Abstract
In this study, the effect of the flavanone naringenin on the growth and genetic expression of the commensal gut microbes, Ruminococcus gauvreauii, Bifidobacterium catenulatum, and Enterococcus caccae, was analyzed. Analysis of growth curves revealed that Ruminococcus gauvreauii was unaffected by naringenin, Bifidobacterium catenulatum was slightly enhanced by naringenin, and Enterococcus caccae was severely inhibited by naringenin. Changes in genetic expression due to naringenin were determined using single-molecule RNA sequencing. Analysis revealed the following responses to naringenin: Ruminococcus gauvreauii upregulated genes involved in iron uptake; Bifidobacterium catenulatum upregulated genes involved in cellular metabolism, DNA repair and molecular transport, and downregulated genes involved in thymidine biosynthesis and metabolism; Enterococcus caccae upregulated pathways involved in transcription and protein transport and downregulated genes responsible for sugar transport and purine synthesis. For the first time, changes in growth and gene expression for commensal gut bacteria in response to naringenin were documented.
Keywords: Naringenin, gut microbiota, single-molecule RNA sequencing
Introduction
Naringenin is a plant polyphenol that belongs to the subgroup flavanone.1 It is found in citrus fruits, predominantly grapefruit, and to some extent in tomatoes, tomato skin, and ketchup.1-3 Due to individual differences in diet, it is difficult to accurately determine the daily intake of naringenin. However, it is estimated that the average dietary intake of polyphenols is between 0.15 and 1.0 g/d.4,5 Similar to other polyphenols, the consumption of naringenin is associated with multiple health benefits.6 Naringenin has been demonstrated to have antioxidant activity,2 and citrus flavanones have been demonstrated to have anti-inflammatory and anticancer properties.7 Naringenin also functions in regulating lipid metabolism,2 and the consumption of orange juice containing flavanones has been indicated in reducing low-density lipoprotein cholesterol8 and increasing high-density lipoprotein concentrations.9
Naringenin is an aglycone; however, it is typically stored as a glycoside in plants, most commonly attached to either d-glucose or l-rhamnose.3,10 After ingestion, naringenin and its glycosides will pass through the small intestine unabsorbed and enter the large intestine, which is host to the gut microbiota.6 Bioavailability of naringenin depends on removal of the sugar groups by the gut microbiota to produce the aglycone naringenin, which can then be absorbed in the large intestine.3,10,11 It has been previously demonstrated that the gut microbiota can not only remove the sugar moieties from the naringenin glycosides but also convert naringenin to phenolic acids through cleavage of the C-ring.1,10 However, although it is known that naringenin has both the opportunity and ability to interact with the gut microbiota, the effect of naringenin itself on the gut microbiota community remains unclear.
Research looking at the interaction of polyphenols and the gut microbiota has mainly focused on metabolism of polyphenols by the gut bacteria,12 although there have been some studies demonstrating that polyphenols can affect bacterial viability by stimulating beneficial microbes and inhibiting pathogens.4,6 For example, the growth of pathogenic bacteria Clostridium perfringens, Clostridium difficile, and some Bacteroides spp was significantly inhibited by tea polyphenols.5 Several polyphenols have been demonstrated to suppress adhesion of the pathogen Salmonella typhimurium and enhance adhesion of the probiotic Lactobacillus rhamnosus to human gut cells.13 However, these studies predominantly analyzed the effect of polyphenols in combination, and not individually, making it difficult to elucidate which polyphenols are responsible for the effect.
Previously, a study of the effect of naringenin on the growth of human intestinal bacteria was performed.12 However, this study only analyzed the effect of naringenin on bacterial growth without exploring the underlying cause. It is possible that naringenin affects bacteria at the genetic level, resulting in upregulation or downregulation of specific genes.14,15 In this case, the bacterial growth pattern may not change but the pattern of gene expression will become altered.14,15 Based on this hypothesis, to fully understand the effect of naringenin, both changes in growth and genetic expression need to be evaluated.
In this study, the effect of naringenin on the growth and genetic regulation of the commensal gut microbes Ruminococcus gauvreauii, Bifidobacterium catenulatum, and Enterococcus caccae was evaluated. These strains represent classical types of commensal microbes that would be found in the human large intestine and were all isolated from human feces.16-18 First, changes in growth due to the addition of naringenin were evaluated by culturing each strain with different concentrations of naringenin for 24 hours. During this time, the turbidity was measured at 0, 4, 8, 12, and 24 hours post inoculation using a densitometer to formulate the growth pattern of each species of bacteria. Second, gene expression profile of each species of bacteria treated with naringenin and the control group, with no naringenin added, was determined through single-molecule RNA sequencing via Helicos Technology and compared with identified changes in gene expression due to naringenin. This study provides evidence that the consumption of naringenin may result in modifications to the gut microbiota composition and gene expression. This effect may contribute to the overall health benefits associated with a diet high in naringenin.
Methods
Bacterial strains
All bacterial strains were ordered as freeze-dried ampoules from Deutsche SammLung von Mikroorganismen und Zellkulturen GmbH (DSMZ) in Germany: type strain Ruminococcus gauvreauii CCRI-16110 (19829), type strain Enterococcus caccae SS-1777 (19114), and type strain Bifidobacterium catenulatum B669 (16992). Each species of bacteria was cultured in strain-specific broth as defined by DSMZ. Prior to being used for experimentation, the bacteria were revived from frozen stock by growing overnight (16 hours) in strain-specific broth under anaerobic conditions at 37°C. Bacteria were inoculated and cultured overnight at least 2 times in sequence prior to its use.
Anaerobic broth preparation
The strain-specific broth was prepared according to the composition supplied by DSMZ and autoclaved at 120°C for 30 minutes using the liquid cycle. Oxygen was then removed from the autoclaved broth by heating while under negative pressure (nitrogen gas) for 10 minutes. The broth was then moved into a Bactron anaerobic chamber, where it was able to cool to room temperature while under anaerobic conditions. All broths were stored in the anaerobic chamber, at room temperature, for up to 3 weeks.
Peptone Yeast Glucose Broth (modified) was used to culture R gauvreauii and comprised the following ingredients in a final volume of 1 L deionized/distilled water: trypticase peptone 5.00 g, peptone 5.00 g, yeast extract 10.00 g, beef extract 5.00 g, glucose 5.00 g, K2HPO4 2.00 g, Tween 80 1.00 mL, cysteine-HCl × H2O 0.50 g, resazurin 1.00 mg, salt solution 40 mL (salt solution composition: CaCl2 × 2 H2O 0.25 g, MgSO4 × 7 H2O 0.50 g, K2HPO4 1.00 g, KH2PO4 1.00 g, NaHCO3 10.00 g, NaCl 2.00 g, distilled water up to 1000.00 mL), hemin solution 10 mL (50 mg hemin in 1 mL 1 N NaOH; final volume of 100 mL using distilled water, stored at 4°C) and vitamin K1 solution 200 µL (0.1 mL of vitamin K1 in 20 mL 95% ethanol and filter sterilized, stored at 4°C). The final pH was adjusted to 7.2 using 10 M NaOH or 37% HCl. Bifidobacterium media was used to culture B catenulatum and contains the following ingredients in a final volume of 1 L deionized/distilled water: casein peptone, tryptic digest 10.00 g, yeast extract 5.00 g, meat extract 5.00 g, Bacto Soytone 5.00 g, glucose 10.00 g, K2HPO4 2.00 g, MgSO4 × 7 H2O 0.20 g, MnSO4 × H20 0.05 g, Tween 80 1.00 mL, NaCl 5.00 g, cysteine-HCl × H2O 0.50 g, salt solution 40 mL (salt solution composition: CaCl2 × 2 H2O 0.25 g, MgSO4 × 7 H2O 0.50 g, K2HPO4 1.00 g, KH2PO4 1.00 g, NaHCO3 10.00 g, NaCl 2.00 g, distilled water up to 1000.00 mL), and resazurin 1.00 mg. The final pH was adjusted to 6.8 using 10 M NaOH or 37% HCl. Trypticase Soy Yeast Extract Medium was used to culture Enterococcus caccae and consisted of the following ingredients in a final volume of 1 L deionized/distilled water: trypticase soy broth 30.0 g and yeast extract 3.0 g. The final pH was adjusted to 7 to 7.2 using either 10 M NaOH or 37% HCl.
Preparation of naringenin
For each experiment, the polyphenol naringenin (W530098-500G; Sigma Aldrich, St. Louis, MO, USA) was dissolved in dimethyl sulfoxide (DMSO) to make a stock solution. Stock solutions were made to contain 100, 75, 50, and 25 mg/mL naringenin. For all testing, 10 µL DMSO stock solution of naringenin was added per 5 mL of anaerobic broth. This resulted in a final concentration of 200, 150, 100, and 50 µg/mL of naringenin in strain-specific broth containing 1% DMSO. For each experiment, the negative control consisted of naringenin added to the broth without bacteria, and the positive control consisted of bacteria cultured in anaerobic broth containing 1% DMSO without the addition of naringenin.
Evaluating changes in growth
To ensure oxygen-free conditions, all work was performed using a Bactron anaerobic chamber. To test the effects of naringenin on each bacterial strain, the following steps were performed, in the manner described previously14,15: to begin, strain-specific anaerobic broth was aliquoted into hungate tubes at a volume of 5 mL per tube.
Each individual hungate tube was sealed using a rubber septa and screw cap (Chemglass, Vineland, NJ, USA) and then stored at room temperature in the anaerobic chamber.
Each bacterial strain was inoculated the night before beginning each experiment and grown to confluency at 37°C. To start the experiment, the DMSO stock solution of naringenin was injected into the hungate tubes containing the aliquoted broth (1 mL needle; 25 gauge syringe). Next, the confluent culture of bacteria was diluted, in the broth specific to that type of bacteria, to 0.5 McFarland units (MU) and 100 µL of this culture was injected into each 5-mL hungate tube (1-mL needle; 25-gauge syringe). To ensure proper distribution after injection, each hungate tube was briefly vortexed and a densitometer was used to determine the MU (time 0 read). The hungate tubes were then incubated at 37°C in an anaerobic chamber. Finally, at 4, 8, 12, and 24 hours post inoculation a densitometer was used to determine the MU for each hungate tube. For each concentration of naringenin tested, 6 hungate tubes of broth containing the desired polyphenol were included. Three of these tubes were used as the negative control and were not inoculated with bacteria. The other 3 tubes were designated as the experimental group and were inoculated with bacteria. Changes in growth were considered statistically significance when P ≤ .05, as determined using a 2-tailed, Student t test.
Evaluating gene expression
RNA extraction
Total RNA was extracted from bacteria cultured with naringenin or DMSO only, at the concentration determined to have the most influence on growth pattern. Ruminococcus gauvreauii was cultured in the presence of 200 µg/mL of naringenin and 1% DMSO for 16 hours. Bifidobacterium catenulatum was cultured in the presence of 200 µg/mL of naringenin and 1% DMSO for 8 hours. Enterococcus caccae was cultured in the presence of 50 µg/mL naringenin and 1% DMSO for 10 hours. After culturing for the determined amount of time, 15 mL of each were centrifuged at 5000g for 10 minutes. The bacterial pellet was then resuspended in 1 mL Trizol and stored at −80°C.
To extract total RNA, the following Zymo Direct-zol RNA Miniprep protocol as previously described14,15: 250 µL of Trizol was added to 100 µL of sample followed by 250 µL of pure ethanol. This mixture was vortexed to homogenize. The entire mixture was loaded onto a Zymo-Spin II C column, centrifuged for 30 seconds at 16 000g, washed with 400 µL of RNA wash buffer, and then centrifuged again for 30 seconds. About 80 µL of DNase I reaction mixture (5 µL DNAse I and 75 µL of DNA digestion buffer) was added and the column was incubated for 15 minutes at room temperature. After incubation, 400 µL of Direct-zol RNA prewash was added to the column and then the column was centrifuged for 30 seconds. The flow through was discarded, and this wash step was repeated. About 700 µL of RNA wash buffer was then added to the column and then the column was centrifuged for 2 minutes. This step was repeated twice. The total RNA was then eluted from the column by adding 30 µL of RNase (ribonuclease)-free water to the column and centrifuged for 30 seconds. The final product was stored at −80°C until needed.
RNA sequencing and data analysis
Single-molecule RNA sequencing was carried out using a Helicos sequencer by SeqLL (Boston, MA, USA) and the results were used to generate gene expression profiles, as previously described.14,15 The gene expression for each species of bacteria treated with either DMSO or naringenin was quantified following these steps. First, the fully assembled genomes were downloaded from the National Center of Biological Information (NCBI; https://www.ncbi.nlm.nih.gov/nuccore/NC013198).19 Next, using UCLUST,20 reads were mapped to their corresponding genomes. Any reads aligning to multiple locations were assigned to their corresponding best match so that the genes are presented by their number of unique reads. Then, using RPKM21 metrics, the reads were normalized according to abundance and length. Changes in gene expression were determined through the comparison of the experimental group (naringenin treated) with the control group (DMSO treated). Any gene that had a statistically significant higher number of reads compared with the DMSO-only control was considered upregulated and, conversely, any gene that had a lower number of reads was considered downregulated.
Results
Effects of naringenin on the growth and genetic regulation of R gauvreauii
The phenotypic effect of naringenin on R gauvreauii was determined by comparing the rate and pattern of growth for cultures treated with different concentrations of naringenin to the control group, which had no naringenin. There was a recorded enhancement of growth for R gauvreauii at 12 hours post inoculation for all concentrations of naringenin tested, although this enhancement did not reach statistical significance (Figure 1A). This is also demonstrated in the percent of control, where at 12 hours post inoculation, culture density was greater than 100% for all concentrations of naringenin (Figure 1B). In fact, R gauvreauii treated with 150 µg/mL naringenin resulted in 127.5% of growth when compared with the control at 12 hours and 105.9% of growth when compared with the control at 24 hours post inoculation (Figure 1B). Although these differences were not statistically significant, they were evident and indicated that the addition of naringenin may not be immaterial. Based on this supposition, it was hypothesized that even though there was no significant change in phenotype, the effects of naringenin on R gauvreauii may be evident in the pattern of gene expression.
Figure 1.
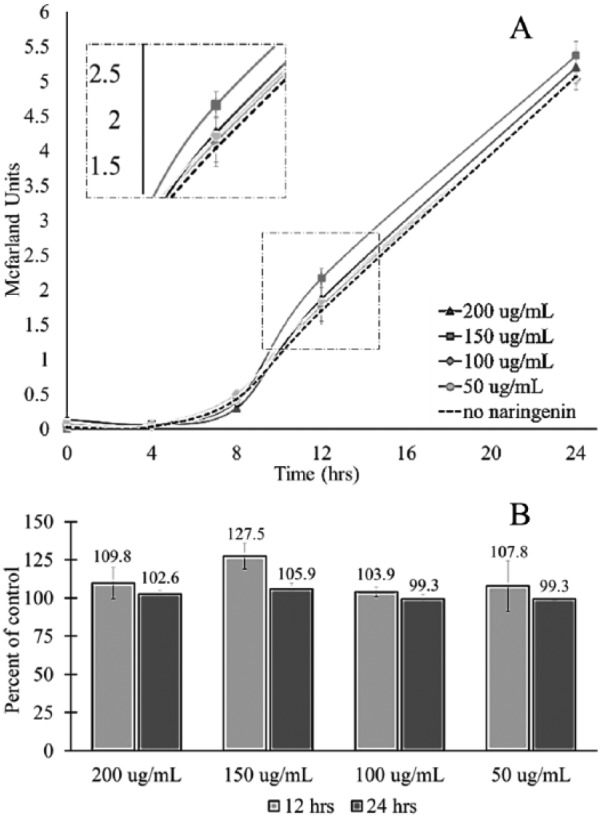
The effect of naringenin on the growth of Ruminococcus gauvreauii. Ruminococcus gauvreauii was cultured for 24 hours in the presence of increasing concentrations of naringenin. A densitometer was used to measure the McFarland units at designated time points. (A) The 24-hour growth curve of R gauvreauii supplemented with increasing concentrations of naringenin. The dotted line represents bacteria grown without naringenin. (B) The percent of control for R gauvreauii cultured in each concentration of naringenin at 12 and 24 hours post inoculation.
The gene expression profile of R gauvreauii grown in the presence of 200 µg/mL of naringenin was assembled and compared with the expression profile of the control, no naringenin added (Tables 1 and 2). For R gauvreauii treated with naringenin, 210 separate genes were identified. Out of the 210 genes, only 12 were found to have a greater than 1.25-fold increase in expression (Table 1) and only 6 produced a greater than 1.25-fold decrease in expression (Table 2). For brevity, only genes that had a greater than 1.25-fold change in expression were considered in the discussion.
Table 1.
Genes upregulated by Ruminococcus gauvreauii in response to naringenin.
| Gene ID | Gene description | Fold↑ | Function |
|---|---|---|---|
| H604_RS0100855 | Hypothetical protein | 1.6 | Unknown |
| H604_RS0100345 | Iron transporter FeoA | 1.4 | Iron uptake |
| H604_RS0100045 | Hypothetical protein | 1.4 | Unknown |
| H604_RS0100920 | Proton-coupled thiamine transporter YuaJ | 1.3 | Thiamine transport |
| H604_RS0100110 | Ribose ABC transporter permease | 1.3 | Molecular translocation |
| H604_RS0100455 | Hypothetical protein | 1.3 | Unknown |
| H604_RS19840 | Hypothetical protein | 1.3 | Unknown |
| H604_RS0100765 | Hypothetical protein | 1.3 | Unknown |
| H604_RS0100350 | Iron transporter FeoA | 1.3 | Iron uptake |
| H604_RS0100290 | ATP-binding protein | 1.3 | Molecular translocation |
| H604_RS0100260 | Hypothetical protein | 1.3 | Unknown |
| H604_RS0101200 | Phosphatidic acid phosphatase | 1.3 | Metabolism |
Table 2.
Genes downregulated by Ruminococcus gauvreauii in response to naringenin.
| Gene ID | Gene description | Fold↓ | Function |
|---|---|---|---|
| H604_RS0100940 | Hypothetical protein | 1.4 | Unknown |
| H604_RS0100620 | Nucleotidyltransferase | 1.3 | DNA synthesis/repair |
| H604_RS0101055 | d-ribose transporter ATP-binding protein | 1.3 | Molecular translocation |
| H604_RS0100980 | Hypothetical protein | 1.3 | Unknown |
| H604_RS0100360 | Hypothetical protein | 1.3 | Unknown |
| H604_RS0100485 | tRNA (N(6)-l-threonylcarbamoyladenosine(37)-C(2))-methylthiotransferase MtaB | 1.3 | Translation |
Effects of naringenin on the growth and genetic regulation of B catenulatum
The rate and pattern of growth for B catenulatum treated with different concentrations of naringenin were compared with a control group, which had no naringenin. For B catenulatum, the addition of naringenin produced no significant difference in growth at 0, 4, 8 and 24 hours post inoculation (Figure 2A). However, at 12 hours post inoculation, there was a significant increase in growth for B catenulatum treated with all concentrations of naringenin (Figure 2A). This is best demonstrated in the percent of control, where at 12 hours post inoculation, B catenulatum treated with naringenin produced growth greater than 100% of the control (Figure 2B). Although the observed increase in growth for B catenulatum treated with naringenin was significant, it was not extensive, only ranging from 106.4% to 109.4% of control (Figure 2B). There was also no distinct connection observed between an increase in dose and culture density, indicating that the observed enhancement is not dose dependent (Figure 2A and B). Although the detected phenotypic change of B catenulatum due to naringenin was small, it provided evidence that this polyphenol does exert an effect on B catenulatum. Based on this conclusion, it was hypothesized that the underlying cause of this phenotypic change could be determined by analyzing changes to gene expression.
Figure 2.
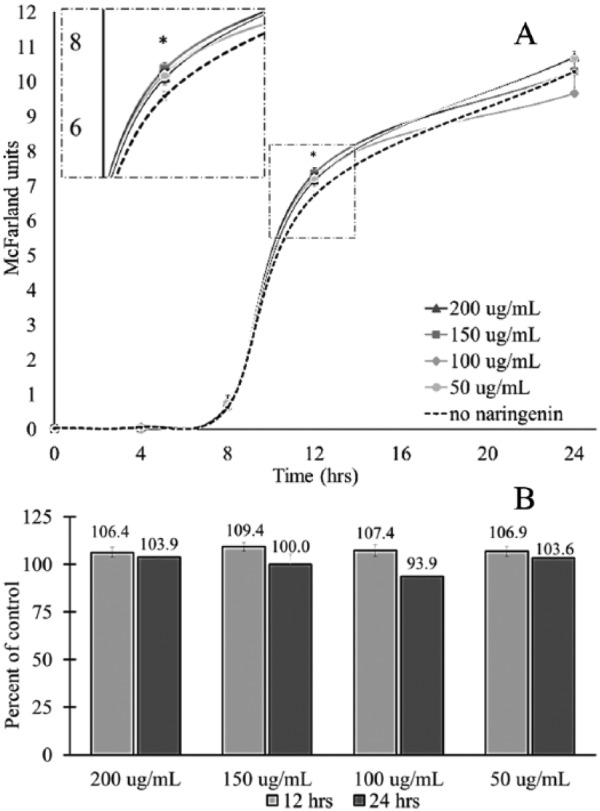
The effect of naringenin on the growth of Bifidobacterium catenulatum. Bifidobacterium catenulatum was cultured for 24 hours in the presence of increasing concentrations of naringenin. A densitometer was used to measure the McFarland units at designated time points. The * mark indicates that there was a statistically significant difference between the control and experimental groups at that time point, according to a 2-tailed, Student t test (P < .05). (A) The 24-hour growth curve of B catenulatum supplemented with increasing concentrations of naringenin. The dotted line represents bacteria grown without naringenin. (B) The percent of control for B catenulatum cultured in each concentration of naringenin at 12 and 24 hours post inoculation.
To elucidate the genotypic effect of naringenin on B catenulatum, the gene expression profile of B catenulatum grown in the presence of 200 µg/mL of naringenin was compared with the expression profile of the control (Tables 3 and 4). A total of 745 genes were identified, 108 genes produced over a 1.25-fold increase in expression, with 38 producing over a 1.5-fold increase in expression (Table 3). Of the genes identified, 44 produced a greater than 2.0-fold decrease in expression, and 17 had a greater than 2.5-fold decrease in expression (Table 4). For concision, only genes with a greater than 1.5-fold increase and 2.5-fold decrease in gene expression were considered for discussion.
Table 3.
Genes upregulated by Bifidobacterium catenulatum in response to naringenin.
| Gene ID | Gene description | Fold↑ | Function |
|---|---|---|---|
| BBCT_0807 | Hypothetical protein | 3.7 | Unknown |
| BBCT_146 | Conserved hypothetical protein | 2.8 | Unknown |
| BBCT_0952 | Conserved hypothetical protein | 2.2 | Unknown |
| BBCT_0976 | Conserved hypothetical protein | 2.1 | Unknown |
| BBCT_1237 | Putative lipoprotein signal peptidase | 2.0 | Protein secretion |
| BBCT_0743 | Conserved hypothetical protein | 2.0 | Unknown |
| BBCT_0923 | Conserved hypothetical protein | 2.0 | Unknown |
| BBCT_1231 | Conserved hypothetical protein | 1.9 | Unknown |
| BBCT_0715 | Hypothetical protein | 1.9 | Unknown |
| BBCT_1471 | ATP synthase epsilon subunit | 1.8 | ATP synthesis |
| BBCT_0629 | Truncated conserved hypothetical protein | 1.8 | Unknown |
| BBCT_0233 | Conserved hypothetical protein | 1.7 | Unknown |
| BBCT_0204 | Phosphopantetheine adenylyltransferase | 1.7 | Metabolism |
| BBCT_0970 | Holliday junction DNA helicase RuvA | 1.7 | DNA repair |
| BBCT_0948 | Conserved hypothetical protein | 1.7 | Unknown |
| BBCT_1598 | Truncated conserved hypothetical protein | 1.6 | Unknown |
| BBCT_1557 | Conserved hypothetical protein | 1.6 | Unknown |
| BBCT_1216 | Conserved hypothetical protein | 1.6 | Unknown |
| BBCT_0983 | Tryptophan synthase alpha subunit | 1.6 | Protein synthesis |
| BBCT_0680 | Transcriptional regulator | 1.6 | Transcription |
| BBCT_1093 | Putative acetyltransferase | 1.5 | Metabolism |
| BBCT_1624 | Conserved hypothetical protein | 1.5 | Unknown |
| BBCT_1013 | Hypothetical protein | 1.5 | Unknown |
| BBCT_1342 | Copper-transporting ATPase | 1.5 | Copper transport |
| BBCT_1688 | Conserved hypothetical protein | 1.5 | Unknown |
| BBCT_1169 | Dipeptide ABC transporter permease component | 1.5 | Molecular transport |
| BBCT_1680 | Galactoside transport protein | 1.5 | Molecular transport |
| BBCT_0013 | Conserved hypothetical protein | 1.5 | Unknown |
| BBCT_1574 | N-acetylglucosamine-6-phosphate deacetylase | 1.5 | Metabolism |
| BBCT_0175 | 6-phosphogluconate dehydrogenase-like protein | 1.5 | Metabolism |
| BBCT_0364 | Conserved hypothetical protein | 1.5 | Unknown |
| BBCT_0688 | Conserved hypothetical protein | 1.5 | Unknown |
| BBCT_0180 | Conserved hypothetical protein | 1.5 | Unknown |
| BBCT_1635 | Hypothetical protein | 1.5 | Unknown |
| BBCT_1478 | ATP synthase subunit A | 1.5 | ATP synthesis |
| BBCT_0202 | Conserved hypothetical protein | 1.5 | Unknown |
| BBCT_1379 | Conserved hypothetical protein | 1.5 | Unknown |
| BBCT_0529 | Conserved hypothetical protein | 1.5 | Unknown |
Table 4.
Genes downregulated by Bifidobacterium catenulatum in response to naringenin.
| Gene ID | Gene description | Fold ↓ | Function |
|---|---|---|---|
| BBCT_1116 | Deoxyuridine 5′-triphosphate nucleotidohydrolase | 3.8 | Thymidine biosynthesis |
| BBCT_0098 | Transcriptional regulator | 3.3 | Transcription |
| BBCT_1543 | UDP-galactopyranose mutase | 3.3 | Cell wall synthesis |
| BBCT_0990 | Ribosome recycling factor | 3.2 | Protein synthesis |
| BBCT_1179 | Nicotinate-nucleotide pyrophosphorylase | 3.1 | Metabolism |
| BBCT_0954 | Conserved hypothetical protein | 3.1 | Unknown |
| BBCT_1033 | Arginine repressor | 2.9 | Transcription |
| BBCT_1640 | Hypothetical protein | 2.8 | Unknown |
| BBCT_1011 | Carbohydrate kinase | 2.8 | Metabolism |
| BBCT_1078 | Conserved hypothetical protein | 2.8 | Unknown |
| BBCT_1329 | Putative transposase | 2.7 | Stress response |
| BBCT_1062 | DNA ligase | 2.6 | DNA repair |
| BBCT_0095 | Putative glutaredoxin | 2.6 | Stress response |
| BBCT_1163 | Conserved hypothetical protein | 2.5 | Unknown |
| BBCT_1455 | l-ribulose-5-phosphate 4-epimerase | 2.5 | Metabolism |
| BBCT_0034 | Putative phosphoprotein phosphatase | 2.5 | Metabolism |
| BBCT_0008 | Glutamate dehydrogenase | 2.5 | Metabolism |
Effects of naringenin on the growth and genetic regulation of E caccae
The pattern and rate of growth for E caccae cultured with increasing concentrations of naringenin was compared with E caccae cultured with no naringenin (Figure 3). For E caccae, at 4, 8, 12, and 24 hours post inoculation, growth was severely and statistically inhibited at all concentrations of naringenin examined (Figure 3A). At 24 hours post inoculation compared with the control, treatment with 200 µg/mL naringenin produced only 49.3% growth, treatment with 150 µg/mL naringenin resulted in 48.7% growth, treatment with 100 µg/mL naringenin produced 60.5% growth, and treatment with 50 µg/mL of naringenin resulted in 75.0% growth (Figure 3B). These results indicate that the inhibition of growth is dose dependent and that while suppression of growth is severe, E caccae is able to partially recover from this inhibition over time.
Figure 3.
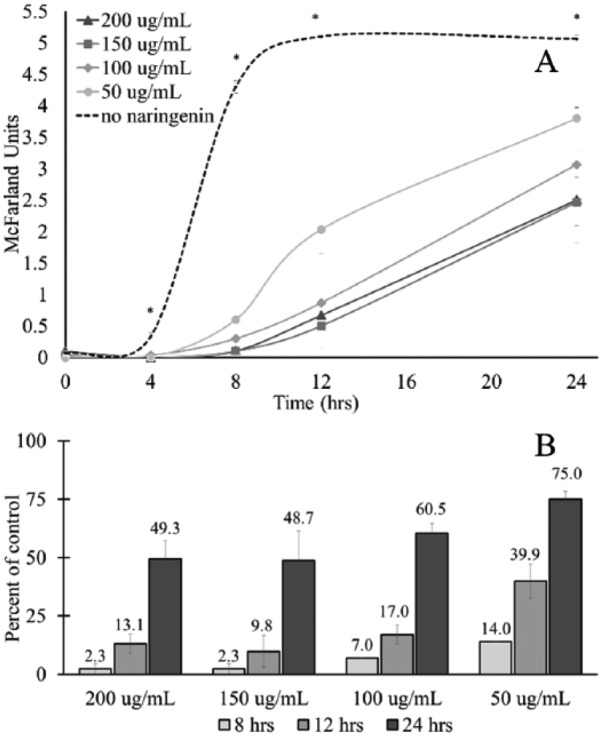
The effect of naringenin on the growth of Enterococcus caccae. Enterococcus caccae was cultured for 24 hours in the presence of increasing concentrations of naringenin. A densitometer was used to measure the McFarland units at designated time points. The * mark indicates that there was a statistically significant difference between the control and experimental groups at that time point, according to a 2-tailed, Student t test (P < .05). (A) The 24-hour growth curve of E caccae supplemented with increasing concentrations of naringenin. The dotted line represents bacteria grown without naringenin. (B) The percent of control for E caccae cultured in each concentration of naringenin at 8, 12, and 24 hours post inoculation.
Not only did naringenin affect the rate of growth but it also changed the pattern of growth for E caccae. Without naringenin, the exponential growth phase occurred between 4 and 8 hours post inoculation, with growth leveling off at 12 hours post inoculation with subsequent entrance into the stationary phase (Figure 3A). The addition of naringenin suppressed entrance into the exponential phase until 8 hours post inoculation for 50 µg/mL naringenin (Figure 3A). However, for E caccae treated with 100, 150, or 200 µg/mL naringenin, there was no obvious entrance into either exponential or stationary phase (Figure 3A). Based on these results, it was hypothesized that the observed change in phenotype of E caccae in response to naringenin would be apparent at the genotypic level.
The gene expression profile of E caccae cultured in the presence of 50 µg/mL of naringenin was assembled and compared with the expression profile of the control. For E caccae treated with naringenin, 759 separate genes were identified (Tables 5 and 6). Of the 759 genes, 343 were upregulated (45.2% of total), of which 132 were hypothetical proteins. A total of 79 genes produced a greater than 1.25-fold increase in expression, with 28 producing a greater than 1.5-fold increase in expression compared with the control (Table 5). Out of the 759 genes, 416 were downregulated (54.8% of total), of which 156 were hypothetical proteins. Of the downregulated genes, 145 presented a greater than 1.25-fold decrease in expression and 49 genes produced a greater than 1.5-fold decrease in expression compared with the control (Table 6). For brevity, only genes with a greater than 1.5-fold increase and 2.5-fold decrease in gene expression were considered for discussion.
Table 5.
Genes upregulated by Enterococcus caccae in response to naringenin.
| Gene ID | Gene description | Fold↑ | Function |
|---|---|---|---|
| UC7_RS13995 | Hypothetical protein | 2.1 | Unknown |
| UC7_RS15645 | Hypothetical protein | 2.1 | Unknown |
| UC7_RS13400 | Hypothetical protein | 2.1 | Unknown |
| UC7_RS13840 | Hypothetical protein | 1.9 | Unknown |
| UC7_RS13985 | Hypothetical protein | 1.8 | Unknown |
| UC7_RS13845 | Hypothetical protein | 1.8 | Unknown |
| UC7_RS14745 | Transcriptional regulator | 1.7 | Transcription |
| UC7_RS12590 | Hypothetical protein | 1.7 | Unknown |
| UC7_RS15085 | Hypothetical protein | 1.7 | Unknown |
| UC7_RS14600 | DNA helicase | 1.7 | DNA replication |
| UC7_RS14805 | ATP-grasp domain-containing protein | 1.7 | Energy utilization |
| UC7_RS13975 | Hypothetical protein | 1.6 | Unknown |
| UC7_RS14295 | Preprotein translocase subunit SecG | 1.6 | Protein translocation |
| UC7_RS16575 | Hypothetical protein | 1.6 | Unknown |
| UC7_RS13125 | Hypothetical protein | 1.6 | Unknown |
| UC7_RS12740 | Hypothetical protein | 1.6 | Unknown |
| UC7_RS15375 | Transcriptional regulator | 1.6 | Transcription |
| UC7_RS16265 | RNA-binding protein | 1.6 | Transcription |
| UC7_RS14730 | Nucleotide sugar dehydrogenase | 1.6 | Metabolism |
| UC7_RS12975 | Hypothetical protein | 1.6 | Unknown |
| UC7_RS13675 | Hypothetical protein | 1.6 | Unknown |
| UC7_RS15295 | Hypothetical protein | 1.5 | Unknown |
| UC7_RS13470 | Hypothetical protein | 1.5 | Unknown |
| UC7_RS16155 | Hypothetical protein | 1.5 | Unknown |
| UC7_RS14625 | Hypothetical protein | 1.5 | Unknown |
| UC7_RS12585 | Cell division protein ZapA | 1.5 | Cell division |
| UC7_RS16570 | Hypothetical protein | 1.5 | Unknown |
| UC7_RS16330 | Acyl carrier protein | 1.5 | Protein synthesis |
Table 6.
Genes downregulated by Enterococcus caccae in response to naringenin.
| Gene ID | Gene description | Fold↓ | Function |
|---|---|---|---|
| UC7_RS15835 | Peroxiredoxin | 2.0 | Stress response |
| UC7_RS13735 | Hypothetical protein | 2.0 | Unknown |
| UC7_RS13805 | Hypothetical protein | 1.9 | Unknown |
| UC7_RS13955 | Protein-(glutamine-N5) methyltransferase, release factor-specific | 1.9 | Methylation |
| UC7_RS14195 | PTS β-glucoside transporter subunit EIIBCA | 1.9 | Sugar transport |
| UC7_RS15880 | Hypothetical protein | 1.9 | Unknown |
| UC7_RS14150 | Serine hydrolase | 1.8 | Metabolism |
| UC7_RS16020 | Hypothetical protein | 1.8 | Unknown |
| UC7_RS14330 | Teichoic acid glycosylation protein | 1.8 | Cell wall synthesis |
| UC7_RS14840 | Hypothetical protein | 1.8 | Unknown |
| UC7_RS13720 | Hypothetical protein | 1.8 | Unknown |
| UC7_RS11285 | Hypothetical protein | 1.8 | Unknown |
| UC7_RS13055 | Amidophosphoribosyltransferase | 1.8 | Purine synthesis |
| UC7_RS11075 | Holliday junction resolvase RecU | 1.7 | DNA repair |
| UC7_RS16300 | β-hydroxyacyl-ACP dehydratase | 1.7 | Protein synthesis |
| UC7_RS11225 | Hypothetical protein | 1.7 | Unknown |
| UC7_RS12935 | Phosphopantetheine adenylyltransferase | 1.7 | Metabolism |
| UC7_RS11045 | Hypothetical protein | 1.7 | Unknown |
| UC7_RS14715 | Hypothetical protein | 1.7 | Unknown |
| UC7_RS15840 | Hypothetical protein | 1.7 | Unknown |
| UC7_RS15355 | Hypothetical protein | 1.7 | Unknown |
| UC7_RS13565 | Hypothetical protein | 1.7 | Unknown |
| UC7_RS14510 | PTS mannose/fructose/sorbose family, IIA component | 1.6 | Sugar transport |
| UC7_RS11425 | Histidinol-phosphate aminotransferase | 1.6 | metabolism |
| UC7_RS15565 | PTS mannose/fructose/sorbose family, IID component | 1.6 | Sugar transport |
| UC7_RS10845 | Hypothetical protein | 1.6 | Unknown |
| UC7_RS15915 | Hypothetical protein | 1.6 | Unknown |
| UC7_RS12815 | Hypothetical protein | 1.6 | Unknown |
| UC7_RS15525 | Ureidoglycolate dehydrogenase | 1.6 | Purine metabolism |
| UC7_RS11535 | Hypothetical protein | 1.6 | Unknown |
| UC7_RS11490 | FMN reductase | 1.6 | Metabolism |
| UC7_RS14490 | Hypothetical protein | 1.6 | Unknown |
| UC7_RS11520 | Hypothetical protein | 1.6 | Unknown |
| UC7_RS10980 | Hypothetical protein | 1.6 | Unknown |
| UC7_RS14385 | Hypothetical protein | 1.6 | Unknown |
| UC7_RS15235 | Hypothetical protein | 1.6 | Unknown |
| UC7_RS13770 | CAAX amino protease | 1.5 | Protein synthesis |
| UC7_RS15515 | Hypothetical protein | 1.5 | Unknown |
| UC7_RS14435 | Hypothetical protein | 1.5 | Unknown |
| UC7_RS12990 | 6-phosphofructokinase | 1.5 | Glycolysis |
| UC7_RS15570 | PTS IIC component | 1.5 | Sugar transport |
| UC7_RS12210 | DNA-directed RNA polymerase subunit delta | 1.5 | Transcription |
| UC7_RS13285 | PTS trehalose IIBC component | 1.5 | Sugar transport |
| UC7_RS15260 | Hypothetical protein | 1.5 | Unknown |
| UC7_RS14935 | Ribosomal RNA large subunit methyltransferase A | 1.5 | Protein synthesis |
| UC7_RS14065 | Hypothetical protein | 1.5 | Unknown |
| UC7_RS15925 | ABC transporter ATP-binding protein | 1.5 | Molecular transport |
| UC7_RS14080 | ModE molybdate transport repressor domain-containing protein | 1.5 | Molecular transport |
| UC7_RS13105 | Xanthine phosphoribosyltransferase | 1.5 | Purine metabolism |
Discussion
Analysis of the rate and pattern of growth for R gauvreauii, B catenulatum, and E caccae revealed that the addition of naringenin variably affected bacterial phenotype. For R gauvreauii, treatment with naringenin resulted in no significant changes in phenotype, whereas for B catenulatum, treatment resulted in an increase in growth at 12 hours post inoculation. For E caccae, naringenin dramatically inhibited growth at 4, 8, 12, and 24 hours post inoculation and altered the pattern of growth by suppressing entrance into exponential phase. Although R gauvreauii, B catenulatum, and E caccae were all exposed to the same amount of naringenin for the same length of time, the outcomes were incongruous. It was hypothesized that these differences in observed phenotype would be detected in the pattern of gene expression.
Analysis of the gene expression profiles for R gauvreauii revealed that the addition of naringenin produced only minimal changes (Tables 1 and 2). There was only one gene identified with a greater than 1.5-fold increase in expression; unfortunately, this gene codes for a hypothetical protein and therefore its function is unknown (Table 1). Interestingly, the addition of naringenin resulted in the upregulation of 2 genes identified as iron transporter FeoA genes, with a 1.4- and 1.3-fold increase (Table 1). The FeoA gene is part of the Feo system, a ubiquitous system in bacteria that is responsible for ferrous iron transport.22 This system is crucial for anaerobic bacteria, playing a role in multiple cellular mechanisms, stress response, and also contributing to virulence.23 Although both of these genes exhibit only a small increase in expression, the fact that there are multiple genes in this pathway upregulated makes this observation more relevant. It is possible that the observed enhancement of growth at 12 hours, where growth was over 100% of control for all concentrations, is related to this increase in iron uptake. If this is the case, it would indicate that the gene expression pathways activated by R gauvreauii in response to naringenin not only counteract any negative effects of the polyphenol but also are ultimately beneficial to the bacteria itself.
There were only a few genes identified that were downregulated by R gauvreauii in response to naringenin (Table 2). Similar to the profile of upregulation, the gene with the highest decrease coded for a hypothetical protein. The gene with the second largest decrease in expression was identified as coding for a nucleotidyltransferase, which is a type of enzyme involved in DNA synthesis and repair (Table 2).24 The other genes identified are involved in pathways such as molecular translocation and translation (Table 2). However, none of the genes downregulated by R gauvreauii in response to naringenin exhibited a greater than 1.5-fold change in gene expression. This makes it difficult to say whether the observed change is meaningful or if the difference in expression is simply due to variation.
Ultimately, R gauvreauii was able to maintain a normal phenotype under all concentrations of naringenin tested (Figure 1A and B). Although it was hypothesized that naringenin may affect gene expression of R gauvreauii, it appears that this effect is minimal (Table 1). From these data, it seems logical to conclude that naringenin is unable to affect R gauvreauii to an extent that can be observed through phenotypic or genotypic evaluation.
The addition of naringenin to B catenulatum resulted in 38 genes with a greater than 1.5-fold increase in expression (Table 3). There was an observed upregulation of multiple genes involved in protein synthesis and protein secretion, which is demonstrated by a 2-fold increase in the putative lipoprotein signal peptidase and a 1.6-fold increase in the tryptophan synthase α-subunit gene (Table 3). The lipoprotein signal peptidase is responsible for removing the signal peptide prior to secretion.25 Upregulation of this gene indicates that B catenulatum is producing and secreting more proteins, which may be related to the documented enhancement of growth for all concentrations of naringenin at 12 hours post inoculation.
In response to naringenin, B catenulatum also upregulated multiple genes associated with cell transport. This is demonstrated by a 1.5-fold increase in the dipeptide ABC transporter permease component, the galactoside transport protein, and the copper-transporting ATPase (adenosine triphosphatase) (Table 3). The ABC transporter system is ubiquitous in bacteria and transports small molecules into and out of the cell.26,27 The copper-transporting ATPase is an ATP-dependent, transmembrane protein that traffics copper into and out of the cells.28 Copper is essential for multiple metabolic pathways, including detoxification of free radicals.28 An increase in the copper-transporting ATPase would require additional ATP, which may explain the upregulation of 2 genes involved in ATP synthesis (Table 3). It is possible that the increase in cellular transport is a way for B catenulatum to maintain homeostasis in the presence of naringenin.
There were 17 genes identified that had a greater than 2.5-fold decrease in expression for B catenulatum in response to naringenin treatment (Table 4). The largest decrease was observed in the deoxyuridine 5′-triphosphate nucleotidohydrolase gene, with a 3.8-fold change in expression. The deoxyuridine 5′-triphosphate nucleotidohydrolase, or dUTPase, is an enzyme responsible for converting dUTP to dUMP, which is then used to synthesis thymidine.29 The dUTPases are ubiquitous to all life-forms and without it, uracil is incorporated into DNA, leading to DNA breaks and ultimately cell death.29 Interestingly, there was a 3.3-fold decrease in the uridine diphosphate (UDP)-galactopyranose mutase, an enzyme that converts UDP-Galp to UDP-Galf, which is a main constituent of the bacterial cell wall and surface.30
There are also 5 separate genes downregulated that are involved with metabolism, including a 2.5-fold decrease in expression of l-ribulose-5-phosphate 4-epimerase (Table 4). This epimerase functions to interconvert l-ribulose 5-phosphate and d-xylulose 5-phosphate.31,32 This conversion is the final step in l-arbinose production, which is then used as a carbon source.33 A decrease in genes involved with metabolism would indicate a drop in catabolism and/or energy production. Taken together, these results indicate that in response to naringenin, B catenulatum downregulates genes associated with metabolism, catabolism, and cell growth.
Despite limited changes to the phenotype of B catenulatum, gene expression analysis revealed that the addition of naringenin produced substantial changes in gene regulation. The addition of naringenin resulted in upregulation of protein synthesis and protein secretion, molecular transport, and ATP synthesis (Table 4). However, there was downregulation of multiple genes involved in critical pathways, such as thymidine biosynthesis, cell wall construction, and metabolism (Table 4). Downregulation of these genes does seem counterintuitive and would indicate a decrease in growth and cell replication. However, this downregulation was not observed in the phenotype. In fact, primary results demonstrated that for all concentrations of naringenin tested, growth of B catenulatum was enhanced at 12 hours, with no difference from the control at 24 hours post inoculation. Therefore, it can only be concluded that the genes downregulated were expendable, at least temporarily, or that they were functionally redundant. In the end, B catenulatum was able to maintain a normal phenotype for all concentrations of naringenin tested. This would indicate that the changes in gene expression were necessary, and provided what was needed to overcome any deleterious effects caused by naringenin.
The addition of naringenin to E caccae resulted in 28 genes with a greater than 1.5-fold increase in expression. However, 19 of these were hypothetical proteins, which unfortunately do not provide any relative information on their function (Table 5). Besides hypothetical proteins, there are 3 genes upregulated that are involved in transcription: 2 transcriptional regulators with a 1.7- and 1.6-fold increase and 1 RNA-binding protein with a 1.6-fold increase (Table 5). This indicates an overall increase in transcription for E caccae in response to naringenin. There is also an observed 1.6-fold increase in the preprotein translocase subunit SecG. SecG is a part of the SecYEG complex, which is located in the membrane and functions to actively transport proteins across the membrane.34 This indicates an increase in protein transport and is possibly used as a mechanism for the cell to maintain homeostasis.
There were 49 genes with a greater than 1.5-fold decrease in activity for E caccae in response to naringenin. The gene that demonstrates the most decrease, a 2.0-fold change, was identified as a peroxiredoxin gene (Table 6). This gene codes for a thiol-specific antioxidant that functions using peroxidase activity.35 Importantly, there was a decrease in 5 individual genes involved in the phosphotransferase system (PTS) pathway. The PTS β-glucoside transporter subunit EIIBCA, with a 1.9-fold decrease; a PTS mannose/fructose/sorbose family IIA and a IID component, both with a 1.6-fold decrease; a PTS IIC component, with a 1.5-fold decrease; and a PTS trehalose IIBC component, with a 1.5-fold decrease in expression (Table 6). All of these are components of the carbohydrate PTS system and function to bring in sugar and carbohydrates into the cell.36 A decrease in so many of the genes responsible for this pathway would most likely limit the ability for E caccae to maintain energy production and normal growth. This is supported by a 1.8-fold decrease in the teichoic acid glycosylation protein, teichoic acid is a component of gram-positive cell membrane, wall, and capsule,37 a 1.5-fold decrease in the 6-phosphofructokinase, which is part of the glycolysis pathway38 and the downregulation of 3 other genes involved in metabolism (Table 6). Ultimately, the growth of E caccae was severely inhibited due to the addition of naringenin. Therefore, it is logical to say that any changes in the pattern of gene expression due to the addition of naringenin were only able to partially counteract any negative effects or were ineffectual.
Conclusions
The results of these experiments clearly show that each bacterial strain tested responded to naringenin differently, both in terms of growth pattern and gene expression. Treatment with naringenin did not measurably affect the growth of R gauvreauii, for B catenulatum there was an increase in growth at 12 hours post inoculation, and for E caccae, a drastic inhibition of growth for each time point tested. The addition of naringenin was determined to produce changes to the gene expression for all 3 of these strains. Ruminococcus gauvreauii had an increase in genes involved with iron uptake and B catenulatum had an upregulation in genes involved in DNA repair and molecular transport and decrease in expression of genes involved in metabolism and thymidine biosynthesis. Enterococcus caccae responded to naringenin by upregulating transcription pathways and protein transport, whereas downregulating sugar transport and purine synthesis.
Taken together, this provides valuable information regarding the way that different types of microbes may respond to the same stimuli. In the context of the gut microbiota, there are 500 to 1000 species of bacteria present at any given time.
The data shown here indicate that each species may respond in a different manner, and the interaction between bacteria and naringenin can be considered somewhat dynamic.
R gauvreauii, B catenulatum, and E caccae were used in this study as representatives of different strains that compose the gut microbiota. It would be valuable, in future work, to see whether the changes in R gauvreauii, B catenulatum, and E caccae are representative of the effect that would occur at a genus or class level. It would also be interesting to see whether or not naringenin would have similar effects on a gut microbiota community. It is quite possible that the addition of polyphenols such as naringenin has a direct hand in shaping the community structure of the gut microbiota, by affecting both growth and gene expression of the bacteria present.
Footnotes
Declaration of Conflicting Interests:The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding:The author(s) received no financial support for the research, authorship, and/or publication of this article.
Author Contributions: JF, LSL, MW, and WX conceived and designed the experiments. LZ and GAA analyzed the data. JF wrote the first draft of the manuscript. PT, SP, and MK contributed to the writing of the manuscript. JF, LSL, MW, WX, LZ, GAA, PT, SP, and MK agree with manuscript results and conclusions. JF, LSL, MW, WX, and SP jointly developed the structure and arguments for the paper. JF, LSL, MW, WX, LZ, GAA, PT, SP, and MK made critical revisions and approved final version. All authors reviewed and approved the final manuscript.
Disclosures and Ethics: As a requirement of publication, authors have provided to the publisher signed confirmation of compliance with legal and ethical obligations including but not limited to the following: authorship and contributorship, conflicts of interest, privacy and confidentiality, and (where applicable) protection of human and animal research subjects. The authors have read and confirmed their agreement with the ICMJE authorship and conflict of interest criteria. The authors have also confirmed that this article is unique and not under consideration or published in any other publication, and that they have permission from rights holders to reproduce any copyrighted material. The external blind peer reviewers report no conflicts of interest.
References
- 1. Felgines C, Texier O, Morand C, Manach C. Bioavailability of the flavanone naringenin and its glycosides in rats. Am J Physiol Gastrointest Liver Physiol. 2000;27:1148–1154. [DOI] [PubMed] [Google Scholar]
- 2. Erlund I. Review of the flavonoids quercetin, hesperetin, and naringenin. Dietary sources, bioactivities, bioavailability, and epidemiology. Nutr Res. 2004;24:851–874. [Google Scholar]
- 3. Pandey R, Gurung R, Sohng J. Apigenin and naringenin natural sources, pharmacology and role in cancer prevention. In: Stacks NM, ed. Biochemistry Research Trends. New York, NY: Nova Science Publishers Inc.; 2015:151–172. [Google Scholar]
- 4. Parkar S, Trower T, Stevenson D. Fecal microbial metabolism of polyphenols and its effects on human gut microbiota. Anaerobe. 2013;23:12–19. [DOI] [PubMed] [Google Scholar]
- 5. Lee HC, Jenner AM, Low CS, Lee YK. Effect of tea phenolics and their aromatic fecal bacterial metabolites on intestinal microbiota. Res Microbiol. 2006;157:876–884. [DOI] [PubMed] [Google Scholar]
- 6. Ozdal T, Sela D, Xiao J, Boyacioglu D. The reciprocal interactions between polyphenols and gut microbiota and effects on bioaccessibility. Nutrients. 2016;8:78–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Manthey J, Grohmann K, Guthrie N. Biological properties of citrus flavonoids pertaining to cancer and inflammation. Curr Med Chem. 2001;8:135–153. [DOI] [PubMed] [Google Scholar]
- 8. Kurowska E, Borradaile N, Spence J, Carroll K. Hypocholesterolemic effects of dietary citrus juices in rabbits. Nutr Res. 2000;20:121–120. [Google Scholar]
- 9. Kurowska E, Spence J, Jordan J, Wetmore S. HDL-cholesterol-raising effect of orange juice in subjects with hypercholesterolemia. Am J Clin Nutr. 2000;72:1095–1100. [DOI] [PubMed] [Google Scholar]
- 10. Kanaze FI, Bounartzi MI, Georgarakis M, Niopas I. Pharmacokinetics of the citrus flavanone aglycones hesperetin and naringenin after single oral administration in human subjects. Eur J Clin Nutr. 2007;61:472–477. [DOI] [PubMed] [Google Scholar]
- 11. Vallejo F, Larrosa M, Escudero E, Zafrilla M. Concentration and solubility of flavanones in orange beverages affect their bioavailability in humans. J Agri Food Chem. 2010;58:6516–6524. [DOI] [PubMed] [Google Scholar]
- 12. Duda-Chodak A. The inhibitory effect of polyphenols on human gut microbiota. J Physiol Pharmacol. 2012;63:497–503. [PubMed] [Google Scholar]
- 13. Parkar S, Stevenson D, Skinner M. The potential influence of fruit polyphenols on colonic microflora and human gut health. Int J Food Microbiol. 2008;124:295–298. [DOI] [PubMed] [Google Scholar]
- 14. Firrman J, Liu L, Zhang L, et al. The effect of quercetin on genetic expression of the commensal gut microbes Bifidobacterium catenulatum, Enterococcus caccae and Ruminococcus gauvreauii. Anaerobe. 2016;42:130–141. [DOI] [PubMed] [Google Scholar]
- 15. Liu L, Firrman J, Arango Argoty G, Tomasula P. Genetic expression profile analysis of the temporal inhibition of quercetin and naringenin on Lactobacillus rhamnosus GG. J Probiot Health. 2016;4:139. [Google Scholar]
- 16. Scardovi V, Crociani F. Bifidobacterium catenulatum, Bifidobacterium dentium, and Bifidobacterium angulatum: three new species and their deoxyribonucleic acid homology relationship. Int J Syst Evol Microbiol. 1974;24:6–20. [Google Scholar]
- 17. Domingo MC, Huletsky A, Boissinot M, Bernard KA, Picard FJ, Bergeron MG. Ruminococcus gauvreauii sp. nov., a glycopeptide-resistant species isolated from a human faecal specimen. Int J Syst Evol Microbiol. 2008;58:1393–1397. [DOI] [PubMed] [Google Scholar]
- 18. Carvalho Mda G, Shewmaker PL, Steigerwalt AG, et al. Enterococcus caccae sp. nov., isolated from human stools. Int J Syst Evol Microbiol. 2006;56:1505–1508. [DOI] [PubMed] [Google Scholar]
- 19. Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:D61–D65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. [DOI] [PubMed] [Google Scholar]
- 21. Mortazavi A, Williams BA, McCue K, Schaeffer L. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. [DOI] [PubMed] [Google Scholar]
- 22. Lau CK, Ishida H, Liu Z, Vogel HJ. Solution structure of Escherichia coli FeoA and its potential role in bacterial ferrous iron transport. J Bacteriol. 2013;195:46–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lau CK, Krewulak KD, Vogel HJ. Bacterial ferrous iron transport: the Feo system. FEMS Microbiol Rev. 2016;40:273–298. [DOI] [PubMed] [Google Scholar]
- 24. Aravind L, Koonin EV. DNA polymerase β-like nucleotidyltransferase superfamily: identification of three new families, classification and evolutionary history. Nucleic Acids Res. 1999;27:1609–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Von Heijne G. The structure of signal peptides from bacterial lipoproteins. Protein Eng. 1989;2:531–534. [DOI] [PubMed] [Google Scholar]
- 26. Higgins C. ABC transporters: physiology, structure and mechanism—an overview. Res Microbiol. 2001;152:205–210. [DOI] [PubMed] [Google Scholar]
- 27. Young J, Holland IB. ABC transporters: bacterial exporters-revisited five years on. Biochim Biophys Acta. 1999;1461:177–200. [DOI] [PubMed] [Google Scholar]
- 28. Migocka M. Copper-transporting ATPases: the evolutionarily conserved machineries for balancing copper in living systems. IUBMB. 2015;67:737–734. [DOI] [PubMed] [Google Scholar]
- 29. Vértessy BG, Tóth J. Keeping uracil out of DNA: physiological role, structure and catalytic mechanism of dUTPases. Acc Chem Res. 2009;42:97–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tanner JJ, Boechi L, McCammon JA, Sobrado P. Structure, mechanism, and dynamics of UDP-galactopyranose mutase. Arch Biochem Biophys. 2014;15:128–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Englesberg E, Anderson Rl, Weinberg R, Lee N. L-Arabinose-sensitive, L-ribulose 5-phosphate 4-epimerase-deficient mutants of Escherichia coli. J Bacteriol. 1962;84:137–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chang C, Tesar C, Li X, Kim Y. A novel transcriptional regulator of L-arabinose utilization in human gut bacteria. Nucleic Acids Res. 2015;43:10546–10559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Samuel J, Luo Y, Morgan PM, Strynadka NC. Catalysis and binding in L-ribulose-5-phosphate 4-epimerase: a comparison with L-fuculose-1-phosphate aldolase. Biochemistry. 2001;40:14772–14780. [DOI] [PubMed] [Google Scholar]
- 34. Veenendaal AK, van der Doe C, Driessen AJ. The protein-conducting channel SecYEG. Biochim Biophys Acta. 2004;1694:81–95. [DOI] [PubMed] [Google Scholar]
- 35. Wood Z, Schroder E, Harris JR, Poole L. Structure, mechanism, and regulation of peroxiredoxins. Trends Biochem Sci. 2003;28:32–40. [DOI] [PubMed] [Google Scholar]
- 36. Postma PW, Lengeler JW, Jacobson GR. Phosphoenolpyruvate: carbohydrate phosphotransferase systems of bacteria. Microbiol Rev. 1993;57:543–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Archibald AR. The structure, biosynthesis and function of teichoic acid. Adv Microb Physiol. 1974;11:53–95. [Google Scholar]
- 38. Mertens E. Pyrophosphate-dependent phosphofructokinase, an anaerobic glycolytic enzyme? FEBS Lett. 1991;258:1–5. [DOI] [PubMed] [Google Scholar]


