Abstract
In recent years, production of fatty acid derivatives has attracted much attention because of their wide range of applications in renewable oleochemicals. Microorganisms such as Saccharomyces cerevisiae provided an ideal cell factory for such chemical synthesis. In this study, an efficient strategy for the synthesis of fatty alcohols based on enhanced supply of free fatty acids (FFAs) was constructed. The FAA1 and FAA4 genes encoding two acyl-CoA synthetases in S. cerevisiae were deleted, resulting in the accumulation of FFAs with carbon chain length from C8 to C18. The coexpression of the carboxylic acid reductase gene (car) from Mycobacterium marinum and the phosphopantetheinyl transferase gene (sfp) from Bacillus subtilis successfully converted the accumulated FFAs into fatty alcohols. The concentration of the total fatty alcohols reached 24.3 mg/L, which is in agreement with that of the accumulated FFAs. To further increase the supply of FFAs, the DGAI encoding the acyl-CoA:diacylglycerol acyltransferase involved in the rate-limiting step of triacylglycerols storage was codeleted with FAA1 and FAA4, and the acyl-CoA thioesterase gene (acot) was expressed together with car and sfp, resulting in an enhanced production of fatty alcohols, the content of which increased to 31.2 mg/L. The results herein demonstrated the efficiency of the engineered pathway for the production of fatty acid derivatives using FFAs as precursors.
Introduction
Known as important precursors for valuable chemicals, fatty acids have attracted tremendous attention for biological production of a variety of fatty acid derivatives, including biodiesel, fatty aldehydes, fatty alcohols, and alkanes/alkenes, some of which are promising substitutes for fossil fuels.1−3 A great deal of efforts have been made in the microbial production of fatty derivatives from fatty acids in recent years,4,5 and particularly, Saccharomyces cerevisiae is regarded to be a potential natural cell factory based on its rapid growth rate, genetic traceability, robust genetic tools, high product yield, and good tolerance to harsh fermentation conditions.6,7
In S. cerevisiae, biosynthesis of fatty acids is initiated by condensing acetyl-CoA and malonyl-CoA, and the final step is catalyzed by the transferase activity, which substitutes CoA for acyl carrier protein (ACP), releasing fatty acyl-CoAs.8 The generated fatty acyl-CoAs are then fed into lipid storage, which mainly existed in the form of triacylglycerols (TAGs) or phospholipids. In addition, the free fatty acids (FFAs) in S. cerevisiae could be rapidly converted to fatty acyl-CoAs by fatty acyl-CoA synthetases, encoded by FAA genes.9 The biosynthesis of fatty acid is in turn to be feedback inhibited by the fatty acyl-CoAs with an extremely low intracellular titer.10,11 As such, the production level of both FFAs and fatty acyl-CoAs in S. cerevisiae is quite low.
Although fatty acids are important building blocks for fatty acid derivatives, their storage forms of TAGs or phospholipids are unable to be directly converted to the target components. A variety of converting enzymes, including wax ester synthase (WS)/acyltransferase (AT) for the synthesis of biodiesel,12,13 fatty acyl-CoA reductase (FAR) for the synthesis of fatty alcohols/aldehydes,14,15 aldehyde decarbonylase (AD)/carboxylic acid reductase (CAR) for the synthesis of fatty aldehyde,16−18 and others such as lipoxygenase (LOX) and hydroperoxide lyase (HPL),19 all use FFAs or fatty acyl-CoAs as precursors. Thus, the activation of fatty acids through engineering the yeast cells is necessary for the efficient biosynthesis of the desired compounds.
In our previous study, the acyl-CoA-dependent TAG biosynthesis pathway was engineered by deleting the rate-limiting enzyme acyl-CoA:diacylglycerol acyltransferase (DAGAT) encoded by dga1, to accumulate fatty acyl-CoAs for the production of fatty acid derivatives. The subsequent overexpression of an alcohol-forming FAR successfully converted the accumulated fatty acyl-CoAs to fatty alcohols, resulting in the production of fatty alcohols in S. cerevisiae at a concentration of 45 mg/L.15 This strategy ideally solved the bottleneck using the storage forms of fatty acids for the production of target derivatives. Because fatty acyl-CoA regulated the biosynthesis of fatty acids, the blockage of TAG synthesis pathway limited the metabolic flux toward the accumulation of fatty acyl-CoA. As such, further increase in fatty acid derivatives in the above-engineered strain is difficult to achieve.
To overproduce target derivatives, the regulatory inhibition by acyl-CoA should be relieved and the flux from fatty acyl-CoAs to FFAs is regarded to be a common strategy. Therefore, another strategy is then focused on the utilization of FFAs as precursors. We previously reported that deletion of FAA1 and FAA4 encoding two acyl-CoA synthetases together with the expression of acyl-CoA thioesterase (acot) successfully accumulated FFAs to a much higher level.20 Among the converting enzymes, the CAR was discovered to be a versatile enzyme that can directly use FFAs to synthesize fatty acid derivatives, such as fatty aldehydes.18 Therefore, S. cerevisiae was designed for metabolic engineering from this perspective in this study. On the basis of the FAA1/FAA4 deletion strain, the FFAs were expectedly accumulated and the CAR was then introduced into the mutant strain, to convert the accumulated FFAs to fatty acid derivatives. The final product produced in the engineered strain was compared with the previously engineered strains with modified TAG synthesis pathway, to investigate the productive capacity of fatty acid derivatives. The study herein aims to look for an alternative strategy realizing the efficient biosynthesis of fatty acid derivatives from the fatty acids in S. cerevisiae.
Results and Discussion
Pathway Engineering for the Production of Enhanced Fatty Acid Derivatives
To realize the successful de novo production of fatty acid derivatives, a fatty acid-dependent pathway was metabolically engineered to supply efficient precursors. Differing from our previous strategy, which increased the production levels of fatty acyl-CoAs, the strategy herein aims to enhance the supply of FFAs. The FAA1 and FAA4 genes encoding the acyl-CoA synthetases were deleted according to the previous study, and the CAR was coexpressed with 4′-phosphopantetheinyl transferase (SFP) to convert the FFAs to their derivatives. To further increase the yield of fatty acid derivatives, the acyl-CoA:DAGAT was also deleted and the acyl-CoA thioesterase fragment was integrated into the genome of the engineered yeast, to block the formation of TAGs, resulting in the release of more FFAs (Figure 1).
Figure 1.
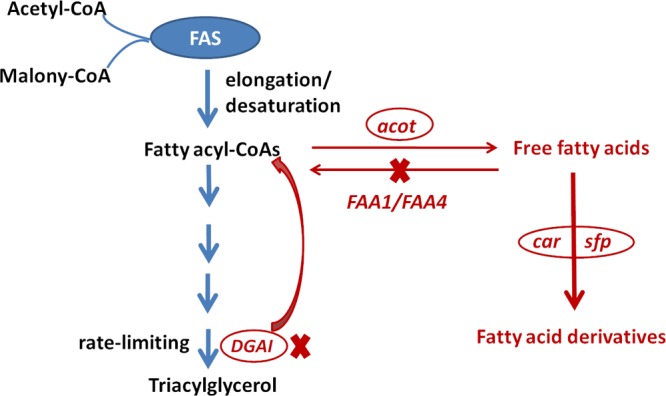
Workflow of engineered pathway in S. cerevisiae for efficient biosynthesis of fatty acid derivatives based on the enhanced supply of FFAs as precursors. FAA1/FAA4: genes encoding the two acyl-CoA synthetases; DGAI: gene encoding the diglyceride acyltransferase; acot: gene encoding the acyl-CoA thioesterase from M. musculus; car: gene encoding the CAR from M. marinum; and sfp: gene encoding the SFP from B. subtilis. The designed pathway was indicated by red color.
Efficient Release of FFAs Based on the Deletion of FAA1, FAA4, and DGAI and Introduction of acot
According to the above strategy, the release of FFAs was investigated. The deletion of FAA1 and FAA4 encoding the acyl-CoA synthetases effectively increased the production level of FFAs. The FFAs mainly detected in ΔFAA1/ΔFAA4 strain were myristic acid (C14:0 fatty acid), cetyl acid (C16:0 fatty acid), palmitoleyl acid (C16:1 fatty acid), stearic acid (C18:0 fatty acid), and oleic acid (C18:1 fatty acid), and small amounts of octanoic acid (C8:0 fatty acid), capric acid (C10:0 fatty acid), and lauric acid (C12:0 fatty acid) was also observed. The codeletion of DGAI and the integration of acot further enhanced the production level of FFAs including C14:0, C16:0, C16:1, C18:0, and C18:1, with maximum yields of 18.3, 83.3, 92.1, 36.8, and 38.4 mg/L, which were increased by 43, 18, 15, 19, and 24%, respectively, compared with those of ΔFAA1/ΔFAA4 strain. However, the yield of medium-chain FFAs was not further increased (Figure 2). In the wild-type strain, even though small amounts of C16:0 and C18:0 were detected, other FFAs were not detectable. The C16:0 and C18:0 FFAs in the wild-type strain reached only about 24.2 and 10.6 mg/L, respectively, and the increase in C16:0 and C18:0 FFAs in the ΔFAA1/ΔFAA4/ΔDGAI-acot strain compared with the wild-type strain reached 244 and 247%, respectively.
Figure 2.
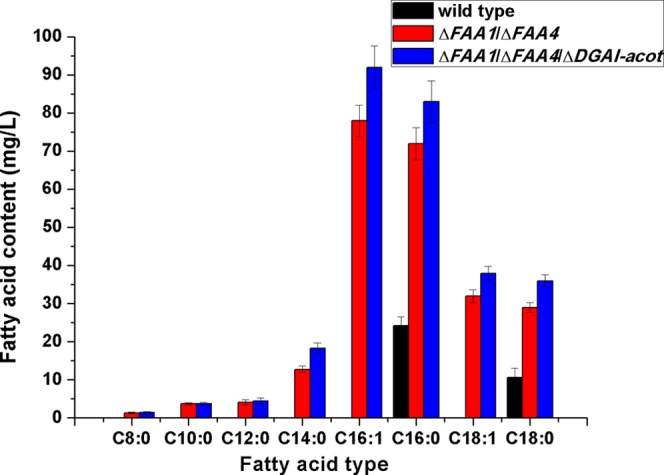
Fatty acid contents among the wild-type, ΔFAA1/ΔFAA4, and ΔFAA1/ΔFAA4/DGAI-acot strains. The standard deviation was calculated from the three independent experimental results.
Efficient Synthesis of Fatty Acid Derivatives Based on Enhanced Supply of FFAs in ΔFAA1/ΔFAA4-car-sfp-Engineered Strain
On the basis of the enhanced supply of FFAs, the coexpression of CAR and SFP successfully converted the individual FFAs into the corresponding fatty alcohols while no fatty aldehydes were detected. In the ΔFAA1/ΔFAA4-car-sfp-engineered strain, total fatty alcohols reached a concentration of 24.3 mg/L and the fatty alcohols mainly detected included 1-tetradecanol (C14:0 fatty alcohol), 1-hexadecanol (C16:0 fatty alcohol), cis-9-hexadecenol (C16:1 fatty alcohol), 1-octadecanol (C18:0 fatty alcohol), and cis-9-octadecenol (C18:1 fatty alcohol) (Figure 3), among which the cis-9-hexadecenol (C16:1) was found to be the most abundant, accounting for approximately 40.7% of the total fatty alcohols produced, followed by 1-hexadecanol (C16:0), 1-tetradecanol (C14:0), 1-octadecanol (C18:0), and cis-9-hexadecenol (C18:1), the concentrations of which accounted for 16.5, 11.4, 11.3, and 12.8%, respectively. Although detectable, 1-octanol (C8:0 fatty alcohol), 1-decanol (C10:0 fatty alcohol), and 1-dodecanol (C12:0 fatty alcohol) represented only less than 1.3, 3.1, and 2.7% of the total fatty alcohols, respectively (Table 2), which was consistent with the profile of the accumulated FFAs in the ΔFAA1/ΔFAA4 double-deletion strain. Although the CAR and SFP were also coexpressed in the wild-type strain, only extremely small amounts of fatty alcohols were detected, including C16:0 fatty alcohol of 0.76 mg/L and C18:0 fatty alcohol of 0.33 mg/L, indicating the limited amount of FFAs produced in the wild-type strain.
Figure 3.
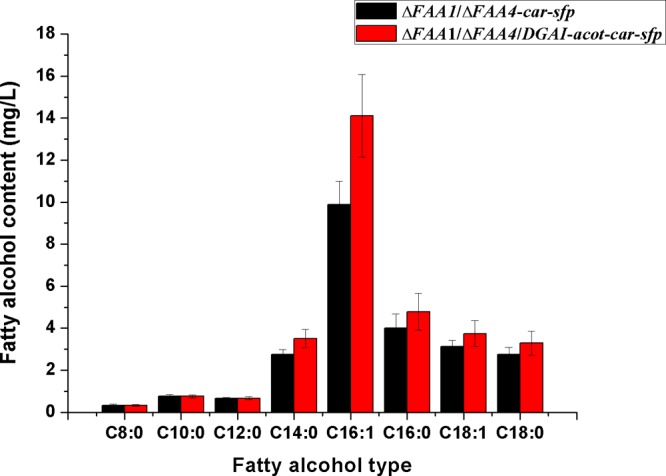
Production levels of fatty alcohol between the engineered strains ΔFAA1/ΔFAA4-car-sfp and ΔFAA1/ΔFAA4/DGAI-acot-car-sfp. The standard deviation was calculated from the three independent experimental results.
Table 2. Free Fatty Alcohol Compositions between the Engineered Strains ΔFAA1/ΔFAA4-car-sfp (Engineered Strain 1) and ΔFAA1/ΔFAA4/ΔDGAI-acot-car-sfp (Engineered Strain 2).
| compositions |
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| strain type | total fatty alcohols (mg/L) | C8:0 (%) | C10:0 (%) | C12:0 (%) | C14:0 (%) | C16:1 (%) | C16:0 (%) | C18:1 (%) | C18:0 (%) |
| engineered strain 1 | 24.3 | 1.3 | 3.1 | 2.7 | 11.4 | 40.7 | 16.5 | 12.8 | 11.3 |
| engineered strain 2 | 31.2 | 1.1 | 2.3 | 2.1 | 11.3 | 45.3 | 15.4 | 12.0 | 10.5 |
From our study, it was obvious that the CAR exhibited activity toward FFAs with a broad range of chain length, from C8 to C18, and the efficient conversion of FFAs to the fatty acid derivatives also depended on the presence of SFP. The results herein were in accordance with the previous report, in which the CAR was characterized and shown to accept a broad range of aliphatic fatty acids.18 The SFP was necessary for the maximum activity of CAR, and as a result, the coexpression of CAR and SFP in this study converted the accumulated FFAs into fatty acid derivatives successfully.
Until now, the CAR has been exploited from limited sources such as Mycobacterium marinum(18,21) and found to convert the FFAs to the corresponding aldehydes. However, in this study, no fatty aldehydes were detected in the engineered strain, but only fatty alcohols with different chain lengths were accumulated. It was reported that S. cerevisiae contained various endogenous aldehyde reductases (ALRs),22 which could effectively convert the synthesized fatty aldehydes into the corresponding fatty alcohols. As such, the majority of FFAs were flux to the formation of fatty alcohols and the trace amounts of fatty aldehydes generated were not detectable.
Enhanced Production of Fatty Alcohols in ΔFAA1/ΔFAA4/ΔDGAI-acot-car-sfp-Engineered Strain
Whereas the accumulated FFAs that resulted from the engineered pathway were secreted into the culture broth, the rest of the fatty acids were stored mainly in the form of TAGs and sterol esters in S. cerevisiae. According to our previous study, the acyl-CoA:DAGAT was the rate-limiting enzyme involved in the synthesis of TAGs from fatty acyl-CoAs and its deletion resulted in the release of fatty acyl-CoAs.15 In this study, the dgaI gene encoding the DAGAT was codeleted with FAA1 and FAA4, and the acyl-CoA thioesterase responsible for the conversion of fatty acyl-CoAs to FFAs was introduced in the modified yeast strain, to supply more FFAs for the production of fatty acid derivatives. For ΔFAA1/ΔFAA4/ΔDGAI-acot, because no “converting enzymes” of car and sfp were expressed, no fatty alcohols were detected. On the other hand, in the ΔFAA1/ΔFAA4/ΔDGAI-acot-car-sfp strain, similar to the ΔFAA1/ΔFAA4-car-sfp-engineered strain, only fatty alcohols were detected and the profiles of the fatty alcohols were in agreement with those in the ΔFAA1/ΔFAA4-car-sfp strain (Figure 3). The concentration of total fatty alcohols increased from 24.3 to 31.2 mg/L, and that of the most abundant cis-9-hexadecenol (C16:1 fatty alcohol) reached 14.1 mg/L, an increase of 42.7%. 1-Tetradecanol (C14:0 fatty alcohol), 1-hexadecanol (C16:0 fatty alcohol), 1-octadecanol (C18:0 fatty alcohol), and cis-9-octadecenol (C18:1 fatty alcohol) increased by 27.2, 19.8, 19.6, and 19.2%, respectively (Table 3). For the medium-chain fatty alcohol including 1-octanol (C8:0 fatty alcohol), 1-decanol (C10:0 fatty alcohol), and 1-dodecanol (C12:0 fatty alcohol), no further increase in yield was observed. In our previous study, the deletion of DAGAT resulted in no increase in medium-chain fatty acyl-CoAs,15 and as a result, no more medium-chain FFAs were further supplied for the synthesis of fatty alcohols in this study. The production level of fatty alcohol in the ΔFAA1/ΔFAA4/DGAI-car-sfp was also investigated. The deletion of DGAI was previously demonstrated to accumulate fatty acyl-CoAs effectively. However, without the expression of acyl-CoA thioesterase, the accumulated fatty acyl-CoAs could not be efficiently converted to FFAs and thus cannot be used as precursors for the CAR. As a result, no more fatty alcohols were detected compared with ΔFAA1/ΔFAA4-car-sfp-engineered strain.
Table 3. Increase in Fatty Alcohols in the Engineered Strain ΔFAA1/ΔFAA4/ΔDGAI-acot-car-sfp (Strain 2) Compared with That in the Engineered Strain ΔFAA1/ΔFAA4-car-sfp (Strain 1)a.
| increase level | total (%) | C8:0 (%) | C10:0 (%) | C12:0 (%) | C14:0 (%) | C16:1 (%) | C16:0 (%) | C18:1 (%) | C18:0 (%) |
|---|---|---|---|---|---|---|---|---|---|
| strain 2–strain 1 | 28.4 | 1.2 | 1.3 | 2.1 | 27.2 | 42.7 | 19.8 | 19.2 | 19.6 |
The increase in the production levels in the engineered strain ΔFAA1/ΔFAA4/ΔDGAI-acot-car-sfp compared with those in the ΔFAA1/ΔFAA4-car-sfp was reflected as “%”.
The comparison of the fatty acid content between ΔFAA1/ΔFAA4/DGAI-acot deletion strain with empty pESC plasmid and that with coexpressed CAR and SFP (ΔFAA1/ΔFAA4/DGAI-acot-car-sfp) found that the FFA content decreased significantly in the ΔFAA1/ΔFAA4/DGAI-acot-car-sfp-engineered strain, which accounted for only 26.2% of that produced in the ΔFAA1/ΔFAA4/DGAI-acot strain; especially, the myristyl acid, cetyl acid, palmitoleyl acid, stearic acid, and oleic acid decreased to 33, 37, 31, 43, and 43%, respectively (Figure 4), demonstrating the successful conversion of FFAs to fatty acid derivatives by the coexpressed CAR and SFP.
Figure 4.
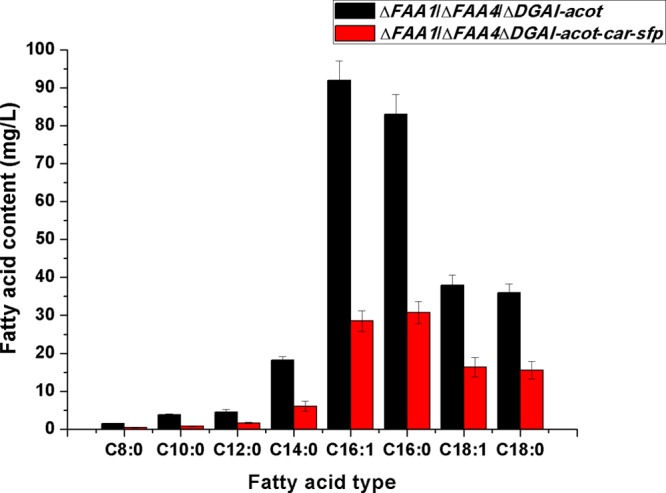
Comparison of fatty acid contents between ΔFAA1/ΔFAA4/DGAI-acot strain and ΔFAA1/ΔFAA4/DGAI-acot-car-sfp strain. The standard deviation was calculated from the three independent experimental results.
Comparison of Production of Fatty Alcohols between Engineered Strains Based on an Increased Supply of FFAs and Fatty Acyl-CoAs
In S. cerevisiae, both fatty acyl-CoAs and FFAs are precursors of fatty acid derivatives, which can be directly converted to the corresponding component either by enzymes using acyl-CoAs as precursors such as FAR or by those with FFAs as precursors such as CAR. Herein, the comparison of the production levels of fatty alcohols between engineered strains based on an increased supply of FFAs (in this study) and fatty acyl-CoAs (previous study) was performed. Both the strategies resulted in the production of fatty alcohols without forming fatty aldehydes, which was due to the substrate specificity of FAR, belonging to the alcohol-forming type14 and the existence of endogenous ALRs in S. cerevisiae,22 converting the fatty aldehydes to the corresponding fatty alcohols. The production level of fatty alcohols in the two engineered strains was comparative, but interestingly, in the engineered strain based on increased supply of fatty acyl-CoAs, the mainly accumulated fatty alcohols were saturated fatty alcohols, especially 1-hexadecanol (C16:0 fatty alcohol), accounting for approximately 80% of the total fatty alcohols. On the other hand, in the engineered strain with increased supply of FFAs, the most abundant fatty alcohol was unsaturated fatty alcohol, especially cis-9-hexadecenol (C16:1 fatty alcohol), representing approximately 45% of the total fatty alcohols. The cis-9-octadecenol (C18:1 fatty alcohol) accounted for approximately 13% in the fatty acyl-CoA-accumulated strain, but it was not detectable in the FFA-accumulated strain. In addition, the medium-chain fatty alcohols with the carbon chain length from C8 to C12 were not detected in the acyl-CoA-accumulated strain but detectable in the FFA-accumulated strain.
While investigating the fatty acid profile in the two engineered strains, it was found that the deletion of FAA1 and FAA4 resulted in the accumulation of medium-chain FFAs including C8:0, C10:0, and C12:0; unsaturated FFAs including C16:1 and C18:1; and saturated FFAs including C14:0, C16:0, and C18:0 (Figure 2). Among them, the C16:1 and C16:0 FFAs were found to be the most abundant and the medium-chain FFAs accounted for only a small part of the total FFAs. As a result, the expression of CAR with a broad substrate specificity resulted in the production of fatty alcohols, the profile of which was in agreement with that of the accumulated FFAs. In our previous study, the DGAI deletion strain resulted in no release of medium-chain fatty acyl-CoAs from TAGs and the FAR was demonstrated to exhibit the highest catalytic efficiency for C16:0 fatty acyl-CoA. As such, the profiles of fatty alcohols produced between engineered strains based on an increased supply of FFAs and fatty acyl-CoAs with expressed CAR and FAR, respectively, reasonably differ from each other.
In the past few years, tremendous efforts have been focused on the study of microbial production of fatty acids and their derivatives. Similarly to our work, Runguphan and Keasling directly overexpressed the FAR from Mus musculus to produce fatty alcohols in S. cerevisiae and followed by the replacement of strong constitutive promoter PTEF1 to the original promoter of genes involved in fatty acid synthesis; the production level of fatty alcohol reached more than 90 mg/L.23 Zhou et al. engineered the yeast strain step by step based on the increased supply of FFAs.24 The initial production of fatty alcohol reached 23.2 mg/L, which was comparable with our results. However, after the deletion of genes involved in endogenous conversion of fatty alcohols to fatty aldehyde and fatty acids, blocking the fatty acid degradation pathway, and coexpression of FAR and CAR, the fatty alcohol yield significantly increased. With further fed-batch strategies, the production of fatty alcohol reached 1.5 g/L. The current results already demonstrated the feasibility of the production of fatty acid derivatives based on the two different strategies and by a further combination of other strategies such as promoter replacement, by-pathway blockage, multiple enzyme coexpression, and fermentation optimization, large-scale production of fatty acid derivatives become realizable.
Conclusions
In this study, an alternative strategy to supply FFAs as precursors for the synthesis of fatty acid derivatives was constructed. The codeletion of FAA1 and FAA4 together with DGAI, followed by the integration of acyl-CoA thioesterase, successfully enhanced the supply of FFAs with the carbon chain length from C8 to C18. The coexpression of the CAR and SFP further converted the accumulated FFAs into their corresponding fatty acid derivatives. Because of the existence of endogenous ALRs in S. cerevisiae, the engineered strain mainly produced fatty alcohols but not fatty aldehydes. The results obtained herein not only demonstrated the efficiency of the constructed pathway for the production of fatty alcohol but also solved the bottleneck involved in the utilization of fatty acids as precursors for the synthesis of fatty acid derivatives directly.
Methods
Strains and Cultivation
The S. cerevisiae BY4741 (ura3Δ, ΔMAT, his3Δ, leu2Δ, met15Δ) was used as a parental strain in this study and cultivated in yeast extract peptone dextrose (YPD) complete medium [1% yeast extract, 2% peptone, and 2% d-glucose (w/v)]. The engineered strains harboring LEU2-based plasmids (pESC-leu) were selected on agar plates of the yeast nitrogen base dextrose (YNBD) minimal medium without leucine [6.7 g/L yeast nitrogen base, 5 g/L ammonium sulfate, 2% d-glucose (w/v), and 0.67 g/L leucine dropout amino acid mixture]. The gene deletion strains with His5-based cassettes were selected on agar plates of YNBD minimal medium plates without histidine [6.7 g/L yeast nitrogen base, 5 g/L ammonium sulfate, 2% d-glucose (w/v), and 0.65 g/L histidine dropout amino acid mixture], and those with KanMX cassette were selected on agar plates of YPD medium containing 200 mg/L Geneticin (G418). All cultivations were performed at 30 °C and 200 rpm (if necessary).
When cultivating the strains for the investigation of fatty acid and fatty acid derivatives, single colonies were selected from the corresponding agar plate. The wild-type strain and engineered gene deletion strains with empty pESC plasmid or recombinant pESC plasmids were cultivated overnight in 5 mL YNBD minimal medium without leucine. After that, the seed liquid was inoculated into a 50 mL YNB minimal medium without leucine, and 1% raffinose (w/v) together with 2% (w/v) galactose was added. The expression of target genes under GAL1/GAL10 promoters was induced by the initial addition of galactose in the absence of glucose at 30 °C and 200 rpm. The cell concentration was measured according to the value of OD600, and after the galactose induction for approximately 24 h and the cell growth reached a stationary phase, the cells were centrifuged and collected for further applications.
Construction of Engineered Strains
The ΔFAA1/ΔFAA4 double gene disruption strain was constructed according to the homologous recombination strategies:25 the polymerase chain reaction (PCR)-amplified fragment containing the loxP-His5-loxP gene disruption cassette and 45 nucleotide stretches that are homologous to the sequences upstream of the start codon of FAA1 gene and downstream of the stop codon were used for the homologous recombination of the FAA1 gene. The deletion of FAA4 gene was based on the FAA1 single gene disruption strain, and the generated PCR fragment containing the loxP-KanMX-loxP and 45 nucleotide stretches that are homologous to the sequences upstream of the start codon of FAA4 gene and downstream of the stop codon was used for the homologous recombination of the FAA4 gene. Once the ΔFAA1/ΔFAA4 double gene disruption strain was obtained, the Kanr marker was rescued according to the method described previously: The mutant strain was transformed with the plasmid pSH47, which carries the cre recombinase gene and URA3 marker gene. After growing in the YPD medium, the cells were shifted to the YPG (galactose) medium to remove the Kanr marker gene. The colonies were detected on both YPD and replica-YPD plus G418 plate. The marker-rescued mutant strain was further used for the deletion of dgaI gene with the same strategy, and the Kanr marker was rescued again.
The carboxylic acid reductase gene (car) from M. marinum (accession no. WP_012393886) and 4′-phosphopantetheinyl transferase gene (sfp) from Bacillus subtilis (accession no. Q9F4F7) were synthesized by GenScript and codon-optimized for S. cerevisiae. The two synthesized gene fragments were flanked with a pair of SpeI/PacI and BamHI/XhoI restriction enzyme sites, which were digested by the corresponding endonucleases and inserted in the two multicloning sites of the predigested pESC-leu plasmid. The recombinant plasmid and empty pESC-leu were then transformed into the ΔFAA1/ΔFAA4 mutant strain separately to construct the engineered strains ΔFAA1/ΔFAA4-pESC and ΔFAA1/ΔFAA4-car-sfp. The acyl-CoA thioesterase gene (acot) fragment from M. musculus (accession NM_145444.3) lacking the C-terminal peroxisomal targeting peptide was obtained in our previous work and was inserted into the pMRI plasmid provided by Prof. Yu26 between the NotI and BglII endonuclease sites. The pMRI recombinant plasmid containing acot was then digested by sfiI endonuclease and transformed into the ΔFAA1/ΔFAA4/ΔDGAI deletion strain. The functional module was integrated into the HO locus, generating the engineered strain ΔFAA1/ΔFAA4/ΔDGAI-acot. The above-constructed pESC-car-sfp plasmid was then transformed into the ΔFAA1/ΔFAA4/ΔDGAI-acot strain, resulting in ΔFAA1/ΔFAA4/ΔDGAI-acot-car-sfp. The primers used in this study are listed in Table 1.
Table 1. Primers Used in This Studya.
| gene | primer | description |
|---|---|---|
| ΔFAA1-F | 5′CAATAAAAACTAGAACAAACACAAAAGACAAAAAAAGACAACAATCAGCTGAAGCTTCGTACGC3′ | forward primer for FAA1 gene deletion |
| ΔFAA1-R | 5′TGCTTTAGTATGATGAGGCTTTCCTATCATGGAAATGTTGATCCAGCATAGGCCACTAGTGGATCTG3′ | reverse primer for FAA1 gene deletion |
| ΔFAA4-F | 5′TCTGTTCTTCACTATTTCTTGAAAAACTAAGAAGTACGCATCAAACAGCTGAAGCTTCGTACGC3′ | forward primer for FAA4 gene deletion |
| ΔFAA4-R | 5′GTGTTTATGAAGGGCAGGGGGGAAAGTAAAAAACTATGTCTTCCTGCATAGGCCACTAGTGGATCTG3′ | reverse primer for FAA4 gene deletion |
| ΔDGAI-F | 5′ATACATAAGGAAACGCAGAGGCATACAGTTTGAACAGTCACATAACAGCTGAAGCTTCGTACGC3′ | forward primer for DGAI gene deletion |
| ΔDGAI-R | 5′CCTTATTTATTCTAACATATTTTGTGTTTTCCAATGAATTCATTAGCATAGGCCACTAGTGGATCTG3′ | reverse primer for DGAI gene deletion |
| acot-F | 5′GAAGAAGCGGCCGCATGCTAGCCAAAGGTAAGCC3′ | forward primer for acot integrated in pMRI plasmid |
| acot-R | 5′CCTTAGATCTTCAAGGACTAGGTCTCTTGTCACC3′ | reverse primer for acot integrated in pMRI plasmid |
The car gene encoding CAR and sfp gene encoding SFP were artificially synthesized with SpeI/PacI and BamHI/XhoI endonuclease sites, respectively, and inserted into the pESC-leu plasmid directly.
Metabolites Extraction and Derivatization
Cells (5 mL) growing in the corresponding culture medium at the stationary phase were centrifuged and collected. After washing twice with distilled water, the cells were resuspended in 400 μL of ethyl acetate, and 5 μL of heptadecanoic acid (10 mg/mL) was added as internal control. The cell samples were added with the same volume of glass beads of diameter 425–600 μm and vortexed in a bead mill with a vortex frequency of 4.0 m/s and a vortex time of 20 s for one cycle. Ten cycles for each sample was performed to break the cells, and the obtained cell lysis was centrifuged at room temperature at 10 000 rpm for 10 min. The supernatants containing the extracted metabolites were collected for derivatization.
For the derivatization of fatty acids, 40 μL of N,O-bistrifluoroacetamide (BSTFA) with 1% chlorotrimethylsilane (TMCS) was added to each of 60 μL samples and reacted at 70 °C for 30 min. The reaction samples were then shaken at room temperature for another 30 min and then transferred to vials for GC–MS detection. Various concentrations of fatty alcohol standards with carbon chain length from C8 to C18 were taken for the derivatization following the same protocol and analyzed using GC–MS under the same conditions. Quantitative calculation of fatty alcohols in the samples was carried out according to the standard curve. Three biological replicates were performed. For the derivatization of fatty acids, the extracted metabolites were evaporated to dryness. Boron trifluoride (BF3) solution (500 μL) with 14% methyl alcohol (MeOH) was added into each of tubes containing the dried samples and was reacted at 95 °C for 20 min. After that, 50 μL of saturated sodium chloride (NaCl) solution was added to terminate the reaction. n-Hexane solution (300 μL) was then added, and the sample tubes were shaken at room temperature for 1 h. The n-hexane layer containing the derived fatty acids (fatty acid methyl esters, FAMEs) was transferred to the vials for GC–MS detection. Standard curves of the FAME standard mix (with C8–C24 carbon chain length, Sigma) were used as references for the quantitative calculation of FFAs in the extracted samples. Three biological replicates were performed.
GC–MS Procedure and Data Analysis
The GC–MS system (Agilent 5975C) equipped with a DB-5 capillary column with 30 m length, 0.25 μm thickness, and 250 μm inner diameter was used to analyze the samples. Samples measuring 1 μL were injected into the GC–MS equipment and separated by the DB-5 capillary column in a splitless mode. The GC–MS program was performed as follows: The temperature of the inlet and the MS source was set at 250 and 230 °C, respectively. The initial temperature of the oven was set at 80 °C, and then the gradient was increased to 250 °C with a ramp of 7 °C/min. The carrier gas was helium, and its flow rate was set at 1.0 mL/min. The chromatogram and identification of mass spectra were acquired using the Shimadzu GC–MS solution software. The final concentrations of the fatty acid derivatives and the fatty acids were calculated with fatty alcohol standards and FAME standards as references. All experiments were performed in three biological replicates.
Acknowledgments
The authors thank Singapore National Research Foundation (grant NRF-CRP5-2009-03) for funding support.
The authors declare no competing financial interest.
References
- Steen E. J.; Kang Y.; Bokinsky G.; Hu Z.; Schirmer A.; McClure A.; del Cardayre S. B.; Keasling J. D. Microbial production of fatty-acid-derived fuels and chemicals from plant biomass. Nature 2010, 463, 559–562. 10.1038/nature08721. [DOI] [PubMed] [Google Scholar]
- Lennen R. M.; Pfleger B. F. Microbial production of fatty acid-derived fuels and chemicals. Curr. Opin. Biotechnol. 2013, 24, 1044–1053. 10.1016/j.copbio.2013.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kung Y.; Runguphan W.; Keasling J. D. From fields to fuels: Recent advances in the microbial production of biofuels. ACS Synth. Biol. 2012, 1, 498–513. 10.1021/sb300074k. [DOI] [PubMed] [Google Scholar]
- Xu Y.; Chu H.; Gao C.; Tao F.; Zhou Z.; Li K.; Li L.; Ma C.; Xu P. Systematic metabolic engineering of Escherichia coli for high-yield production of fuel bio-chemical 2,3-butanediol. Metab. Eng. 2014, 23, 22–33. 10.1016/j.ymben.2014.02.004. [DOI] [PubMed] [Google Scholar]
- Pfleger B. F.; Gossing M.; Nielsen J. Metabolic engineering strategies for microbial synthesis of oleochemicals. Metab. Eng. 2015, 29, 1–11. 10.1016/j.ymben.2015.01.009. [DOI] [PubMed] [Google Scholar]
- Tang X.; Chen W. N. Investigation of fatty acid accumulation in the engineered Saccharomyces cerevisiae under nitrogen limited culture condition. Bioresour. Technol. 2014, 162, 200–206. 10.1016/j.biortech.2014.03.061. [DOI] [PubMed] [Google Scholar]
- Nielsen J. Systems biology of lipid metabolism: From yeast to human. FEBS Lett. 2009, 583, 3905–3913. 10.1016/j.febslet.2009.10.054. [DOI] [PubMed] [Google Scholar]
- Tehlivets O.; Scheuringer K.; Kohlwein S. D. Fatty acid synthesis and elongation in yeast. Biochim. Biophys. Acta, Mol. Cell Biol. Lipids 2007, 1771, 255–270. 10.1016/j.bbalip.2006.07.004. [DOI] [PubMed] [Google Scholar]
- Black P. N.; DiRusso C. C. Yeast acyl-CoA synthetases at the crossroads of fatty acid metabolism and regulation. Biochim. Biophys. Acta, Mol. Cell Biol. Lipids 2007, 1771, 286–298. 10.1016/j.bbalip.2006.05.003. [DOI] [PubMed] [Google Scholar]
- Faergeman N. J.; Knudsen J. Role of long-chain fatty acyl-CoA esters in the regulation of metabolism and in cell signalling. Biochem. J. 1997, 323, 1–12. 10.1042/bj3230001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beopoulos A.; Nicaud J.-M.; Gaillardin C. An overview of lipid metabolism in yeasts and its impact on biotechnological processes. Appl. Microbiol. Biotechnol. 2011, 90, 1193–1206. 10.1007/s00253-011-3212-8. [DOI] [PubMed] [Google Scholar]
- Shi S.; Valle-Rodríguez J. O.; Khoomrung S.; Siewers V.; Nielsen J. Functional expression and characterization of five wax ester synthases in Saccharomyces cerevisiae and their utility for biodiesel production. Biotechnol. Biofuels 2012, 5, 7. 10.1186/preaccept-1932279820621895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong B. W.; Shi S.; Siewers V.; Nielsen J. Improved production of fatty acid ethyl esters in Saccharomyces cerevisiae through up-regulation of the ethanol degradation pathway and expression of the heterologous phosphoketolase pathway. Microb. Cell Fact. 2014, 13, 39. 10.1186/1475-2859-13-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellenbrand J.; Biester E.-M.; Gruber J.; Hamberg M.; Frentzen M. Fatty acyl-CoA reductases of birds. BMC Biochem. 2011, 12, 64. 10.1186/1471-2091-12-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X.; Chen W. N. Enhanced production of fatty alcohols by engineering the TAGs synthesis pathway in Saccharomyces cerevisiae. Biotechnol. Bioeng. 2015, 112, 386–392. 10.1002/bit.25356. [DOI] [PubMed] [Google Scholar]
- Bernard A.; Domergue F.; Pascal S.; Jetter R.; Renne C.; Faure J.-D.; Haslam R. P.; Napier J. A.; Lessire R.; Joubès J. Reconstitution of plant alkane biosynthesis in yeast demonstrates that Arabidopsis ECERIFERUM1 and ECERIFERUM3 are core components of a very-long-chain alkane synthesis complex. Plant Cell 2012, 24, 3106–3118. 10.1105/tpc.112.099796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirmer A.; Rude M. A.; Li X.; Popova E.; del Cardayre S. B. Microbial biosynthesis of alkanes. Science 2010, 329, 559–562. 10.1126/science.1187936. [DOI] [PubMed] [Google Scholar]
- Akhtar M. K.; Turner N. J.; Jones P. R. Carboxylic acid reductase is a versatile enzyme for the conversion of fatty acids into fuels and chemical commodities. Proc. Natl. Acad. Sci. U.S.A. 2013, 110, 87–92. 10.1073/pnas.1216516110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X.; Chen W. N. Proteomics analysis of metabolically engineered yeast cells and medium-chained hydrocarbon biofuel precursors synthesis. AMB Express 2014, 4, 61. 10.1186/s13568-014-0061-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L.; Zhang J.; Lee J.; Chen W. N. Enhancement of free fatty acid production in Saccharomyces cerevisiae by control of fatty acyl-CoA metabolism. Appl. Microbiol. Biotechnol. 2014, 98, 6739–6750. 10.1007/s00253-014-5758-8. [DOI] [PubMed] [Google Scholar]
- He A.; Li T.; Daniels L.; Fotheringham I.; Rosazza J. P. N. Nocardia sp. carboxylic acid reductase: Cloning, expression, and characterization of a new aldehyde oxidoreductase family. Appl. Environ. Microbiol. 2004, 70, 1874–1881. 10.1128/aem.70.3.1874-1881.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buijs N. A.; Zhou Y. J.; Siewers V.; Nielsen J. Long-chain alkane production by the yeast Saccharomyces cerevisiae. Biotechnol. Bioeng. 2015, 112, 1275–1279. 10.1002/bit.25522. [DOI] [PubMed] [Google Scholar]
- Runguphan W.; Keasling J. D. Metabolic engineering of Saccharomyces cerevisiae for production of fatty acid-derived biofuels and chemicals. Metab. Eng. 2014, 21, 103–113. 10.1016/j.ymben.2013.07.003. [DOI] [PubMed] [Google Scholar]
- Zhou Y. J.; Buijs N. A.; Zhu Z.; Qin J.; Siewers V.; Nielsen J. Production of fatty acid-derived oleochemicals and biofuels by synthetic yeast cell factories. Nat. Commun. 2016, 7, 11709. 10.1038/ncomms11709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegemann J. H.; Güldener U.; Köhler G. J.. Gene disruption in the budding yeast Saccharomyces cerevisiae. In Methods in Molecular Biology; Humana Press, 2006; Vol. 313, pp 129–144. [DOI] [PubMed] [Google Scholar]
- Xie W.; Liu M.; Lv X.; Lu W.; Gu J.; Yu H. Construction of a controllable β-carotene biosynthetic pathway by decentralized assembly strategy in Saccharomyces cerevisiae. Biotechnol. Bioeng. 2014, 111, 125–133. 10.1002/bit.25002. [DOI] [PubMed] [Google Scholar]


