Abstract
Plants produce a wide variety of secondary metabolites in response to adverse environmental factors. Z,Z-Farnesyl diphosphate (Z,Z-FPP), synthesized by Z,Z-farnesyl diphosphate synthase (zFPS), supports the formation of phytochemicals in wild tomatoes. Here, the crystal structure of N-terminal truncated zFPS (ΔzFPS) was determined. Irregular products including lavandulyl diphosphate and an unknown compound were surprisingly found. Apart from the truncated N-terminus as a functional regulator, structure-based analysis and mutagenesis assays revealed a residue H103 in ΔzFPS as one of the key elements to this irregular function. A series of substrate–enzyme complex structures were obtained from ΔzFPS-H103Y by co-crystallizing with isopentenyl diphosphate, dimethylallyl thiolodiphosphate, or both. Various substrate-binding modes were revealed. The catalytic mechanisms of both the head-to-tail and head-to-middle reactions in ΔzFPS were proposed. Functional switch between the two mechanisms in this enzyme and the essential role played by the flexible C-terminus were elucidated as well.
Introduction
Terpenoids are among the most important metabolites in plants. They play various roles, such as mediating defense or beneficial interactions.1−6 Prenyltransferases (PTSs), which are also known as isoprenyl diphosphate synthases, catalyze the formation of the upstream substrates for terpene synthases (TPSs).3 The reaction begins with dimethylallyl diphosphate (DMAPP) and isopentenyl diphosphate (IPP), which are synthesized from acetyl-CoA via the mevalonate pathway in most animal and fungal cells or from carbohydrate derivatives via the methylerythritol phosphate pathway in bacteria and plant chloroplasts.6−9 PTSs catalyze consecutive prenyl chain elongation via nucleophilic attack to the initiatory substrate, resulting in trans (E) or cis (Z) products,10,11 which differ in their molecular geometry. Regarding the three-dimensional folds of the enzymes, the α-helical bundle is generally adopted by trans-PTS.12,13 For cis-PTS, a Rossmannlike fold is presented,14,15 in which the dimeric enzyme takes the shape of a butterfly composed of a pairing domain and a catalytic domain.
The terpenoids identified to date are mostly derived from the trans-type substrates. Knowledge about cis-PTS is rather limited. The zFPS protein in this study is a short-chain cis-PTS from the glandular trichomes of the tomato species Solanum habrochaites. Its major product, Z,Z-FPP, is an important precursor for the formation of phytotoxins with vital defense roles in wild tomato.16,17 The crystal structure of zFPS, lacking the transit peptide and an additional 27 residues on the N-terminus, was determined, named ΔzFPS. To obtain substrate–enzyme complex structures, the ΔzFPS-H103Y mutant was designed by structure-based sequence alignment with other homologous enzymes. From this mutant, a series of substrate–enzyme complex structures were obtained and the irregular head-to-middle function was also revealed. Both the head-to-tail and head-to-middle condensation reactions are elucidated from the complex structures. The results provide new insights into the study of the highly diverse cis-PTS family.
Results and Discussion
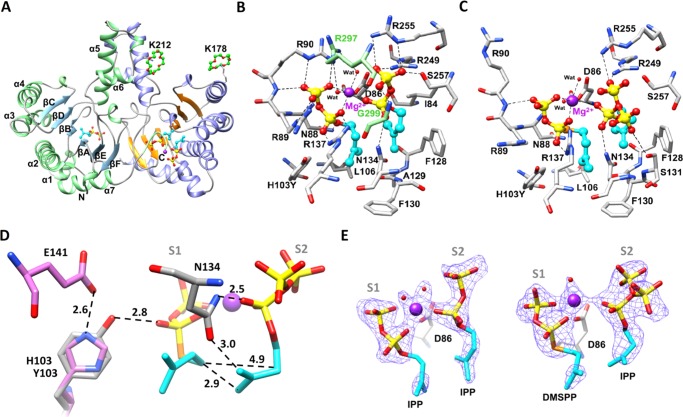
The full-length enzyme includes an N-terminal chloroplast-targeting peptide predicted to be 45 amino acids in length.16 The N-terminal 45-residue truncated clone, which was expressed and purified, was named zFPS directly. Because no crystal was obtained from this clone, an additional N-terminal 27-residue truncation was made and named ΔzFPS, which was successfully crystallized. This apo-form ΔzFPS crystal (ΔzFPS-APO) belonged to the P43212 space group, with one protein molecule in an asymmetric unit (Table 1; see crystallographic information files in the Supporting Information). Similar to the structures of other cis-PTSs, the overall fold of ΔzFPS is Rossmannlike (Figure 1A). Two dyad-related monomers are associated into a dimer via hydrogen bonds and hydrophobic interactions. This dimeric structure is consistent with the results of the sedimentation velocity (SV) analysis (Figure S1). However, we were unable to obtain the complex structure after testing a series of substrates or substrate analogues, including IPP, ISPP (isopentenyl S-thiolodiphosphate), DMSPP, GPP (geranyl diphosphate), and ibandronate.
Table 1. Data Collection and Refinement Statisticsa,b.
| name/PDB code | ΔzFPS-APO/5HXN | ΔzFPS-M/5HXO | ΔzFPS-MI/5HXP | ΔzFPS-MD/5HXQ | ΔzFPS-MDI/5HXT |
|---|---|---|---|---|---|
| data collection | |||||
| space group | P43212 | P43212 | P212121 | P212121 | P212121 |
| cell dimensions (Å) | a = 51.89 | a = 51.68 | a = 58.31 | a = 59.75 | a = 59.98 |
| b = 51.89 | b = 51.68 | b = 64.55 | b = 66.80 | b = 66.32 | |
| c = 173.29 | c = 173.56 | c = 127.16 | c = 128.73 | c = 128.9 | |
| resolution (Å) | 25.0–2.05 | 25.0–2.05 | 25.0–1.95 | 25.0–1.95 | 20.0–2.15 |
| (2.12–2.05) | (2.12–2.05) | (2.02–1.95) | (2.02–1.95) | (2.23–2.15) | |
| unique reflections | 15 386 | 15 682 | 35 453 | 38 064 | 28 515 |
| Rmerge (%) | 4.4 (49.5) | 4.3 (41.2) | 4.1 (54.5) | 4.3 (76.4) | 5.5 (47.7) |
| I/σ(I) | 52.3 (4.3) | 42.8 (5.1) | 33.0 (3.0) | 42.4 (2.8) | 37.4 (3.9) |
| completeness | 97.4 (94.6) | 99.9 (99.9) | 99.7 (100.0) | 99.3 (99.8) | 99.8 (98.5) |
| redundancy | 11.0 (10.0) | 10.6 (9.6) | 4.0 (4.1) | 5.9 (5.7) | 7.1 (6.5) |
| refinement | |||||
| resolution (Å) | 25.0–2.05 | 25.0–2.05 | 25.0–1.95 | 25.0–1.95 | 20.0–2.15 |
| no. of reflections | 14 337/751 | 14 812/777 | 31 558/1761 | 31 779/1679 | 25 353/1286 |
| Rwork/Rfree | 19.2/24.1 | 19.1/23.7 | 19.9/24.3 | 19.4/24.1 | 19.7/24.9 |
| no. of nonhydrogen atoms/avg B factor (Å2) | |||||
| protein | 1771/27.9 | 1755/27.5 | 3587/23.1 | 3643/30.2 | 3602/28.6 |
| water | 154/38.8 | 118/35.5 | 204/32.3 | 315/38.7 | 320/39.3 |
| ligand | 42/36.0 | 14/34.6 | 56/39.7 | ||
| Mg2+; crown-ether | 1/28.1; 36/37.2 | na; 36/58.5 | 2/37.9; 18/52 | ||
| RMSD bond lengths (Å) | 0.007 | 0.007 | 0.009 | 0.008 | 0.007 |
| RMSD bond angles (deg) | 1.16 | 1.21 | 1.43 | 1.28 | 1.18 |
| Ramachandran plot (%) | |||||
| most favored | 94.9 | 94.9 | 94.2 | 95.8 | 94.8 |
| additionally allowed | 4.6 | 5.1 | 5.5 | 4.2 | 5.0 |
| generously allowed | 0.0 | 0.0 | 0.0 | 0.0 | 0.2 |
| disallowed | 0.5 | 0.0 | 0.3 | 0.0 | 0.0 |
Values corresponding to the highest-resolution shells are shown in parentheses.
The stereochemistry of the model was validated with PROCHECK.
Figure 1.
Substrate–enzyme complex structures of ΔzFPS-H103Y. (A) General structure of ΔzFPS-MI. The cyclic crown-ether molecules are shown as balls and sticks. (B, C) The side chains of the ligand-interacting residues of ΔzFPS-MI and ΔzFPS-MDI, respectively. The Mg2+ ion (purple globe), the C-terminal residues from the neighboring monomer (light green), the hydrocarbon tails (cyan), water molecules (red spheres), and sulfur atom of the ligand (orange). (D) Interactions between the ligands and the key catalytic residues in ΔzFPS-MDI and the side chains of E141 and H103 from ΔzFPS-APO (pink sticks). (E) Fo-Fc OMIT maps calculated for the ligand(s) bound in ΔzFPS-MI (middle) and ΔzFPS-MDI (right). Both were contoured at the 2.0 level.
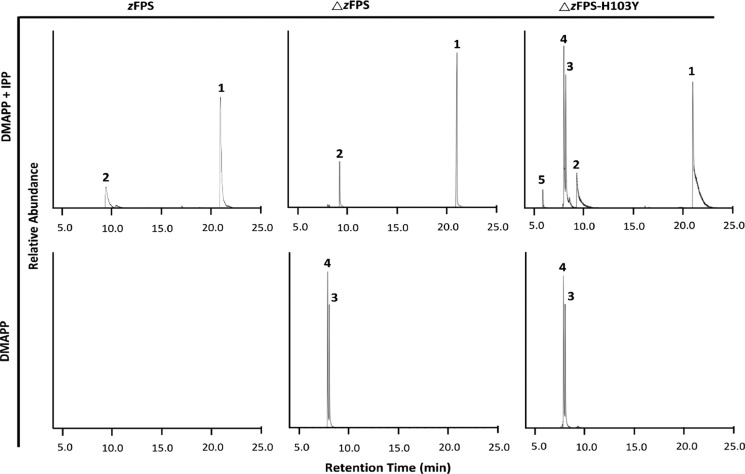
To obtain complex structures, catalytic residues around the active site were examined. Four catalytic residues including N134, H103, S131, and R137 of ΔzFPS, corresponding to N74, H43, S71, and R77 from an undecaprenyl diphosphate synthase (UPPS),18 potentially serve as general bases or acids for the chemical reaction. These catalytic residues are highly conserved in the cis-type PTS family, except the position ΔzFPS-H103. Indeed, this positon is frequently replaced by Tyr in other homologous enzymes (Figure S2). Using the ΔzFPS-H103Y mutant, complex structures were successfully obtained (see below) and the enzyme function was altered as well. Irregular products including LPP (Peak-4), an unidentified Peak-3 compound, and the monoterpene limonene (Peak-5) were found, with the native function retained (Figures 2 and S4). On the basis of the NIST Standard Database, the most reasonable candidate for Peak-3 is the monoterpene “menthol”, which showed the same retention time as that for Peak-3; however, the similarity of the mass spectra is too low (around 30%). The Peak-5 limonene is a cyclized terpenoid compound.
Figure 2.
GC chromatograms of the enzyme products from zFPS (without the signal peptide), ΔzFPS, and mutant ΔzFPS-H103Y. The peaks are identified as follows: 1, Z,Z-farnesol; 2, nerol; 3, unidentified irregular product; 4, lavandulol; and 5, limonene. RT, retention time (in minutes).
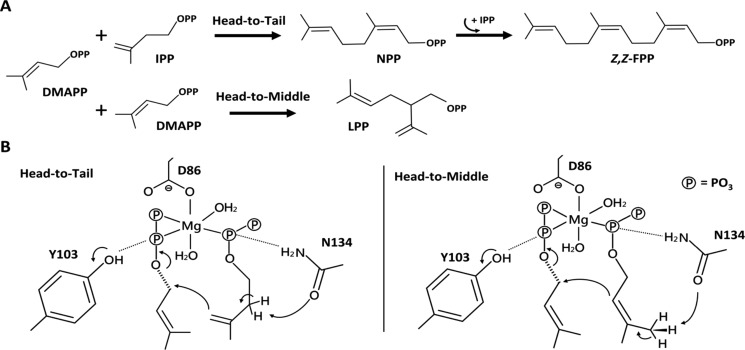
Most TPS structures share a common α-helical TPS fold.19,20 However, di-TPS Rv3378c from Mycobacterium tuberculosis was found to adopt the same Rossmannlike fold as that by cis-PTS.21 Cyclization function in cis-PTS is rare, but it was reported in a cyclolavandulyl diphosphate synthase22 (CLDPS). A structure-based sequence comparison revealed the conserved character among the same corresponding positions Rv3378-Y51, CLDPS-Y26, and Y103 from ΔzFPS-H103Y. On the other hand, the Peak-4 LPP is an irregular prenyl diphosphate, synthesized by the head-to-middle condensation reaction from DMAPP molecules.22,23 We supposed that a similar irregular reaction is adopted for the formation of unknown Peak-3. Unlike ΔzFPS-H103Y, the two irregular products were significantly reduced in ΔzFPS when IPP was provided. However, without IPP, ΔzFPS produced the two irregular products dominantly (Figure 2).
Three complex structures were successfully determined, including ΔzFPS-MI (ΔzFPS-H103Y/Mg2+/IPP), ΔzFPS-MDI (ΔzFPS-H103Y/DMSPP/Mg2+/IPP), and ΔzFPS-MD (ΔzFPS-H103Y/DMSPP). All complex structures belonged to the P212121 space group, with one protein dimer per asymmetric unit and crown-ether molecule(s) bound to the surface lysine (Figures 1A and S5A,B).
Two substrate-binding sites can be discerned: an S1 site for the binding of the initiatory substrate and an S2 site for the successive isoprene units.18 The ligand in the S1 site interacts with the side chains of N88, R89, R90, Y103, L106, and R137. Among these, L106, which regulates the product chain length in zFPS,17 is also crucial for ligand positioning. Two alternative binding modes potentially representing the transient binding states of the β-phosphate group from the S2 ligand were observed (Figure 1C), which may be regulated by the side chains of S131, R249, R255, and S257. The side chain from N134 interacts with both the α-phosphate group and the allylic tail from the S2 ligand.
The ΔzFPS-MI and ΔzFPS-MDI structures suggest that N134 can serve as a general base for proton removal from the S2 ligand (Figure 1D). The N atom and the O atom of the side chain from N134 are close to the diphosphate group and the C2/C4 atom of the ligand, respectively. Nucleophilic attack at the C1 atom of the S1 ligand by the π-electrons from the S2 ligand results in condensation. In ΔzFPS-MDI, with a distance of 2.9 Å between the C4 atom from the S2 ligand and the C1 atom from the S1 ligand, head-to-tail condensation occurs. For the head-to-middle condensation reaction, the distance between the C2 atom of the S2 ligand and the C1 atom of the S1 ligand is 4.9 Å. ΔzFPS-MI shows an even longer distance. The ligand positioning seems to be more favored for the head-to-tail condensation by the short reacting distance. The occurrence of the head-to-middle condensation function may need a greater potential in the ionization of the S1 ligand.
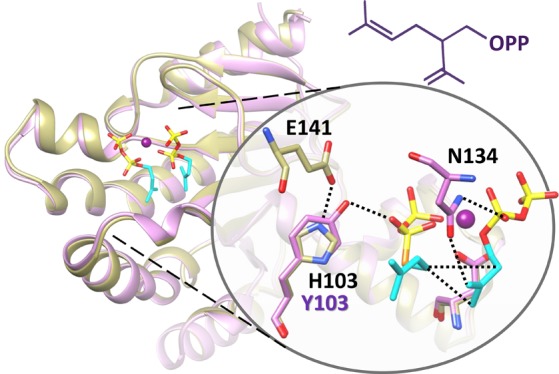
The catalytic mechanism of head-to-middle condensation was proposed previously in a lavandulyl diphosphate synthase24 (LPPS, PDB ID: 5HC6), a cis-PTS with 39% amino acid sequence identity to zFPS. Superposition of LPPS and ΔzFPS-MDI showed a highly conserved active site, with most substrate-binding residues identical (Figure S6A). The corresponding position of zFPS-H103 is LPPS-H93. Two catalytic residues in the S1 site are different: zFPS-R90/LPPS-L80 and zFPS-N88/LPPS-H78 (Scheme 1). According to our complex structures, zFPS-R90 potentially regulates the opening/closing of S1 site (Figure 1B,C). LPPS-L80 may play a similar role. On the other hand, as proposed previously, LPPS-H78 is the key catalytic residue for the head-to-middle reaction in LPPS, facilitating the diphosphate release from the S1 ligand.24 However, in ΔzFPS, the corresponding position of LPPS-H78 is ΔzFPS-N88. Alternatively, the Y103 residue of the ΔzFPS-H103Y mutant can play a crucial role in the ionization reaction of the diphosphate group (Scheme 1). In contrast, H103 in the ΔzFPS structure (ΔzFPS-APO) forms a hydrogen bond with E141, which inhibits the bonding between H103 and the diphosphate group from the S1 ligand (Figure 1D). The head-to-middle reaction is much less favored under this situation. As the enzymatic assays showed, ΔzFPS catalyzed head-to-middle condensation only in the absence of IPP (Figure 2). The elimination function served by Y103 in ΔzFPS-H103Y may also contribute to the small amounts of limonene found in the product profile. Revealed by structural comparison, the residue served for the diphosphate elimination in di-TPS Rv3378c is Y5121 and the corresponding position is close to Y103 in ΔzFPS-MI (Figure S7).
Scheme 1. Catalytic Mechanisms of ΔzFPS.
The head-to-tail condensation mechanism in cis-PTS, as proposed previously,18 starts with the binding of an allylic ligand in the S1 site. The homoallylic ligand then binds to the S2 site in the form of a MgIPP complex for the successive reaction. However, from the ΔzFPS-MD complex structure, only the S2 site was occupied by one DMSPP molecule, which may represent the starting configuration for head-to-middle catalysis. This binding was initially assisted by the side chains of E133 and Y266, and C-terminal R296 from the neighboring monomer (Figure S5C). A second DMAPP molecule accompanied by Mg2+ then moves into the S1 site.
The adverse turn of the binding order for the head-to-middle reaction may be attributed to the S1 ligand (Figure S8), in which ready ionization of the diphosphate group is crucial. The reactive nature of the S1 ligand would result in an unstable binding; thus, the binding sequence is opposite compared to that in the head-to-tail reaction. We suppose that both of the binding sequences for head-to-tail and head-to-middle condensation reactions can be adopted in ΔzFPS.
The final coverage of the catalytic pocket is accomplished by the extended C-terminal segment from the neighboring monomer (Figure 1B). The backbone of G299 on the terminus stabilizes the S2 ligand, and the side chain of R297 binds to two water molecules of the Mg2+-coordinated complex. The Mg2+ ion will eventually migrate to the diphosphate group of the S1 ligand and then leave as a MgPPi complex.18 The elongated product then occupies the S1 site for the subsequent reaction. The C-terminal segment is crucial because the ΔzFPS-H103Y/R297A mutant was inactivated, a tremendous loss of activity in ΔzFPS-H103Y/R296A was also observed (Figure S3E,F).
Most PTSs reported to date are classified with a specific functional mechanism. However, a cis-PTS from Methanosarcina acetivorans was identified that naturally synthesizes both head-to-tail and head-to-middle reaction products.25 Using a structural approach, the potential of the multifunctional character of ΔzFPS was brought to light. The 103 position in zFPS is not the only factor to regulate the functional specificity. With the N-terminal segment retained, the enzymatic assay showed no irregular products in zFPS (Figure 2). On the basis of the crystal structures of ΔzFPS-MI and ΔzFPS-MDI, the 23-residue N-terminus might be able to extend to the active site of the neighboring monomer near the C-terminus, and this N-terminal segment is one of the important functional regulators of zFPS, apparently inhibiting the occurrence of the head-to-middle condensation reaction. However, traceable amounts of the irregular products can still be detected when DMAPP was provided in excessive concentration (Figure S3B). The reason why zFPS could not be crystallized without the N-terminal truncation is likely the flexibility of this segment, which probably restricts the binding of IPP in the S2 site.
Conclusions
In the present study, the head-to-middle condensation function was observed in the N-terminal truncated ΔzFPS. High substrate specificity was demonstrated in this enzyme as the irregular function only occurs without the presence of IPP. The regulatory nature of this enzyme was characterized on the basis of the structural information obtained from complex structures accompanied by mutagenesis and enzymatic assays. Catalytic mechanisms of both the typical head-to-tail and irregular head-to-middle condensation reactions were proposed. The regulatory role played by the residue at positon 103 in zFPS was elucidated as well. These findings contribute to furthering our understanding about the formation of highly diverse prenyl phytochemicals.
Methods
Protein Expression and Purification
The nucleotide sequence of ΔzFPS from tomato lacking the N-terminal segment (residues 1–70) with the double mutations D71M and E75A was synthesized on a codon-optimized basis of the Escherichia coli translational system (http://www.genscript.com). As the native hydrophilic N-terminal is flexible, which may have a negative effect on crystallization, we had mutated the two residues to hydrophobic methionine and alanine to improve the crystal packing. The obtained nucleotides were then cloned into the pET-32 Xa/LIC vector (www.merckmillipore.com), which includes a Factor Xa cleavage site between the upstream His-tag and the downstream coding sequence. The same expression strategy was applied to the mutants in this study. The mutants obtained from site-directed mutagenesis were produced using PCR-based methods by the following primers (3′–5′): ΔzFPS-H103Y, forward primer: CGTTTCGGAAGGTTATAAACACCTG; reverse primer: CAGGTGTTTATAACCTTCCGAAACG. ΔzFPS-H103Y/R296A, forward primer: GCAGCGTCATGCTCGTTTTGGTGGTCACACG; reverse primer: CGTGTGACCACCAAAACGAGCATGACGCTGC. ΔzFPS-H103Y/R297A, forward primer: GCAGCGTCATCGTGCTTTTGGTGGTCACACG; reverse primer: CGTGTGACCACCAAAAGCACGATGACGCTGC. All PCR reactions were performed using high-fidelity DNA polymerase KOD-Plus following the manufacturer’s instructions (www.toyobo-global.com). The obtained PCR products were then digested by DpnI (www.neb.com) at 37 °C for 1 h and transformed into E. coli DH5α cells for plasmid production.
Each constructed clone was transformed into E. coli BL21 (DE3) cells for protein expression. The cells were grown to A600 of 0.6 at 37 °C in LB medium containing 50 μg mL–1 of ampicillin, induced with 0.5 mM isopropyl-1-thio-β-d-galactopyranoside, and grown for 16 h at 16 °C. They were then harvested by centrifugation at 6000g at 4 °C, resuspended in 50 mL of extraction buffer (25 mM tris-base, pH 7.8, 500 mM NaCl, 10 mM imidazole), and broken by a French Press (Constant Systems Ltd). For protein purification, the lysates were recovered by centrifugation at 20 000g for 40 min at 4 °C. ΔzFPS or ΔzFPS mutants with N-terminal thioredoxin and His-tag fusion in supernatants were then applied to binding columns with nickel-nitrilotriacetic acid (Ni-NTA) agarose resin (https://www.qiagen.com). The imidazole concentration of the wash buffer was increased to 40 mM, and the recombinant protein fractions were eluted at an imidazole concentration between 50 and 70 mM. After processing by Factor Xa protease digestion to remove the N-terminal fusion tags (www.neb.com), the mixtures were then passed through a second Ni-NTA column to remove the uncleaved fusion protein and free fusion tags. The purified protein was concentrated to 10 mg mL–1 in a buffer solution of 25 mM tris-base, pH 8.0, 150 mM NaCl. SDS-polyacrylamide gel electrophoresis analyses were performed to demonstrate the protein purity. The purified proteins were frozen with liquid nitrogen and stored at −80 °C.
Crystallization and Data Collection
Crystals of ΔzFPS (ΔzFPS-APO) and ΔzFPS-H103Y (ΔzFPS-M) were grown by mixing 2 μL of protein with 2 μL of reservoir solution using the sitting-drop vapor diffusion method at 18 °C. The ΔzFPS crystals were obtained in a reservoir solution of 70% 2-methyl-2,4-pentanediol (MPD), 0.1 M Tris, pH 8.0. The ΔzFPS-H103Y mutant was crystallized in a reservoir solution of 5% (w/v) PEG 8000, 40% MPD, 0.1 M cacodylate, pH 7.0. Complex crystals of ΔzFPS-MI (ΔzFPS-H103Y/Mg2+/IPP), ΔzFPS-MD (ΔzFPS-H103Y/DMSPP), and ΔzFPS-MDI (ΔzFPS-H103Y/Mg2+/DMSPP/IPP) were obtained with reservoir solution of 13% PEG10k, 20 mM 18-crown-6,26 0.1 M MES, pH 6.5. All diffraction data were collected using X-rays of wavelength 1.000 Å, recorded at cryogenic temperatures with a cryoprotectant solution of 20–25% glycerol. Data of ΔzFPS-APO were collected at the beamline BL5-A of the Photon Factory in Japan, with an ADSC Quantum 315r CCD detector. Data for the apo-form ΔzFPS-H103Y crystal were collected at the beamline BL12B2 of SPring-8 in Japan, using an ADSC Quantum 210 CCD detector. Data for other complex crystals including ΔzFPS-MI and ΔzFPS-MD were collected using the beamline BL15A1 of the National Synchrotron Radiation Research Center (NSRRC) in Taiwan. The ΔzFPS-MDI complex data was collected at the NSRRC Taiwan Photon Source (TPS) beamline TPS-05A with a Rayonix MX300-HS CCD detector. All diffraction data were processed and scaled using the HKL2000 program.27
Structure Determination and Refinement
All crystal structures in this study were determined by molecular replacement using the MOLREP program of the CCP4 program suite.28 The E. coli UPPS (PDB code: 1JP3) was used as the initial search model. Both ΔzFPS and ΔzFPS-H103Y crystals belong to space group P43212, with one protein molecule in an asymmetric unit. The complex structures, ΔzFPS-MDI, ΔzFPS-MI, and ΔzFPS-MD, were crystallized in space group P212121, with two molecules per asymmetric unit. Throughout the refinement, 5% of randomly selected data were set aside for cross-validation with Rfree values. Manual modifications of the models were performed using the program Coot.29 Difference Fourier (Fo-Fc) maps were calculated to locate the solvent molecules. All crystal structures were refined using Refmac5,30 including individual isotropic B-factor refinement. The molecular figures were produced using UCSF Chimera.31
In Vitro Enzymatic Assays
Enzymatic assays were performed in 1 mL of 50 mM HEPES, pH 7.8, containing 100 mM KCl, 7.5 mM MgCl2, 5% (w/v) glycerol, and 5 mM DTT. For each assay, 50 μg of zFPS, ΔzFPS or ΔzFPS mutants was incubated at 32 °C for 2 h, with different combinations of DMAPP, IPP, neryl diphosphate (NPP), and GPP (www.echelon-inc.com) at 65 μM. The reaction products were then dephosphorylated by adding 20 units of alkaline phosphatase (www.neb.com) and incubated at 37 °C for 1 h. The final products were extracted three times, each by vortexing with 2 mL of pentane. After separating the water phase by centrifugation, the obtained organic phase was then concentrated to 10 μL. The products were analyzed on an Agilent 5975C Series GC/MSD. The samples in pentane were injected directly into the GC–MS system. Separation was assured with a DB-5 capillary column (J&W Scientific; 30 m, 0.25 mm inside diameter). The temperature program used was isothermal 70 °C for 2 min, followed by a linear gradient (8 °C min–1) to 250 °C. Mass spectra from NIST Mass Spectral Library and authentic standards were used for product confirmation. Authentic standards including lavandulol, farnesol, limonene, and α-terpineol were purchased from Sigma (www.sigmaaldrich.com), and Z,Z-farnesol was purchased from Echelon (www.echelon-inc.com).
Analytical Ultracentrifugation
The SV experiments were performed at 50 000 rpm using a 4-hole AnTi60 rotor at 20 °C in a Beckman Optima XL-I AUC equipped with absorbance optics. The purified ΔzFPS samples were collected and diluted to different concentrations: 1, 0.5, and 0.1 mg mL–1, in a protein storage buffer (25 mM tris-base, 150 mM NaCl, 1 mM DTT, pH 8.0). Standard 12 mm aluminum double-sector centerpieces were filled with protein solution, with a reference cell containing the blank buffer. Quartz windows were used with absorbance optics (OD 280) in a continuous mode without averaging. No time interval was set between scans. The data were analyzed with a c(s) distribution of the Lamm equation solutions calculated using SEDFIT Version 14.1 (http://analyticalultracentrifugation.com). Software Sednterp (http://www.jphilo.mailway.com) was used to estimate the protein partial specific volume (Vbar), buffer density (1.0051 g mL–1), and buffer viscosity (0.0102331 P) at 20 °C.
Acknowledgments
We thank many groups at Academia Sinica: Yu-Ching Wu from the Small Molecule Metabolomics Core Lab, Institute of Plant and Microbial Biology, for use of GC–MS instrument; Hui-Ling Shih from Core Facilities for protein structural analysis, Institute of Biological Chemistry, for crystallization screening processing; and Meng-Ru Ho from the Biophysics Core Facility, Scientific Instrument Center, for AUC analysis. We are also grateful to the National Synchrotron Radiation Research Center (NSRRC) of Taiwan for the synchrotron beam time allocations.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsomega.6b00562.
Experimental materials, methods, supporting tables, and figures (PDF)
Crystallographic information file for 5HXN (CIF)
Crystallographic information file for 5HXO (CIF)
Crystallographic information file for 5HXP (CIF)
Crystallographic information file for 5HXQ (CIF)
Crystallographic information file for 5HXT (CIF)
This work was supported by grants from the Taiwan Protein Project (MOST105-0210-01-12-01 and MOST106-0210-01-15-04), the Academia Sinica-Industrial Technology Research Institute Collaboration Project (23yy-y110505), and Academia Sinica and Ministry of Science and Technology (MOST106-0210-01-15-02).
The authors declare no competing financial interest.
Supplementary Material
References
- Pichersky E.; Gang D. R. Trends Plant Sci. 2000, 5, 439–445. 10.1016/S1360-1385(00)01741-6. [DOI] [PubMed] [Google Scholar]
- Gershenzon J.; Dudareva N. Nat. Chem. Biol. 2007, 3, 408–414. 10.1038/nchembio.2007.5. [DOI] [PubMed] [Google Scholar]
- Christianson D. W. Chem. Rev. 2006, 106, 3412–3442. 10.1021/cr050286w. [DOI] [PubMed] [Google Scholar]
- Bohlmann J.; Meyer-Gauen G.; Croteau R. Proc. Natl. Acad. Sci. U.S.A. 1998, 95, 4126–4133. 10.1073/pnas.95.8.4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coates R. M.; Denissen J. F.; Juvik J. A.; Babka B. A. J. Org. Chem. 1988, 53, 2186–2192. 10.1021/jo00245a012. [DOI] [Google Scholar]
- Kasahara H.; Hanada A.; Kuzuyama T.; Takagi M.; Kamiya Y.; Yamaguchi S. J. Biol. Chem. 2002, 277, 45188–45194. 10.1074/jbc.M208659200. [DOI] [PubMed] [Google Scholar]
- Rohmer M.; Knani M.; Simonin P.; Sutter B.; Sahm H. Biochem. J. 1993, 295, 517–524. 10.1042/bj2950517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charon L.; Hoeffler J. F.; Pale-Grosdemange C.; Lois L. M.; Campos N.; Boronat A.; Rohmer M. Biochem. J. 2000, 346, 737–742. 10.1042/bj3460737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seemann M.; Bui B. T. S.; Wolff M.; Miginiac-Maslow M.; Rohmer M. FEBS Lett. 2006, 580, 1547–1552. 10.1016/j.febslet.2006.01.082. [DOI] [PubMed] [Google Scholar]
- Liang P. H.; Ko T. P.; Wang A. H. J. Eur. J. Biochem. 2002, 269, 3339–3354. 10.1046/j.1432-1033.2002.03014.x. [DOI] [PubMed] [Google Scholar]
- Vandermoten S.; Haubruge É.; Cusson M. Cell. Mol. Life Sci. 2009, 66, 3685–3695. 10.1007/s00018-009-0100-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z.; Zhou J.; Wu R.; Xu J. J. J. Chem. Theory Comput. 2014, 10, 5057–5067. 10.1021/ct500607n. [DOI] [PubMed] [Google Scholar]
- Köksal M.; Jin Y.; Coates R. M.; Croteau R.; Chris-tianson D. W. Nature 2011, 469, 116–120. 10.1038/nature09628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko T. P.; Chen Y. K.; Robinson H.; Tsai P. C.; Gao Y. G.; Chen A. P. C.; Wang A. H. J.; Liang P. H. J. Biol. Chem. 2001, 276, 47474–47482. 10.1074/jbc.M106747200. [DOI] [PubMed] [Google Scholar]
- Fujihashi M.; Zhang Y. W.; Higuchi Y.; Li X. Y.; Koyama T.; Miki K. Proc. Natl. Acad. Sci. U.S.A. 2001, 98, 4337–4342. 10.1073/pnas.071514398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallaud C.; Rontein D.; Onillon S.; Jabés F.; Duffé P.; Giacalone C.; Thoraval S.; Escoffier C.; Herbette G.; Leonhardt N.; Causse M.; Tissier A. Plant Cell. 2009, 21, 301–317. 10.1105/tpc.107.057885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J. H.; Gonzales-Vigil E.; Matsuba Y.; Pichersky E.; Barry C. S. Plant Physiol. 2014, 164, 80–91. 10.1104/pp.113.230466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo R. T.; Ko T. P.; Chen A. P. C.; Kuo C. J.; Wang A. H. J.; Liang P. H. J. Biol. Chem. 2005, 280, 20762–20774. 10.1074/jbc.M502121200. [DOI] [PubMed] [Google Scholar]
- Lesburg C. A.; Zhai G.; Cane D. E.; Christianson D. W. Science 1997, 277, 1820–1824. 10.1126/science.277.5333.1820. [DOI] [PubMed] [Google Scholar]
- Whittington D. A.; Wise M. L.; Urbansky M.; Coates R. M.; Croteau R. B.; Christianson D. W. Proc. Natl. Acad. Sci. U.S.A. 2002, 99, 15375–15380. 10.1073/pnas.232591099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan H. C.; Feng X.; Ko T. P.; Huang C. H.; Hu Y.; Zheng Y.; Bogue S.; Nakano C.; Hoshino T.; Zhang L.; Lv P.; Liu W.; Crick D. C.; Liang P. H.; Wang A. H. J.; Oldfield E.; Guo R. T. J. Am. Chem. Soc. 2014, 136, 2892–2896. 10.1021/ja413127v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozaki T.; Zhao P.; Shinada T.; Nishiyama M.; Kuzuyama T. J. Am. Chem. Soc. 2014, 136, 4837–4840. 10.1021/ja500270m. [DOI] [PubMed] [Google Scholar]
- Demissie Z. A.; Erland L. A. E.; Rheault M. R.; Mahmoud S. S. J. Biol. Chem. 2013, 288, 6333–6341. 10.1074/jbc.M112.431171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M.; Chen C. C.; Chen L.; Xiao X.; Zheng Y.; Huang J. W.; Liu W.; Ko T. P.; Cheng Y. S.; Feng X.; Oldfield E.; Guo R. T.; Ma Y. Angew. Chem. 2016, 128, 4799–4802. 10.1002/ange.201600656. [DOI] [Google Scholar]
- Ogawa T.; Emi K. I.; Koga K.; Yoshimura T.; Hemmi H. FEBS J. 2016, 283, 2369–2383. 10.1111/febs.13749. [DOI] [PubMed] [Google Scholar]
- Lee C. C.; Maestre-Reyna M.; Hsu K. C.; Wang H. C.; Liu C. I.; Jeng W. Y.; Lin L. L.; Wood R.; Chou C. C.; Yang J. M.; Wang A. H. J. Angew. Chem., Int. Ed. 2014, 53, 13054–13058. 10.1002/anie.201405664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z.; Minor W. Methods Enzymol. 1997, 276, 307–326. 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Bailey S. Acta Crystallogr., Sect. D: Biol. Crystallogr. 1994, 50, 760–763. 10.1107/S0907444993011898. [DOI] [PubMed] [Google Scholar]
- Emsley P.; Cowtan K. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2004, 60, 2126–2132. 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Murshudov G. N.; Vagin A. A.; Dodson E. J. Acta Crystallogr., Sect. D: Biol. Crystallogr. 1997, 53, 240–255. 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Pettersen E. F.; Goddard T. D.; Huang C. C.; Couch G. S.; Greenblatt D. M.; Meng E. C.; Ferrin T. E. J. Comput. Chem. 2004, 25, 1605–1612. 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.