Abstract
A number of amphiphilic difatty acyl linear and cyclic R5K2 peptide conjugates were synthesized by solid-phase peptide methods to enhance the interaction with the hydrophobic cellular phospholipid bilayer and to improve siRNA delivery and silencing. Binding to siRNA molecules was significantly less for the cyclic peptide conjugates. A gradual decrease was observed in the particle size of the complexes with increasing peptide/siRNA ratio for most of the synthesized peptides, suggesting the complex formation. Most of the complexes showed a particle size of less than 200 nm, which is considered an appropriate size for in vitro siRNA delivery. A number of fatty acyl-conjugated peptides, such as LP-C16 and LP-C18, displayed near complete protection against serum degradation. Flow cytometry studies demonstrated significantly higher internalization of fluorescence-labeled siRNA (FAM-siRNA) in the presence of LP-C16, LP-C18, and CP-C16 with 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE) addition. Confocal microscopy confirmed the cellular internalization of fluorescence-labeled siRNA in the presence of LP-C16 and LP-C18 with DOPE when compared with cells exposed to DOPE/FAM-siRNA. While C16- and C18-conjugated peptides (especially linear peptides) showed silencing against kinesin spindle protein (KSP) and janus kinase 2 (JAK2) proteins, the addition of DOPE enhanced the silencing efficiency significantly for all selected peptides, except for CP-C16. In conclusion, C16 and C18 difatty acyl peptide conjugates were found to enhance siRNA delivery and generate silencing of targeted proteins in the presence of DOPE. This study provides insights for the design and potential application of optimized difatty acyl peptide/lipid nanoparticles for effective siRNA delivery.
Introduction
RNA interference (RNAi) is an alternative approach to traditional medicine, which usually uses small molecules to target specific molecules and/or aims to control symptoms of a disorder. RNAi research has attracted significant interest since the dawn of the century. This approach is based on post-transcriptional interference with the expression of a specific protein (or in some cases, a family of proteins) that could be involved in the pathology of the disorder. The possibility of this “silencing” mechanism was reported in late 1990s by Fire et al. who reported interference with the protein expression at the mRNA level by delivering an exogenous double-stranded RNA.1 Among different approaches to RNAi, small-interfering RNA (siRNA) is the closest to the small-molecule approach because it does not alter the genetic information of the cell, does not require incorporation into the chromosome, and creates a dose-dependent and transient effect.
Although siRNA silencing is temporary and the effect is limited to the number of RNA strands delivered to the cell, the siRNA approach to RNAi offers several advantages, which include the following: (i) unlike short hairpin RNAs and micro-RNAs, siRNA is ready to use and does not require intracellular processing; (ii) it is only needed in the cytoplasm and not in the nucleus, which eliminates the need for intranuclear delivery; (iii) it is highly specific, which is specifically advantageous in basic research, by eliminating off-target effect and enabling researchers to evaluate the functionality of the targeted protein; and (iv) as mentioned before, among different RNAi approaches, siRNA is the closest to a traditional “drug”, which is feasible in vivo and can be applicable to clinical settings. Despite these advantages, however, the use of siRNA in clinical settings has been marred by a few challenges for efficient siRNA delivery. Whereas in vivo delivery of siRNA is shown to be a daunting task, even in vitro siRNA delivery has been challenging and inconsistent because of the hydrophilic nature of the siRNA molecule (which minimizes its interaction with the cell membrane) and its negative electronic charge (due to the presence of phosphate groups on the backbone of the nucleotides, which is repulsed by the negatively charged cell membrane).2 In vitro siRNA delivery has been studied extensively for a variety of purposes, including evaluating the efficiency of a novel delivery system, investigating the intracellular functions of a specific protein, and determining the potential therapeutic effect of silencing specific proteins.
Many different categories of carriers have been studied for siRNA delivery, which include polymers, lipids, and peptides. Short sequences of amino acids (less than 30) have been studied as delivery systems since the late 20th century. Specifically, cell-penetrating peptides (CPPs) are 5–40 amino acid-long peptides that are known for their ability to internalize their cargo into the cell and have been used for siRNA delivery as well.3,4 The exact mechanism of cellular uptake has been a subject of debate; however, macropinocytosis, signal-activated endocytosis, and “inverted micelle” model have been suggested.5 Positively charged amino acids, for example, arginine and lysine, are required for complex formation with siRNA,6 and it has been suggested that intracellular delivery could be optimized by adjusting the number of arginines in the peptide.7 Recent reports indicate the in vitro use of folate-poly(ethylene glycol) PEG-appended dendrimer/α-cyclodexdrin conjugates for delivering siRNA to keratin-forming tumor cell line, HeLa (KB) cells (ATCC CCL-17),8 polyelectrolyte-gold nanoassemblies to deliver β-site APP cleaving enzyme1 siRNA to murine neuronal cell lines,9 polycation liposome-encapsulated calcium phosphate nanoparticles for vascular endothelial growth factor (VEGF) siRNA delivery to the MCF-7 breast cancer cell line,10 and folic acid-modified mesoporous silica nanocarriers to deliver VEGF siRNA to HeLa cells over-expressing folic acid.11 The most recent developments in siRNA delivery include stable nucleic acid lipid particles (SNALPs) and N-acetylgalactosamine (GalNAc). SNALPs are multicomponent liposome-like particles that use neutral and cationic lipids for a more effective siRNA delivery12,13 and has reached clinical trials for a few formulations.14 GalNAc, on the other hand, is a liver-targeting moiety with high affinity for asialoglycoprotein receptor, which has been studied as a direct conjugation to siRNA or a component in the delivery system.15,16
We have reported the application of cyclic CPPs containing arginine (R) and tryptophan (W) residues or fatty acyl chains for delivery of different cargo molecules including negatively charged phosphopeptides.17,18 Cyclic peptides are more rigid and expected to be more stable toward degradation. The cyclic nature of the peptides provides higher stability versus the linear counterparts and a more efficient cell penetration. Recently, we also reported that [WR]5 and [WR]5-capped gold nanoparticles improved the cellular delivery of siRNA in HeLa cells by 2- and 3.8-fold, respectively.19
Our earlier work also showed that short linear peptide amphiphiles containing arginine and lysine that conjugated with two fatty acyl groups of hexadecanoyl groups were able to act as a molecular transporter of negatively charged phosphopeptide (PEpYLGLD) intracellularly.17 We have previously reported monofatty acyl conjugates of cyclic peptides containing five and six arginine residues.20 Two conjugates, dodecanoyl-[R5] and dodecanoyl-[R6], improved the cellular uptake of a cell-impermeable negatively charged phosphopeptide (GpYEEI) by 3.4-fold and 5.5-fold, respectively, in human ovarian cancer cells, suggesting that a combination of peptide cyclization and fatty acylation can be used to improve the intracellular uptake of compounds with limited cell permeability.
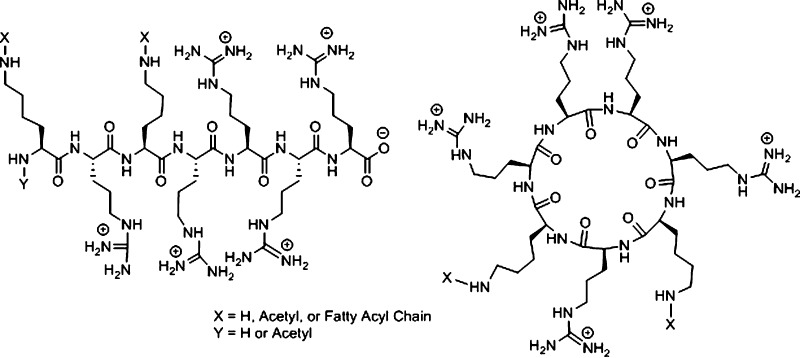
On the basis of the previous data on the importance of the presence of fatty acyl chains and positively charged peptides for cellular delivery efficiency, we report here the design, synthesis, characterization, and evaluation of linear and cyclic peptides conjugated with two fatty acyl chains for siRNA delivery. To the best of our knowledge, no cyclic and linear peptides containing two fatty acyl chains and five arginine residues have been reported previously for siRNA delivery. We hypothesized that the combination of two fatty acids and positively charged peptides will enhance the interaction with the two hydrophobic chains and negatively charged phosphate in the cellular phospholipid bilayer, respectively, and enhance the cellular delivery of siRNA and siRNA silencing. Two lipid tails of the peptide are expected to increase the hydrophobic interior interactions with the cellular membrane. One of the objectives of this research was to determine whether the side chain length of the fatty acyl conjugates and the linear or cyclic nature of the peptides can affect the cellular internalization. For this purpose, a variety of linear and cyclic peptides were synthesized with various fatty acyl conjugations with different lengths (from C2 to C18) and degrees of saturation. Both linear and cyclic peptides had the same amino acid sequence of R5K2 as the backbone (Figure 1). (R5K2) and [R5K2] represent the number and presence of five arginine and two lysine residues in linear and cyclic peptides, respectively. The sequences are KRKR4 and [KRKR4] for linear and cyclic peptides, respectively. For simplicity, the backbone of both group of peptides is shown as R5K2. For convenience, square brackets [ ] and parentheses ( ) were used to represent cyclic and linear peptides, respectively.
Figure 1.
Chemical structures of difatty acyl linear and cyclic peptide conjugates.
The diversity in hydrophobicity of the compounds allows studying the interactions with siRNA and transfection efficiency. We analyzed the amount of peptide required for 50% binding to siRNA (BC50, as an indirect method of estimating “binding affinity”) to determine the effect of lipid conjugation and the size of the conjugate on the interionic interaction. The ability of the peptides in protecting siRNA against early enzymatic degradation was investigated by exposing the complexes to fetal bovine serum (FBS), and the toxicity of the peptides was explored in different human cancer cell lines. We characterized the size and surface electrical charge of the complexes formed via ionic interaction between the peptides and siRNA to confirm complex formation and neutralization of siRNA negative charge and to evaluate the physical characteristics of the complex. Finally, the efficiency of the peptide libraries in internalizing siRNA into different human cancer cells was studied, which was confirmed by the silencing efficiency for a model protein.
Results
Chemistry
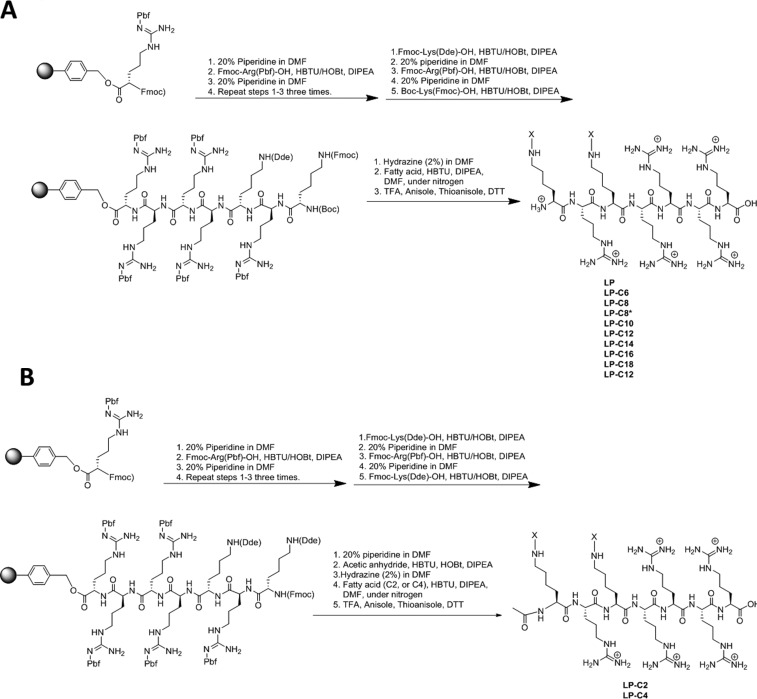
Fmoc solid-phase peptide synthesis was used to synthesize the cyclic and linear difatty acyl conjugates of the designed peptides with five arginines and two lysines (R5K2) (Schemes 1 and 2). Protected Arg(Pbf)-Wang resin was used as a solid support for the linear peptide synthesis, and Fmoc-Arg(Pbf)-OH and Fmoc-Lys(Dde)-OH or Boc-Lys(Fmoc)-OH were used as building block amino acids. After deprotection of the Fmoc group and activation of the amino group, Dde groups of lysine residues were selectively deprotected in the presence of hydrazine in dimethylformamide (DMF, 2:98 v/v). Prior to the final cleavage of the linear peptide conjugates, the fatty acid conjugations were carried out with the resin-attached linear peptide. Fatty acid conjugation was performed in the presence of 2-(1H-benzotriazole-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate (HBTU) and N,N-diisopropylethylamine (DIPEA). The final cleavage was performed in the presence of trifluoroacetic acid (TFA)/anisole/thioanisole/dithiothreitol (DTT) (94:2:2:2 v/v/v/v) (Scheme 1).
Scheme 1. (A) Synthesis of Linear N-Free Amino Difatty Acyl Conjugates of R5K2; (B) Synthesis of Linear N-Acetylamino Difatty Acyl Conjugates of R5K2.
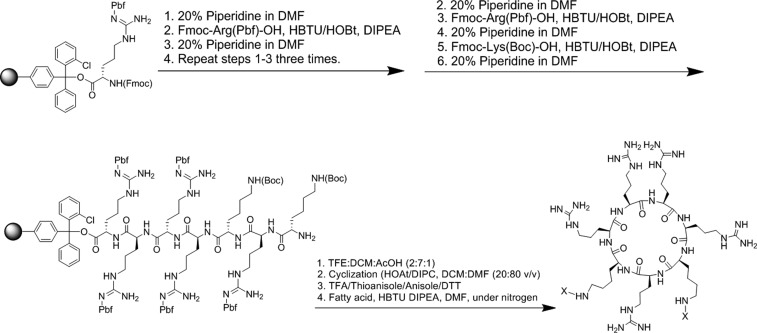
Scheme 2. Synthesis of Cyclic Difatty Acyl Conjugates of R5K2.
2-Chlorotrityl arginine resin was used as a solid support for the synthesis of difatty acyl cyclic peptide conjugates. Fmoc-Arg(Pbf)-OH and Fmoc-Lys(Boc)-OH were used as the building block amino acids. After assembly of the peptide on the solid phase, the resin was cleaved in the presence of dichloromethane (DCM)/trifluoroethanol/AcOH (1:2:7 v/v/v). N- to C-cyclization was accomplished in the solution phase and in the presence of 1-hydroxy-7-azabenzotriazole (HOAT)/DIPEA. After deprotection of side chains in the presence of TFA/anisole/thioanisole/DTT (94:2:2:2 v/v/v/v), fatty acid conjugation was accomplished in the presence of HBTU and DIPEA to afford difatty acyl cyclic peptide conjugates (Scheme 2).
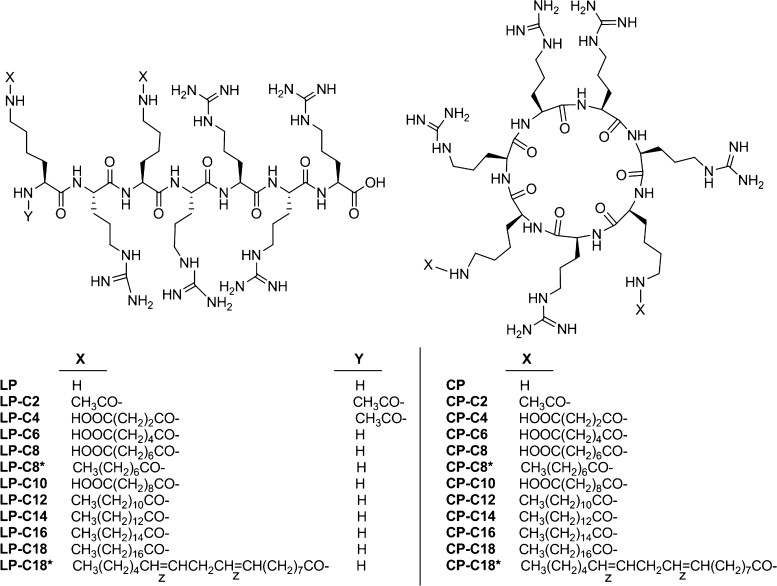
Fatty acids were (CH3(CH2)n–COOH) and dicarboxylic acids were (HOOC(CH2)n–COOH), where n varied from 2 to 16. Also, linoleic acid was chosen as the fatty acid to incorporate an analogue with an unsaturated bond in the library. The structures of all conjugated linear and cyclic peptides can be found in Figures S1 and S2 (Supporting Information), respectively. The crude linear and cyclic difatty acyl peptide conjugates were purified by reversed-phase high-performance liquid chromatography (HPLC) and characterized by matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF). During the purification process, it was discovered that the polarity of the solvents affected the purification process of the conjugates. For example, isopropanol alcohol and methanol worked more efficiently when purifying the hydrophobic C18 linear and cyclic conjugates.
BC50 for siRNA Binding
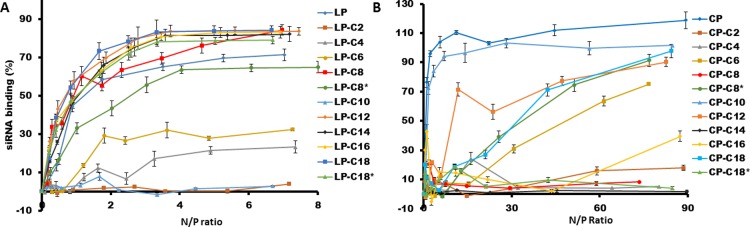
To evaluate the capability of linear and cyclic peptides to form complexes with siRNA via interionic interaction, a binding experiment was designed based on the quantification of bound siRNA using SYBR Green II, and the data were used to calculate BC50 for each peptide. Figure 2 summarizes the siRNA-binding of both libraries. The fluorescent signal quantified for the positive control was not significantly different from the signal recorded for SYBR Green added to siRNA-free saline (data not shown), which confirmed the reliability of the binding data. Among linear peptides, the unmodified linear peptide (R5K2) showed the lowest BC50 for binding to siRNA (BC50 = 0.7; n = 3). Hydrophobically modified linear peptides showed two different patterns for siRNA binding; whereas most modified peptides showed a strong siRNA-binding affinity, LP-C2, LP-C4, LP-C6, and LP-C10 did not follow this trend, and in fact, complete complex formation was not observed even with the highest ratio studied for these modified peptides. Among this group of peptides, LP-C10 showed the lowest BC50 to form complexes with siRNA, with almost no siRNA binding detected at any of the studied peptide/siRNA ratios. The common theme among the fatty acid-conjugated peptides with low affinity for siRNA binding is the terminal −COOH group at the end of the dicarboxylic fatty acid used for conjugation. The only other modified peptide with the −COOH group at the end of the conjugated fatty acid was LP-C8, which showed ∼84% complex formation at an nitrogen/phosphate (N/P) ratio of 7.0, with a relatively lower BC50 (1.18) compared to other peptides (Figure 2A). The final ranking of the BC50 values calculated for peptides with near-complete complex formation was LP (0.70) > LP-C12 (0.97) = LP-C18 (1.0) > LP-C8 (1.2) = LPC16 (1.2) > LP-C14 (1.4) = LP-C18* (1.4) > LP-C8* (2.6).
Figure 2.
BC50 of difatty acyl linear and cyclic peptides to siRNA. Representative line graph of siRNA binding of linear (A) and cyclic peptides (B) to scrambled siRNA, indicating the percentage of siRNA bond to the peptide as a function of peptide/siRNA N/P ratios of 0–8 and 0–90 (compared to free siRNA as control), respectively.
The cyclic-modified peptides showed a more versatile profile. Only unmodified cyclic [R5K2] and four modified peptides achieved complete complexation with siRNA at the highest N/P ratio. The BC50 value of the unmodified cyclic peptide was very similar to that of the linear counterpart (BC50 = 0.73); however, modified cyclic peptides generally showed a higher BC50. Whereas CP-C2, CP-C4, CP-C8, CP-C14, and CP-C18* showed negligible affinity to siRNA (less than 20% complex formation at a w/w ratio of 80:1), CP-C6 and CP-C16 showed some affinity and achieved ∼77 and ∼40% complex formation at the highest studied peptide/siRNA ratio, respectively (Figure 2B). The final ranking of BC50 values among the peptides that did show complete complexation was CP (0.73) > CP-C10 (1.3) > CP-C12 (18) > CP-C8* (37) > CP-C18 (38).
Size and Electrical Charge of Peptide/siRNA Complexes
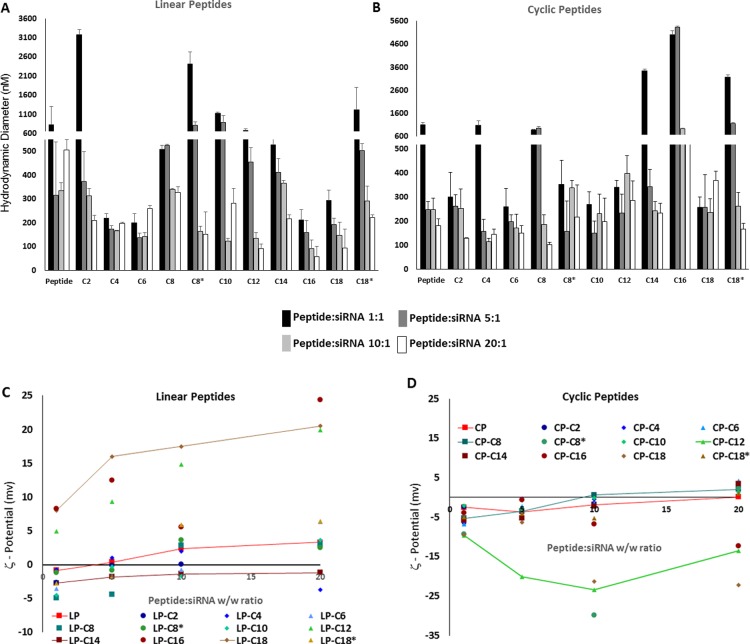
The in vitro and in vivo performances of siRNA delivery systems largely depends on the size and overall surface electrical charge of the siRNA-associated carrier. We used a dynamic light scattering method to determine the size of the peptide/siRNA complexes at four different w/w ratios. For most of the linear and cyclic peptides, a decline in the size of the peptide/siRNA complexes was observed on increasing the peptide/siRNA w/w ratio. This was specifically significant for LP-C2 (3171 vs 210 nm for ratios of 1:1 [N/P ratio of 1.12] and 20:1 [N/P ratio of 22.4], respectively), LP-C8* (2590 vs 152 nm), CP-C14 (3349 vs 400 nm), CP-C16 (5020 vs 646 nm), and CP-C18 (2633 vs 168 nm). On the other hand, some of the other peptides showed a rather steady size regardless of the peptide/siRNA ratio (including LP, LPC4, LP-C6, CP-C2, CP-C6, CP-C10, and CP-C12). Overall and at the highest ratio studied in this experiment, most of the peptide/siRNA ratios showed a hydrodynamic diameter of 200 nm or less. Among these peptides, LP-C16 (57.47 nm), LP-C18 (92.78 nm), and CP-C8 (103.12 nm) were the smallest complexes observed (all sizes were measured in triplicates). However, some of the peptide/siRNA complexes showed a hydrodynamic diameter larger than 400 nm at this ratio, which included LP, CP-C12, CP-C16, and CP-C18 (Figure 3A,B).
Figure 3.
Particle size and electrical surface charge of peptide/siRNA complexes. Hydrodynamic diameter of the linear (A) and cyclic (B) peptides/siRNA complexes at different w/w ratios. The particle size for most of the complexes at the peptide/siRNA w/w ratio of 20:1 (N/P ratios of 10.2–29.5 for modified peptides) was about 200 nm or less. ζ-Potential of linear (C) and cyclic (D) peptides/siRNA complexes at different w/w ratios. Whereas most of the linear peptides showed a steady increase in the ζ-potential with a positive value for the 20:1 peptide/siRNA w/w ratio, many cyclic peptides failed to show a similar trend, and a few reached a positive ζ-potential at the highest peptide/siRNA ratio studied in this experiment. The error bars represent the standard deviation.
The ζ-potential of the nonmodified peptide/siRNA nanoparticles showed a steady increase on increasing the peptide/siRNA w/w ratio (−0.83, 0.371, 2.4, and 3.3 for 1:1, 5:1, 10:1, and 20:1 ratios equivalent to N/P ratios of 2.2, 11.0, 21.9, and 43.9, respectively). Most of the fatty acid-conjugated linear peptides showed a similar pattern with a gradual increase in the ζ-potential of the complexes. The only exception was LP-C4, which after an initial increase (−0.701, 1.01, and 1.93 for peptide/siRNA ratios of 1:1, 5:1, and 10:1 equivalent to N/P ratios of 0.51, 2.55, and 5.1, respectively) demonstrated a negative ζ-potential for the 20:1 peptide/siRNA w/w ratio (N/P ratio of 10.2). LP-C14 was the only other modified peptide that (despite a gradual increase in ζ-potential values) demonstrated a negative value at the w/w ratio of 20:1 (equivalent to N/P ratio of 22.4; ζ-potential = −1.21). LP-C12, LP-C16, LP-C18, and LP-C18* were the only peptides that showed positive ζ-potential values for all studied peptide/siRNA ratios and a significant increase in the ζ-potential compared to the unmodified linear peptide (Figure 3C). The unmodified cyclic peptide also showed an increase in the ζ-potential on increasing the peptide/siRNA ratio (−5.39, −3.7, −1.9, and 0.101 for ratios of 1:1, 5:1, 10:1, and 20:1 equivalent to N/P ratios of 2.23, 11.2, 22.3, and 44.6, respectively). A similar pattern, however, was not observed for all of the peptides in this library. CP-C12, CP-C16, and CP-C18 demonstrated negative ζ-potential values for all peptide/siRNA w/w ratios studied and showed a drop in the ζ-potential with the highest ratio studied (e.g., −3.90, −0.58, −6.77, and −12.40 were recorded as the average ζ-potential at different N/P ratios studied). Other modified cyclic peptides showed a gradual increase in the ζ-potential; however, they never achieved any values more than 4.5 (ζ-potential = 4.22 for CP-C6 at a w/w ratio of 20:1, equivalent to an N/P ratio of 15.3; Figure 3D).
Serum Stability of Complexes
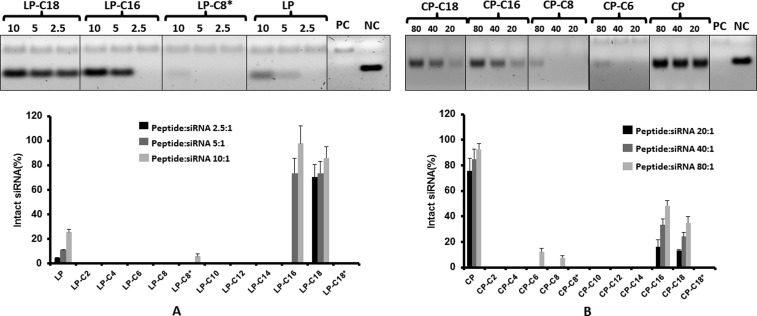
To determine the capability of the complexes to protect the siRNA against enzymatic degradation, we exposed different study groups to a diluted FBS solution, with “naked” siRNA as the positive control and siRNA exposed to saline as the negative control, and the results are summarized in Figure 4. Although in vitro experiments are performed in cell culture media containing 10% FBS, a higher concentration of FBS (25%) was used to determine the stability of siRNA under a harsher condition and a more biologically relevant environment. After 24 h exposure to serum, we did not observe any bands for siRNA in the positive control groups, which indicated complete degradation of “naked” siRNA under these conditions and validated the method. Considering the negative control band as 100% intact siRNA, we calculated the percentage of siRNA that remained intact in each study group. Although only ∼25% of the siRNA was protected by the unmodified linear peptide at the peptide/siRNA w/w ratio of 10:1, LP-C16 and LP-C18 were able to protect siRNA almost completely after 24 h of incubation with the serum in 37 °C. The level of protection was not significantly different for the three different peptide/siRNA w/w ratios studied for LP-C18; however, it showed significant improvement for LP-C16 on increasing the w/w ratio from 2.5:1 to 5:1. Other modified linear peptides showed minimum to no protection against enzymatic degradation (Figure 4A). The cyclic unmodified peptide, on the other hand, showed significant protection that approached 100% on increasing the peptide/siRNA w/w ratio to 80:1. The only modified peptides that showed noticeable protection against enzymatic degradation were CP-C16 and CP-C18, with 48 and 35% intact siRNA detected after incubation of the complexes formed at a peptide/siRNA ratio w/w of 80:1 (Figure 4B).
Figure 4.
Serum stability of siRNA after complex formation. Representative image of gel electrophoresis and a bar graph summarizing the percentage of intact siRNA after 24 h of exposure to serum at 37 °C for linear (A) and cyclic (B) peptide/siRNA complexes at peptide/siRNA w/w ratios of 2.5–10 and 20–80, respectively. PC and NC represent positive control (scrambled siRNA exposed to saline for 24 h at 37 °C, quantified as 100%) and negative control (representing “naked” siRNA incubated with serum for 24 h at 37 °C), respectively. Linear and cyclic peptides conjugated with C16 and C18 fatty acids as well as the unconjugated cyclic peptide showed the most protection against enzymatic degradation of siRNA. Data are presented as mean, n = 3, and the error bars represent standard deviation.
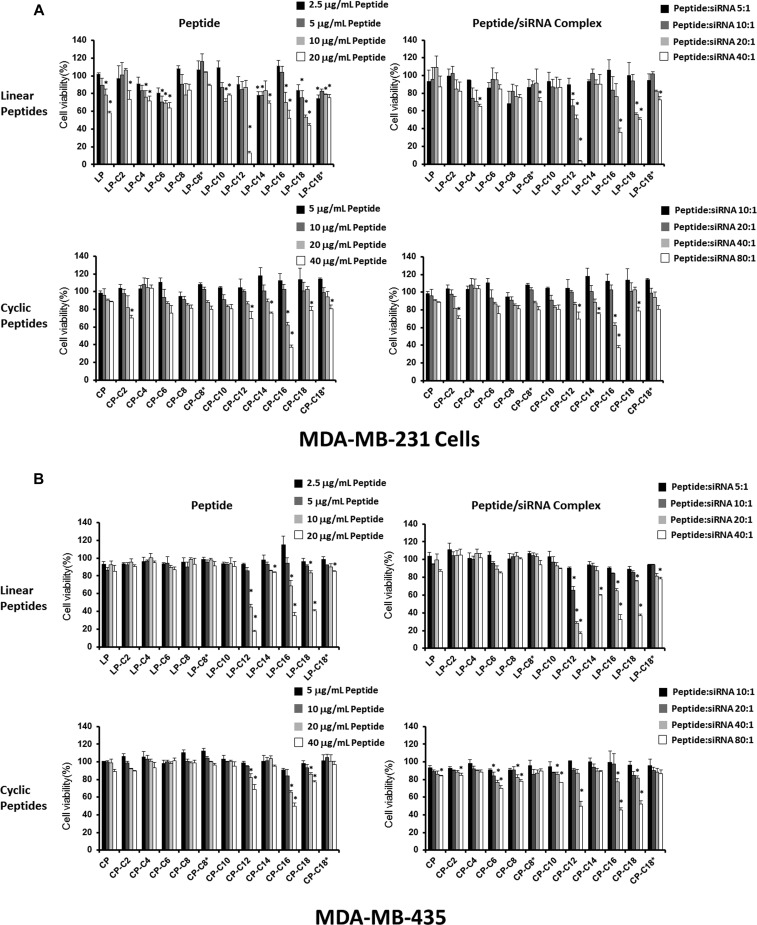
Cytotoxicity
The effect of peptide libraries on the viability of human cell lines was evaluated using two approaches: exposing the cell lines to different concentrations of peptide solution and exposing the cells to peptide/siRNA complexes prepared with different peptides at various peptide/siRNA ratios. We duplicated this study in human cancer cell lines MDA-MB-231 and MDA-MB-435 (Figure 5). The viability of cells followed a similar pattern after exposure to peptide solutions and peptide/siRNA complexes in both the cell lines. In MDA231 cells, LP-C12 showed significant cytotoxicity at a peptide/siRNA w/w ratio of 20:1 (N/P ratio of 23.3), both alone and incorporated in the siRNA complex. Some toxicity was also observed with LP-C16 and LP-C18 at this peptide/siRNA ratio, which was more significant with the peptide/siRNA complexes for LP-C16 (52.9% viability after peptide exposure vs 35.8% after peptide/siRNA exposure). Interestingly, a similar toxicity was not observed for C12-conjugated cyclic peptide in this cell line; however, a similar toxicity was observed for CP-C16 both as peptide solution and peptide/siRNA complex (Figure 5A). LP-C12, LP-C16, and LP-C18 showed similar toxicity levels in MDA-MB-435 cells as well. For some of the fatty acid-conjugated cyclic peptides, however, the toxicity was more significant when the MDA-MB-435 cells were treated with peptide/siRNA complexes as compared to the peptide solution (68.6% vs 50.1% viability for CP-C12, 77.5% vs 52.4% viability for CP-18; Figure 4B). The other peptides showed minimum or no significant toxicity in any of the study groups in either of the cell lines.
Figure 5.
Safety profiles of the peptide libraries and the peptide/siRNA complexes in human cell lines. The viability of MDA-MB-231 (A) and MDA-MB-435 (B) cell lines after 72 h of exposure to different peptide concentrations (left panels) and peptide/siRNA complexes in a range of peptide/siRNA ratios (right panels) are presented for linear (upper panels) and cyclic (lower panels) peptide libraries. Linear peptide conjugated with C12 fatty acid showed the most significant toxicity in both cell lines. Data are presented as mean, n = 3, and the error bars represent standard deviation. The asterisks represent a significant difference (p < 0.05) compared to “no treatment” cells (representing 100% viability; data for which is not included in the figure).
Cellular Internalization
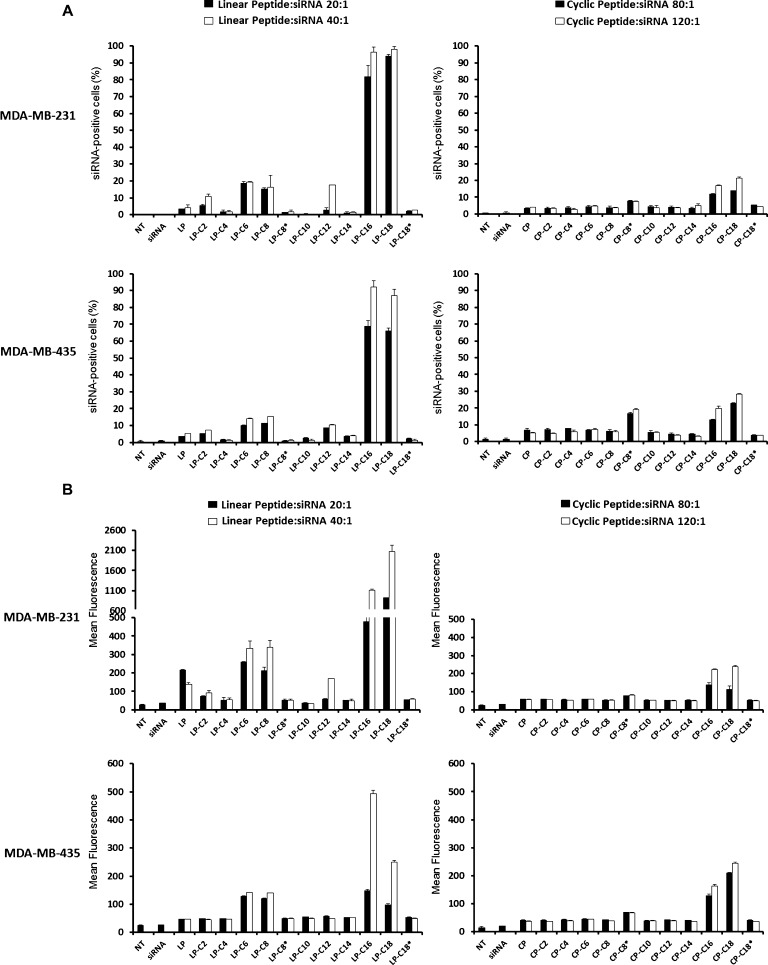
The ability of peptide libraries to deliver siRNA into the cells was evaluated using FAM-labeled scrambled siRNA and analysis by flow cytometry and fluorescent microscopy. The siRNA uptake pattern was similar for both MDA-MB-231 and MDA-MB-435 cell lines. The internalization of siRNA was minimum with most of the linear library in both cell lines. However, LP-C16 and LP-C18 were exceptions and showed a significant siRNA uptake, which was significantly higher at a peptide/siRNA ratio of 20:1 (N/P ratios of 21.6 and 20.8, respectively) compared to the ratio of 10:1 (N/P ratios of 10.8 and 10.4, respectively) (43.8 vs 17.1% siRNA-positive cells for LP-C16 and 52.4 vs 19.1% for LP-C18) in MDA-MB-231 cells. The siRNA internalization followed a similar trend in MDA-MB-435 cells for these two modified linear peptides; however, a higher level of efficiency was achieved for LP-C16 at both peptide/siRNA ratios studied compared to the MDA-MB-231 cells (82.3% vs 43.8% siRNA-positive cells at a w/w ratio of 20:1 [N/P ratio of 21.6] and 48.7% vs 17.1% at a ratio of 10:1; Figure 6A, left panels). However, a different pattern was observed for cyclic peptides. None of the cyclic peptides achieved siRNA internalization in more than 20% of the cells in either of the cell lines included in the study. In the MDA-MB-231 cells, CP-C18, CP-C16, and CP-C8* internalized siRNA into 12.5, 11.7, and 9.8% of the cells, respectively, at a peptide/siRNA w/w ratio of 120:1 (N/P ratios of 126, 131, and 154, respectively). The level of siRNA uptake was slightly higher in MDA-MB-435 cells, and the same peptides achieved 19.3, 14.9, and 14.2% siRNA-positive cells at the same ratios(Figure 6A, right panels). The mean fluorescence results confirmed a similar pattern in both cell lines (Figure 6B). The siRNA uptake with other peptides was not statistically different from “naked siRNA”.
Figure 6.
Cellular uptake of FAM-siRNA by human cancer cell lines. The internalization of FAM-labeled siRNA into MDA-MB-231 (the upper panels in each set) and MDA-MB-435 (the lower panels in each set) cells for linear (left panels in each set) and cyclic (right panels in each set) peptides is presented as percentage of the cells positive for siRNA uptake (A) and average of the fluorescence in the cell population (B). The cells were transfected by two different peptide/siRNA w/w ratios (20:1 and 40:1 for linear and 80:1 and 120:1 for cyclic peptides). Linear peptide conjugated with C16 and C18 fatty acids demonstrated the most efficient intracellular siRNA delivery. Data are presented as mean, n = 3, and the error bars represent standard deviation.
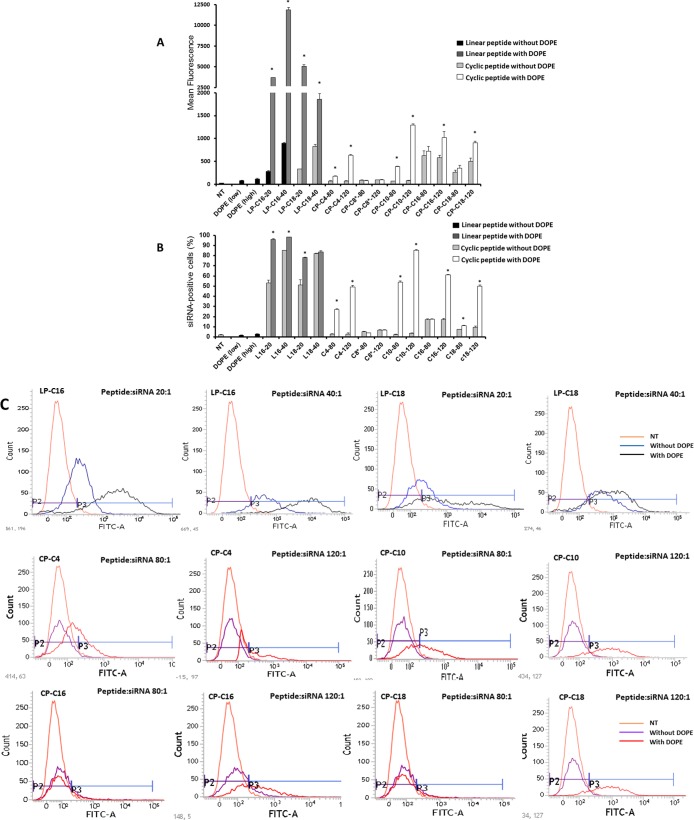
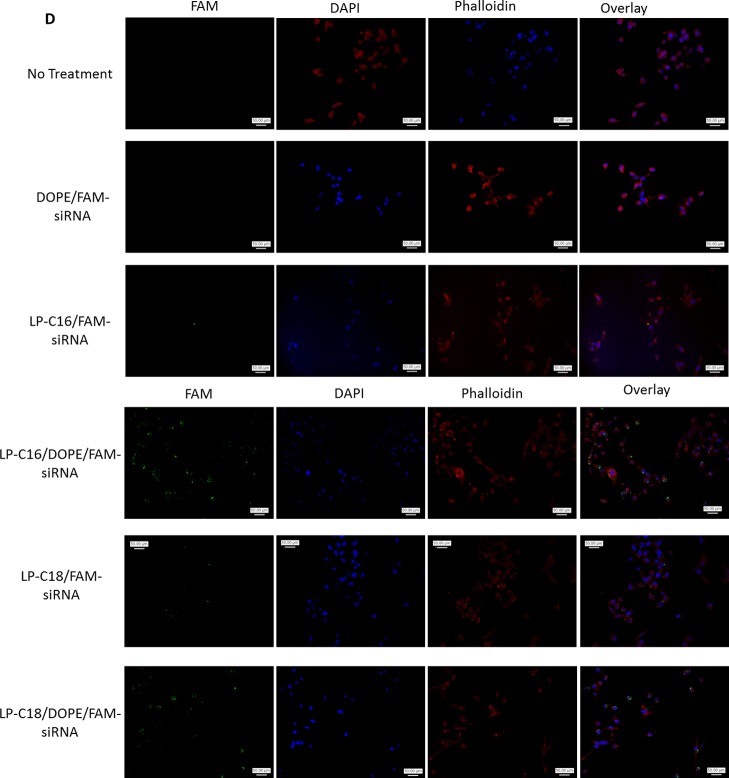
To enhance the siRNA internalization capacity, we added another lipophilic component to the delivery system and compared the siRNA uptake in MDA-MB-231 cells with and without incorporating 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE) in selected peptide/siRNA complexes using peptide/siRNA w/w ratios of 20:1 and 40:1 for linear peptides and 80:1 and 120:1 for cyclic peptides (Figure 7A,B). The addition of DOPE to the delivery system increased the siRNA-positive cells from 53 to 96% and from 85 to 98% for 20:1 and 40:1 ratios of LP-C16/siRNA (N/P ratios of 21.6 and 43.2), respectively. A similar increase (from 51 to 78%) was observed for the LP-C18/siRNA w/w ratio of 20:1 (N/P ratio of 20.8). The increase was not statistically significant for the w/w ratio of 40:1 (N/P ratio of 41.6) for this peptide (82–84%; Figure 7B). The role of DOPE in improving the delivery performance of the selected linear peptides, however, was more noticeable with analyzing the mean fluorescence of the cells treated with peptide/DOPE/siRNA complexes. The mean fluorescence of the treated cells increased from 281 to 3693 and from 893 to 11 861 for LP-C16/siRNA w/w ratios of 20:1 and 40:1, respectively. A similar pattern was observed for the LP-C18-modified peptide (increasing the mean fluorescence from 329 to 5040 and from 824 to 1855 for ratios of 20:1 and 40:1, respectively, Figure 7A). Different fatty acid-conjugated cyclic peptides responded slightly differently to DOPE addition. Whereas CP-C4, CP-C10, and CP-C18 demonstrated a significant increase in the percentage of cells internalizing siRNA at both peptide/siRNA ratios, CP-C16 only showed a significant improvement in efficacy with DOPE addition at a w/w ratio of 120:1 (N/P ratio of 131) (at a w/w ratio of 80:1 [N/P ratio of 87.4], 17.6 and 17.1% of cells internalized siRNA with and without DOPE addition to the delivery system). Adding DOPE did not improve siRNA delivery at either of the ratios for CP-C8* (Figure 7B). Mean fluorescence results confirmed the same trends for cyclic peptides, except CP-C16, which showed a significant improvement in siRNA internalizing efficiency at a CP-C16/siRNA w/w ratio of 120:1 (582–1016 without and with DOPE, respectively, Figure 7A). Delivering FAM-labeled siRNA using only DOPE (with no peptide) with the lowest and highest final concentration used in this study did not result in a significant cellular internalization of siRNA. Figure 7C illustrates examples of the fluorescence recordings of flow cytometry for different peptides with and without DOPE compared to nontreated (NT) cells. The fluorescent microscope images confirmed our findings using flow cytometry. Although DOPE/FAM-labeled siRNA did not show any internalization into MDA-MB-231 cells, both LP-C16 and LP-C18 peptides demonstrated cell uptake that increased significantly with the addition of DOPE.
Figure 7.
Effect of incorporation of DOPE in the siRNA complexes on siRNA intracellular delivery. The cellular uptake studies were repeated with selected peptides to investigate the effect of addition of an additional lipophilic component, and the results are presented as the average of quantified fluorescence (A) and percentage of the cells positive for FAM-labeled siRNA uptake (B). Addition of DOPE to the siRNA complexes enhanced intracellular delivery of siRNA significantly for most of the selected peptides. Data are presented as mean, n = 3, and the error bars represent standard deviation. The asterisks indicate significant difference in cellular internalization as a result of DOPE incorporation into each complex. DOPE (low) and (high) indicate the siRNA uptake of the cells exposed to complexes of siRNA and DOPE (with no peptide) using the lowest (for complexes with peptide/siRNA w/w ratio of 20:1 and the highest (for complexes with peptide/siRNA w/w ratio of 120:1) DOPE final concentration used in the other study groups included in the experiment. (C) Samples of flow cytometry results for siRNA uptake: the uppermost row represents the fluorescence pattern of cells transfected with linear peptide/siRNA w/w ratio of 20:1 or 40:1, with or without DOPE incorporated into the carrier. The middle and bottom rows represent samples of fluorescence pattern of cells transfected with cyclic peptide/siRNA w/w ratio of 80:1 or 120:1, with or without DOPE. The gate P2 represents fluorescence of cells considered siRNA-free, whereas the cells in the P3 region are positive for siRNA uptake. (D) Fluorescent microscope images showing individual channels (green, blue, and red), and an overlay for each treatment group as well as “No treatment” cells as the negative control.
Silencing Efficiency
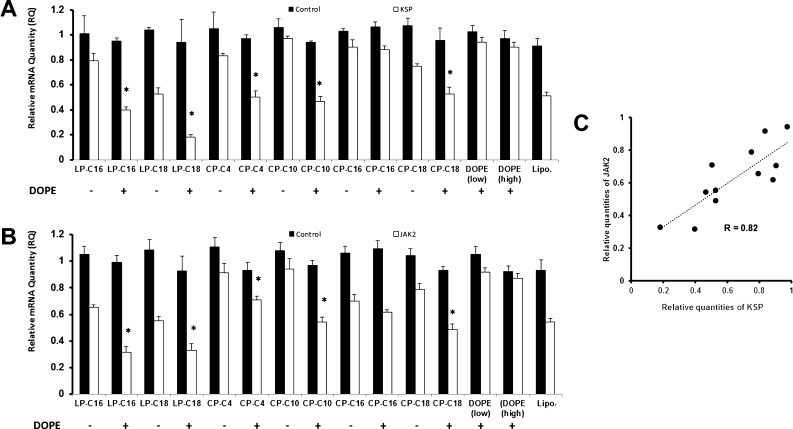
Two model proteins were selected to demonstrate the in vitro silencing efficiency of the designed modified peptides. LP-C16, LP-C18, CP-C4, CP-C10, CP-C16, and CP-C18 were selected for delivering siRNAs targeting kinesin spindle protein (KSP) or Janus kinase 2 (JAK2), and the mRNA levels were directly compared to cells treated with scrambled siRNA. Each peptide was tested with and without incorporation of DOPE into the carrier. Between the selected linear peptides, LP-C18 showed a higher efficiency [relative quantity (RQ) of 0.52 compared to 0.79 for LB-C16] in silencing KSP (Figure 8A). The efficiency difference between the two peptides was not as much against JAK2 (0.55 vs 0.65; Figure 8B). None of the cyclic peptides showed an RQ lower than 0.75 for KSP without DOPE. However, delivering JAK2 siRNA, CP-C18 demonstrated an RQ of ∼0.7. Overall, without adding DOPE to the carriers, the highest silencing efficiency was achieved with the selected linear peptides delivering JAK2 siRNA. Incorporating DOPE to the carrier had a similar enhancing effect observed in cellular internalization studies. All selected peptides (except CP-C16) showed a higher silencing efficiency against both selected proteins with the addition of DOPE. The DOPE enhancing effect was more noticeable in silencing KSP, where the CP-C10 (0.47 with DOPE vs 0.97 without it), LP-C16 (0.40 with DOPE vs 0.79 without it), and LP-C18 (0.18 with DOPE vs 0.53 without it) showed the most significant response to the addition of DOPE to the delivery system.
Figure 8.
Quantification of downregulation of mRNA levels of selected targeted proteins. MDA-MB-231 cells were treated with peptide/siRNA complexes with peptide/siRNA w/w ratios of 40:1 and 120:1 for linear and cyclic peptides, respectively, while the effect of incorporation of DOPE in the delivery system was also evaluated. Lipofectamine 2000 (Lipo.) was used as the positive control. The mRNA levels of KSP (A) and JAK2 (B) was quantified after 48 h of exposure to siRNA complexes using quantitative PCR. Data are presented as mean of RQ compared to cells exposed to saline (normalized using mRNA levels of β-actin), n = 3, and the error bars represent standard deviation. The asterisks identify the study groups that demonstrated a significant increase in silencing efficiency as a result of DOPE incorporation. DOPE (low) and (high) indicate the siRNA uptake of the cells exposed to DOPE (with no peptide) using the lowest and highest DOPE final concentrations used in the other study groups included in the experiment. (C) Correlation between silencing efficiency for the two selected targets: RQs presented in (A) and (B) are plotted on x and y axes, respectively, and the correlation factor (R) was calculated. The silencing data show significant correlation (p < 0.01) between the two selected targets.
Overall, with the addition of DOPE, the highest silencing efficiency against KSP and JAK2 was achieved by LP-C18 (RQ of 0.18) and LP-C16 (RQ of 0.32), respectively (Figure 8A,B). DOPE alone (with no peptide) did not induce significant silencing for either of the selected proteins, even with the highest final concentration of DOPE used in this study. The silencing efficiency of delivered siRNA against KSP and JAK2 showed a strong correlation (R = 0.82; p < 0.01), as illustrated in Figure 8C.
Discussion
The therapeutic possibilities with RNAi via siRNAs have not been utilized to the full potential, mostly because of the underperformance of the carriers. Cationic lipids or polymers are among the commonly studied nonviral carriers for nucleic acids because of their spontaneous tendency to interact with the cargo via interionic interaction. Neutralization of the siRNA electric charge in the process also enhances cell internalization by compacting nucleic acid and reducing the hydrophilicity of this type of molecule. Short (around 30 amino acids or less) CPPs have also been used as an alternative approach, either by covalently binding the peptide to siRNA or by complex formation using positively charged peptides.2 In 2013, van Asbeck et al. reported a study on siRNA complex formation with a small library of arginine- or lysine-rich linear peptides and demonstrated that using a stearyl group to replace the acetyl group could enhance the silencing efficiency against the luciferase reporter gene in a leukemia cell line,21 which the authors contributed to induced amphipathic characteristics. In this study, we used a variety of fatty acids with a wide range (C2–C18) to modify linear and cyclic peptides composed of five arginine and two lysines.
Herein, we designed two libraries of linear and cyclic peptides with similar amino acid compositions and studied the effect of hydrophobic modification of synthesized peptides with fatty acids with a wide range of chain lengths. We hypothesized that the cyclic structure of the second library could offer higher cell-penetrating ability and stability and less toxicity over linear counterparts. In our synthesized peptides, no inter- and/or intra-dicarboxylate cross-linked products were observed because 3 equiv of fatty acids was used in the coupling reactions in a very dilute solution in DMF that minimizes coupling of both carboxylic acids in each fatty acid in the intramolecular or intermolecular reactions. Furthermore, all final products were purified by HPLC as described in the manuscript. MALDI spectral data did not exhibit the presence of intermolecular or intramolecular products.
To evaluate the ability of synthesized peptides to bind to siRNA, we performed a binding assay we had previously reported using the SYBR Green dye to quantify the free siRNA.22 Among the linear-modified peptides, LP-C2, LP-C4, LP-C6, and LP-C10 did not achieve a complete siRNA binding in the range of peptide/siRNA w/w ratio studies. In addition to a relatively smaller fatty acid conjugation, these peptides have another common characteristic, the dicarboxylic conjugated fatty acid, which means a −COOH group that exists at the free end of the fatty acid could be partially ionized in a neutral pH and create a negatively charged group. This ionization could potentially interfere with the interionic interaction with negatively charged siRNA and therefore limit the extent of binding. The only other linear peptides conjugated with dicarboxylic fatty acids in the library is LP-C8, which demonstrated one of the highest BC50 values among the linear peptides that achieved complete binding. Cyclic peptides showed a higher BC50. In fact, in addition to the unmodified cyclic peptide, only CP-C8*, CP-C10, CP-C12, and CP-C18 showed complete binding at the highest peptide/siRNA w/w ratio studied (80:1) (Figure 2). Cyclic peptides are more rigid and expected to be more stable toward degradation. However, this rigidity of the molecular structure could hinder the interionic interaction with siRNA molecules, which could explain the higher BC50 for this library. A significant correlation between the BC50 values and the length of the conjugation was not observed for either of the libraries (data not shown).
Difatty acyl peptide conjugates have different physicochemical properties. A number of factors are involved in interactions with siRNA, such as charge, hydrophobicity, size, overall structure, and conformation of the conjugates. The level of hydrophobicity/charge ratio is a key factor for the binding of amphiphilic conjugates to siRNA. The chain length and its flexibility are postulated to generate highly condensed positively charged amphiphilic particles for efficient interactions with siRNA. However, our data indicate that the enhanced hydrophobicity of the delivery system (both by increasing the chain length in the fatty acid structure and by addition of DOPE as an extra hydrophobic component) plays a significant role in the level of interaction of the delivery system with the cell membrane.
The size and electric surface charge of siRNA carriers are crucial characteristics for the efficiency of delivery, and we used a Zetasizer to analyze both the hydrodynamic diameter and ζ-potential of the peptide/siRNA complexes. The ability of a carrier to condense the nucleic acid cargo is considered one of the important factors in protecting siRNA and internalizing it into the target cells.23 We observed a gradual decrease in the particle size of the complexes with increasing peptide/siRNA w/w ratio from 1:1 to 20:1 for most of the synthesized peptides (Figure 2A,B), which could indicate complex formation. Most of the complexes showed a particle size of less than 200 nm, which is considered the proper size for in vitro siRNA delivery.24 We previously reported a similar range of particle sizes for hydrophobically modified low-molecular-weight polyethyleneimine (PEI),22 which was shown effective for in vitro and in vivo delivery of siRNA to a variety of cell lines.25,26 The presence of a positive surface charge seems to be important for the cellular uptake and limits the risk of particle aggregation. It has been suggested that a slightly positive surface charge (+2 to +10) is the ideal surface charge to optimize cell surface interactions while minimizing nontarget carrier binding.27,28 The ζ-potential of siRNA complexes is expected to move toward positive values as the peptide/siRNA ratio is increased. This gradual increase was observed for most of the linear peptides (except LP-C4 and LP-C14, which showed negative values for the peptide/siRNA w/w ratio of 20:1) and a few of the cyclic peptides (Figure 3C,D). A negative value suggests incomplete binding, which confirms our observations in the binding study.
One of the major hurdles in siRNA delivery is the susceptibility to enzymatic degradation, which limits siRNA in vivo half-life to a few minutes.29 A gel electrophoresis method was used to quantify intact siRNA after exposure of peptide/siRNA complexes to serum. Whereas positive and negative controls confirmed the validity of the method, most of the peptides failed to protect siRNA for 24 h. Only C16- and C18-conjugated peptides showed protection against enzymatic degradation, which could be due to the maximum hydrophobicity of these modified peptides. This protection potentially limits the interaction of siRNA with the enzymatic components in the aqueous surrounding medium (Figure 4). A near complete protection was also provided by the unmodified cyclic peptide, which could be explained by the high affinity for siRNA binding (the highest among the cyclic library) and a small positive ζ-potential at a ratio of peptide/siRNA ratio of 20:1 (N/P ratio of 44.7). Higher peptide/siRNA w/w ratios were used for the cyclic library (up to 80:1) because of limited serum stability and BC50 observed in this study and previous experiments.
We selected two approaches to evaluate the toxicity of the delivery system: exposure of the cells to different concentrations of peptides and treatment with peptide/scrambled siRNA with corresponding peptide concentrations. Both approaches were evaluated based on the speculations that positively charged carriers enhance cellular uptake via the whole formation in the cell membrane.30 Because complex formation changes the overall charge of the peptide, the cytotoxicity of the peptide alone and as a complex with siRNA could potentially be different. LP-C12 showed an unusual toxicity compared to other synthesized peptides in both cell lines used (Figure 5). LP-C16 and LP-C18 were the other peptides that showed a higher toxicity compared to other peptides in the highest concentration examined. Whereas the higher toxicity of the C16- and C18-conjugated peptides could be explained by a closer interaction with the cell membrane because of increased hydrophobicity of the peptide, the unusual toxicity of LP-C12 might be due to an incomplete purification process that could expose the cells to impurities remaining in peptide solution. The trend of toxicity was similar for both approaches, which indicates that the toxicity mechanism might be independent of the electrical charge of the peptide. Most synthesized peptides were nontoxic in the selected concentration range, and overall, the cyclic library was less toxic than the linear equivalents.
In general, conjugation with C18* demonstrated a safer toxicity profile compared to conjugation with C18. Whereas these two conjugations showed a comparable safety profile for the cyclic peptide in MDA-MB-231 cells, LP-C18 and CP-C18 showed a more significant effect on cell viability in both selected cell lines both as peptide alone (in the concentration range of 5–20 μg/mL) and as a peptide/siRNA complex, especially at peptide/siRNA w/w ratios of 20:1 and 40:1, compared to LP-C18* and CP-C18*, respectively. These differences suggest that the conjugation of the peptide with unsaturated linoleic acid in C18* (compared to saturated stearic acid in C18) reduces the interaction with the cell membrane, which was confirmed by a significantly higher cellular internalization in the subsequent experiments for C18. The cytotoxicity data also indicated an unusual toxicity for LP-C12, which was not seen in the cyclic peptide counterpart. This might be an indication of an impurity in the synthesis product that was not detected in our analysis of the peptide. However, we did not investigate this unusual effect further because LP-C12 did not show promising siRNA delivery characteristics in the selected cell lines.
Our cellular internalization studies confirmed our results from our particle characterization studies. Linear peptide conjugated with C16 and C18 fatty acids showed the highest efficiency in internalizing FAM-labeled siRNA into both human cancer cell lines studied. The average fluorescence signal, however, was significantly higher in MDA-MB-231 triple-negative breast cancer cells (Figure 6B). This was surprising because in one of our previous studies, we observed the opposite (more efficient delivery to MDA-MB-435 cells using fatty acid-conjugated low-molecular-weight PEI).26 The average fluorescence signal and the percentage of cells showing siRNA uptake were both lower for the cyclic peptides. CP-C16 and CP-C18 showed the highest delivery efficiency in this library. This was not surprising because we perform our uptake studies in serum-rich media, and the binding affinity, ζ-potential of complexes, and protection against enzymatic degradation all play important roles in the cellular internalization of siRNA. Whereas the cyclic peptide library showed an overall lower affinity to bind to siRNA and demonstrated a lower ζ-potential for most of the cyclic peptides and a lower serum stability of siRNA, only the most hydrophobic cyclic peptides (C16- and C18-conjugated peptides) showed some promising results in those characterization experiments. Noticing the role of hydrophobicity of the carriers in internalizing siRNA, we repeated the cell internalization study with selected peptides, adding a new hydrophobic component to the delivery system. DOPE has been commonly used as a component in different liposomal and polymeric siRNA delivery systems.31−34 We investigated the effect of incorporating DOPE into the peptide/siRNA complexes and used DOPE/siRNA as a control group to evaluate the effect of DOPE alone in siRNA delivery. In our studies, DOPE did not show a significant siRNA uptake; however, the addition of DOPE to peptide/siRNA complexes had a significant effect on the siRNA delivery efficiency of all selected peptides (except caprylic acid-conjugated cyclic peptide and CP-C16 at the lower selected peptide/siRNA ratio). The enhancing effect of DOPE was specifically significant on LP-C16, where the average of the fluorescence signal increased more than 10-fold (from 893 to 11861) for a peptide/siRNA w/w ratio of 40:1 (N/P ratio of 43.2) (Figure 6). Lipids have been extensively used as the individual carrier of siRNA (cationic lipids as lipoplexes) or as a hydrophobic modifier or component in other delivery systems. It is speculated that they enhance cellular internalization by enhancing the interaction of the particle with the cell membrane, which in turn could trigger a cellular uptake. On the basis of observations on the effect of fatty acid chain length on the efficiency of siRNA delivery, we believe this hydrophobic interaction is the main mechanism involved in the internalization of these particles. Our microscopic images (Figure 7D) confirmed the impact of DOPE addition to the delivery system.
Cell internalization of siRNA is only the first step in protein silencing. The ability to escape endosomes and the timely release from the complexes are the other necessary steps in successful mRNA targeting and degradation. To demonstrate the silencing efficiency of our hydrophobically modified peptides, we targeted two unrelated proteins that have been extensively studied for their roles in cell proliferation and survival. KSP is a member of kinesin superfamily of microtubule motors involved in mitosis and has been targeted by small-molecule drugs35,36 and siRNA37,38 as an anticancer strategy. JAK2, on the other hand, is a pivotal component of the JAK/STAT pathway that has been studied extensively for its key role in development, proliferation, differentiation, and survival of cancer cells.39 JAK inhibitors, for example, ruxolitinib and tofacitinib, are used in clinics.40 We have previously reported JAK2 silencing via siRNA delivery and its impact on inhibiting the growth of human cancer cells.25,26,41 Our silencing experiments in this project confirmed our observations in the cell internalization studies. Whereas C16- and C18-conjugated peptides (especially linear peptides) showed silencing against both the targeted proteins, the addition of DOPE enhanced the silencing efficiency significantly for all selected peptides, except CP-C16. This is consistent with our observations in the uptake study and confirms that the addition of DOPE not only increases the cellular uptake but this uptake also translates to efficient protein silencing. Maybe even more significantly, the significant correlation between the silencing efficiency against two selected proteins indicates the efficiency of this approach regardless of the expression level of the targeted protein or its role in intracellular mechanisms. Our experiments also demonstrated a silencing efficiency that was comparable, and even superior in some cases, to commercially available Lipofectamine 2000. Whereas the RQ of ∼0.5 was observed for siRNAs delivered by Lipofectamine, LP-C18 reduced the mRNA levels of KSP and JAK2 to RQs of 0.18 and 0.32, respectively (Figure 8).
These data indicate that an optimal fatty acyl chain length is required for efficient interaction with the cell membrane and internalization, as observed for both cyclic and linear peptides. The difatty acyl linear peptides, LP-C16 and LP-C18, are more efficient siRNA carriers when compared with the corresponding difatty acyl cyclic peptides, CP-C16 and CP-C18, possibly because of the more efficient interactions of positively charged amino acids and fatty acyl chains with the cell membrane and siRNA in more flexible linear conjugates versus the cyclic ones. It was also noticed that the presence of more rigid unsaturated linoleic acid in both LP-C18* and CP-C18* significantly reduced siRNA stability and siRNA internalization when compared with the corresponding more flexible stearic acid analogues LP-C18 and CP-C18 peptides, respectively. These data indicate that the fatty acid chain length and peptide and fatty acid flexibility are the determinant factors for optimal interactions with the membrane and siRNA and cellular internalization.
Conclusions
Peptides have shown promising efficiency as cell-penetrating components in siRNA delivery. We hypothesized that synthesizing hydrophobically modified cationic peptides can be an effective approach in forming stable complexes with siRNA that are capable of internalizing into the target cells and target any mRNA in the cytoplasm. Our results showed that the level of hydrophobicity is a key factor in the efficiency of the carriers. In fact, the addition of an extra hydrophobic element significantly enhanced the performance of designed peptides as shown in C16 and C18 peptides. This study reveals the promising potential of a number of peptide/lipid nanoparticles, such as C16 and C18 peptides, for effective siRNA delivery that justifies further evaluation in vivo.
Experimental Section
Materials
For the list of materials used in this project, please see the Supporting Information.
Methods
Assembly of Amino Acids on Resins for the Synthesis of Resin-Attached Linear and Cyclic Peptides
The synthesis of R5K2 peptides was achieved by using 0.50 mmol Fmoc-L-Arg(Pbf)-Wang or H-Arg(Pbf)-2-ClTrt resins for linear and cyclic peptides, respectively. The resin was added to a reusable fritted glass reaction vessel and placed on the Tribute peptide synthesizer. Each vial was filled with 3 equiv of amino acids (Fmoc-Arg(Pbf)-OH and Fmoc-Lys(Dde)-OH for linear peptides and Fmoc-Arg(Pbf)-OH and Fmoc-Lys(Boc)-OH for cyclic peptides) and 2.5 equiv of HBTU as the coupling reagent. All of the solvent bottles were pressurized with nitrogen, while the reaction vessel was also pressurized and mixed by the nitrogen inlet and was vacuum-sealed. The deprotection of the Fmoc group was performed by addition of piperidine in DMF (20 v/v %). The program configured to remove the Fmoc-protecting group of the resin-attached amino acid before coupling the first amino acid residue. Once the first amino acid residue was coupled, the remaining residues were added to the reaction vessel by single coupling. The poor coupling of the hydrophobic fatty acids for linear and cyclic peptides was mostly likely because of some aggregation, possibly due to the interactions between hydrophobic residues during the fatty acid coupling. To overcome the aggregation, double-coupling was used for the linear peptide fatty acid conjugation. In regard to the fatty acid conjugations for the cyclic peptide, the reactions were performed under solution phase air-free peptide synthesis. A percentage yield of approximately 60–70% was observed for most of the linear peptide compared to 40–50% for cyclic peptide conjugations.
Synthesis of Difatty Acyl Linear Peptide Conjugates
Fmoc-protecting group was removed from the Fmoc-L-Arg(Pbf)-Wang resin (0.50 mmol) in the presence of piperidine in DMF (20 v/v %). For the synthesis of linear peptides with capped N-acetyl amino group (LP-C2 and LP-C4), two Fmoc-Lys(Dde)-OH amino acids were used in coupling; then, the N-terminal Fmoc group was removed first in the presence of piperidine (20%) in DMF followed by capping with acetic anhydride before removal of the Dde group and fatty acid (C2 or C4) conjugation as described below. For the remaining linear peptides, two kinds of lysines were used: Fmoc-Lys(Dde)-OH and Boc-Lys(Fmoc)-OH. The Boc group was removed after the fatty acid conjugation described below and after the final cleavage. Once the peptide sequence was assembled on the resin and completed by the Tribute, the side chain Dde of the first lysine and the Fmoc group of the second lysine were removed by addition of hydrazine (2%, 1 mL) and DMF (98%, 49 mL). The mixture of 2% hydrazine was mixed with the resin for 2 h at room temperature under nitrogen. The resin was drained and washed with DMF three times (3 × 3 mL). The coupling of the fatty acid was carried out after the deprotection of lysines. The reaction was accomplished by mixing 3 equiv of the fatty acid with 2.5 equiv of HBTU and 6 equiv of DIPEA in DMF. The fatty acid coupling was repeated to ensure maximum coupling to the unprotected peptide. The fatty acids used for both libraries included acetic anhydride (C2), succinic acid (C4), adipic acid (C6), suberic acid (C8), caprylic acid (saturated eight carbon chain, C8*), sebacic acid (C10), lauric acid (C12), myristic acid (C14), palmitic acid (C16), stearic acid (C18), and linoleic acid (unsaturated 18 carbon chain, C18*). The final cleavage of the linear peptide was performed by adding a cocktail of TFA, anisole, thioanisole, and DTT. A cleavage cocktail (20 mL) was prepared by adding 18 mL of TFA, 0.50 mL of anisole, 0.50 mL of thioanisole, and 50 mg of DTT, and it was added to the linear peptide assembled on the Wang resin. The mixture was then stirred for 4 h to afford crude difatty acyl linear peptide conjugates.
Synthesis of Difatty Acyl Cyclic Peptide Conjugates
H-Arg(Pbf)-2-ClTrt (0.5 mmol) was swelled in DMF. Fmoc-Arg(Pbf)-OH and Fmoc-Lys(Boc)-OH were used as building blocks. After assembly of the peptide sequence, the cleavage of the peptide from the 2-chlorotrityl resin was performed manually from the resin with acetic acid/trifluoroethanol/DCM (1:2:7 v/v/v) for 2 h at room temperature. The solution was filtered and washed with DCM to remove acetic acid. The cleavage solution was diluted with n-hexane and evaporated four times. The remaining acetic acid was evaporated overnight by vacuum. To cyclize the peptide, HOAT (122.50 mg, 0.3 mmol, 3 equiv) was added to the dried linear peptide in a 500 mL flask. The mixture of DCM and DMF (1:4) was added to the flask, followed by addition of 140 μL of N,N′-diisopropylcarbodiimide into the solution under dry nitrogen. The reaction was continued for 24 h to complete. Once the cyclization was complete, deprotection was carried out with reagent R (TFA/anisole/thioanisole/DTT), followed by the coupling of fatty acid in the presence of HBTU and DIPEA, according to the method described above for linear peptides. The same selected fatty acids that were used for the generation of the difatty acyl linear peptide library were also utilized to afford the crude difatty acyl cyclic peptide conjugates.
Analysis and Purification Method
All of the crude peptide fatty acid conjugates were purified by using the preparative HPLC to achieve the minimum purity of 90%. The dicarboxylic fatty acid cyclic and linear conjugates were dissolved in 2:1 water/acetonitrile (v/v), and the solution was filtered before injection to the HPLC. The HPLC system included an XBridge BEH130 reversed-phase C18 column (19 × 250 mm, 10 μm particle size, Waters Corp., Milford, MA, USA) maintained at 50 °C, LC-20AD pumps (Shimadzu Corp., Kyoto, Japan), an FRC-10A fraction collector, and an SPD-M20A diode array detector. For shorter chain difatty acyl conjugates, gradient elution was performed at a flow rate of 10 mL/min with mobile phase composition ranging from 95:5 water/acetonitrile (v/v) to 5:95 water/acetonitrile (v/v) throughout the 60 min run. The mobile phase contained 0.1% TFA throughout the gradient. Hydrophobic difatty acyl peptides, containing longer fatty acid chains, were dissolved in a mixture of methanol/isopropanol alcohol (1:1, v/v), and a gradient system of methanol/isopropanol alcohol with 0.1% TFA was used for dilution. The detector monitored the absorbance of the amide bond at the given wavelength of 215 nm.42,43 Mass spectra were collected from m/z 900 to 3500 using the positive reflective mode of MALDI mass spectrometer to characterize the molecular weight, using freshly prepared α-cyano-4-hydroxycinnamic acid (5 mg/mL) in acetonitrile (0.1% TFA) and water (0.1% TFA) (1:1, v/v) as the matrix. Using the FlexControl software (Bruker Corp.), the retention times for the peak tops of the chromatograph were determined from the ion chromatograms of the protonated difatty acyl peptide conjugates. MALDI-TOF mass spectrometry data for difatty acyl linear and cyclic peptides are shown in Table S1 (Supporting Information).
Complex Formation with Modified Peptides and siRNA
The peptide/siRNA complexes were formed in different w/w ratios for the variety of in vitro studies conducted for this project. The complexes formed spontaneously based on the ion/ion interaction in normal saline. For each formulation, the required amount of siRNA stock solution (10 μM) was added to normal saline, and then the calculated amount of peptide stock solution (1 mg/mL) was added to the solution. The mixture was gently mixed and was incubated at room temperature for 30 min to ensure complete complexation. The peptide/siRNA ratios used in complex formations are reported as weight/weight ratios as well as N/P ratios. The N/P ratios were calculated using the following formula
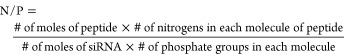 |
All siRNAs used in this project contain 21 base pairs in each strand, with a 2 nucleotide overhang on each strand, which represents 20 phosphodiester linkages for each siRNA molecule. The siRNA targeting JAK2 (Qiagen catalogue # SI02659657) represented the following sequence: sense: 5′-CCAUCAUACGAGAUCUUAATT-3′; antisense: 5′UUAAGAUCUCGUAUGAUGGCT-3′). The sequence for other siRNAs used in this study was not provided to the authors as they were considered proprietary information. The number of protonable nitrogen atoms differed among the peptides, and the net positive charge varied between 2 and 7. Table S2 summarizes the N/P ratios for some of the w/w ratios reported in this manuscript.
Amount of Peptide Required for 50% siRNA Binding (BC50)
To evaluate the effect of hydrophobic modification of the linear and cyclic peptides on their ability to bind to siRNA, a previously reported SYBR Green II dye exclusion assay was performed using a wide range of N/P ratios. This method has been extensively used for DNA44 and has been shown to correlate with the results of the electrophoretic mobility shift assay for quantification of siRNA binding.45 Complexes were formed using scrambled siRNA and all modified peptides in both libraries with N/P ratios up to 8 for linear library and N/P ratios up to 90 for cyclic library. After completion of complex formation, SYBR Green II dye solution (1:10 000 dilution in double-distilled water) was immediately added to each complex solution, and the fluorescence of the solution was quantified in a 96-well plate at a λ excitation of 485 nm and a λ emission of 527 nm. Free siRNA (with similar concentration to complexes) was used as the control to determine the unbound siRNA in each complex formulation, using the following equation
A completely bond siRNA sample was also prepared as a positive control using a previously reported hydrophobically modified, low-molecular-weight PEI.22 To determine the ratio required for 50% siRNA binding (BC50) for each hydrophobically modified peptide, the percentage of siRNA bond to each peptide versus the peptide/siRNA ratios was plotted, and BC50 was calculated based on the linear portion of the curve for each experiment.
Light Scattering for Size and ζ-Potential of siRNA Complexes
The hydrodynamic diameter and surface charge of the complexes were analyzed using a Malvern Nano ZS Zetasizer (Westborough, MA) at 25 °C in disposable cuvettes and folded capillary cells, respectively. The peptide/siRNA complexes were prepared with a variety of peptide/siRNA w/w ratios ranging from 1:1 to 20:1 using scrambled siRNA to study the effect of the composition of the complex on these physical characteristics. The ζ-potential of the complexes was determined at 40 V, using Smoluchowski approximation.
Protection of siRNA against Enzymatic Degradation
To study the capability of the fatty acid-conjugated linear and cyclic peptides to enhance the stability of siRNA in biological environments, samples of unprotected scrambled siRNA (positive control) and peptide/siRNA complexes at different peptide/siRNA ratios, ranging from 2.5:1 to 10:1 for linear and from 20:1 to 80:1 for cyclic peptides, were exposed to FBS solutions. After completion of complex formation for each formulation, each sample was added to a 25 v/v % FBS solution in Hank’s balanced salt solution (HBSS), and the mixture was incubated at 37 °C for 24 h. A sample of unprotected siRNA in HBSS was used as the negative control, representing 100% intact siRNA. After incubation, the complexes were dissociated using a 2:3 mixture of heparin (5% solution in normal saline) and ethylenediaminetetraacetic acid (0.5 mM), and the samples were analyzed using 1% agarose gel (with 1 μg/mL ethidium bromide) at 70 V for 20 min. Ultraviolet illumination (Gel-Doc system, Bio-Rad; Hercules, CA) was used to visualize the gel, and the intensity of the bands (representing remaining intact siRNA) was quantified by Image J software.
Cell Lines
The human breast cancer cell line MDA-MB-231 and the human cell line MDA-MB-435 were purchased from American Type Culture Collection (ATCC; Manassas, VA). MDA-MB-231 and MDA-MB-435 were cultured in Dulbecco’s modified Eagle’s medium and in Roswell Park Memorial Institute 1640 medium, respectively. Both media were supplemented with 10% (v/v) FBS, 100 U/mL penicillin, and 100 μg/mL streptomycin. Cells were maintained under a normal condition of 37 °C and 5% CO2 under a humidified atmosphere and were subcultured when 80–100% confluent.
Toxicity of the Difatty Acyl Peptides and Peptide/siRNA Complexes
The safety profiles of the linear and cyclic peptide libraries were evaluated in two different human cancer cell lines by exposing the cells to the peptides alone and in peptide/siRNA complexes. Confluent cultures (∼800 000 cells/mL) of MDA-MB-231 and MDA-MB-435 human cancer cell lines were seeded in 96-well plates. After 24 h, the cells were exposed to different concentrations of peptide solutions or peptide/siRNA complexes. Linear peptide solutions were added to both cell lines to create final concentrations of 2.5, 5, 10, and 20 μg/mL, whereas cyclic peptides were added to the final concentrations of 5, 10, 20, and 40 μg/mL. Peptide/siRNA complexes were prepared with different peptide/siRNA w/w ratios of 5:1, 10:1, 20:1, and 40:1 (translating to 2.5, 5, 10, and 20 μg/mL peptide final concentration) for linear and 10:1, 20:1, 40:1, and 80:1 (equivalent to 5, 10, 20, and 40 μg/mL peptide final concentration) for cyclic peptides. The final concentration of scrambled siRNA was 36 nM in all cytotoxicity studies performed with peptide/siRNA complexes. The cells were then incubated for 72 h at 37 °C with controlled CO2 and humidity. A standard 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay was performed after the incubation period to determine the cell viability as a percentage of NT cells.
Internalization of siRNA into Human Cancer Cell Lines
The capability of difatty acyl linear and cyclic peptide libraries in internalizing siRNA into human cells was evaluated using FAM-labeled scrambled siRNA and flow cytometry and fluorescent microscopy. Both MDA-MB-231 and MDA-MB-435 cell lines were used for these studies and were seeded in 24-well plates (∼200 000 cells per well). Peptide/siRNA complexes were prepared in ratios of 10:1 and 20:1 for the linear peptide library and 80:1 and 120:1 for the difatty acyl cyclic peptide library, with a final concentration of 36 nM for the fluorescent-labeled scrambled siRNA. After addition of the siRNA complexes to the cell culture media, the cells were incubated in 37 °C and standard growth conditions for 24 h. After the incubation period, for flow cytometry studies, the cells were washed with clear HBSS (×2), trypsinized, and fixed using 3.7% formaldehyde solution. The suspended cells were analyzed with a BD-FACSVerse flow cytometer (BD Biosciences; San Jose, CA) using the fluorescein isothiocyanate (FITC) channel to quantify cell-associated fluorescence. To investigate the effect of the addition of DOPE to the delivery system, DOPE was added to the complexes in a peptide/DOPE molar ratio of 3:2. After each flow cytometry analysis, the percentage of cells with a fluorescence signal and the mean fluorescence of the cell population were calculated based on the calibration of the signal gated with NT cells (as the negative control), so that the autofluorescence would be ∼1% of the population.
To obtain fluorescent images of the siRNA cellular uptake, cells were seeded on a glass slide in a 6-well plate (at ∼400 000 cells/mL), and after 24 h, they were exposed to FAM-labeled siRNA complexes. After 24 h of exposure to FAM-labeled siRNA, the cells were washed with clear HBSS and fixed in 3.7% formaldehyde solution for 30 min at room temperature. The cell membrane and nucleus were stained with Texas Red Phalloidin (Invitrogen) (1:250 in HBSS) and 4′,6-diamidino-2-phenylindole (DAPI), respectively. A KEYENCE BZ-X700 all-in-one fluorescence microscope (KEYENCE Corp. of America, Itasca, IL) was used to produce fluorescence images using the 40× objective and different filters for DAPI, FITC, and Texas Red. NT cells were used as negative controls.
In Vitro Silencing of Selected Proteins
The efficacy of siRNA delivered by selected peptides in silencing specific proteins was studied in the MDA-MB-231 cell line. Confluent cells were seeded in 6-well plates (∼400 000 cells/mL) and were treated with scrambled siRNA (CsiRNA) or targeted siRNAs (final concentration of 54 nM) delivered with selected peptides with or without DOPE. After 48 h of incubation at 37 °C and under normal cell culture conditions, the cells were lysed with TRIzol (1 mL for each 1 × 106 cells) for cDNA synthesis and real-time polymerase chain reaction (RT-PCR). The cell lysates were incubated at room temperature for 5 min, before chloroform (with a 1:5 v/v ratio to TRIzol) was added to the lysates. The tubes were mixed and incubated for 2–3 min at room temperature, before the aqueous phase was collected. Isopropanol was added to the precipitate and pellet RNA using centrifuge (12 000g for 10 min in 4 °C). The pellet was then washed with 75% ethanol, and the extracted RNA was then dissolved in RNase-free water. The total extracted RNA in each sample was determined by BioSpec-Nano spectrophotometer (Shimadzu, Columbia, MD). To synthesize cDNA, 0.5 μg of RNA was reverse-transcribed using an iScriptTM reverse transcription supermix and a C1000 Touch thermocycler (Bio-Rad, Hercules, CA), following the manufacturer’s guidelines. A CFX96TM optical module (Bio-Rad, Hercules, CA) was used for the RT-PCR analysis, where human β-actin was used as the endogenous gene to normalize the mRNA level of targeted proteins. Primers were designed using primer Blast software available at The National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov/) and were synthesized with the following sequences by IDT Technologies (Coralville, Iowa): β-actin (forward: 5′-CCA CCC CAC TTC TCT CTA AGG A-3′; reverse: 5′-AAT TTA CAC GAA AGC AAT GCT-3′), KSP (forward: 5′-TCA CAA AAG CAA TGT GGA AAC CTA-3′; reverse: 5′-TCT GTC CAA AGA TTCA TTA ACT TGC A-3′), and JAK2 (forward: 5′-AAC TGC AGA TGC ACA TCA TTA CCT-3′; reverse: 5′-TCG AAA TTG GGC CAT GAC A-3′). Primers were validated to assure equal efficiency at different cDNA concentrations (−0.1 < slope < 0.1 for the ΔCT vs cDNA dilution graph) and selectivity for the protein of interest (by gel electrophoresis and determination of the length of the resulting DNA). The mRNA levels were determined by calculating ΔCT, ΔΔCT, and RQ using the endogenous gene and “no treatment” group as reference points.
Statistical Analysis
The significance of the differences observed reported values (including siRNA uptake, mRNA levels, and cell viability) were evaluated using Student’s t test (p < 0.05). Standard deviations were calculated for all data and are represented by the error bars in all figures. Pearson’s correlation coefficient was calculated where indicated, and its significance was determined by the t test, according to the following equation
where r and n are the correlation coefficient and the number of samples, respectively. The calculated value of t was compared to p values for each degree of freedom to determine the significance of the correlation.
Acknowledgments
The authors would like to acknowledge the financial support for this research from the Chapman University School of Pharmacy.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsomega.7b00741.
Chemical structures of linear and peptide fatty acid conjugates; MALDI-TOF mass spectrometry data for difatty acyl linear and cyclic peptides; N/P ratio for selected w/w peptide/siRNA ratios used in this project; MALDI-TOF mass spectra; and materials (PDF)
Author Contributions
H.D. and M.S. contributed equally.
The authors declare no competing financial interest.
Supplementary Material
References
- Fire A.; Xu S.; Montgomery M. K.; Kostas S. A.; Driver S. E.; Mello C. C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998, 391, 806–811. 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Aliabadi H. M.; Landry B.; Sun C.; Tang T.; Uludağ H. Supramolecular assemblies in functional siRNA delivery: where do we stand?. Biomaterials 2012, 33, 2546–2569. 10.1016/j.biomaterials.2011.11.079. [DOI] [PubMed] [Google Scholar]
- Lehto T.; Ezzat K.; Wood M. J. A.; Andaloussi S. E. Peptides for nucleic acid delivery. Adv. Drug Delivery Rev. 2016, 106, 172–182. 10.1016/j.addr.2016.06.008. [DOI] [PubMed] [Google Scholar]
- Tai W.; Gao X. Functional peptides for siRNA delivery. Adv. Drug Delivery Rev. 2016, 110–111, 157–168. 10.1016/j.addr.2016.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Järver P.; Mäger I.; Langel Ü. In vivo biodistribution and efficacy of peptide mediated delivery. Trends Pharmacol. Sci. 2010, 31, 528–535. 10.1016/j.tips.2010.07.006. [DOI] [PubMed] [Google Scholar]
- Adami R. C.; Rice K. G. Metabolic stability of glutaraldehyde cross-linked peptide DNA condensates. J. Pharm. Sci. 1999, 88, 739–746. 10.1021/js990042p. [DOI] [PubMed] [Google Scholar]
- Futaki S.; Suzuki T.; Ohashi W.; Yagami T.; Tanaka S.; Ueda K.; Sugiura Y. Arginine-rich peptides. An abundant source of membrane-permeable peptides having potential as carriers for intracellular protein delivery. J. Biol. Chem. 2001, 276, 5836–5840. 10.1074/jbc.m007540200. [DOI] [PubMed] [Google Scholar]
- Ohyama A.; Higashi T.; Motoyama K.; Arima H. In Vitro and In Vivo Tumor-Targeting siRNA Delivery Using Folate-PEG-appended Dendrimer (G4)/α-Cyclodextrin Conjugates. Bioconjugate Chem. 2016, 27, 521–532. 10.1021/acs.bioconjchem.5b00545. [DOI] [PubMed] [Google Scholar]
- Chaudhary A.; Garg S. siRNA delivery using polyelectrolyte-gold nanoassemblies in neuronal cells for BACE1 gene silencing. Mater. Sci. Eng., C 2017, 80, 18–28. 10.1016/j.msec.2017.05.101. [DOI] [PubMed] [Google Scholar]
- Chen J.; Sun X.; Shao R.; Xu Y.; Gao J.-q.; Liang W.-q. VEGF siRNA delivered by polycation liposome-encapsulated calcium phosphate nanoparticles for tumor angiogenesis inhibition in breast cancer. Int. J. Nanomed. 2017, 12, 6075–6088. 10.2147/ijn.s142739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng G.; Shen Y.; Zhao R.; Chen F.; Zhang Y.; Xu A.; Shao J. Dual-Targeting Multifuntional Mesoporous Silica Nanocarrier for Codelivery of siRNA and Ursolic Acid to Folate Receptor Overexpressing Cancer Cells. J. Agric. Food Chem. 2017, 65, 6904–6911. 10.1021/acs.jafc.7b03047. [DOI] [PubMed] [Google Scholar]
- Campani V.; Salzano G.; Lusa S.; De Rosa G. Lipid Nanovectors to Deliver RNA Oligonucleotides in Cancer. Nanomaterials 2016, 6, 131. 10.3390/nano6070131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H.; Kanasty R. L.; Eltoukhy A. A.; Vegas A. J.; Dorkin J. R.; Anderson D. G. Non-viral vectors for gene-based therapy. Nat. Rev. Genet. 2014, 15, 541–555. 10.1038/nrg3763. [DOI] [PubMed] [Google Scholar]
- Titze-de-Almeida R.; David C.; Titze-de-Almeida S. S. The Race of 10 Synthetic RNAi-Based Drugs to the Pharmaceutical Market. Pharm. Res. 2017, 34, 1339–1363. 10.1007/s11095-017-2134-2. [DOI] [PubMed] [Google Scholar]
- Huang Y. Preclinical and Clinical Advances of GalNAc-Decorated Nucleic Acid Therapeutics. Mol. Ther.—Nucleic Acids 2017, 6, 116–132. 10.1016/j.omtn.2016.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smekalova E. M.; Kotelevtsev Y. V.; Leboeuf D.; Shcherbinina E. Y.; Fefilova A. S.; Zatsepin T. S.; Koteliansky V. lncRNA in the liver: Prospects for fundamental research and therapy by RNA interference. Biochimie 2016, 131, 159–172. 10.1016/j.biochi.2016.06.007. [DOI] [PubMed] [Google Scholar]
- Shirazi A. N.; Oh D.; Tiwari R. K.; Sullivan B.; Gupta A.; Bothun G. D.; Parang K. Peptide amphiphile containing arginine and fatty acyl chains as molecular transporters. Mol. Pharmaceutics 2013, 10, 4717–4727. 10.1021/mp400539r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirazi A. N.; Tiwari R. K.; Oh D.; Banerjee A.; Yadav A.; Parang K. Efficient delivery of cell impermeable phosphopeptides by a cyclic peptide amphiphile containing tryptophan and arginine. Mol. Pharmaceutics 2013, 10, 2008–2020. 10.1021/mp400046u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirazi A. N.; Paquin K. L.; Howlett N. G.; Mandal D.; Parang K. Cyclic peptide-capped gold nanoparticles for enhanced siRNA delivery. Molecules 2014, 19, 13319–13331. 10.3390/molecules190913319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh D.; Shirazi A. N.; Northup K.; Sullivan B.; Tiwari R. K.; Bisoffi M.; Parang K. Enhanced cellular uptake of short polyarginine peptides through fatty acylation and cyclization. Mol. Pharmaceutics 2014, 11, 2845–2854. 10.1021/mp500203e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Asbeck A. H.; Beyerle A.; McNeill H.; Bovee-Geurts P. H. M.; Lindberg S.; Verdurmen W. P. R.; Hällbrink M.; Langel Ü.; Heidenreich O.; Brock R. Molecular parameters of siRNA–cell penetrating peptide nanocomplexes for efficient cellular delivery. ACS Nano 2013, 7, 3797–3807. 10.1021/nn305754c. [DOI] [PubMed] [Google Scholar]
- Aliabadi H. M.; Landry B.; Bahadur R. K.; Neamnark A.; Suwantong O.; Uludağ H. Impact of lipid substitution on assembly and delivery of siRNA by cationic polymers. Macromol. Biosci. 2011, 11, 662–672. 10.1002/mabi.201000402. [DOI] [PubMed] [Google Scholar]
- Boussif O.; Lezoualc’h F.; Zanta M. A.; Mergny M. D.; Scherman D.; Demeneix B.; Behr J. P. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proc. Natl. Acad. Sci. U.S.A. 1995, 92, 7297–7301. 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard K. A.; Rahbek U. L.; Liu X.; Damgaard C. K.; Glud S. Z.; Andersen M. Ø.; Hovgaard M. B.; Schmitz A.; Nyengaard J. R.; Besenbacher F.; Kjems J. RNA interference in vitro and in vivo using a novel chitosan/siRNA nanoparticle system. Mol. Ther. 2006, 14, 476–484. 10.1016/j.ymthe.2006.04.010. [DOI] [PubMed] [Google Scholar]
- Aliabadi H. M.; Mahdipoor P.; Bisoffi M.; Hugh J. C.; Uludağ H. Single and Combinational siRNA Therapy of Cancer Cells: Probing Changes in Targeted and Nontargeted Mediators after siRNA Treatment. Mol. Pharmaceutics 2016, 13, 4116–4128. 10.1021/acs.molpharmaceut.6b00711. [DOI] [PubMed] [Google Scholar]
- Aliabadi H. M.; Maranchuk R.; Kucharski C.; Mahdipoor P.; Hugh J.; Uludağ H. Effective response of doxorubicin-sensitive and -resistant breast cancer cells to combinational siRNA therapy. J. Controlled Release 2013, 172, 219–228. 10.1016/j.jconrel.2013.08.012. [DOI] [PubMed] [Google Scholar]
- Cao N.; Cheng D.; Zou S.; Ai H.; Gao J.; Shuai X. The synergistic effect of hierarchical assemblies of siRNA and chemotherapeutic drugs co-delivered into hepatic cancer cells. Biomaterials 2011, 32, 2222–2232. 10.1016/j.biomaterials.2010.11.061. [DOI] [PubMed] [Google Scholar]
- Convertine A. J.; Benoit D. S. W.; Duvall C. L.; Hoffman A. S.; Stayton P. S. Development of a novel endosomolytic diblock copolymer for siRNA delivery. J. Controlled Release 2009, 133, 221–229. 10.1016/j.jconrel.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raemdonck K.; Vandenbroucke R. E.; Demeester J.; Sanders N. N.; De Smedt S. C. Maintaining the silence: reflections on long-term RNAi. Drug Discovery Today 2008, 13, 917–931. 10.1016/j.drudis.2008.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S.; Leroueil P. R.; Janus E. K.; Peters J. L.; Kober M.-M.; Islam M. T.; Orr B. G.; Baker J. R. Jr.; Holl M. M. B. Interaction of polycationic polymers with supported lipid bilayers and cells: nanoscale hole formation and enhanced membrane permeability. Bioconjugate Chem. 2006, 17, 728–734. 10.1021/bc060077y. [DOI] [PubMed] [Google Scholar]
- Dakwar G. R.; Braeckmans K.; Ceelen W.; De Smedt S. C.; Remaut K. Exploring the HYDRAtion method for loading siRNA on liposomes: the interplay between stability and biological activity in human undiluted ascites fluid. Drug Delivery Transl. Res. 2017, 7, 241–251. 10.1007/s13346-016-0329-4. [DOI] [PubMed] [Google Scholar]
- Gu L.; Nusblat L. M.; Tishbi N.; Noble S. C.; Pinson C. M.; Mintzer E.; Roth C. M.; Uhrich K. E. Cationic amphiphilic macromolecule (CAM)–lipid complexes for efficient siRNA gene silencing. J. Controlled Release 2014, 184, 28–35. 10.1016/j.jconrel.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori Y.; Hara E.; Shingu Y.; Minamiguchi D.; Nakamura A.; Arai S.; Ohno H.; Kawano K.; Fujii N.; Yonemochi E. siRNA delivery into tumor cells by cationic cholesterol derivative-based nanoparticles and liposomes. Biol. Pharm. Bull. 2015, 38, 30–38. 10.1248/bpb.b14-00526. [DOI] [PubMed] [Google Scholar]
- Rezaee M.; Oskuee R. K.; Nassirli H.; Malaekeh-Nikouei B. Progress in the development of lipopolyplexes as efficient non-viral gene delivery systems. J. Controlled Release 2016, 236, 1–14. 10.1016/j.jconrel.2016.06.023. [DOI] [PubMed] [Google Scholar]
- Perez-Melero C. KSP inhibitors as antimitotic agents. Curr. Top. Med. Chem. 2014, 14, 2286–2311. 10.2174/1568026614666141130095532. [DOI] [PubMed] [Google Scholar]
- Sarli V.; Giannis A. Targeting the kinesin spindle protein: basic principles and clinical implications. Clin. Cancer Res. 2008, 14, 7583–7587. 10.1158/1078-0432.ccr-08-0120. [DOI] [PubMed] [Google Scholar]
- Marra E.; Palombo F.; Ciliberto G.; Aurisicchio L. Kinesin spindle protein SiRNA slows tumor progression. J. Cell. Physiol. 2013, 228, 58–64. 10.1002/jcp.24103. [DOI] [PubMed] [Google Scholar]
- Tabernero J.; Shapiro G. I.; LoRusso P. M.; Cervantes A.; Schwartz G. K.; Weiss G. J.; Paz-Ares L.; Cho D. C.; Infante J. R.; Alsina M.; Gounder M. M.; Falzone R.; Harrop J.; White A. C. S.; Toudjarska I.; Bumcrot D.; Meyers R. E.; Hinkle G.; Svrzikapa N.; Hutabarat R. M.; Clausen V. A.; Cehelsky J.; Nochur S. V.; Gamba-Vitalo C.; Vaishnaw A. K.; Sah D. W. Y.; Gollob J. A.; Burris H. A. 3rd First-in-humans trial of an RNA interference therapeutic targeting VEGF and KSP in cancer patients with liver involvement. Cancer Discovery 2013, 3, 406–417. 10.1158/2159-8290.cd-12-0429. [DOI] [PubMed] [Google Scholar]
- Kisseleva T.; Bhattacharya S.; Braunstein J.; Schindler C. W. Signaling through the JAK/STAT pathway, recent advances and future challenges. Gene 2002, 285, 1–24. 10.1016/s0378-1119(02)00398-0. [DOI] [PubMed] [Google Scholar]
- Dymock B. W.; See C. S. Inhibitors of JAK2 and JAK3: an update on the patent literature 2010–2012. Expert Opin. Ther. Pat. 2013, 23, 449–501. 10.1517/13543776.2013.765862. [DOI] [PubMed] [Google Scholar]
- Zhu P.; Aliabadi H. M.; Uludağ H.; Han J. Identification of Potential Drug Targets in Cancer Signaling Pathways using Stochastic Logical Models. Sci. Rep. 2016, 6, 23078. 10.1038/srep23078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldfarb A. R.; Saidel L. J.; Mosovich E. The ultraviolet absorption spectra of proteins. J. Biol. Chem. 1951, 193, 397–404. [PubMed] [Google Scholar]
- Zinieris N.; Leondiadis L.; Ferderigos N. Nα-Fmoc removal from resin-bound amino acids by 5% piperidine solution. J. Comb. Chem. 2005, 7, 4–6. 10.1021/cc049872d. [DOI] [PubMed] [Google Scholar]
- Saxena V.Preparation and Physical Characterization of DNA-Binding Cationic Liposomes. Liposomes; Methods in Molecular Biology; Humana Press, 2017; Vol. 1522, pp 245–250. [DOI] [PubMed] [Google Scholar]
- Neamnark A.; Suwantong O.; Bahadur K. C. R.; Hsu C. Y. M.; Supaphol P.; Uludağ H. Aliphatic lipid substitution on 2 kDa polyethylenimine improves plasmid delivery and transgene expression. Mol. Pharmaceutics 2009, 6, 1798–1815. 10.1021/mp900074d. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.