EM data collection was carried out at Harvard Medical School (Boston, MA), using a Polara electron microscope (FEI) operated at 300 kV and equipped with a K2 Summit direct electron detector (Gatan). All images were recorded in super-resolution counting mode using the semi-automated data collection software UCSFImage440 at a nominal magnification of 31,000x (corresponding to a calibrated super-resolution pixel size of 0.62 Å). The defocus was set to range from −1.6 μm to −3.0 μm. The total exposure time of 6 s was dose-fractionated into 30 frames (200 ms per frame), with a dose rate of 8 electrons per pixel per second. A total of 1,628 and 569 image stacks were collected for Cdc48(ADP) and Cdc48(ATPγS), respectively.
For Cdc48 in complex with the UN cofactor in the presence of ADP or ATPγS, all data collection was performed in the Cryo-EM Resource Center at the Rockefeller University using the automated data collection software SerialEM41. Using a Titan Krios electron microscope (FEI) operated at 300 kV, 3,279 and 9,299 image stacks were collected for Cdc48-cofactor(ADP) and Cdc48-cofactor(ATPγS), respectively. The image stacks were recorded at a nominal magnification of 22,500x (yielding a calibrated super-resolution pixel size of 0.65 Å) with a K2 Summit camera in super-resolution counting mode. The defocus was set to range from −1.2 μm to −2.8 μm. The total exposure time of 15 s was dose-fractionated into 50 frames (300 ms per frame), with a dose rate of 10 electrons per pixel per second.
Image processing
For the datasets of Cdc48 alone with ADP or ATPγS, the image stacks were motion-corrected and binned over 2 × 2 pixels with MotionCor42, yielding a pixel size of 1.23 Å. The defocus values were estimated by CTFFIND443. Each image was then manually inspected and rejected if considered of inadequate quality for further image processing (e.g., ice contamination, blurriness, bad CTF fit, etc.). Particles were manually picked from the remaining images using the e2boxer.py command in EMAN244.
For the Cdc48(ADP) dataset, 52,348 particles were manually picked from 1,440 images. The particles were extracted into 256 × 256-pixel boxes in RELION45. A subset of these particles was used to generate 2D class averages using the iterative stable alignment and clustering (ISAC) algorithm46. These were then used to calculate an initial 3D map using the validation of individual parameter reproducibility (VIPER) algorithm, both implemented in SPARX47. Using the map obtained with VIPER as reference, all the particles were subjected to RELION 3D classification into 5 classes. One class containing 9,751 particles (~19% of the dataset) showed the most detailed structural features with apparent six-fold symmetry. This class was subjected to 3D refinement and yielded a density map at 8.9 Å resolution. When the same class was refined with C6 symmetry imposed, an improved density map at 7.2 Å resolution was obtained.
For the Cdc48(ATPγS) dataset, 29,313 particles were manually picked from 354 images and extracted into 200 × 200-pixel boxes in RELION. The particles were used to calculate 2D class averages with ISAC, and the resulting averages were used to calculate an initial 3D map with VIPER. Using this map as reference, the particles were subjected to RELION 3D classification into 10 classes. One class containing 3021 particles (~10% of the dataset) showed the most well-defined structural features. This class was subjected to 3D refinement and yielded a density map at 10.3 Å resolution. Since the structure also exhibited apparent six-fold symmetry, the same class was also refined with C6 symmetry imposed, which resulted in a map with an improved resolution of 8.2 Å.
For the Cdc48-cofactor complex in the presence of ADP or ATPγS, the image stacks were motion-corrected, dose-weighted, and binned over 2 × 2 pixels (yielding a pixel size of 1.3 Å) using MotionCor248. The defocus values were estimated with CTFFIND4. Each image was manually inspected and imperfect images were excluded from further processing. The remaining images were subjected to template-free particle auto-picking using Gautomatch (www.mrclmb.cam.ac.uk/kzhang/Gautomatch/).
For the Cdc48-cofactor(ADP) complex, 145,947 auto-picked particles were extracted from 2,551 images into 256 × 256-pixel boxes in RELION. These particles were subjected to reference-free 2D classification and classes showing poor averages were removed. To obtain an initial 3D model, the remaining 141,422 particles were aligned to the Cdc48(ADP) cryo-EM map (obtained as described above and filtered to 40 Å) using RELION 3D classification with the number of classes set to one. The resulting map showed density not present in the reference map. Using this new map as reference, the particles were sorted into 6 classes by 3D classification. The classes that showed strong cofactor density (class 2, 5 and 6; 82,249 particles; ~58% of the dataset) were combined and subjected to 3D classification into 10 classes. The resulting classes showed that the Cdc48-cofactor(ADP) complex also exhibits conformational variability, most notably in the N domain and the cofactor region. Classes 2, 8, 9 and 10 showing the strongest cofactor density and relatively well-ordered N domains were combined, and the resulting 52,178 particles were subjected to 3D refinement in RELION, yielding a map at 6.7 Å resolution.
For the Cdc48-cofactor(ATPγS) complex, the 808,059 auto-picked particles were binned over 4 × 4 pixels, resulting in a pixel size of 5.2 Å, extracted into 64 × 64-pixel boxes in RELION-249, and subjected to reference-free 2D classification in RELION-2. After removing classes giving poor averages, the remaining 616,772 particles were used as input for cryoSPARC50 to calculate an initial 3D map. Using this map as reference, the particles were sorted into 8 classes by RELION-2 3D classification. Only one of the classes (containing 91,883 particles; ~15% of the dataset) showed strong density for the cofactor bound to Cdc48 and detailed structural features. Refinement of this class yielded a density map at 10.4 Å resolution. The refined particles were then re-extracted from the original micrographs as re-centered and unbinned particles into 256 × 256-pixel boxes (pixel size of 1.3 Å). 3D refinement of the newly extracted particles was performed using the orientation parameters determined from the dataset of 4x binned particles as the starting point for further optimization. The final density map had a resolution of 4.6 Å, according to the gold-standard Fourier shell correlation (FSC) curve and using the FSC = 0.143 cutoff. Because the flexibility of the N domains affects the precision of the particle alignment, the N domains were masked out for the final alignment cycles in RELION-2, which improved the resolution of the remaining map region to 4.3 Å.
Model building and refinement for the Cdc48-cofactor(ATPγS) structure
The structure of human p97 bound to ATPγS (PDB: 5FTN) was used to build a homology model of C. thermophilum Cdc48 using SWISS-MODEL51. The homology model of Cdc48 and the crystal structure of the zinc-finger, MPN, and CTD domains of Npl4 were docked into the cryo-EM density map using UCSF Chimera52. The atomic model was optimized by cycles of real-space refinement using phenix.real_space_refine53 against half-map 1 from RELION-2 and manual re-building in Coot54. FSC curves were calculated between the refined models and half-map 1 (work), half-map 2 (free), and the combined map. The statistics from the structure determination are summarized in Supplementary Table 2.
Protease-protection experiments
Purified C. thermophilum UN was treated with increasing concentrations of trypsin (0, 2.4, 7.3, 22, and 66 μg/mL), incubated at room temperature for 30 min, and subjected to SDS-PAGE and Coomassie blue staining. Bands were subjected to mass spectrometry.
Crystallization
The Zn2+-finger/MPN/CTD fragment (57 mg/mL) was thawed immediately before crystallization setup and centrifuged for 10 min at 20,000xg. Crystals were grown using the hanging drop method at 4°C by mixing 2 μL of protein solution with 2 μL of well solution (0.2 M Na/K phosphate pH 7, 9.2% wt/vol poly-γ-glutamic acid [Molecular Dimensions], 0.2 M potassium thiocyanate, 5% propylene glycol). Crystals appeared in 1–2 days and were harvested, incubated briefly in cryo-protection solution (0.2 M Na/K phosphate pH 7, 10% wt/vol poly-γ-glutamic acid, 0.2 M potassium thiocyanate, 30% propylene glycol), and flash frozen in liquid nitrogen.
X-ray data collection and structure determination
Crystals were screened at NE-CAT beamline 24-ID-C at the Advanced Photon Source (Argonne National Laboratory). An X-ray absorption scan showing a peak at 9665.7 eV confirmed the presence of zinc, and data were accordingly collected at this energy (1.283Å). The dataset used for structure determination (doi:10.15785/SBGRID/565) was gathered from a single crystal at 100K. Data were processed with XDS55 and analyzed with Aimless56. The crystal belonged to the P1211 space group, diffracted to 2.58 Å (outer shell, 2.675-2.582 Å: I/sigma = 0.49, CC1/2 = 0.322) and contained two copies per asymmetric unit. To solve the structure, the cryo-EM density corresponding to the cofactor central tower was used as a molecular replacement (MR) model according to the protocol of Jackson et al.57, using the programs Chimera, Phaser, RESOLVE, and the CCP4i and Phenix crystallographic suites52,53,58,59,60. The central tower density was converted to structure factors and placed at the origin of a P1 unit cell. This model was used to obtain a molecular replacement solution with initial low-resolution phases. Phases were extended to high resolution using density modification as implemented in RESOLVE, taking advantage of the two-fold non-crystallographic symmetry present in the crystal. The MR solution was passed to the MR-SAD module in Phenix. Initial phases were of sufficient quality for the majority of the structure in the asymmetric unit to be assembled using the Autobuild function in Phenix61. The structure was completed by iterative rounds of manual adjustment in Coot54 and refinement in Phenix, with TLS parameter refinement enabled and a riding hydrogen model. Zinc fingers were refined with geometry restraints as suggested in ref. 62. Ramachandran statistics were: 96.03% favored, 3.86% allowed, 0.11% outliers. A composite omit map with simulated annealing was generated with Phenix60. Figures were generated using UCSF Chimera52 and PyMOL (Schrödinger, LLC). Crystallographic software was maintained by SBGrid63.
LC-MS/MS and cross-link mapping
Buffer components for cross-linking were BioUltra grade (Sigma Aldrich). LC-MS/MS was carried out with Thermo Scientific LC-MS grade reagents and solvents. The cross-linker bis[sulfosuccinimidyl] suberate (BS3) was purchased from Thermo Scientific.
Purified protein complexes (0.5 mg/mL in 50 mM HEPES pH 7.5, 200 mM NaCl, 5 mM MgCl2, 0.5 mM TCEP, 0.1 mM ATPγS) were cross-linked with 100, 200, or 400 μM of BS3 (3.5 mM stock in water) for 30 min at room temperature. The reactions were quenched by addition of Tris-HCl pH 7.5 to a final concentration of 10 mM. Samples were dialyzed against 100 mM ammonium bicarbonate, reduced with 50 mM TCEP at 60°C for 10 min, and alkylated with 50 mM iodoacetamide in the dark for 60 min at room temperature. Digestion was carried out at 37°C overnight with 0.5 μg sequencing grade modified trypsin (Promega) in 100 mM ammonium bicarbonate. The resulting peptides were passed though C18 Spin Tips (Thermo Scientific) before elution with 40 μL of 80% acetonitrile (ACN) in 0.1% trifluoroacetic acid. Eluted peptides were dehydrated in vacuum and resuspended in 20 μL 0.1% formic acid for MS analysis.
Peptides were analyzed in an Orbitrap Fusion Lumos mass spectrometer64 (Thermo Scientific) coupled to an EASY-nLC (Thermo Scientific) liquid chromatography system, with a 2 μm, 500 mm EASY-Spray column. The peptides were eluted over a 120-min linear gradient from 96% buffer A (water) to 40% buffer B (ACN), then continued to 98% buffer B over 20 min with a flow rate of 250 nL/min. Each full MS scan (R = 60,000) was followed by 20 data-dependent MS2 (R = 15,000) with high-energy collisional dissociation and an isolation window of 2.0 m/z. Normalized collision energy was set to 35. Precursors of charge state 4–6 were collected for MS2 scans; monoisotopic precursor selection was enabled and a dynamic exclusion window was set to 30.0 s.
Raw LC-MS/MS data files were converted into mgf format using Proteome Discoverer (Thermo Scientific) and searched using pLink65,66 with default FDR<5%, maximum e-value set at =0.001, trypsin digest with up to 3 missed cleavages, constant modification at 1=carbamidomethyl[C], variable modification at 1=oxidation[M]. Cross-linker was set to BS3 ([K [K 138.068 138.068 156.079 156.079]. Mass tolerances for fragments and precursors were left unaltered. mgf files were searched against a database comprising Fasta sequences of Cdc48, Ufd1, and Npl4.
To estimate the dynamic range of the BS3 spacer in solution, we performed a molecular dynamic simulation of BS3 using YASARA Dynamics (YASARA Biosciences GmbH). A target structure in pdb format was generated from the BS3 structural formula. MD simulation was carried out for 25 ns in explicit water with 0.9% NaCl, at 298K with the AMBER14 force field. Spacer length was recorded as C4-C11 distance. See Suppl Data Set 3.
Hydrogen/deuterium exchange mass spectrometry
Five μM stock solutions of C. thermophilum Npl4 residues 129-602 or the full-length UN complex were prepared in equilibration buffer (10 mM HEPES pH 7.5, 100 mM NaCl, H2O). To initiate labeling, 3 μL of each protein were diluted with 45 μL labeling buffer (10 mM HEPES pD 7.5, 100 mM NaCl, 99.8% D2O) and incubated for 10 s, 10 min, 1 hr, or 4 hrs). At the specified time, 48 μL of quench buffer (150 mM potassium phosphate pH 2.4, 0°C) were added. All subsequent steps were performed at 0°C.
Quenched samples were digested online with an immobilized pepsin column (prepared in house according to ref. 67) and directed into a Waters nanoAcquity UPLC with HDX technology68. Peptides were trapped on a Waters UPLC BEH C18 1.7 μm VanGuard BEH column and desalted with 0.1% formic acid in water for 3 min at 100 μL/min. Peptides were separated over 6 min using a 5–35% gradient of water:acetonitrile with 0.1% formic acid at a flow rate of 100 μL/min using a Waters HSS T3 1.8 μm C18, 1.0 mm × 50 mm analytical column. Deuterium incorporation was measured using a Waters Synapt G2Si system equipped with a standard ESI source in HDMSE mode. Mass spectra were acquired over a m/z range of 50–2000, and mass accuracy was confirmed by calibration with 500 fmol/μL of human glu-fibrinopeptide. Peptic peptides were identified using ProteinLynx Global Server (PLGS) 3.0 (Waters) and deuterium incorporation measured using DynamX 3.0 (Waters). The deuterium levels were not corrected for back exchange and are reported as relative69. All experiments were performed in duplicate.
Substrate-unfolding assays
mEos3.2 was purified and polyubiquitinated as described18. Briefly, the protein was expressed as an N-terminal His14-SUMO fusion. After SUMO cleavage, an N-terminal arginine is exposed, facilitating ubiquitination by the purified S. cerevisiae enzymes Uba1, Ubc2, and Ubr1. Eos was separated from the ubiquitination machinery on the basis of a C-terminal streptavidin-binding peptide (SBP) tag, which was then removed by 3C protease. Finally, substrates bearing ubiquitin chains of 5–10 moieties were isolated by gel filtration. The substrate molecules carry a single ubiquitin chain with K48 linkages at lysine 19 (ref. 18).
Substrate unfolding was monitored as described18. Briefly, the substrate (200 nM) was mixed with 300 nM UN variants and 400 nM Cdc48 variants in 50 mM HEPES pH 7.2, 100 mM KCl, 10 mM MgCl2, 0.25 mg/mL protease-free bovine serum albumin. After 10 min incubation at 30°C, ATP (2 mM) was added, and fluorescence (excitation: 540 nm, emission; 580 nm) was monitored in a Spectramax M5 plate reader for 30 min.
Substrate-binding experiments
The C. thermophilum Npl4 Zn2+-finger/MPN/CTD fragments (residues 129-602, 129-519, or 519-602) with an N-terminal SBP tag were incubated with streptavidin agarose beads (Pierce) in binding buffer (50 mM Tris pH 8.0, 150 mM NaCl) for 30 min at room temperature. The beads were washed three times with binding buffer to remove excess bait protein. Next, DyLight 800-labeled, polyubiquitinated superfolder GFP, generated as described for the Eos substrate above and purified by gel filtration, was incubated with the beads for 30 min at a concentration of 50 nM in 100 μL binding buffer. The beads were again washed three times. Bound material was eluted with binding buffer plus 1 mM biotin and subjected to SDS-PAGE and fluorescence scanning on an Odyssey CLx infrared scanner (Licor) followed by Coomassie blue staining.
Yeast experiments
The npl4-1 strain (Mata npl4-1 ura3-52 leu2Δ1 trp1Δ63) was transformed with plasmids derived from pPS402 (gift of Pedro Carvalho, originally generated by the lab of Pamela Silver70). The original plasmid encodes wild-type Npl4 under its endogenous promoter and includes a Ura3 cassette. Initial cultures were grown at room temperature, as the npl4-1 strain grows poorly at 30°C. Yeast were spotted in 10-fold serial dilution on SD-Ura plates and incubated at room temperature, 30°C, or 37°C for 2–3 days.
Data availability
The crystal structure determined in this work has been deposited in the Protein Data Bank (PDB) with accession code 6CDD. The Cdc48/Npl4 model has been deposited in the PDB with accession code 6CHS. The cryo-EM maps have been deposited in the Electron Microscopy Data Bank (EMDB) with accession codes 7476 (Cdc48-Ufd1-Npl4 [ATPγS]), 7477 (Cdc48 [ADP]), 7478 (Cdc48 [ATPγS]), and 7479 (Cdc48-Ufd1-Npl4 [ADP]). X-ray diffraction data are available from SBGrid with the identifier doi:10.15785/SBGRID/565. Source data for Figure 3a, 3c, 8a, and 10b are available with the paper online. Other datasets generated during the study are available from the authors on request.
Supplementary Material
Acknowledgments
We thank X. Wu and L. Li for assistance with crystallography, D. Finley for critical reading of the manuscript, the SBGrid consortium at Harvard Medical School, and the ICCB Longwood for use of equipment. We thank M. Ebrahim and J. Sotiris at the Rockefeller University Evelyn Gruss Lipper Cryo-Electron Microscopy Resource Center for assistance with microscope operation. This work is based on research conducted at the Northeastern Collaborative Access Team beamlines, which are funded by the NIH/NIGMS (P41 GM103403). The Pilatus 6M detector on 24-ID-C beam line is funded by a NIH-ORIP HEI grant (S10 RR029205). This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357. Z.J. is a Howard Hughes Medical Institute Fellow of the Damon Runyon Cancer Research Foundation, DRG-2315-18. This research was supported in part by a Helmsley Postdoctoral Fellowship at The Rockefeller University (to K.H.K.), funding from the Blavatnik Family Foundation (to E.N.), a research collaboration with the Waters Corporation (J.R.E.), and NIGMS grants R01GM052586 and T32GM007753. E.N. and T.A.R. are Howard Hughes Medical Institute investigators.
Footnotes
Accession Codes
The crystal structure determined in this work has been deposited in the Protein Data Bank (PDB) with accession code 6CDD. The Cdc48/Npl4 model has been deposited in the PDB with accession code 6CHS. The cryo-EM maps have been deposited in the Electron Microscopy Data Bank (EMDB) with accession codes 7476 (Cdc48-Ufd1-Npl4 [ATPγS]), 7477 (Cdc48 [ADP]), 7478 (Cdc48 [ATPγS]), and 7479 (Cdc48-Ufd1-Npl4 [ADP]).
Author contributions
N.O.B. purified proteins, solved the crystal structure, and performed biochemical experiments. K.H.K. performed cryo-EM data collection and analysis. Z.J. purified and tested Cdc48 FFF mutants and performed biochemical experiments. T.E.W. performed H/D exchange experiments with supervision from J.R.E. V.S. performed crosslinking mass spectrometry analysis with supervision from E.N. T.W. oversaw the cryo-EM experiments. N.O.B. and T.A.R. wrote the manuscript. T.A.R. supervised the project.
The authors declare no competing financial interests.
References
- 1.Stach L, Freemont PS. The AAA+ ATPase p97, a cellular multitool. Biochem J. 2017;474:2953–2976. doi: 10.1042/BCJ20160783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang X, et al. Structure of the AAA ATPase p97. Mol Cell. 2000;6:1473–1484. doi: 10.1016/s1097-2765(00)00143-x. [DOI] [PubMed] [Google Scholar]
- 3.Meyer HH, Shorter JG, Seemann J, Pappin D, Warren G. A complex of mammalian ufd1 and npl4 links the AAA-ATPase, p97, to ubiquitin and nuclear transport pathways. EMBO J. 2000;19:2181–2192. doi: 10.1093/emboj/19.10.2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lander GC, et al. Complete subunit architecture of the proteasome regulatory particle. Nature. 2012;482:186–191. doi: 10.1038/nature10774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tang WK, et al. A novel ATP-dependent conformation in p97 N-D1 fragment revealed by crystal structures of disease-related mutants. EMBO J. 2010;29:2217–2229. doi: 10.1038/emboj.2010.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Banerjee S, et al. 2.3 Å resolution cryo-EM structure of human p97 and mechanism of allosteric inhibition. Science. 2016;351:871–875. doi: 10.1126/science.aad7974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hänzelmann P, Schindelin H. The Interplay of Cofactor Interactions and Post-translational Modifications in the Regulation of the AAA+ ATPase p97. Front Mol Biosci. 2017;4:21. doi: 10.3389/fmolb.2017.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ye Y, Meyer HH, Rapoport TA. The AAA ATPase Cdc48/p97 and its partners transport proteins from the ER into the cytosol. Nature. 2001;414:652–656. doi: 10.1038/414652a. [DOI] [PubMed] [Google Scholar]
- 9.Skrott Z, et al. Alcohol-abuse drug disulfiram targets cancer via p97 segregase adaptor NPL4. Nature. 2017;552:194–199. doi: 10.1038/nature25016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Isaacson RL, et al. Detailed structural insights into the p97-Npl4-Ufd1 interface. J Biol Chem. 2007;282:21361–21369. doi: 10.1074/jbc.M610069200. [DOI] [PubMed] [Google Scholar]
- 11.McCullough J, Clague MJ, Urbé S. AMSH is an endosome-associated ubiquitin isopeptidase. J Cell Biol. 2004;166:487–492. doi: 10.1083/jcb.200401141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cope GA, et al. Role of predicted metalloprotease motif of Jab1/Csn5 in cleavage of Nedd8 from Cul1. Science. 2002;298:608–611. doi: 10.1126/science.1075901. [DOI] [PubMed] [Google Scholar]
- 13.Maytal-Kivity V, Reis N, Hofmann K, Glickman MH. MPN+, a putative catalytic motif found in a subset of MPN domain proteins from eukaryotes and prokaryotes, is critical for Rpn11 function. BMC Biochem. 2002;3:28. doi: 10.1186/1471-2091-3-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alam SL, et al. Ubiquitin interactions of NZF zinc fingers. EMBO J. 2004;23:1411–1421. doi: 10.1038/sj.emboj.7600114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hänzelmann P, Schindelin H. Characterization of an Additional Binding Surface on the p97 N-Terminal Domain Involved in Bipartite Cofactor Interactions. Structure. 2016;24:140–147. doi: 10.1016/j.str.2015.10.027. [DOI] [PubMed] [Google Scholar]
- 16.Hetzer M, et al. Distinct AAA-ATPase p97 complexes function in discrete steps of nuclear assembly. Nat Cell Biol. 2001;3:1086–1091. doi: 10.1038/ncb1201-1086. [DOI] [PubMed] [Google Scholar]
- 17.Heo JM, et al. A stress-responsive system for mitochondrial protein degradation. Mol Cell. 2010;40:465–480. doi: 10.1016/j.molcel.2010.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bodnar NO, Rapoport TA. Molecular Mechanism of Substrate Processing by the Cdc48 ATPase Complex. Cell. 2017;169:722–735.e9. doi: 10.1016/j.cell.2017.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee C, Prakash S, Matouschek A. Concurrent translocation of multiple polypeptide chains through the proteasomal degradation channel. J Biol Chem. 2002;277:34760–34765. doi: 10.1074/jbc.M204750200. [DOI] [PubMed] [Google Scholar]
- 20.Burton RE, Siddiqui SM, Kim YI, Baker TA, Sauer RT. Effects of protein stability and structure on substrate processing by the ClpXP unfolding and degradation machine. EMBO J. 2001;20:3092–3100. doi: 10.1093/emboj/20.12.3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park S, Isaacson R, Kim HT, Silver PA, Wagner G. Ufd1 exhibits the AAA-ATPase fold with two distinct ubiquitin interaction sites. Structure. 2005;13:995–1005. doi: 10.1016/j.str.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 22.DeLaBarre B, Christianson JC, Kopito RR, Brunger AT. Central pore residues mediate the p97/VCP activity required for ERAD. Mol Cell. 2006;22:451–462. doi: 10.1016/j.molcel.2006.03.036. [DOI] [PubMed] [Google Scholar]
- 23.Prakash S, Tian L, Ratliff KS, Lehotzky RE, Matouschek A. An unstructured initiation site is required for efficient proteasome-mediated degradation. Nat Struct Mol Biol. 2004;11:830–837. doi: 10.1038/nsmb814. [DOI] [PubMed] [Google Scholar]
- 24.Pye VE, et al. Structural insights into the p97-Ufd1-Npl4 complex. Proc Natl Acad Sci USA. 2007;104:467–472. doi: 10.1073/pnas.0603408104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bebeacua C, et al. Distinct conformations of the protein complex p97-Ufd1-Npl4 revealed by electron cryomicroscopy. Proc Natl Acad Sci USA. 2012;109:1098–1103. doi: 10.1073/pnas.1114341109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bruderer RM, Brasseur C, Meyer HH. The AAA ATPase p97/VCP interacts with its alternative co-factors, Ufd1-Npl4 and p47, through a common bipartite binding mechanism. J Biol Chem. 2004;279:49609–49616. doi: 10.1074/jbc.M408695200. [DOI] [PubMed] [Google Scholar]
- 27.Blok NB, et al. Unique double-ring structure of the peroxisomal Pex1/Pex6 ATPase complex revealed by cryo-electron microscopy. Proc Natl Acad Sci USA. 2015;112:E4017–25. doi: 10.1073/pnas.1500257112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ambroggio XI, Rees DC, Deshaies RJ. JAMM: a metalloprotease-like zinc site in the proteasome and signalosome. PLoS Biol. 2004;2:E2. doi: 10.1371/journal.pbio.0020002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lingaraju GM, et al. Crystal structure of the human COP9 signalosome. Nature. 2014;512:161–165. doi: 10.1038/nature13566. [DOI] [PubMed] [Google Scholar]
- 30.Worden EJ, Dong KC, Martin A. An AAA Motor-Driven Mechanical Switch in Rpn11 Controls Deubiquitination at the 26S Proteasome. Mol Cell. 2017;67:799–811.e8. doi: 10.1016/j.molcel.2017.07.023. [DOI] [PubMed] [Google Scholar]
- 31.Sato Y, et al. Structural basis for specific cleavage of Lys 63-linked polyubiquitin chains. Nature. 2008;455:358–362. doi: 10.1038/nature07254. [DOI] [PubMed] [Google Scholar]
- 32.Davies CW, Paul LN, Kim MI, Das C. Structural and thermodynamic comparison of the catalytic domain of AMSH and AMSH-LP: nearly identical fold but different stability. J Mol Biol. 2011;413:416–429. doi: 10.1016/j.jmb.2011.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsuchiya H, et al. In Vivo Ubiquitin Linkage-type Analysis Reveals that the Cdc48-Rad23/Dsk2 Axis Contributes to K48-Linked Chain Specificity of the Proteasome. Mol Cell. 2017;66:488–502.e7. doi: 10.1016/j.molcel.2017.04.024. [DOI] [PubMed] [Google Scholar]
- 34.Worden EJ, Padovani C, Martin A. Structure of the Rpn11-Rpn8 dimer reveals mechanisms of substrate deubiquitination during proteasomal degradation. Nat Struct Mol Biol. 2014;21:220–227. doi: 10.1038/nsmb.2771. [DOI] [PubMed] [Google Scholar]
- 35.Shi Y, et al. Rpn1 provides adjacent receptor sites for substrate binding and deubiquitination by the proteasome. Science. 2016;351:aad9421–aad9421. doi: 10.1126/science.aad9421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matyskiela ME, Lander GC, Martin A. Conformational switching of the 26S proteasome enables substrate degradation. Nat Struct Mol Biol. 2013;20:781–788. doi: 10.1038/nsmb.2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen S, et al. Structural basis for dynamic regulation of the human 26S proteasome. Proc Natl Acad Sci USA. 2016;113:12991–12996. doi: 10.1073/pnas.1614614113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Frey S, Görlich D. A new set of highly efficient, tag-cleaving proteases for purifying recombinant proteins. J Chromatogr A. 2014;1337:95–105. doi: 10.1016/j.chroma.2014.02.029. [DOI] [PubMed] [Google Scholar]
- 39.Ohi M, Li Y, Cheng Y, Walz T. Negative Staining and Image Classification - Powerful Tools in Modern Electron Microscopy. Biol Proced Online. 2004;6:23–34. doi: 10.1251/bpo70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li X, Zheng S, Agard DA, Cheng Y. Asynchronous data acquisition and on-the-fly analysis of dose fractionated cryoEM images by UCSFImage. J Struct Biol. 2015;192:174–178. doi: 10.1016/j.jsb.2015.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 42.Li X, et al. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat Methods. 2013;10:584–590. doi: 10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rohou A, Grigorieff N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J Struct Biol. 2015;192:216–221. doi: 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tang G, et al. EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 45.Scheres SHW. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang Z, Fang J, Chittuluru J, Asturias FJ, Penczek PA. Iterative stable alignment and clustering of 2D transmission electron microscope images. Structure. 2012;20:237–247. doi: 10.1016/j.str.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hohn M, et al. SPARX, a new environment for Cryo-EM image processing. J Struct Biol. 2007;157:47–55. doi: 10.1016/j.jsb.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 48.Zheng SQ, et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kimanius D, Forsberg BO, Scheres SH, Lindahl E. Accelerated cryo-EM structure determination with parallelisation using GPUs in RELION-2. Elife. 2016;5:19. doi: 10.7554/eLife.18722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Punjani A, Rubinstein JL, Fleet DJ, Brubaker M. A cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat Methods. 2017;14:290–296. doi: 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- 51.Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 52.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 53.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 55.Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Evans PR, Murshudov GN. How good are my data and what is the resolution? Acta Crystallogr D Biol Crystallogr. 2013;69:1204–1214. doi: 10.1107/S0907444913000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jackson RN, McCoy AJ, Terwilliger TC, Read RJ, Wiedenheft B. X-ray structure determination using low-resolution electron microscopy maps for molecular replacement. Nat Protoc. 2015;10:1275–1284. doi: 10.1038/nprot.2015.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Terwilliger TC. Maximum-likelihood density modification. Acta Crystallogr D Biol Crystallogr. 2000;56:965–972. doi: 10.1107/S0907444900005072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Terwilliger TC, et al. Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr D Biol Crystallogr. 2008;64:61–69. doi: 10.1107/S090744490705024X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Touw WG, van Beusekom B, Evers JMG, Vriend G, Joosten RP. Validation and correction of Zn-CysxHisy complexes. Acta Crystallogr D Struct Biol. 2016;72:1110–1118. doi: 10.1107/S2059798316013036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Morin A, et al. Collaboration gets the most out of software. Elife. 2013;2:e01456. doi: 10.7554/eLife.01456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zubarev RA, Makarov A. Orbitrap mass spectrometry. Anal Chem. 2013;85:5288–5296. doi: 10.1021/ac4001223. [DOI] [PubMed] [Google Scholar]
- 65.Yang B, et al. Identification of cross-linked peptides from complex samples. Nat Methods. 2012;9:904–906. doi: 10.1038/nmeth.2099. [DOI] [PubMed] [Google Scholar]
- 66.Fan S-B, et al. Using pLink to Analyze Cross-Linked Peptides. Curr Protoc Bioinformatics. 2015;49:8.21.1–19. doi: 10.1002/0471250953.bi0821s49. [DOI] [PubMed] [Google Scholar]
- 67.Wang L, Pan H, Smith DL. Hydrogen exchange-mass spectrometry: optimization of digestion conditions. Mol Cell Proteomics. 2002;1:132–138. doi: 10.1074/mcp.m100009-mcp200. [DOI] [PubMed] [Google Scholar]
- 68.Wales TE, Fadgen KE, Gerhardt GC, Engen JR. High-speed and high-resolution UPLC separation at zero degrees Celsius. Anal Chem. 2008;80:6815–6820. doi: 10.1021/ac8008862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wales TE, Engen JR. Hydrogen exchange mass spectrometry for the analysis of protein dynamics. Mass Spectrom Rev. 2006;25:158–170. doi: 10.1002/mas.20064. [DOI] [PubMed] [Google Scholar]
- 70.DeHoratius C, Silver PA. Nuclear transport defects and nuclear envelope alterations are associated with mutation of the Saccharomyces cerevisiae NPL4 gene. Mol Biol Cell. 1996;7:1835–1855. doi: 10.1091/mbc.7.11.1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The crystal structure determined in this work has been deposited in the Protein Data Bank (PDB) with accession code 6CDD. The Cdc48/Npl4 model has been deposited in the PDB with accession code 6CHS. The cryo-EM maps have been deposited in the Electron Microscopy Data Bank (EMDB) with accession codes 7476 (Cdc48-Ufd1-Npl4 [ATPγS]), 7477 (Cdc48 [ADP]), 7478 (Cdc48 [ATPγS]), and 7479 (Cdc48-Ufd1-Npl4 [ADP]). X-ray diffraction data are available from SBGrid with the identifier doi:10.15785/SBGRID/565. Source data for Figure 3a, 3c, 8a, and 10b are available with the paper online. Other datasets generated during the study are available from the authors on request.