Abstract
Cadmium, a heavy metal pollutant, causes cancer. The existence of cancer stem cells (CSCs) in tumors is widely considered to be the reason for the recurrence and treatment failure of cancer. Increasing evidence has confirmed that under certain conditions non-CSCs could be converted into CSCs. The impact of cadmium on the development of CSC lineage in the bulk tumor cell population is not yet studied. The aim of this study was to evaluate the effect of cadmium on the conversion of non-CSCs to CSCs and the identification of CSCs based on the concurrent monitoring of multiple CSC markers. High-content monitoring of molecular markers was performed using quantum dot (QD) nanoprobes and an acousto-optical tunable filter (AOTF)-based imaging device. Cadmium treatment significantly increased the CSC population in MCF-7 and HepG2 cell lines. The cadmium-induced CSCs were identified by a concurrent analysis of stem-cell markers, namely, CD44, CD24, CD133, and ALDH1. Moreover, increased m-RNA expression of CD44, ALDH1, and CD133 and protein expression of p-Ras, p-Raf-1, p-MEK-1, and p-ERK-1 were observed in the cadmium-treated MCF-7 and HepG2 cells. This study demonstrates that cadmium induces the gene expression of CSC markers in the breast and liver cancer cell lineage and promotes the conversion of non-CSCs to CSCs.
Introduction
Cancer is the second major cause of death worldwide. Several environmental reports indicated that the incidence of cancer increased in proportion to the levels of environmental pollutants.1,2 Heavy metal pollutants have been reported to inflict a wide array of health risks, including cancer, on the human population. Cadmium is one of the major heavy metal pollutants, and it is widely used in the metal industry, paint industry, and plastic industry and in the preparation of rechargeable nickel–cadmium batteries. Improper disposal of heavy metals is a major concern because they cannot be biodegraded and can accumulate in living organisms existing in the food web.3 Many global health reports indicated that continuous exposure to cadmium poses a cancer risk to the human population. Industrial emissions and effluents of a lead–zinc mine are the major source of cadmium contamination. Cigarette smoking is a major exposure to cadmium. The cadmium content in the tobacco ranges between 1 and 2 μg/g dry weight, and the average cadmium content per cigarette ranges between 0.5 and 1 μg.4 It has been reported that blood cadmium levels in smokers are generally twice those of nonsmokers.5 Apart from cigarette smoking, another major exposure to cadmium is the consumption of cadmium-contaminated water and food. Because of its poor metabolic excretion and long half-life (15–30 years), cadmium generally accumulates in the liver and kidney and causes liver, prostate, and lung cancer. Cadmium and its compounds are currently classified by International Agency for Research as a group 1 carcinogen for humans.
Despite the advances in chemotherapy, radiotherapy, and monoclonal antibody therapy in cancer treatment, the occurrence of treatment failure is still a major concern. The inherent drug-resistance mechanism of cancer reduces the survival chances of patients.6 One of the well-proven and accepted hypotheses for the treatment failure is the existence of cancer stem cells (CSCs) in tumor population. CSCs are pluripotent cells, which exhibit a high level of drug resistance, metastatic, and self-renewal capabilities as compared with normal cancer cells.7 Targeted therapies against CSCs still remain a challenge. Conventional therapies can effectively eradicate the rapidly proliferating cancer cell in tumor but leaves the drug-resistant CSCs; the latter has the ability to generate a pool of drug-resistant proliferating cells. Hence, a rapid identification and targeted therapy against CSCs are required to effectively treat cancer, but marker identification still poses a challenge.
Even though several studies reported the carcinogenicity of cadmium, till date, no studies have reported the impact of cadmium on the CSC marker expression. The present study addressed the role of cadmium in the generation of CSCs in the tumor cell population. CSCs are generally identified based on the expression of a unique set of markers; till date, simultaneous identification of multiple markers with greater accuracy remains a challenge. At present, serum marker analysis and diagnostic enzyme analysis are widely used for the cancer diagnosis. However, serum marker-based cancer diagnosis provides false positive results.8 In addition, serum marker diagnosis cannot provide details of the phenotype of cancer cells and CSCs. Even though techniques such as immunohistochemistry and magnetic resonance imaging enable the detection of CSCs,9 the success of these techniques mostly relied on the expertise of the physician. Deciphering the phenotype of CSCs is highly helpful for the prognosis of cancer.10 However, successful identification of the CSC phenotype depends on the accurate determination of positiveness or negativeness of many biomarkers. Quantum dot (QD)-based concurrent detection of multiple tumor markers is a suitable option for the analysis of CSCs. QD exhibits broad excitation and narrow emission spectra, which enables the concurrent monitoring of multiple QDs through excitation at a single wavelength. In addition, QDs possess strong photostability.11 These excellent imaging properties of QDs are more suitable for multiplex imaging of CSC markers. Moreover, the acousto-optical tunable filter (AOTF) used in the imaging device is an electronically tunable filter and facilitates the scanning of QD-conjugated nanoprobes at a single wavelength, which avoids the spectral overlap among the QDs. In this work, cadmium-induced breast and hepatocellular CSCs were reported by concurrent determination of molecular markers such as CD133, CD44, CD24, and aldehyde dehydrogenase1 (ALDH1). An AOTF and QD nanoprobe-based hypermulticolor cellular imaging system was used for the identification of molecular markers. In addition, the present high-content cellular assay devoid of cell lysis may provide new insights into a mechanistic study on environmental carcinogen-induced CSC formation, including cadmium dealt with in this work. The correlation among proteins involved in signal transduction can be expected to be observed at the single cell level without interruption arising from cell lysis. In this work, the role of Ras/Raf/MEK/ERK signaling cascade in the cadmium-induced CSC markers was also analyzed.
Results and Discussion
CSC Markers of Breast and Hepatic CSCs
CSCs are considered to be responsible for the generation of highly proliferative bulk tumor cell population.12 However, recently, the differentiation of pancreatic cancer cells into CSCs has been reported.13 Mutation, epigenetic changes, and tumor microenvironment-induced alteration in the metabolism and regulatory pathway are key factors for the generation of CSCs in the tumor.14,15 Environmental pollutants could be one of the causative factors for the generation of the CSCs. Earlier, Chang et al. reported that sublethal stress induced by arsenic reprogrammed the human bronchial epithelial cells to CD61– CSCs.16,17 Similarly, benzopyrene has been reported to enhance the stemness property in MCF-7 cells.18,19 This indicates a possible role of carcinogens in the expression of stemness genes in cancer. In the present study, the effect of cadmium on the generation of CSCs among bulk tumor cells of MCF-7 and HepG2 cells was studied. A schematic illustration of the present study is represented in Figure 1. Identification of CSCs based on the markers is a challenging task because the confirmation of CSCs requires an analysis of expression of multiple stem-cell markers. In breast cancer, CD44 and CD24 are the major stem-cell markers. It has been reported that CD44+ CD24–/low CSC population can induce palpable tumors in nonobese diabetic/severe combined immune deficiency mice with a 100-fold efficacy than CD44+ CD24+ cancer cells.20 Recently, ALDH1 has also been reported as an important breast CSC marker. It has been evidenced that ALDH1+ CD44+ CD24–/low cells were highly tumorigenic than ALDH1– CD44+ CD24–/low cells.21 On the basis of the previous reports, cells with the phenotype of ALDH1+ CD44+ CD24–/low are considered to be breast CSCs. Hepatic CSCs can be identified with a number of markers, namely, CD44, CD24, CD133, CD90, CD13, ALDH1, and epithelial cell adhesion molecule (EpCAM).22 CD133 was reported to be associated with invasive features of breast, liver, brain, and lung cancer.23 CD44 has been proven, in many types of cancer, to facilitate cell migration and promotes metastasis process.24 ALDH1 expression has been reported to enrich the stamens property that provides chemo- and radioresistance characteristics to CSCs.25,26 On the basis of the previous reports, HepG2 cells expressing CD133, CD44, and ALDH1 are considered to be hepatic CSCs.
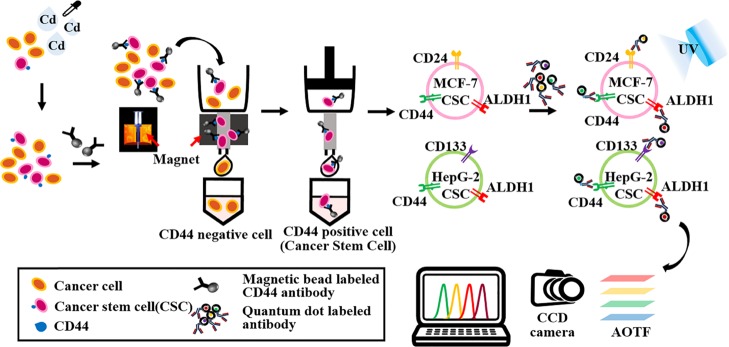
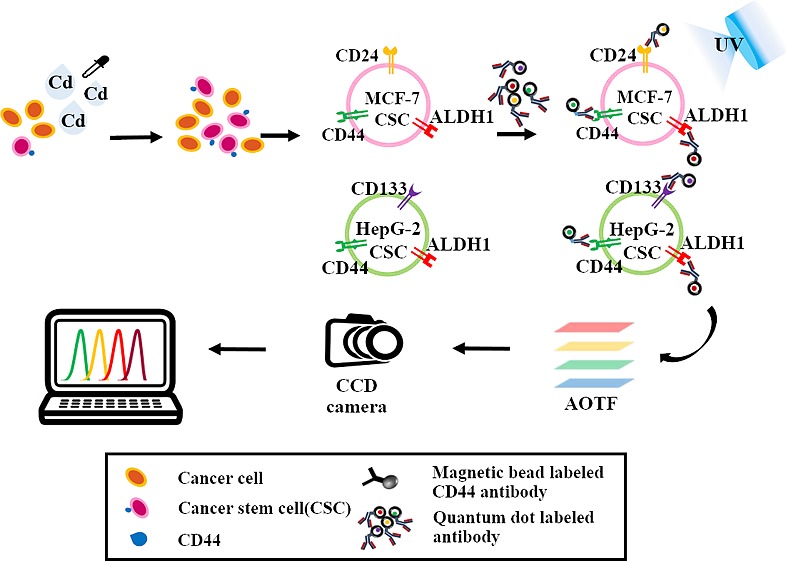
Figure 1.
Schematic illustration of the monitoring of cadmium-induced CSCs based on the concurrent identification of multiple CSC markers using QD and AOTF-based cellular imaging. MCF-7 and HepG2 cells were treated with cadmium for 72 h, followed by the separation of CSCs using a magnetic sorter. Then, hepatic CSCs were identified based on the expression of QD-conjugated CD133, CD44, and ALDH1, and breast CSCs were identified based on the expression of CD44, CD24, and ALDH1. The AOTF scans the cellular images at a single wavelength emitted from the QD-based nanoprobes without spectral overlap.
High-Content Monitoring of CSC Population in Cultures Exposed to Cadmium Using AOTF-Based Cellular Imaging
At present, conventional methods such as western blot, immunohistochemistry, immunofluorescence, and flow cytometry are used for the identification of stem-cell markers. However, each of these techniques has its own merits and demerits. Western blot involves cell lysis, separation of proteins in sodium dodecyl sulfate (SDS) gel, transfer of proteins into a membrane, and a detection process; all of these processes need high technical expertise, including proper maintenance of pH and temperature, buffer preparation, and homogenization.27 Dye-labeled antibodies used in immunohistochemistry and immunofluorescence analyses for the detection of target proteins also have their own limitations. Most of the dyes exhibit broad emission spectra,28 which considerably limits the concurrent probing of different CSC markers. Simultaneous monitoring of multiple markers existing in a single cell is highly advantageous because the stemness property of CSCs is determined by the combination of the presence or absence of many markers. Moreover, it has to be considered that the tumor population is not a phenotypically homogeneous population.29 Even though fluorescence-activated cell sorting is a widely used tool to monitor the molecular markers, concurrent analysis of multiple CSC markers was hardly reported. In this work, for the successful identification of multiple markers, QD-tagged antibodies and AOTF-based cellular imaging was used. QDs have narrow spectral ranges; hence, multiple QDs can be excited at a single wavelength. This phenomenon enables the use of multiple QDs to obtain multicolor imaging for simultaneous monitoring of multiple markers.30 QDs exhibit multifold brightness, photostability, and photobleaching thresholds as compared with the conventional dyes.31 Moreover, the AOTF filter used in the fluorescence microscopic detection system is an electronically tunable spectral bandpass filter. AOTF can produce beams at a single wavelength, and this property enables the detection of multiple CSC markers without overlapping their emission wavelength among each other. Hence, the combination of AOTF and QD-based constructed detection system can provide the facility to concurrently monitor more than four CSC markers at a single cell level.
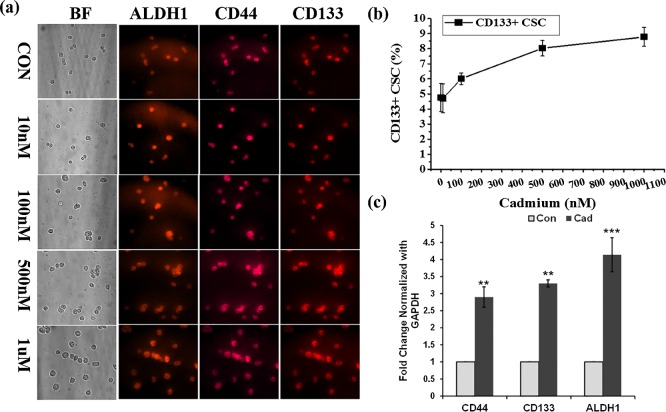
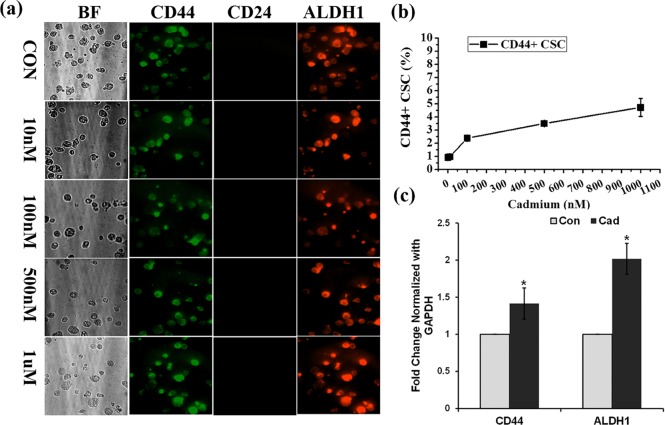
In the present study, HepG2 cells were treated with or without different concentrations of cadmium for 72 h and then CSCs were isolated using CD133 microbeads. To further confirm the sorted-out cells as CSCs, the CD133 positive cells were immunostained with a mixture of QD-conjugated CD133, CD44, and ALDH1 antibodies. As shown in Figure 2a, all of the isolated CD133 positive hepatic CSCs were positive for both CD44 and ALDH1, which correlated well with the phenotype of hepatocellular CSCs. Before the magnetic sorting of CSCs, the total number of live HepG2 cells was counted using a hemocytometer. Similarly, the total number of CD133 positive cells obtained via the sorting was counted (Table 1). The line graph in Figure 2b represents the percentage of CSCs obtained from HepG2 cells after treatment with cadmium: 4.76, 4.71, 6, 8.03, and 8.78% of CSCs were observed at 0 nM, 10 nM, 100 nM, 500 nM, and 1 μM cadmium, respectively. As compared with the control, cadmium-induced CSCs were determined to be 1.24, 3.27, and 4.01%, respectively, at 100 nM, 500 nM, and 1 μM cadmium. A significant change was not observed between the control and 10 nM concentration of cadmium. However, a significant change was observed in cell cultures treated with a high concentration of cadmium. In addition, the gene expression of CSC markers CD133, CD44, and ALDH1 in HepG2 cells was analyzed with the presence and absence of cadmium. Quantitative real-time (RT) polymerase chain reaction (PCR) results displayed in Figure 2c show that cadmium significantly increased the gene expression of CD133, CD44, and ALDH1 (p < 0.001, 0.01, and 0.01, respectively) in HepG2 cell lines. To confirm the breast CSCs, antibodies that are specific for stem-cell markers CD44, CD24, and ALDHI were conjugated with QD525, QD565, and QD605, respectively. All isolated CD44 positive breast CSCs are CD44 and ALDH1 positive and CD24 negative (Figure 3a). A total number of MCF-7 cells used for the breast CSCs were counted using a hemocytometer, and after sorting, isolated CSCs were also counted (Table 2). The percentage of CSCs isolated from the MCF-7 cell culture after treating with cadmium is represented in Figure 3b. The observed CSC percentages were 0.93, 0.97, 2.39, 3.50, and 4.73%, respectively, at 0 nM, 10 nM, 100 nM, 500 nM and 1 μM cadmium. As compared with the control, the percentages of cadmium-induced CSCs were 0.03, 1.42, 2.87, and 3.80%, respectively, at 10 nM, 100 nM, 500 nM, and 1 μM cadmium. To further confirm the cadmium-induced gene expression of CSC markers, a semiquantitative RT-PCR analysis was performed. RT-PCR results displayed in Figure 3c show that cadmium significantly increased the gene expression of CD44 and ALDH1 (p < 0.01) in the breast cancer cell line MCF-7. From the observed results, we can conclude that the CSC population increased in correlation with the concentration of cadmium exposure. Herein, the results observed in the present study clearly indicated that cadmium treatment significantly increased the number of CSC population in the cultures of both breast and hepatic cancer cell lines. The AOTF-based cellular imaging results observed in the present study show that all isolated CSCs from MCF-7 cells were positive for CD44 and ALDH1 and negative for CD24. Similarly, most of the CSCs isolated from the HepG2 cells were positive for CD133, CD44, and ALDH1 markers.
Figure 2.
(a) High-content detection of hepatocellular CSCs in the cadmium-treated HepG2 cell culture. CSC markers CD133, CD44, and ALDH1 were concurrently monitored using the QD-conjugated antibodies. (b) Line graph represents the quantitative determination of hepatocellular CSCs obtained from the HepG2 cell culture in response to the incremental concentrations of cadmium. (c) Semiquantitative RT-PCR analysis of CD133, CD44, and ALDH1 expression in HepG2 cells treated with and without cadmium (1 μM) over the period of 72 h. The values are means ± SD from three independent experiments.
Table 1. Number of CD133 Positive Cells Increased as a Result of Treatment of HepG2 Cells with Cadmium and the Percentage of Hepatic CSCs.
| group | total cell (cell/mL) | CD133 + CSC cell (cell/mL) | CD133 + CSC (%) | average of CSC (%) | cadmium-induced CSC (%) |
|---|---|---|---|---|---|
| 0 nM | 8.4 × 106 | 3.8 × 105 | 4.52 | 4.76 | 0 |
| 4.2 × 106 | 2.44 × 105 | 5.8 | |||
| 6.8 × 106 | 2.7 × 105 | 3.97 | |||
| 10 nM | 7.6 × 106 | 3.3 × 105 | 4.34 | 4.71 | 0 |
| 4.6 × 106 | 2.67 × 105 | 5.8 | |||
| 6.2 × 106 | 2.48 × 105 | 4 | |||
| 100 nM | 8.0 × 106 | 4.8 × 105 | 6 | 6 | 1.24 |
| 5.1 × 106 | 3.26 × 105 | 6.39 | |||
| 6.5 × 106 | 3.65 × 105 | 5.62 | |||
| 500 nM | 9.4 × 106 | 7.6 × 105 | 8.09 | 8.03 | 3.27 |
| 5.0 × 106 | 4.26 × 105 | 8.52 | |||
| 5.9 × 106 | 4.42 × 105 | 7.49 | |||
| 1 μM | 9.0 × 106 | 8.4 × 105 | 9.33 | 8.78 | 4.02 |
| 4.6 × 106 | 4.1 × 105 | 8.9 | |||
| 6.3 × 106 | 5.11 × 105 | 8.11 |
Figure 3.
(a) Breast CSCs obtained from the concurrent monitoring of CD44, CD24, and ALDH1 in the cadmium-treated MCF-7 cell culture. The emission wavelength of QD-conjugated CD44, CD24, and ALDH1 antibodies was measured at 525, 565, and 625 nm, respectively. (b) Quantitative determination of breast CSCs in the MCF-7 cell culture treated at the functional concentration of cadmium. (c) Semiquantitative RT-PCR analysis of CD44, CD24, and ALDH1 expression in MCF-7 cells treated with and without cadmium (1 μM) over the period of 72 h. The values are means ± SD from three independent experiments.
Table 2. Total Number and Percentage of CSCs in the Control and Cadmium-Treated MCF-7 Cell Culture.
| group | total cell (cell/mL) | CD44 + CSC cell (cell/mL) | CD44 + CSC (%) | average of CSC (%) | cadmium-induced CSC (%) |
|---|---|---|---|---|---|
| 0 nM | 9.80 × 106 | 1.00 × 105 | 1.02 | 0.93 | 0 |
| 1.215 × 107 | 1.00 × 105 | 0.82 | |||
| 5.20 × 106 | 0.50 × 105 | 0.96 | |||
| 10 nM | 1.045 × 107 | 1.00 × 105 | 0.96 | 0.97 | 0.04 |
| 1.11 × 107 | 1.00 × 105 | 0.90 | |||
| 9.45 × 106 | 1.00 × 105 | 1.06 | |||
| 100 nM | 8.80 × 106 | 2.00 × 105 | 2.27 | 2.39 | 1.46 |
| 1.095 × 107 | 2.50 × 105 | 2.28 | |||
| 9.55 × 106 | 2.50 × 105 | 2.61 | |||
| 500 nM | 1.185 × 107 | 4.00 × 105 | 3.38 | 3.50 | 2.57 |
| 1.075 × 107 | 3.50 × 105 | 3.26 | |||
| 9.05 × 106 | 3.50 × 105 | 3.87 | |||
| 1 μM | 4.90 × 106 | 2.00 × 105 | 4.08 | 4.73 | 3.8 |
| 5.30 × 106 | 3.00 × 105 | 5.66 | |||
| 5.60 × 106 | 2.50 × 105 | 4.46 |
Flow Cytometric Analysis of Breast and Hepatic CSCs
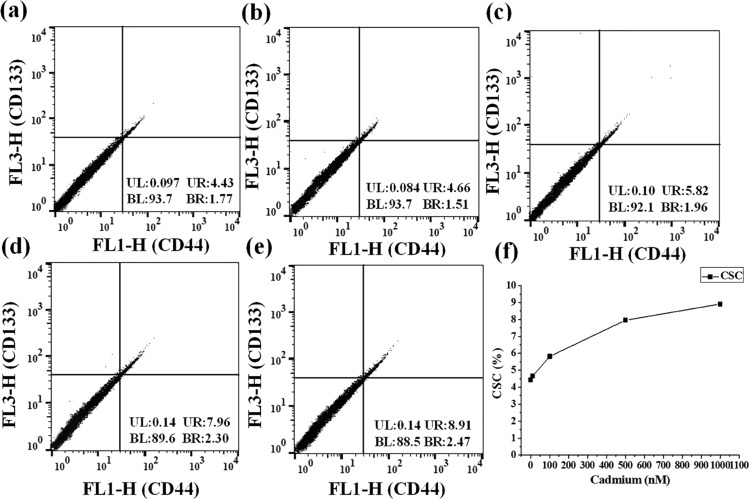
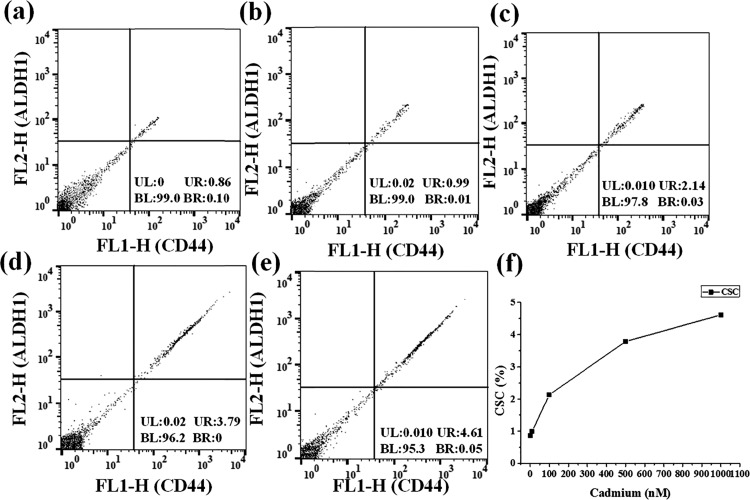
The results observed in the multicolor cellular imaging analysis were verified using flow cytometry. The flow cytometry method was widely accepted for the detection of cellular markers. The CD44 and CD133 markers were used for the identification of hepatic CSCs. Similarly, CD44 and ALDH1 were used to monitor the breast CSC population existing in the MCF-7 cell line. In Figure 4a–e, the x-axis represents the fluorescence emission of CD44 and the y-axis represents the fluorescence emission of CD133. Fluorescence observed near the x-axis represents CD44 positive cells. Similarly, fluorescence observed near the y-axis represents CD133 positive cells. Cell population observed in the upper-right section in the flow-cytometry results corresponds to CSCs, expressing both CD44 and CD133 markers. A total of 30 000 HepG2 cells were used for the flow cytometric analysis. The percentage of hepatocellular CSCs was found to be 4.43, 4.66, 5.82, 7.96, and 8.91%, respectively, at 0 nM, 10 nM, 100 nM, 500 nM, and 1 μM cadmium (Figure 4a–e). These values were plotted as a line graph in Figure 4f. Breast CSCs were also analyzed using FACS, and the fluorescence emission of CD44 and ALDH1 was plotted on the x-axis and y-axis, respectively. As shown in Figure 5a–e, the upper-right section corresponds to the population of breast CSCs, expressing both CD44 and ALDH1 markers. Flow cytometric measurements were recorded for a total of 10 000 MCF-7 cells. The percentages of CSCs found at 0 nM, 10 nM, 100 nM, 500 nM, and 1 μM cadmium treatment were 0.86, 0.99, 2.14, 3.79, and 4.61%, respectively. These values were plotted as a line graph in Figure 5f.
Figure 4.
(a–e) Detection of hepatocellular CSCs in the cadmium-treated HepG2 cell culture based on the expression of CD133 and CD44 markers using a flow cytometer. The upper-right section corresponds to the hepatocellular CSCs, expressing both CD133 and CD44. (f) Flow cytometry-based quantitative determination of hepatocellular CSCs obtained from the HepG2 cell culture treated at the functional concentration of cadmium.
Figure 5.
(a–e) FACS analysis for the breast CSC population induced by cadmium in the MCF-7 cell culture. The x-axis represents CD44 emission, and the y-axis represents ALDH1 emission. For the x-axis, the right side indicates CD44 positive; for the y-axis, the upper side indicates ALDH1 positive. (f) Quantitative determination of breast CSCs obtained using flow cytometry.
High-Content Immunofluorescence and Western Blot Analysis of Ras Signaling Cascade in Cadmium-Treated MCF-7 and HepG2 Cell Lines
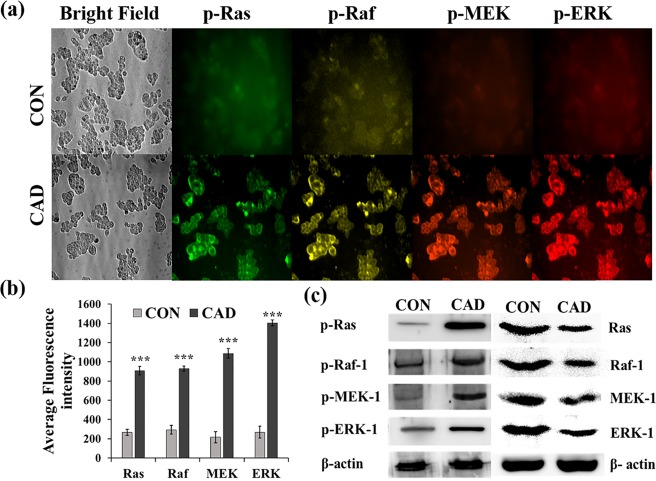
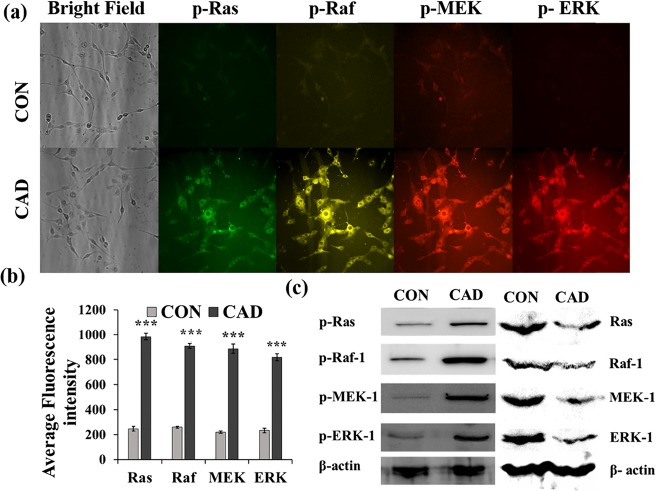
Tumor initiation is a complex multistep process, which causes mutation in a number of genes that are involved in cell regulation and proliferation. It has been reported that a 6 h exposure of cadmium to the HepG2 cells results in a vast change in the gene expression pattern.32,33 Epigenetic modifications are one of the major causes of mutation, which involves acetylation of histones and methylation of genes, and these modifications can alter gene expression. Cartularo et al., reported that 1.0 μM cadmium chloride for 24 h or 0.1 μM cadmium chloride for 3 weeks in HepG2 cells results in the methylation of a number of genes that are involved in cell regulation.34 Earlier, it has been reported that cadmium uptake in HepG2 cells significantly increased during the 24 h exposure period and mostly accumulated in cytoplasm, nuclei, and mitochondria.35 The present study investigated the impact of cadmium on the activation of Ras/Raf/MEK/ERK signaling cascade and its role in the gene expression of CSC markers. The expression pattern of p-Ras, p-Raf, p-MEK, and p-ERK at a single cell level was simultaneously analyzed using AOTF and respective QD-conjugated antibodies. Antip-Ras, p-Raf, p-MEK, and p-ERK antibodies were tagged with QD525, QD565, QD605, and QD655, respectively. The MCF-7 and HepG2 cells were treated with 1 μM cadmium for 72 h, and the fluorescence intensities emitted from probes specific for p-Ras, p-Raf, p-MEK, and p-ERK were measured at 525, 565, 605, and 655 nm, respectively. The representative images are depicted in Figures 6a and 7a. The observed fluorescence intensities were quantitatively evaluated using the Metamorph software and are represented in Figures 6b and 7b. The observed results show that the active forms of p-Ras, p-Raf, p-MEK, and p-ERK were significantly (p < 0.001) increased in the cadmium-treated MCF-7 and HepG2 cells as compared with the control cells. Furthermore, to confirm the immunofluorescence analysis, western blot analysis was carried out; the obtained results show that the protein expressions of p-Ras, p-Raf, p-MEK, and p-ERK were increased as compared with the control (Figures 6c and 7c).
Figure 6.
(a) Effect of cadmium on the levels of phosphorylated Ras, Raf-1, MEK-1, and ERK-1 in HepG2 cells was concurrently monitored over a period of 72 h by high-content immunofluorescence analysis. (b) Bar graph represents the quantitative expression of phosphorylated Ras, Raf-1, MEK-1, and ERK-1 in HepG2 cells based on the average fluorescence intensity observed in the microscopic image. (c) Representative western blot image shows the expression of phosphorylated and unphosphorylated Ras, Raf-1, MEK-1, and ERK-1 in HepG2 cells with the presence and absence of cadmium. The values are means ± SD from three independent experiments.
Figure 7.
(a) High-content monitoring of phosphorylated Ras, Raf-1, MEK-1, and ERK-1 in MCF-7 cells treated with cadmium over a period of 72 h. The fluorescence intensities emitted from QD probes specific for p-Ras, p-Raf-1, p-MEK-1, and p-ERK-1 were measured at 525, 565, 605, and 655 nm, respectively. (b) Bar graph represents the quantitative expression of phosphorylated Ras, Raf-1, MEK-1, and ERK-1 in MCF-7 cells based on the average fluorescence intensity observed in the microscopic image. Metamorph software was used for the image analysis. (c) Representative western blot image shows the expression of phosphorylated and unphosphorylated Ras, Raf-1, MEK-1, and ERK-1 in MCF-7 cells in the presence and absence of cadmium. Values are means ± SD from three independent experiments.
Ras/Raf/MEK/ERK signaling cascade is mainly attributed to the cell survival and proliferation of cells. Ras is the upstream regulator of this pathway, and several studies reported that the constitutive activation of Ras due to mutation results in cancer.36 Ras signaling cascade was reported in the prevention of apoptosis.37 Activation of the MAPK pathway is closely related to the progression of metastasis of cancer. Extensive preclinical data support that MEK inhibitor could be a potential anticancer agent to treat human cancer.38 The inhibitory potential of MEK inhibitor against apoptosis and cell proliferation was reported in the HepG2 cell culture.39 The overexpression of Raf has been considered to be a prognostic marker for the recurrence of hepatocellular carcinoma.40 Estrogen receptor-mediated Ras activation by cadmium has been reported in the MCF-7 cell line.41 Interaction of cadmium with a G-protein-coupled estrogen receptor and the activation of MAPK/ERK signaling cascade were reported in human lung adenocarcinoma cells.42 In the present study, elevated levels of phosphorylated Raf, MEK, and ERK were observed in MCF-7 and HepG2 cells in response to the treatment of cadmium. High-content multicolor imaging used in the current study monitored the concurrent expression of MAPK components at the single cell level; it clearly implicates the cadmium-induced activation of MAPK pathway, and the study was further confirmed using western blot analysis. Recent reports implicated that an overexpression of various components of Ras pathway in CD44 positive cells,43 vice versa the inhibition of Ras signaling, has been reported in the suppression of the stemness property of CSCs.44
Ras Inhibitor Salirasib Suppresses the Cadmium-Induced Gene Expression of CD44, CD133, and ALDH1
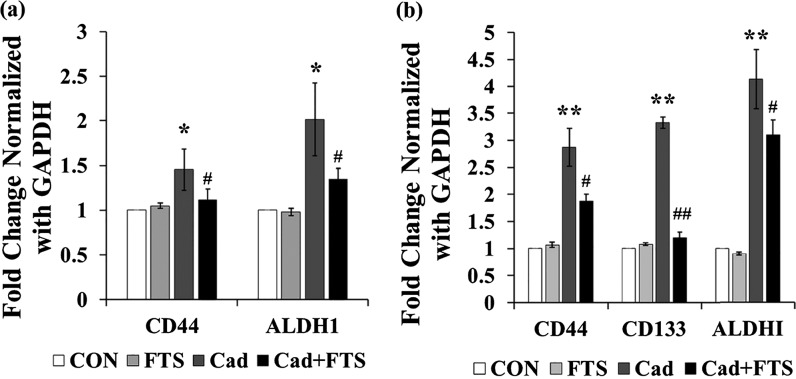
To confirm the possible involvement of Ras signaling cascade in the cadmium-induced CSC marker expressions, HepG2 and MCF-7 cells were treated with 25 μM Ras inhibitor salirasib (farnesylthiosalicylic acid; Sigma-Aldrich, USA) and 1 μM cadmium for 72 h. As shown in Figure 8, the gene expressions of CD44, CD133, and ALDH1 were not significantly changed in MCF-7 and HepG2 cell lines treated with salirasib alone as compared with the control. However, in HepG2 cells, salirasib cotreatment with cadmium significantly suppressed the cadmium-induced gene expression of CD44, CD133, and ALDH1 (p < 0.05, 0.01, and 0.5, respectively). Similarly, salirasib co-treatment with cadmium significantly suppressed the cadmium-induced gene expression of CD44 and ALDH1 (p < 0.05) in MCF-7 cells. The results of the present study show that elevated activation of Ras signaling cascade in response to the cadmium-induced stress plays a partial role in the expression of stemness genes in MCF-7 and HepG2 cells.
Figure 8.
Gene expression of CSC markers in HepG2 cells and MCF-7 cells treated with 25 μM Ras inhibitor and 1 μM cadmium over the period of 72 h. The mRNA expression was analyzed by RT-PCR. (a) mRNA expression of CD44 and ALDH1 in MCF-7 cells. (b) mRNA expression of CD44, CD133, and ALDH1 in HepG2 cells. Values are means ± SD from three independent experiments.
From the results observed in this study, it can be concluded that the CSC population was significantly increased in MCF-7 and HepG2 cell lines in response to the cadmium treatment. In addition, cadmium stimulates the gene expression of CSC markers CD44 and ALDH1 in MCF-7 cells and CD44, CD133, and ALDH1 in HepG2 cells through Ras signaling cascade.
Materials and Methods
Cell Culture and Cadmium Treatment
Breast cancer cell line MCF-7 and hepatic cell line HepG2 were purchased from the Korean Cell Line Bank. Both cell lines were cultured in Dulbecco’s modified Eagle’s medium (Invitrogen, 11995-073) supplemented with 10% heat-inactivated fetal bovine serum (16000-044, Gibco), at 37 °C under a 5% CO2 atmosphere. The cells were treated with different concentrations of cadmium, ranging from 10 nM to 1 μM (Sigma-Aldrich, 356107) for 72 h at 37 °C under 5% CO2.
Microbead-Based CSC Sorting from MCF-7 and HepG2 Cell Lines
The molecular marker-based magnetic cell sorting technique was used to isolate the CSCs. Magnetic beads conjugated with anti-CD44 antibody were used to separate the breast CSCs from MCF-7 cell lines. Similarly, magnetic beads conjugated with anti-CD133 were used to separate the hepatic CSCs from HepG2 cell lines. Briefly, after treating with cadmium for 72 h, the cell cultures were washed twice with 1× PBS and incubated with Accutase solution (Invitrogen, Carlsbad, CA, USA) for 20 min for the detachment of cells from the culture plate. The detached cells were centrifuged briefly at 1250 rcf for 5 min. Then, the cell pellet was briefly resuspended in sorting buffer and then centrifuged at 300g for 10 min. The collected MCF-7 and HepG2 cells were incubated with 20 μL of CD44 and 20 μL of CD133 microbead solution, respectively, for 15 min at 4 °C (CD44 microbead kit human and CD133 microbead kit human, MACS Miltenyi Biotec). The cells were then resuspended in 1 mL of sorting buffer and centrifuged at 300g for 10 min. The obtained cell pellet was resuspended in 500 μL of sorting buffer. The LS column provided in the kit was (130-042-401; MACS Miltenyi Biotec) fixed in the magnetic field of MACS separator, and then preseparation filter (130-041-407; MACS Miltenyi Biotec) was kept on the LS column. The filter and column setup was rinsed thoroughly using sorting buffer, and then the resuspended cells were applied on the preseparation filter. CD44/CD133 negative cells were eluted using sorting buffer. To elute the attached CD44 positive and CD133 positive cells from the column, the column was kept on a collecting tube and rinsed thoroughly using 5 mL of sorting buffer. The collected CD44 positive fraction of MCF-7 cells, and the CD133 positive fraction of HepG2 cells were used for further analysis.
Preparation of Antibody–QD Conjugates for CSC Screening
Antibodies specific for CSC markers CD133 (ab-19898; Abcam, USA), CD44 (sc-65265), CD24 (sc-19585), and ALDH1 (sc-374149) (Santa Cruz biotechnology, Santa Cruz, USA) were conjugated to QD705, QD525, QD565, and QD625, respectively, according to the manufacturer’s instructions (Quantum Dot Conjugation Kit; Invitrogen, Carlsbad, CA, USA). Disulfide bonds in the antibodies were reduced by dithiothreitol and then incubated with maleimide-functionalized QD for 1 h at room temperature. The maleimide group in the QD–antibody conjugates was removed by treating with 2-mercaptoethanol. The unconjugated QDs were eluted using a column provided in the kit. The QD–antibody conjugate solution was diluted at 1:200 with 1% bovine serum albumin. The cancer cells isolated from the magnetic bead-sorting procedure were fixed in 4% formaldehyde for 10 min and then washed twice with 1× PBS. The formaldehyde-fixed cells were centrifuged at 230g for 3 min, and the cell pellet was collected. The collected cell pellet was treated with 0.2% saponin for 10 min at room temperature and then washed twice with 1× PBS. The cells processed using the above-mentioned steps were incubated with the diluted QD–antibody conjugates for 2 h at room temperature. The following mixture of antibody–QD conjugates containing QD605–ALDH1, QD655–CD133, and QD705–CD44 was used for the HepG2 cells, and a mixture of antibody–QD conjugates containing QD525–CD44, QD565–CD24, and QD605–ALDH1 was used for the MCF-7 cells. After incubation, the cells were washed twice with 1× PBS and centrifuged at 230g for 3 min. The obtained cell pellet was resuspended again in 50 μL of 1× PBS solution, and 10 μL of cells was placed in the 1.5 μ slide vi for imaging (1.5 μ-slide vi; ibide GmbH, Am Klopferspitz, 82152 Martinsried, Germany). The antibody–QD conjugates QD525–CD44, QD565–CD24, QD605–ALDH1, QD655–CD133, and QD705–CD44 were scanned at 705, 525, 565, and 625 nm, respectively, using an AOTF-enabled detection system.
Flow Cytometry
CD44 and CD133 antibody–QD conjugates were used to detect hepatocellular CSCs. Similarly, CD44 and ALDH1 antibody–QD conjugates were used to detect the CSCs of MCF-7 cell lines. Briefly, the MCF-7 and HepG2 cells were treated with different concentrations of cadmium (10 nM, 100 nM, 500 nM, and 1 μM) for 72 h. Then, the cells were detached from the culture flask using the cell detachment solution Accutase (Thermo Fisher Scientific, USA) for 20 min at 37 °C. The detached cells were treated with 4% formaldehyde and then incubated with 0.2% saponin for 10 min at room temperature. The cells were then treated with QD-conjugated antibodies for 2 h at room temperature. After incubation, the cells were washed with phosphate-bufferedsaline (PBS) solution and centrifuged at 230g for 3 min. The obtained cell pellet was resuspended in 2 mL of 1× PBS solution, and then the cells were subjected to the flow cytometric analysis using an FACS Calibur (FACS Calibur; BD Bioscience).
High-Content Screening of Ras/Raf/MEK/ERK Pathway
For the simultaneous quantitative expression of Ras signaling cascade, antibodies specific for p-Ras (sc-130215), p-Raf-1 (sc-101791), p-MEK-1 (sc-271914), and p-ERK (sc-7383) (Santa Cruz Biotechnology, USA) were conjugated with QD525, QD565, QD625, and QD655, respectively. The MCF-7 and HepG2 cells were treated with 1 μM cadmium for 72 h. After treating with cadmium, the cells were washed twice with 1× PBS and then fixed using 4% formaldehyde for 10 min at room temperature. Then, the cells were washed with PBS and incubated with 0.2% saponin for 10 min at room temperature. After that, the cells were washed thrice with PBS and incubated with QD-conjugated antibodies for 2 h at room temperature. After washing thrice with PBS buffer, the cells were scanned at 525, 565, 625, and 655 nm using an AOTF-based detection system.
Western Blot
The cadmium-induced activation of phosphorylated and unphosphorylated forms of Ras (sc-224), Raf-1 (sc-7267), MEK-1 (sc-6250), and ERK-1 (sc-94) (Santa Cruz Biotechnology, USA) in MCF-7 and HepG2 cells was analyzed using the western blot. The MCF-7 and HepG2 cells were treated with 1 μM cadmium for 72 h. After the cadmium treatment, cell lysates were prepared using radioimmunoprecipitation assay buffer (RIPA Buffer; R0278, Sigma-Aldrich). The protein concentration of cell lysates was quantified using Bradford protein assay (BioRod, USA). Thirty micrograms of homogenized protein samples was separated using 10% SDS-polyacrylamide gel electrophoresis and transferred to the nitrocellulose membrane (Sigma, USA). Then, the membranes were probed with primary antibodies. Reactive proteins were detected using horseradish peroxidase-tagged secondary antibodies (Santa Cruz Biotechnology, USA) and visualized using the enhanced chemiluminescence detection (Amersham). The images were taken using LAS4000 (Image Quant LAS4000; GE Healthcare).
Quantitative RT-PCR
Total RNA from the cadmium-treated MCF-7 and HepG2 cells was extracted, as described by the manufacturer’s protocol (Dynabeads MRNA Purification Kit, Invitrogen, USA). The quality of isolated RNA was assessed by the ratio of absorbance at 260:280 nm and then by 1% agarose RNA gel electrophoresis. The concentration of RNA was measured at 260 nm using a Nanodrop (Thermo Scientific, USA). From the total RNA, cDNAs specific for GAPDH, CD44, CD133, and ALDH1 were prepared using QuantiTect Reverse Transcription Kit (Qiagen, USA), as described by the manufacturer’s protocol. Two picomoles of primers was used for the amplification of target m-RNA. The starting template quantity of the samples was determined using GAPDH expression, and then quantitative PCRs were carried out. The following primers were used: CD44 sense TGCCGCTTTGCAGGTGTATT, antisense CCGATGCTCAGAGCTTTCTCC; Cd133 sense GCATTGGCATCTTCTATGGTT, antisense CGCCTTGTCCTTGGTAGTGT; ALDH1 sense CCTGTCCTACTCACCGATTTG, antisense TCCTCCTTATCTCCTTCTTCTACC; and GAPDH sense CATGAGAAGTATGACAACAGCCT, antisense AGTCCTTCCACGATACCAAAGT. A SYBR green method was adopted to perform the RT-PCR (SYBR green qPCR kit, Thermo Scientific, USA). An RT-PCR instrument (7300, Applied Biosystems) was used to execute the RT-PCR. Data were normalized with GAPDH and analyzed using the 2–ΔΔCT method.
Statistical Analysis
All experiments were performed in triplicates (n = 3). The data were expressed as mean ± standard deviation. One-way analysis of variance with a Tukey–Kramer post-test was used to compare the data between groups. The results obtained in the present study were considered to be statistically significant when p ≤ 0.05.
Acknowledgments
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Ministry of Education, Science and Technology (MEST) (2015R1A2A1A05001842 and 2016R1A4A1010796). We are grateful to the Research Institute of Pharmaceutical Sciences at Seoul National University for providing experimental equipment.
Author Contributions
† H.J. and P.A. contributed equally.
The authors declare no competing financial interest.
References
- Raaschou-Nielsen O.; Beelen R.; Wang M.; Hoek G.; Andersen Z. J.; Hoffmann B.; Stafoggia M.; Samoli E.; Weinmayr G.; Dimakopoulou K. Particulate matter air pollution components and risk for lung cancer. Environ. Int. 2016, 87, 66–73. 10.1016/j.envint.2015.11.007. [DOI] [PubMed] [Google Scholar]
- Bode A. M.; Dong Z.; Wang H. Cancer prevention and control: Alarming challenges in China. Natl. Sci. Rev. 2016, 3, 117–127. 10.1093/nsr/nwv054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchounwou P. B.; Yedjou C. G.; Patlolla A. K.; Sutton D. J.. Heavy metal toxicity and the environment. In Molecular, Clinical and Environmental Toxicology; Springer, 2012; pp 133–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satarug S.; Moore M. R. Adverse health effects of chronic exposure to low-level cadmium in foodstuffs and cigarette smoke. Environ. Health Perspect. 2004, 112, 1099–1103. 10.1289/ehp.6751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Järup L.; Berglund M.; Elinder C. G.; Nordberg G.; Vahter M. Health effects of cadmium exposure–A review of the literature and a risk estimate. Scand. J. Work, Environ. Health 1998, 24, 1–51. [PubMed] [Google Scholar]
- Medema J. P. Cancer stem cells: The challenges ahead. Nat. Cell Biol. 2013, 15, 338–344. 10.1038/ncb2717. [DOI] [PubMed] [Google Scholar]
- Borah A.; Raveendran S.; Rochani A.; Maekawa T.; Kumar D. S. Targeting self-renewal pathways in cancer stem cells: Clinical implications for cancer therapy. Oncogenesis 2015, 4, e177 10.1038/oncsis.2015.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madu C. O.; Lu Y. Novel diagnostic biomarkers for prostate cancer. J. Cancer 2010, 1, 150–177. 10.7150/jca.1.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennedige T.; Venkatesh S. K. Advances in computed tomography and magnetic resonance imaging of hepatocellular carcinoma. World J. Gastroenterol. 2016, 22, 205. 10.3748/wjg.v22.i1.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju H.; Shim Y.; Arumugam P.; Song J. M. Crosstalk-eliminated quantitative determination of aflatoxin B1-induced hepatocellular cancer stem cells based on concurrent monitoring of CD133, CD44, and aldehyde dehydrogenase1. Toxicol. Lett. 2016, 243, 31–39. 10.1016/j.toxlet.2015.12.006. [DOI] [PubMed] [Google Scholar]
- Pisanic T. R. II; Zhang Y.; Wang T. H. Quantum dots in diagnostics and detection: Principles and paradigms. Analyst 2014, 139, 2968–2981. 10.1039/c4an00294f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattabiraman D. R.; Weinberg R. A. Tackling the cancer stem cells—What challenges do they pose?. Nat. Rev. Drug Discovery 2014, 13, 497–512. 10.1038/nrd4253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning X.; Du Y.; Ben Q.; Huang L.; He X.; Gong Y.; Gao J.; Wu H.; Man X.; Jin J.; Xu M.; Li Z. Bulk pancreatic cancer cells can convert into cancer stem cells (CSCs) in vitro and 2 compounds can target these CSCs. Cell Cycle 2016, 15, 403–412. 10.1080/15384101.2015.1127471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auffinger B.; Tobias A. L.; Han Y.; Lee G.; Guo D.; Dey M.; Lesniak M. S.; Ahmed A. U. Conversion of differentiated cancer cells into cancer stem-like cells in a glioblastoma model after primary chemotherapy. Cell Death Differ. 2014, 21, 1119–1131. 10.1038/cdd.2014.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks M. D.; Burness M. L.; Wicha M. S. Therapeutic implications of cellular heterogeneity and plasticity in breast cancer. Cell Stem Cell 2015, 17, 260–271. 10.1016/j.stem.2015.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Q.; Chen B.; Thakur C.; Lu Y.; Chen F. Arsenic-induced sub-lethal stress reprograms human bronchial epithelial cells to CD61– cancer stem cells. Oncotarget 2014, 5, 1290. 10.18632/oncotarget.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokar E. J.; Diwan B. A.; Waalkes M. P. Arsenic exposure transforms human epithelial stem/progenitor cells into a cancer stem-like phenotype. Environ. Health Perspect. 2010, 118, 108–115. 10.1289/ehp.0901059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pluchino L. A.; Wang H.-C. R. Chronic exposure to combined carcinogens enhances breast cell carcinogenesis with mesenchymal and stem-like cell properties. PLoS One 2014, 9, e108698 10.1371/journal.pone.0108698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim Y.; Song J. M. Spectral overlap-free quantum dot-based determination of benzo[a]pyrene-induced cancer stem cells by concurrent monitoring of CD44, CD24 and aldehyde dehydrogenase 1. Chem. Commun. 2015, 51, 2118–2121. 10.1039/c4cc08953g. [DOI] [PubMed] [Google Scholar]
- Hurt E. M.; Kawasaki B. T.; Klarmann G. J.; Thomas S. B.; Farrar W. L. CD44+CD24– prostate cells are early cancer progenitor/stem cells that provide a model for patients with poor prognosis. Br. J. Cancer 2008, 98, 756–765. 10.1038/sj.bjc.6604242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginestier C.; Hur M. H.; Charafe-Jauffret E.; Monville F.; Dutcher J.; Brown M.; Jacquemier J.; Viens P.; Kleer C. G.; Liu S.; Schott A.; Hayes D.; Birnbaum D.; Wicha M. S.; Dontu G. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell 2007, 1, 555–567. 10.1016/j.stem.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung P. F.; Cheung T. T.; Yip C. W.; Ng L. W.; Fung S. W.; Lo C. M.; Fan S. T.; Cheung S. T. Hepatic cancer stem cell marker granulin-epithelin precursor and β-catenin expression associate with recurrence in hepatocellular carcinoma. Oncotarget 2016, 7, 21644–21657. 10.18632/oncotarget.7803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irollo E.; Pirozzi G. CD133: To be or not to be, is this the real question?. Am. J. Transl. Res. 2013, 5, 563–581. [PMC free article] [PubMed] [Google Scholar]
- Hou Y.; Zou Q.; Ge R.; Shen F.; Wang Y. The critical role of CD133+CD44+/high tumor cells in hematogenous metastasis of liver cancers. Cell Res. 2012, 22, 259–272. 10.1038/cr.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLeo A. B. Targeting cancer stem cells with ALDH1A1-based immunotherapy. OncoImmunology 2012, 1, 385–387. 10.4161/onci.18826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B.; Lee J. K.; Choi S. Continuous sorting and washing of cancer cells from blood cells by hydrophoresis. BioChip J. 2015, 10, 81–87. 10.1007/s13206-016-0201-0. [DOI] [Google Scholar]
- Aebersold R.; Burlingame A. L.; Bradshaw R. A. Western blots versus selected reaction monitoring assays: Time to turn the tables?. Mol. Cell. Proteomics 2013, 12, 2381–2382. 10.1074/mcp.e113.031658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resch-Genger U.; Grabolle M.; Cavaliere-Jaricot S.; Nitschke R.; Nann T. Quantum dots versus organic dyes as fluorescent labels. Nat. Methods 2008, 5, 763–775. 10.1038/nmeth.1248. [DOI] [PubMed] [Google Scholar]
- Kreso A.; Dick J. E. Evolution of the cancer stem cell model. Cell Stem Cell 2014, 14, 275–291. 10.1016/j.stem.2014.02.006. [DOI] [PubMed] [Google Scholar]
- Kim M.; Rangasamy S.; Shim Y.; Song J. Cell lysis-free quantum dot multicolor cellular imaging-based mechanism study for TNF-α-induced insulin resistance. J. Nanobiotechnol. 2015, 13, 4. 10.1186/s12951-015-0064-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovtun O.; Arzeta-Ferrer X.; Rosenthal S. J. Quantum dot approaches for target-based drug screening and multiplexed active biosensing. Nanoscale 2013, 5, 12072–12081. 10.1039/c3nr02019c. [DOI] [PubMed] [Google Scholar]
- Kawata K.; Yokoo H.; Shimazaki R.; Okabe S. Classification of heavy-metal toxicity by human DNA microarray analysis. Environ. Sci. Technol. 2007, 41, 3769–3774. 10.1021/es062717d. [DOI] [PubMed] [Google Scholar]
- Fabbri M.; Urani C.; Sacco M. G.; Procaccianti C.; Gribaldo L. Whole genome analysis and microRNAs regulation in HepG2 cells exposed to cadmium. ALTEX-Alternatives to Animal Experimentation 2012, 29, 173. 10.14573/altex.2012.2.173. [DOI] [PubMed] [Google Scholar]
- Cartularo L.; Laulicht F.; Sun H.; Kluz T.; Freedman J. H.; Costa M. Gene expression and pathway analysis of human hepatocellular carcinoma cells treated with cadmium. Toxicol. Appl. Pharmacol. 2015, 288, 399–408. 10.1016/j.taap.2015.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehn P. F.; White C. M.; Conners D. E.; Shipkey G.; Cumbo T. A. Characterization of the human hepatocellular carcinoma (hepg2) cell line as an in vitro model for cadmium toxicity studies. In Vitro Cell. Dev. Biol.: Anim. 2004, 40, 172–182. . [DOI] [PubMed] [Google Scholar]
- McCubrey J. A.; Steeman L. S.; Chappell W. H.; Abrams S. L.; Montalto G.; Cervello M.; Nicoletti F.; Fagone P.; Malaponte G.; Mazzarino M. C.; Candido S.; Libra M.; Bäsecke J.; Mijatovic S.; Maksimovic-Ivanic D.; Milella M.; Tafuri A.; Cocco L.; Evangelisti C.; Chiarini F.; Martelli A. M. Mutations and deregulation of Ras/Raf/MEK/ERK and PI3K/PTEN/Akt/mTOR cascades which alter therapy response. Oncotarget 2012, 3, 954. 10.18632/oncotarget.652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adjei A. A. Blocking oncogenic Ras signaling for cancer therapy. J. Natl. Cancer Inst. 2001, 93, 1062–1074. 10.1093/jnci/93.14.1062. [DOI] [PubMed] [Google Scholar]
- Stewart A.; Thavasu P.; de Bono J. S.; Banerji U. Titration of signalling output: Insights into clinical combinations of MEK and AKT inhibitors. Ann. Oncol. 2015, 26, 1504–1510. 10.1093/annonc/mdv188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein P. J.; Schmidt C. M.; Wiesenauer C. A.; Choi J. N.; Gage E. A.; Yip-Schneider M. T.; Wiebke E. A.; Wang Y.; Omer C.; Sebolt-Leopold J. S. The effects of a novel MEK inhibitor PD184161 on MEK-ERK signaling and growth in human liver cancer. Neoplasia 2006, 8, 1–8. 10.1593/neo.05373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L.; Shi Y.; Jiang C.-Y.; Wei L.-X.; Wang Y.-L.; Dai G.-H. Expression and prognostic role of pan-Ras, Raf-1, pMEK1 and pERK1/2 in patients with hepatocellular carcinoma. Eur. J. Surg. Oncol. 2011, 37, 513–520. 10.1016/j.ejso.2011.01.023. [DOI] [PubMed] [Google Scholar]
- Brama M.; Gnessi L.; Basciani S.; Cerulli N.; Politi L.; Spera G.; Mariani S.; Cherubini S.; d’Abusco A. S.; Scandurra R.; Migliaccio S. Cadmium induces mitogenic signaling in breast cancer cell by an ERα-dependent mechanism. Mol. Cell. Endocrinol. 2007, 264, 102–108. 10.1016/j.mce.2006.10.013. [DOI] [PubMed] [Google Scholar]
- Huff M. O.; Todd S. L.; Smith A. L.; Elpers J. T.; Smith A. P.; Murphy R. D.; Bleser-Shartzer A. S.; Hoerter J. E.; Radde B. N.; Klinge C. M. Arsenite and cadmium activate MAPK/ERK via membrane estrogen receptors and G-protein coupled estrogen receptor signaling in human lung adenocarcinoma cells. Toxicol. Sci. 2016, 152, 62–71. 10.1093/toxsci/kfw064. [DOI] [PubMed] [Google Scholar]
- Dang H.; Steinway S. N.; Ding W.; Rountree C. B. Induction of tumor initiation is dependent on CD44s in c-Met+ hepatocellular carcinoma. BMC Cancer 2015, 15, 1. 10.1186/s12885-015-1166-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccarelli C.; Vulcano F.; Milazzo L.; Gravina G. L.; Marampon F.; Macioce G.; Giampaolo A.; Tombolini V.; Di Paolo V.; Hassan H. J.; Zani B. M. Key role of MEK/ERK pathway in sustaining tumorigenicity and in vitro radioresistance of embryonal rhabdomyosarcoma stem-like cell population. Mol. Cancer 2016, 15, 1. 10.1186/s12943-016-0501-y. [DOI] [PMC free article] [PubMed] [Google Scholar]