Abstract
DNA aptamers are single-stranded oligonucleotides that are generated by an in vitro selection method to bind targets with high affinity and specificity. Understanding molecular recognition by DNA aptamers is of fundamental importance in the development of biosensor applications. The small molecule ochratoxin A (OTA) is a fungal-derived food toxin, and OTA DNA aptamers have been established for the development of rapid detection platforms required for food safety. One such OTA aptamer (OTAA) is a guanine-rich DNA oligonucleotide that folds into an antiparallel G-quadruplex (GQ) upon OTA binding, although structural details of the GQ fold and its interaction with OTA are currently unknown. In the present study, the fluorescent nucleobase analogue, 8-thienyl-2′-deoxyguanosine (ThdG), was inserted into various G sites of OTAA to determine the probe impact on GQ folding and OTA binding affinity. Our results suggest that OTAA contains three lateral (l) loops connecting two stacked G-tetrads with an anticlockwise loop progression to afford a −(lll) GQ topology. The phenolic ring system of OTA undergoes π-stacking interactions with the G-tetrads of OTAA. Our results also demonstrate aptamer sites that can be modified with ThdG to afford a fluorescent light-up signal upon OTA binding.
Introduction
Aptamers are single-stranded oligonucleotides that bind a variety of targets, including small molecules, proteins, and cells, with high affinity and specificity.1,2 DNA aptamers have the potential to replace antibodies as recognition elements due to their ease of synthesis on a large-scale level at low cost and their higher chemical stability.3,4 They are also much easier to modify than antibodies,5 and efforts have been focused on the utility of chemically modified aptamers to improve chemical diversity for enhanced target binding affinity6−11 and therapeutic potential.12,13
Aptamer base modifications can create fluorescent nucleobase analogues (FBAs) that facilitate sensitive biophysical measurements14,15 and are ideal candidates for characterizing local binding events.16,17 One simple strategy for preparation of a novel FBA series is arylation of the 8-position of 2′-deoxyguanosine (dG) to afford a fluorescent 8-aryl-dG base.18−25 Such FBAs have proven useful for monitoring G-quadruplex (GQ) folding because the bases exhibit intense fluorescence in the GQ compared to their emission in a duplex structure. Furthermore, depending on their site of incorporation, they minimally perturb the native GQ. This feature can be exploited for aptamer detection by G-rich DNA aptamers that frequently fold into a GQ upon target binding.18,20,23 A classic example is the 15-mer thrombin binding aptamer (TBA, 5′-GGTTGGTGTGGTTGG26) that produces an antiparallel GQ consisting of two G-tetrads connected through lateral (TGT and two TT) loops.27 The guanosine residues within the tetrads display an alternating syn- and anti-conformation (see G-tetrad in Figure 1A) and interact through Hoogsteen hydrogen bonds stabilized by a central potassium ion and provide a scaffold for thrombin binding to the loop residues.27
Figure 1.
(A) Schematic illustration of the G-tetrad with alternating syn- and anti-G bases with a central K+ ion (purple ball); (B) the structure of ochratoxin A (OTA); and (C) the structure of the FBA 8-thienyl-2′-dG (ThdG) and sequence of the 31-mer OTA aptamer (OTAA) with possible G-tetrad-Gs underlined in bold, indicating sites of ThdG incorporation.
Recently, we incorporated 8-thienyl-dG (ThdG,28Figure 1) into TBA and demonstrated the utility of the probe for monitoring thrombin binding using fluorescence spectroscopy.23 The ThdG probe favors a syn-conformation28 and stabilizes the antiparallel GQ when placed in a syn-G-tetrad position but destabilizes the GQ in an anti-G-tetrad position, as evidenced by UV thermal melting experiments.23 The fluorescent nature of ThdG (λex = 315 nm, λem = 411 nm in TBA,23 ϕfl = 0.79 for free nucleoside in water28) also provides a diagnostic energy-transfer excitation peak at 290 nm for probe stacking with a G-tetrad,23 which can distinguish G-tetrad guanosines from those in loop and tail positions. Finally, ThdG exhibits quenched emission in the duplex that lights-up in the GQ, providing emission switching properties for monitoring thrombin binding using duplex → GQ exchange.23
DNA aptamers have also been generated for the small molecule ochratoxin A (OTA, Figure 1).29−31 OTA is a common food-contaminating mycotoxin produced by the species Aspergillus and Penicillium.32 It is detected in a wide variety of foodstuffs, including cereals, coffee, dried fruits, wine, and beers, and is a potent renal carcinogen.33 Given its toxicity and world-wide occurrence, rapid OTA detection platforms are of great interest for food safety.34 Previously developed platforms required an OTA antibody for immunoreaction-based assays.35−37 The discovery of OTA aptamers29−31 has opened the door for the development of DNA-based detection platforms,38−40 and the OTA–aptamer (OTAA) interaction is fast becoming a proof-of-concept system for small molecule detection.41
The first described in vitro selection for an OTA aptamer produced a 61-mer that binds OTA in a 1:1 fashion with an apparent dissociation constant (Kd) of 0.36 μM.29 Aptamer truncation afforded a G-rich 31-mer (Figure 1C) as the shortest sequence, with a Kd less than 1 μM.29 The analysis of the OTAA–OTA interaction using circular dichroism (CD) identified an antiparallel GQ aptamer topology upon OTA binding;38 however, structural details of the GQ topology and its interaction with OTA are unknown. The knowledge of an aptamer–target structure is critical for optimizing analytical potential, aptamer length for ease of synthesis, establishing preferred site(s) of modification for enhanced aptamer performance and expanding our understanding of self-assembly and molecular recognition. Unfortunately, determination of a high-resolution GQ-target structure using NMR or X-ray crystallography is not a trivial task. Both require expensive instrumentation to yield data that are challenging for interpretation from relatively large quantities of the aptamer–target complex, often involving 13C,15N-enriched DNA for NMR analysis. This suggested the potential utility of ThdG to probe the OTAA–OTA interaction using dilute aptamer solutions. Our studies provide a model for the overall antiparallel GQ topology of OTAA and demonstrate aptamer sites that can be modified by ThdG to afford a fluorescent light-up signal upon OTA binding. Furthermore, we have established that the intrinsic fluorescence of OTA is a sensitive marker for measuring aptamer–target binding affinities.
Results and Discussion
Probe Influence on GQ Folding
Analysis of the 31-mer OTAA using the on-line QGRS mapper for predicting GQ formation42 suggests that eight GQ structures with two G-tetrads and three loops are possible. The central 11 Gs (highlighted in bold and underlined for native OTAA in Figure 1C) can participate in GQ formation. Common to all eight GQs are the involvement of G15 and G16, which must be present in the G-tetrad. The other G-tetrad sites are derived from guanosine residues in a three-base cluster (G5–G7, G11–G13, and G23–G25) indicating that the first or last two guanosines of each cluster participate in G-tetrad formation. To distinguish guanosine residues in G-tetrad positions from those in loops and tail regions, the ThdG probe was inserted into the three three-base clusters of the aptamer (Figure 1C) via solid-phase synthesis to yield nine modified strands (ThdG-phosphoramidite has been previously described;23 see Supporting Information (SI) for mass spectrometry (MS) of mOTAA samples). The mOTAA oligonucleotides were analyzed using UV–vis thermal melting, fluorescence, and CD spectroscopy to determine probe influence on GQ topology.
The UV thermal melting (Tm) values of the mOTAA samples (6 μM) were recorded in a previously optimized OTA binding buffer (10 mM Tris pH 8.0, 20 mM CaCl2, 120 mM NaCl, and 5 mM KCl)29 by monitoring the change in absorbance at 295 nm as a function of temperature. Under these conditions the native OTAA antiparallel GQ had an average Tm of 46.6 °C from the five heating and cooling ramps. The change in Tm (ΔTm) induced by ThdG (Figure 2) in G5–G7 and G11–G13 displayed positive ΔTm values at G6 and G11 that were followed by strongly negative values at G7 and G12, as expected for ThdG placement within the G-tetrad at syn-G (G6 and G11) versus anti-G (G7 and G12) positions.23 In contrast, placement of the ThdG probe at positions G5 and G13 reduced Tm of the GQ to only a minor extent, suggesting that these guanosine residues are not in the G-tetrad. For G23–G25, positive ΔTm values were observed at both G23 and G24, whereas it was not possible to record a Tm value with the probe at G25. The lack of Tm for the ThdG-25 suggested probe placement at an anti-G-tetrad position that completely hindered GQ folding, as observed previously for TBA, with certain 8-aryl-dG derivatives inserted into anti-G-tetrad sites.19 For the modified sites that afforded Tm values, the error bars presented in Figure 2 represent the range in ΔTm due to differences in the melting and annealing curves (hysteresis). Representative melting curves for native OTAA and ThdG-24 are provided in the SI (Figure S1).
Figure 2.
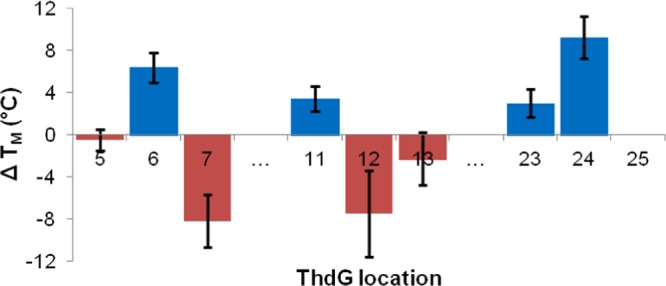
Change in UV thermal melting temperature (ΔTm) of the antiparallel GQ produced by OTAA upon site-specific incorporation of ThdG. Oligonucleotide solutions (6 μM) were monitored at 295 nm over five ramps at a rate of 0.5 °C min–1 in the OTA binding buffer. ΔTm = Tm (mOTAA GQ) – Tm (native OTAA GQ). Error bars represent the range in ΔTm due to differences in Tm values between the melting and annealing curves (hysteresis).
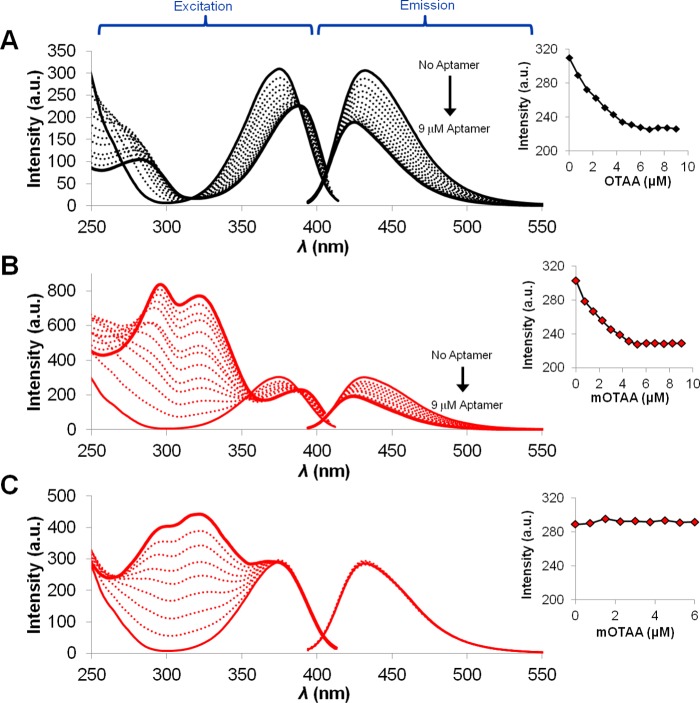
The influence of the ThdG probe on the antiparallel GQ topology, as monitored by CD, and the fluorescence response of the probe (λex = 316 nm and λem = 411 nm) is highlighted in Figure 3. For the first three-base cluster (G5–G7, Figure 3A,B), only ThdG-6 generated the characteristic antiparallel CD spectrum (red-dashed trace, Figure 3A) produced by native OTAA (black trace, Figure 3A) with positive peaks at 290 and 240 nm and a negative peak at 260 nm. The CD spectrum of ThdG-7 exhibited a broad positive peak at 275 nm and a negative peak at 246 nm (red dotted trace), suggesting a perturbed GQ topology. ThdG-5 displayed the positive peak at 290 nm for the antiparallel topology, but failed to exhibit the strong negative peak at 260 nm (solid red trace, Figure 3A). The fluorescence response of these mOTAA samples all displayed the diagnostic 290 nm peak in the excitation spectrum signifying probe stacking with the G-tetrad (Figure 3B).
Figure 3.
Circular dichroism (A, C, and E) and fluorescence excitation/emission (B, D, and F) spectra of native OTAA (solid black traces in (A), (B), and (E)) and mOTAA GQ (solid, dashed, and dotted red traces in (A)–(F)). All spectra of oligonucleotides (6 μM) were recorded at 10 °C in the OTA binding buffer.
At the G11–G13 sites, both ThdG-11 (solid red trace) and ThdG-13 (dotted red trace) produced the antiparallel GQ observed for the native OTAA (Figure 3C). The CD spectrum for ThdG-12 (dashed red trace, Figure 3C) resembled the CD spectrum of ThdG-7 (dotted red trace, Figure 3A), implying similar perturbation to the GQ topology. The fluorescence spectra of ThdG-11 (solid red trace) and ThdG-12 (dashed red trace) displayed the 290 nm peak in the excitation spectrum for G-tetrad stacking (Figure 3D). The ThdG-13 mOTAA exhibited relatively bright emission with no peak at 290 nm in the excitation spectrum (dotted trace, Figure 3D), suggesting no probe stacking with the G-tetrad.
In the final G23–G25 three-base cluster, only ThdG-24 (dashed red trace, Figure 3E) produced the antiparallel GQ topology. This mOTAA sample also displayed a strong positive ΔTm value for GQ folding (Figure 2), suggesting probe placement within the G-tetrad at a syn-G position. The probe also exhibited a strong prominent 290 nm peak in the excitation spectrum, signifying probe stacking with the G-tetrad (dashed red trace, Figure 3F). In contrast, the CD spectrum for ThdG-23 (that also displayed a positive ΔTm value, Figure 2) exhibited a broad positive band peaking at 265 nm with a negative peak at 245 nm (solid red trace, Figure 3E), suggesting formation of GQ with a parallel topology.21 The ThdG-25 sample exhibited a broad positive peak at 275 nm and a negative peak at 246 nm (red dotted trace) suggesting a perturbed GQ topology. The fluorescence spectra of the ThdG-23 and ThdG-25 samples displayed the 290 nm peak in the excitation spectra, although the peak was not particularly prominent, especially for the ThdG-25 sample (dotted red trace, Figure 3F). Overall, the CD analysis confirmed that only four mOTAA samples (ThdG-6, -11, -13, and -24) fold into the antiparallel GQ topology, as noted for native OTAA. Three of the four oligonucleotides (ThdG-6, -11, and -24) also strongly stabilized the GQ structure (Figure 2), suggesting probe placement in syn-G-tetrad positions. ThdG-13 also produced the antiparallel fold yet slightly decreased GQ stability (Figure 2), suggesting probe placement in a loop, where the syn-conformation of the probe can be tolerated.
Probe Influence on OTA Binding
To determine the impact of the ThdG probe on target binding, titrations were performed by monitoring changes in the intrinsic fluorescence of OTA upon aptamer addition. In the OTA binding buffer, the free toxin has an excitation (λex) of 375 nm and emission (λem) at 432 nm. This fluorescence was found to be sensitive to aptamer binding, and changes in excitation/emission wavelength and intensity were employed to measure OTA binding affinity. Previously reported OTA binding studies with the 31-mer OTAA were carried out using equilibrium dialysis and afforded an apparent Kd of 0.5 μM.29 Differences in Kd values can occur when the method used to determine it changes;41 therefore, a fluorescence titration was performed with the native OTAA to afford a standard Kd value.
When the native OTAA was added to the toxin in the binding buffer, the OTA excitation underwent a bathochromic (red) shift, which was accompanied by loss in intensity, suggesting π-stacking interactions of the phenolic group of OTA with the antiparallel GQ (Figure 4A).43 The OTA emission underwent a hypsochromic (blue) shift with reduced intensity, suggesting placement of the OTA in a more nonpolar environment upon OTAA binding.44 Through monitoring the changes in excitation at 375 nm, a binding isotherm was produced (inset, Figure 4A) that afforded a 1:1 binding interaction with an apparent Kd of 1.1 μM.
Figure 4.
Fluorescence titrations of OTA (3 μM) with (A) native 31-mer OTAA, (B) ThdG-11 mOTAA, and (C) ThdG-12 mOTAA at 20 °C. Inset: intensity change in fluorescence excitation (λex = 375 nm) with increasing aptamer concentration.
To confirm that OTA binding to the 31-mer OTAA was sequence and structure specific, negative control titrations using the 15-mer TBA and the 22-mer human telomeric repeat sequence (HTelo22)21 that can also form antiparallel GQ structures were carried out (Figure S2A,B, SI). In each case, the fluorescence response of OTA indicated no change in λex or λem wavelengths or intensities, suggesting no binding of OTA to GQ structures produced by TBA or HTelo22.
Given the sensitivity of OTA fluorescence to aptamer binding, the same titration conditions were used to determine OTA affinity for the mOTAA strands. Representative titrations (Figure 4B,C) emphasize the drastic influence of the probe location on the OTA–aptamer interaction. For addition of the ThdG-11 sample (Figure 4B), OTA excitation and emission underwent similar changes noted for addition of native OTAA and a Kd of 2.1 μM was determined, suggesting that the ThdG probe at G11 minimally impacts OTA binding. The additional excitation peaks in Figure 4B at 290 and 315 nm are due to the ThdG probe within the mOTAA sample and suggest GQ formation due to the strong energy-transfer peak at 290 nm. In sharp contrast, the OTA fluorescence displayed no response to addition of the ThdG-12 sample over the course of the titration (Figure 4B), suggesting that OTA did not bind to the mOTAA. Fluorescence titrations for the other mOTAA samples are given in the SI (Figure S3). Negative control fluorescence titrations were also performed with the nonchlorinated ochratoxin B (OTB) derivative, which is less toxic than OTA and is frequently found as a co-contaminant of food sources.33 Neither the native OTAA nor the ThdG-11 mOTAA samples displayed OTB binding affinity (Figure S4, SI).
The results from ThdG impact on GQ folding and OTA binding are summarized in Table 1. Due to disruption of the antiparallel GQ, no OTA binding was observed with ThdG placed at predicted anti-G-tetrad sites (G7, G12, and G25), as anticipated. At predicted syn-G-tetrad sites (G6, G11, and G24), probe placement at G11 was an optimal location for FBA insertion to monitor OTA binding, whereas G24 insertion surprisingly inhibited toxin binding. For probe placements at G5, G13, and G23, which are not predicted to reside in the G-tetrads, G5 and G23 were disruptive to OTA binding, whereas G13 had minimal impact.
Table 1. Summary of ThdG Impact on GQ Folding, OTA Binding, and Predicted Location of the ThdG Probe within the Antiparallel GQ.
| ID | Tm (ΔTm)a | λex (290)b | GQc | location | Kdd |
|---|---|---|---|---|---|
| OTAA | 46.6 | yes | 1.1 ± 0.2 | ||
| ThdG-5 | (−0.5) | yes | no | 5′-tail | 19.9 ± 2.7 |
| ThdG-6 | (+6.4) | yes | yes | syn-tetrad | 10.5 ± 2.2 |
| ThdG-7 | (−8.4) | yes | no | anti-tetrad | NBe |
| ThdG-11 | (+3.4) | yes | yes | syn-tetrad | 2.1 ± 0.3 |
| ThdG-12 | (−7.5) | yes | no | anti-tetrad | NB |
| ThdG-13 | (−2.3) | no | yes | loop | 3.3 ± 0.4 |
| ThdG-23 | (+3.0) | yes | no | loop | NB |
| ThdG-24 | (+9.2) | yes | yes | syn-tetrad | NB |
| ThdG-25 | no Tm | yes | no | anti-tetrad | NB |
Tm of GQ in degree Celsius, ΔTm = Tm (mOTAA GQ) – Tm (native OTAA).
Presence of the excitation peak at 290 nm for probe-G-tetrad stacking interactions.
Formation of antiparallel GQ, as evidenced by CD.
Dissociation constant (μM) for OTA binding measured by changes in the intrinsic fluorescence of OTA upon aptamer additions.
No binding (NB) observed from fluorescence titration.
Model for the OTA–OTAA Interaction
The unimolecular antiparallel GQ produced by OTAA contains two stacked tetrads connected by three loops. According to the geometric formalism for DNA GQ folding presented by da Silva45 and co-workers,46 the characteristic antiparallel CD spectrum produced by OTAA places it in group III that contains four possible antiparallel GQ topologies with two stacked tetrads (Figure 5).46 Starting from the 5′-end of OTAA, the spectral data with the mOTAA samples suggest that G5 is present in the 5′-tail, placing G6 (syn), G7 (anti), G11 (syn), and G12 (anti) in the tetrad. This indicates that the first loop contains three-residues (T8G9T10) starting with a pyrimidine, which commonly form diagonal loops (d) and lateral loops (l) with wide grooves.45 However, none of the four possible antiparallel GQ topologies start with a diagonal loop and thus the first TGT loop of OTAA is a lateral loop stretching across a wide groove. It is noteworthy that TBA also contains a lateral wide TGT loop as the second loop in the sequence.27
Figure 5.
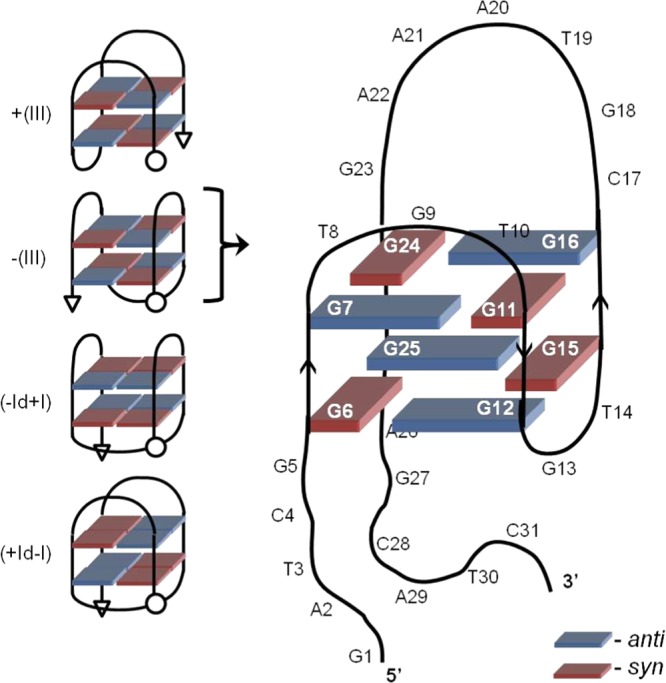
Schematic structures of antiparallel GQs containing two stacked tetrads connected by three loops and predicted model for −(lll) GQ topology for OTAA.
The spectral data also suggests that G13 is part of a two-residue loop (G13T14), which represents the central loop in the antiparallel GQ. Two-residue loops are too short to form a diagonal loop and typically occupy a lateral narrow loop.45 This restriction rules out (−ld + l) and (+ld – l) topologies (Figure 5) for the antiparallel GQ produced by OTAA and indicates that all three loops must be lateral. The final loop is predicted to be an unusual seven-residue lateral wide loop in which there is a possibility for the looping bases to form hydrogen-bond alignments. The optical data strongly suggests that G24 (syn) and G25 (anti) are present in the tetrad with G23 in the seven-residue loop. Overall, OTAA is predicted to form the −(lll) GQ topology with an anticlockwise loop progression, which is stereoisomeric of the clockwise +(lll) topology produced by TBA.45 For TBA, the central lateral loop spans a wide groove, whereas the first and third loops span narrow grooves.27 For OTAA, the first and third loops span wide grooves, whereas the central two-residue loop is restricted to spanning a narrow groove. The −(lll) topology predicted for OTAA was first reported in 2009 for the chair-type GQ formed by a Bombyx mori telomeric sequence (d[TAGG(TTAGG)3]) that contains three lateral TTA loops, with the central loop spanning across a narrow groove.47
The optical data for OTA binding to the −(lll) GQ topology for OTAA suggests that the phenolic moiety of the toxin undergoes π-stacking interactions (Figure 4). Although toxin placement within the −(lll) GQ topology is uncertain, it is noteworthy that ThdG insertion at G6, G11, G13, and G24 all produced the antiparallel GQ topology of the native OTAA (Table 1). Of these mOTAA samples, G6, G11, and G13 had relatively little impact on OTA binding, suggesting that these guanosine residues do not interact directly with the toxin. In contrast, ThdG insertion at G24 inhibited toxin binding, suggesting that this guanosine residue interacts directly with OTA and the thiophene ring of the probe inhibits insertion of the toxin into its binding site. Probe placement at G5 also perturbed OTA binding (Table 1), and previous studies have demonstrated the importance of the 5′- and 3′-tails for OTA binding affinity.29 This information suggests that the toxin may insert between the G-tetrad residues G24 and G25, which is distant from G6, G11, and G13 and would permit interaction of the toxin with the tail residues. Further efforts are ongoing to refine this model for OTA binding to OTAA using additional FBAs.
Duplex–GQ Exchange for OTA Detection
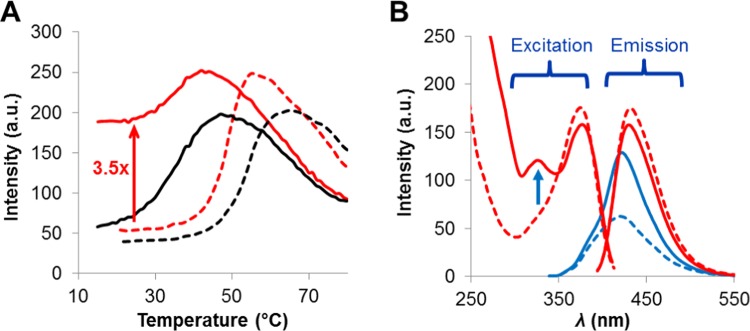
Although the ThdG probe is not practical for OTA detection strategies in the low nanomolar regime because its emission at 411 nm overlaps with the emission of OTA and it lacks sufficient brightness in low nanomolar concentrations, it was still employed in duplex → GQ exchange studies to demonstrate the potential of an internal 8-aryl-dG probe for OTA detection. For these studies, the ThdG-11 mOTAA sample was initially annealed to a 12-mer complementary strand 5′-CCATCCACACCC that produces a G13-T mismatch in the duplex with the aptamer and quenches the fluorescence emission of the ThdG probe at G11. Upon addition of 1.2 equiv of OTA at room temperature, no change in the ThdG probe fluorescence was observed, suggesting that the toxin was unable to facilitate denaturation of the duplex. Thus, a thermal ramp was employed to melt the duplex (heat to 80 °C) and promote OTA binding to mOTAA upon slow cooling to 20 °C (Figure 6A). Under these conditions, the aptamer sample containing OTA exhibited a 3.5-fold increase in emission intensity of the ThdG probe, with λex = 290 nm (solid red trace vs dashed red trace, Figure 6A) upon slow cooling to 20 °C. In contrast, the sample lacking the toxin regenerated the quenched emission of the ThdG probe upon slow cooling to 20 °C (black traces, Figure 6A). These observations suggested that the toxin was able to bind to the mOTAA sample and facilitate GQ formation, which accounted for the 3.5-fold increase in emission intensity of the ThdG probe.
Figure 6.
(A) Emission response of the ThdG probe (411 nm), with λex = 290 nm, following heating (dashed lines) and cooling (sold lines) ThdG-11 mOTAA-12-mer duplex (3 μM) in the presence (1.2 equiv, red traces) and absence (black traces) of OTA. (B) Fluorescence spectra of the sample containing OTA prior (dashed lines) and after (solid lines) the thermal experiment. Red traces represent changes in the excitation and emission of OTA, whereas blue traces represent changes in the emission of the ThdG probe.
Full fluorescence spectra of the samples were also obtained before and after completion of the thermal experiment (Figure 6B). A 1.9-fold increase in emission of the ThdG probe at 411 nm was observed between the beginning and end of the thermal experiment (dashed blue trace vs solid blue trace, Figure 6B), and the increase in intensity of the 290 nm peak in the excitation spectrum are positive indicators of GQ formation. In addition, the observed excitation of OTA at 375 nm displayed characteristic quenching and red-shifting, indicative of π-stacking and binding to the GQ (dashed red trace vs solid red trace, Figure 6B). The spectral data presented in Figure 6B confirmed that the 3.5-fold increase in emission intensity in the thermal melting experiment (Figure 6B) was due to OTA binding to the mOTAA sample with GQ formation and displacement of the complementary strand.
Conclusions
The studies presented here demonstrate the utility of internal emissive nucleobase probes for monitoring small molecule–aptamer interactions. The information gained from this study indicates that FBAs can serve as structural probes to determine the topology of an antiparallel GQ motif critical for target binding. The internal ThdG probe also provided insight into target binding location, as certain probe locations that afforded the antiparallel GQ topology were found to inhibit OTA binding, suggesting that the thiophene ring system of the probe blocked toxin insertion into the aptamer binding site. The structural ThdG probe was also employed as a sensor in a duplex → GQ fluorescent light-up system to demonstrate the potential utility of internal FBAs for OTA detection methods. Although the ThdG probe itself is not practical for OTA detection methods in food samples in low nanomolar concentrations, it successfully inferred structural information on the nature of the OTA–OTAA interaction. This information is currently being utilized to guide probe placement of new FBA sensors that may be practical for rapid OTA aptasensor detection strategies required for food safety.
Experimental Section
Materials
OTA was supplied as a 99.5% pure powder from Dr. Hans-Ulrich Humpf at the University of Münster, Germany, and quantified in MeOH via UV–vis at 333 nm, with extinction coefficient of 6400 mol–1 cm–1.48 The nonchlorinated OTB derivative was purchased from Toronto Research Chemicals Canada and quantified in MeOH via UV–vis at 318 nm, with extinction coefficient of 6900 mol–1 cm–1.49 Unmodified oligonucleotides including the TBA and HTelo22 were purchased from Sigma-Aldrich Ltd. and purified via reverse phase cartridge purification.
Oligonucleotide Synthesis
The unmodified OTAA and ThdG–mOTAA strands were synthesized via standard phosphoramidite synthesis with a 5′-dimethoxytrityl (DMT) label and were carried out on a 1 μmol scale on a MerMade 12 solid-phase oligonucleotide synthesizer. The ThdG-phosphoramidite was synthesized according to previously published protocols.23 DNA was purified via high-performance liquid chromatography and the DMT was removed in the presence of mildly acidic high purity water. Masses of the modified OTAA strands were acquired on a Bruker AmaZon ion trap SL spectrometer in a solution of 90% high purity water and 10% methanol, with 0.1 mM ammonium acetate.
UV Thermal Denaturation
Solutions were prepared to a concentration of 6 μM OTAA in OTA binding buffer consisting of 20 mM CaCl2, 120 mM NaCl, 10 mM trisphosphate, and 5 mM KCl, with pH 8.0.29 Samples were monitored at 295 nm on a Cary 300-Bio Spectrometer with a 6 × 6 multicell block with Peltier thermal controller. The samples were heated and cooled at a rate of 0.5 °C min–1, and an average of five ramps was reported. Thermal melting temperatures (Tm values) were calculated by determining the first derivative of the melting curve through the Varian Thermal software.
Circular Dichroism
CD spectra were recorded on a Jasco J-815 CD spectrophotometer equipped with a thermal controlled 1 × 6 multicell block. The annealed samples obtained from the Tm studies were measured at 10 °C in quartz cells (110-QS), with a light path of 1 mm, and monitored between 200 and 400 nm at a bandwidth of 1 nm and scanning speed of 100 nm min–1. A minimum of three scans were collected and averaged. Data was corrected against a blank measurement of the appropriate buffer and smoothed using the Jasco software.
OTA Binding Titrations
OTA was prepared as a 100 μL solution with concentration of 3 μM in a Hellma Analytics ultra-micro cuvette 105.253-QS, with path length of 10 × 2 mm2, with temperature control at 20 °C. Strands of mOTAA were titrated until a final concentration of 2–3 equiv of aptamer to OTA was reached. Scans were taken directly after manual mixing of the solution. A plot of the fraction of OTA bound versus concentration of mOTAA generated a binding isotherm that was analyzed with SigmaPlot 11.0 using the one site saturation ligand binding macro to obtain Kd values and standard errors.
Acknowledgments
This research was funded in part through Growing Forward 2 (GF2), a federal–provincial–territorial initiative. The Agricultural Adaptation Council assists in the delivery of GF2 in Ontario. This research was also funded in part through Grain Farmers of Ontario. R.A.M. also thanks Dr. Hans-Ulrich Humpf from the University of Münster, Germany, for the sample of OTA.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsomega.7b00782.
UV thermal melting curves; titration of OTA and OTB (Figures S1–S4); ESI-MS spectra of ThdG–mOTAA (Figures S5–S7); ESI-MS analysis of modified OTAA (Table S1) (PDF)
Author Contributions
† K.L.F. did the complete experimental work.
The authors declare no competing financial interest.
Supplementary Material
References
- Ellington A. D.; Szostak J. W. In vitro selection of RNA molecules that bind specific ligands. Nature 1990, 346, 818–822. 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- Tuerk C.; Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 1990, 249, 505–510. 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- Jayasena S. D. Aptamers: an emerging class of molecules that rival antibodies in diagnostics. Clin. Chem. 1999, 45, 1628–1650. [PubMed] [Google Scholar]
- Bunka D. H. J.; Stockley P. G. Aptamers come of age – at last. Nat. Rev. Microbiol. 2006, 4, 588–596. 10.1038/nrmicro1458. [DOI] [PubMed] [Google Scholar]
- Diafa S.; Hollenstein M. Generation of aptamers with an expanded chemical repertoire. Molecules 2015, 20, 16643–16671. 10.3390/molecules200916643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaught J. D.; Bock C.; Carter J.; Fitzwater T.; Otis M.; Schneider D.; Rolando J.; Waugh S.; Wilcox S. K.; Eaton B. E. Expanding the chemistry of DNA for in vitro selection. J. Am. Chem. Soc. 2010, 132, 4141–4151. 10.1021/ja908035g. [DOI] [PubMed] [Google Scholar]
- Kimoto M.; Yamashige R.; Matsunaga K.-i.; Yokoyama S.; Hirao I. Generation of high-affinity DNA aptamers using an expanded genetic alphabet. Nat. Biotechnol. 2013, 31, 453–457. 10.1038/nbt.2556. [DOI] [PubMed] [Google Scholar]
- Imaizumi Y.; Kasahara Y.; Fujita H.; Kitadume S.; Ozki H.; Endoh T.; Kuwahara M.; Sugimoto N. Efficacy of base-modification on target binding of small molecule DNA aptamers. J. Am. Chem. Soc. 2013, 135, 9412–9419. 10.1021/ja4012222. [DOI] [PubMed] [Google Scholar]
- Zhao J.; Katsube S.; Yamamoto J.; Yamasaki K.; Miyagishi M.; Iwai S. Analysis of ATP and AMP binding to a DNA aptamer and its imidazole-tethered derivatives by surface plasmon resonance. Analyst 2015, 140, 5881–5884. 10.1039/C5AN01347J. [DOI] [PubMed] [Google Scholar]
- Tsvetkov V. B.; Varizhuk A. M.; Pozmogova G. E.; Smirnov I. P.; Kolganova N. A.; Timofeev E. N. A universal base in a specific role: tuning up a thrombin aptamer with 5-nitroindole. Sci. Rep. 2015, 5, 16337 10.1038/srep16337. [DOI] [Google Scholar]
- Cservenyi T. Z.; Van Riesen A. J.; Berger F. D.; Desoky A.; Manderville R. A. A simple molecular rotor for defining nucleoside environment within a DNA aptamer–protein complex. ACS Chem. Biol. 2016, 11, 2576–2582. 10.1021/acschembio.6b00437. [DOI] [PubMed] [Google Scholar]
- Ng E. W. M.; Shima D. T.; Calias P.; Cunningham E. T. Jr.; Guyer D. R.; Adamis A. P. Pegaptanib, a targeted anti-VEGF aptamer for ocular vascular disease. Nat. Rev. Drug Discovery 2006, 5, 123–132. 10.1038/nrd1955. [DOI] [PubMed] [Google Scholar]
- Astakhova I. K.; Wengel J. Scaffolding along nucleic acid duplexes using 2′-amino-locked nucleic acids. Acc. Chem. Res. 2014, 47, 1768–1777. 10.1021/ar500014g. [DOI] [PubMed] [Google Scholar]
- Sinkeldam R. W.; Greco N. J.; Tor Y. Fluorescent analogs of biomolecular building blocks: design, properties, and applications. Chem. Rev. 2010, 110, 2579–2619. 10.1021/cr900301e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matarazzo A.; Hudson R. H. E. Fluorescent adenosine analogs: a comprehensive survey. Tetrahedron 2015, 71, 1627–1657. 10.1016/j.tet.2014.12.066. [DOI] [Google Scholar]
- Schmidt O. P.; Mata G.; Luedtke N. W. Fluorescent base analogue reveals T-HgII-T base pairs have high kinetic stabilities that perturb DNA metabolism. J. Am. Chem. Soc. 2016, 138, 14733–14739. 10.1021/jacs.6b09044. [DOI] [PubMed] [Google Scholar]
- Burns D. D.; Teppang K. L.; Lee R. W.; Lokensgard M. E.; Purse B. W. Fluorescence turn-on sensing of DNA duplex formation by a tricyclic cytidine analogue. J. Am. Chem. Soc. 2017, 139, 1372–1375. 10.1021/jacs.6b10410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manderville R. A.; Wetmore S. D. C-Linked 8-aryl guanine nucleobase adducts: biological outcomes and utility as fluorescent probes. Chem. Sci. 2016, 7, 3482–3493. 10.1039/C6SC00053C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sproviero M.; Fadock K. L.; Witham A. A.; Manderville R. A.; Sharma P.; Wetmore S. D. Electronic tuning of fluorescent 8-aryl-guanine probes for monitoring DNA duplex–quadruplex exchange. Chem. Sci. 2014, 5, 788–796. 10.1039/C3SC52625A. [DOI] [Google Scholar]
- Sproviero M.; Manderville R. A. Harnessing G-tetrad scaffolds within G-quadruplex forming aptamers for fluorescence detection strategies. Chem. Commun. 2014, 50, 3097–3099. 10.1039/C3CC49560D. [DOI] [PubMed] [Google Scholar]
- Sproviero M.; Fadock K. L.; Witham A. A.; Manderville R. A. Positional impact of fluorescently modified G-tetrads within polymorphic human telomeric G-quadruplex structures. ACS Chem. Biol. 2015, 10, 1311–1318. 10.1021/cb5009926. [DOI] [PubMed] [Google Scholar]
- Blanchard D. J. M.; Cservenyi T. Z.; Manderville R. A. Dual fluorescent deoxyguanosine mimics for FRET detection of G-quadruplex folding. Chem. Commun. 2015, 51, 16829–16831. 10.1039/C5CC07154B. [DOI] [PubMed] [Google Scholar]
- Fadock K. L.; Manderville R. A.; Sharma P.; Wetmore S. D. Optimization of fluorescent 8-heteroaryl-guanine probes for monitoring protein-mediated duplex→G-quadruplex exchange. Org. Biomol. Chem. 2016, 14, 4409–4419. 10.1039/C6OB00474A. [DOI] [PubMed] [Google Scholar]
- Dumas A.; Luedtke N. W. Cation-mediated energy transfer in G-quadruplexes revealed by an internal fluorescent probe. J. Am. Chem. Soc. 2010, 132, 18004–18007. 10.1021/ja1079578. [DOI] [PubMed] [Google Scholar]
- Dumas A.; Luedtke N. W. Highly fluorescent guanosine mimics for folding and energy transfer studies. Nucleic Acids Res. 2011, 39, 6825–6834. 10.1093/nar/gkr281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock L. C.; Griffin L. C.; Latham J. A.; Vermaas E. H.; Toole J. J. Selection of single-stranded DNA molecules that bind and inhibit human thrombin. Nature 1992, 355, 564–566. 10.1038/355564a0. [DOI] [PubMed] [Google Scholar]
- Krauss I. R.; Merlino A.; Randazzo A.; Novellino E.; Mazzarella L.; Sica F. High-resolution structures of two complexes between thrombin and thrombin-binding aptamer shed light on the role of cations in the aptamer inhibitory activity. Nucleic Acids Res. 2012, 40, 8119–8128. 10.1093/nar/gks512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rankin K. M.; Sproviero M.; Rankin K.; Sharma P.; Wetmore S. D.; Manderville R. A. C8-Heteroaryl-2′-deoxyguanosine adducts as conformational fluorescent probes in the NarI recognition sequence. J. Org. Chem. 2012, 77, 10498–10508. 10.1021/jo302164c. [DOI] [PubMed] [Google Scholar]
- Cruz-Aguado J. A.; Penner G. Determination of ochratoxin A with a DNA aptamer. J. Agric. Food Chem. 2008, 56, 10456–10461. 10.1021/jf801957h. [DOI] [PubMed] [Google Scholar]
- Barthelmebs L.; Jonca J.; Hayat A.; Prieto-Simon B.; Marty J.-L. Enzyme-linked aptamer assays (ELAAs), based on a competition format for a rapid and sensitive detection of ochratoxin A in wine. Food Control 2011, 22, 737–743. 10.1016/j.foodcont.2010.11.005. [DOI] [Google Scholar]
- McKeague M.; Velu R.; Hill K.; Bardóczy V.; Mészáros T.; DeRosa M. C. Selection and characterization of a novel DNA aptamer for Label-free fluorescence biosensing of ochratoxin A. Toxins 2014, 6, 2435–2452. 10.3390/toxins6082435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohland A. E.; Nesheim S.; Friedman L. Ochratoxin A: A review. Pure Appl. Chem. 1992, 64, 1029–1046. 10.1351/pac199264071029. [DOI] [Google Scholar]
- Pfohl-Leszkowicz A.; Manderville R. A. Ochratoxin A: An overview on toxicity and carcinogenicity in animals and humans. Mol. Nutr. Food Res. 2007, 51, 61–99. 10.1002/mnfr.200600137. [DOI] [PubMed] [Google Scholar]
- Ha T. H. Recent advances for the detection of ochratoxin A. Toxins 2015, 7, 5276–5300. 10.3390/toxins7124882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieto-Simón B.; Campàs M.; Marty J.-L.; Noguer T. Novel highly-performing immunosensor-based strategy for ochratoxin A detection in wine samples. Biosens. Bioelectron. 2008, 23, 995–1002. 10.1016/j.bios.2007.10.002. [DOI] [PubMed] [Google Scholar]
- Liu B.-H.; Tsao Z.-J.; Wang J.-J.; Yu F.-Y. Development of a monoclonal antibody against ochratoxin A and its application in enzyme-linked immunosorbent assay and gold nanoparticle immunochromatographic strip. Anal. Chem. 2008, 80, 7029–7035. 10.1021/ac800951p. [DOI] [PubMed] [Google Scholar]
- Li T.; Jeon K.-S.; Suh Y. D.; Kim M.-G. A label-free, direct and noncompetitive FRET immunoassay for ochratoxin A based on intrinsic fluorescence of an antigen and antibody complex. Chem. Commun. 2011, 47, 9098–9100. 10.1039/c1cc12604k. [DOI] [PubMed] [Google Scholar]
- Yang C.; Wang Y.; Marty J.-L.; Yang X. Aptamer-based colorimetric biosensing of ochratoxin A using unmodified gold nanoparticles indicator. Biosens. Bioelectron. 2011, 26, 2724–2727. 10.1016/j.bios.2010.09.032. [DOI] [PubMed] [Google Scholar]
- Lv Z.; Chen A.; Liu J.; Guan Z.; Zhou Y.; Xu S.; Yang S.; Li C. A simple and sensitive approach for ochratoxin A detection using a label-free fluorescent aptasensor. PLoS One 2014, 9, e85968 10.1371/journal.pone.0085968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S.; Zhang Y.; Pang G.; Zhang Y.; Guo S. Tuning the aggregation/disaggregation behavior of graphene quantum dots by structure-switching aptamer for high-sensitivity fluorescent ochratoxin A sensor. Anal. Chem. 2017, 89, 1704–1709. 10.1021/acs.analchem.6b03913. [DOI] [PubMed] [Google Scholar]
- McKeague M.; De Girolamo A.; Valenzano S.; Pascale M.; Ruscito A.; Velu R.; Frost N. R.; Hill K.; Smith M.; McConnell E. M.; DeRosa M. C. Comprehensive analytical comparison of strategies used for small molecule aptamer evaluation. Anal. Chem. 2015, 87, 8608–8612. 10.1021/acs.analchem.5b02102. [DOI] [PubMed] [Google Scholar]
- Kikin O.; D’Antonio L.; Bagga P. S. QGRS Mapper: a web-based server for predicting G-quadruplexes in nucleotide sequences. Nucleic Acids Res. 2006, 34, W676–W682. 10.1093/nar/gkl253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaulage M.; Maji B.; Bhat J.; Iwasaki Y.; Chatterjee S.; Bhattacharya S.; Muniyappa K. Discovery and structural characterization of G-quadruplex DNA in human acetyl-CoA carboxylase gene promoters: Its role in transcriptional regulation and as a therapeutic target for human disease. J. Med. Chem. 2016, 59, 5035–5050. 10.1021/acs.jmedchem.6b00453. [DOI] [PubMed] [Google Scholar]
- Frenette C.; Paugh R. J.; Tozlovanu M.; Juzio M.; Pfohl-Leszkowicz A.; Manderville R. A. Structure–activity relationships for the fluorescence of ochratoxin A: Insight for detection of ochratoxin A metabolites. Anal. Chim. Acta 2008, 617, 153–161. 10.1016/j.aca.2007.12.030. [DOI] [PubMed] [Google Scholar]
- da Silva M. W. Geometric formalism for DNA quadruplex folding. Chem. – Eur. J. 2007, 13, 9738–9745. 10.1002/chem.200701255. [DOI] [PubMed] [Google Scholar]
- Karsisiotis A. I.; Hessari N. M.; Novellino E.; Spada G. P.; Randazzo A.; da Silva M. W. Topological characteriaztion of nucleic acid G-quadruplexes by UV absorption and circular dichroism. Angew. Chem., Int. Ed. 2011, 50, 10645–10648. 10.1002/anie.201105193. [DOI] [PubMed] [Google Scholar]
- Amrane S.; Ang R. W. L.; Tan Z. M.; Li C.; Lim J. K. C.; Lim J. M. W.; Phan A. T.; Lim K. W. A novel chair-type G-quadruplex formed by a Bombyx mori telomeric sequence. Nucleic Acids Res. 2009, 37, 931–938. 10.1093/nar/gkn990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Merwe K. J.; Steyn P. S.; Fourie L.; Scott D. B.; Theron J. J. Ochratoxin A, a toxic metabolite produced by Aspergillus ochraceus Wilh.. Nature 1965, 205, 1112–1113. 10.1038/2051112a0. [DOI] [PubMed] [Google Scholar]
- Steyn P. S.; Holzapfel C. W. The synthesis of ochratoxins A and B: Metabolites of Aspergillus ochraceus Wilh.. Tetrahedron 1967, 23, 4449–4461. 10.1016/S0040-4020(01)88843-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.