Abstract
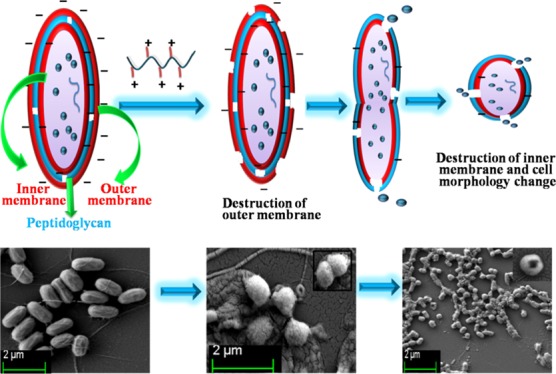
Synthetic polymer-based antimicrobial materials destroy conventional antibiotic resistant microorganisms. Although these antibacterial polymers imitate the properties of antimicrobial peptides (AMPs), their effect on bacterial cell morphology has not been studied in detail. To investigate the morphology change of a bacterial cell in the presence of antimicrobial polymer, herein we have designed and synthesized side-chain amino acid-based cationic polymers, which showed efficient antibacterial activity against Gram-negative (Escherichia coli), as well as Gram-positive (Bacillus subtilis) bacteria. Morphological switching from a rod shape to a spherical shape of E. coli cells was observed by field emission-scanning electron microscopy analysis due to cell wall disruption, whereas the B. subtilis cell structure and size remained intact, but stacks of the cells formed after polymer treatment. The zone of inhibition experiment on an agar plate for E. coli cells exhibited drastic morphological changes at the vicinity of the polymer-treated portion and somewhat less of an effect at the periphery of the plate.
Introduction
Multidrug-resistant pathogenic microorganisms have created a serious problem in the medical sciences.1,2 They cannot be destroyed by conventional antibiotics and cause several diseases and infections in humans.3,4Escherichia coli XL10 (E. coli XL10) is a well-known Gram-negative bacterium, and is such a multidrug-resistant microorganism that causes half of the infections in humans.5 Recently, antimicrobial peptides (AMPs)6,7 have been considered as a promising alternative to conventional antibiotics.8 Antibiotics preserve the bacterial cell morphology, whereas AMPs efficiently show bactericidal properties by physically disrupting the bacterial cytoplasmic membrane instead of targeting mammalian cells.9,10 They attack the bacterial cell membrane directly, and the disruption is mediated by forming electrostatic interactions between the cationic charge of the AMPs and anionic charge of the phosphate headgroups on the membrane surface, which in turn disrupt the membrane by insertion of hydrophobic components into the plasma membrane.11,12 AMPs selectivity attack microorganisms over mammalian cells as zwitterionic phospholipids provide a net neutral charge on the surface of mammalian cells.13,14
Currently, an alternative approach to develop new antimicrobial agents utilizing synthetic polymer chemistry has become popular due to the difficulty and cost of large scale synthesis of AMPs, and also the rapid degradation of AMPs by the protease enzyme inside the human body.15,16 Different classes of polymers such as polyethers,17 polycarbonates,18 polymethacrylates,19 polynorbornenes,20 poly-β-lactams,21 and so forth, have been synthesized as AMP mimics. When designing antimicrobial polymers,22 sufficient cationic charge has to be incorporated into the macromolecule to undergo electrostatic adhesion to the negatively charged microbial cell wall. Further, introduction of a hydrophobic moiety into the polymeric system can lead to disruption of the cellular membrane.23,24
Recently, the antimicrobial efficacy of cationic or hydrophobic polymeric substances and their cell penetration have been investigated extensively.25,26 For example, Haldar and co-workers studied the antibacterial properties of novel hydrolyzable cationic amphiphiles bearing one, two, and three trimethylammonium headgroups and pyridinium headgroups and observed that the incorporation of multiple headgroups led to improved antibacterial activity.27 Their group developed a set of cationic dimeric amphiphiles (bearing cleavable amide linkages between the head group and the hydrocarbon tail with different methylene spacers) with high antibacterial activity against human pathogenic bacteria (E. coli and Staphylococcus aureus) and low cytotoxicity.4 Interaction of cationic amphiphiles with the negatively charged bacterial cell membrane and disruption of the bacterial membrane leading to cell death were observed by field emission-scanning electron microscopy (FESEM) and fluorescence spectroscopy. Zhou et al. investigated the selective antibacterial activities and action mechanism of oligomeric surfactants bearing amide moieties through isothermal titration microcalorimetry, SEM and zeta potential measurements.9 A very recent investigation on polypeptide-based macroporous cryogels, prepared through a polycationic polylysine-b-polyvaline block copolymer with glutaraldehyde as the cross-linker under cryogenic conditions showed inherent antimicrobial properties.28 The key findings were a 95.6% reduction of viable E. coli cells after a brief 1 h incubation and a very interesting “trap and kill” mechanism due to the macroporous structure of the cryogels. Chen et al. investigated a quantitative cell wall disruption mechanism, similar to AMPs, through analyzing the interaction between lipid bilayers acting as a model for a cellular membrane with synthetic antimicrobial polymers by sum frequency generation vibrational spectroscopy.29 Recently, the design of antimicrobial polymers has been extended to the use of primary ammonium groups to mimic the amphiphilic property and cationic functionality of natural AMPs.30,31
Although significant progress has been made in the area of cationic antibacterial polymers, very little attention has been paid to the morphological switching of the bacterial cell. Therefore, we became interested in investigating the morphological switching of Gram-negative bacteria (E. coli) during side-chain amino acid-based cationic polymer treatment. To further our investigation, we studied the effect of these compounds on Gram-positive (Bacillus subtilis) bacteria. The double membrane structures of E. coli (Gram-negative bacterium) are well established.32,33 According to this model, the cell membrane is more difficult to disrupt compared to the single membrane structure of B. subtilis (Gram-positive bacterium).34 However, B. subtilis has a very thick outer cell wall composed of a negatively charged peptidoglycan layer (polysaccharide with amino acid side chains), whereas E. coli has a thin peptidoglycan layer sandwiched between the outer and inner membrane (IM) composed of lipopolysaccharide.35 Because of the difference in cell wall structure, the cell penetration ability of any antimicrobial polymer for two different types of bacteria should be different. In Gram-positive bacteria, the antimicrobial polymer rather easily interacts with the loosely packed porous peptidoglycan layer and attacks the inner cytoplasmic membrane, whereas for Gram-negative bacteria, the additional outer membrane (OM) protects the IM to some extent.34 Hence, the cell wall penetration ability of any antimicrobial agent is expected to be greater in the case of Gram-positive bacteria than for that of Gram-negative bacteria.36 To understand this, three homopolymers with controlled molecular weight and narrow dispersity, composed of alanine, leucine, and phenylalanine-based monomers, and two block copolymers with methyl methacrylate (MMA) and poly(ethylene glycol) methyl ether methacrylate (PEGMA) using an alanine-based macro chain transfer agent (CTA) were prepared via reversible addition–fragmentation chain transfer (RAFT) polymerization. Herein, we selected side-chain amino acid-based polymers due to their biocompatibility and cationic nature.37,38 The antimicrobial effect was more prominent with increasing hydrophobicity of the −R group of the amino acid-based cationic homopolymers and correlated with their cell penetrating ability. The drastic switching (rod shape to spherical shape) of cell morphology of the polymer-treated bacterial cell was observed using FESEM analysis and the Gram staining approach, and was most conspicuous at the vicinity of the polymer-treated region. Cell death resulting from cell membrane disruption and stacking of cells was observed by FESEM and the Gram staining approach. The antimicrobial effect of the above polymers on Gram-negative (E. coli) and Gram-positive (B. subtilis) bacterial cell morphology was investigated systematically with distance from the zone of inhibition. A mechanism of cell morphology switching for Gram-negative bacteria is proposed in the presence of side-chain amino acid-based cationic polymers.
Results and Discussion
Synthesis of Side-Chain Amino Acid-Based Homopolymers and Block Copolymers
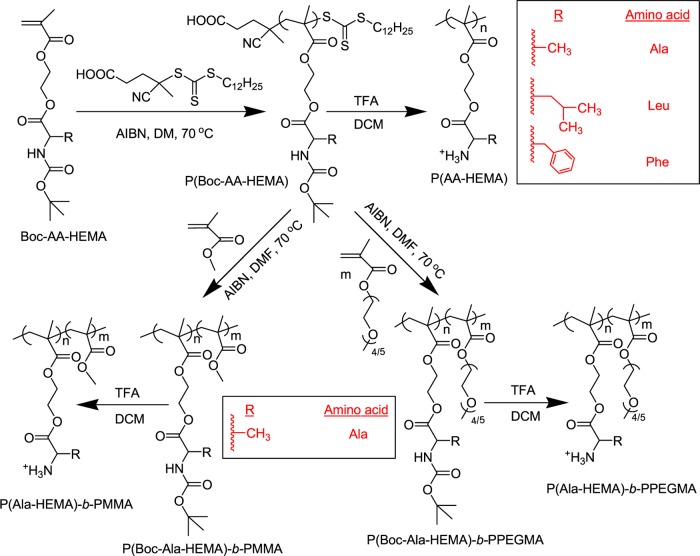
Side-chain amino acid containing methacrylate monomers (Boc-AA-HEMA, Scheme 1, AA = amino acid, i.e., alanine (Ala) or leucine (Leu) or phenylalanine (Phe)) were polymerized via the RAFT technique in dimethylformamide (DMF) at 70 °C using AIBN as the radical initiator and CDP as CTA (Scheme 1) at a constant Boc-AA-HEMA to CDP to AIBN ratio of [Boc-AA-HEMA]/[CDP]/[AIBN] = 25:1:0.1 (Table 1). For the polymer synthesis, we used the RAFT technique to obtain polymers with controlled molecular weight, narrow dispersity (Đ), and defined chain ends. The gel permeation chromatography (GPC) refractive index (RI) traces for all of the homopolymers (P(Boc-AA-HEMA)) indicate a unimodal distribution (Figure S1). Number-average molecular weights (Mn,GPC) and Đ values (1.10–1.18) were determined from the GPC analysis and the results are shown in Table 1. P(Boc-AA-HEMA) was characterized by 1H NMR spectroscopy in CDCl3 (Figures S2–S4). Typical resonance signals for the different protons in the repeating unit of the polymer are assigned on the spectrum. The number-average degree of polymerization (DPn) for P(Boc-Ala-HEMA) was determined by comparing the integration areas of the signals at 4.1–4.5 ppm from the Boc-Ala-HEMA repeating unit (5H from O—CH2—CH2—O— and chiral proton) in the main chain of P(Boc-Ala-HEMA) and at 2.4–2.6 ppm from the terminal —CH2—CH2— protons (4H) from the HOOC—CH2—CH2—C(CN)(CH3)— chain ends (Figure S2A). The DPn values of the other two homopolymers, P(Boc-Leu-HEMA) and P(Boc-Phe-HEMA), were also determined from NMR signal comparison. The DPn of each polymer is denoted by subscripts; for example, P(Boc-Ala-HEMA)14 represents the homopolymer of Boc-Ala-HEMA with DPn = 14. From the NMR chain-end analysis,42 the molecular weights (Mn,NMR) were determined (Table 1). Also, Table 1 summarizes the theoretical molecular weight (Mn,theo) values, which were calculated based on conversion (Conv.) for different homopolymers using the equation: Mn,theo = (([monomer]/[CDP] × average molecular weight (MW) of monomer × conversion) + (MW of CDP)). A nice agreement between Mn,theo, Mn,GPC, and Mn,NMR is observed in Table 1, thus indicating that we have used well-defined polymers for further study.
Scheme 1. Synthesis of Amino Acid-Based Homopolymers and Block Copolymers by RAFT Polymerization, Followed by Deprotection of Side-Chain Boc Groups.
Table 1. Results from the Synthesis of P(Boc-AA-HEMA) Homopolymers and Two Block Copolymers of PEGMA and MMA Using P(Boc-Ala-HEMA) as Macro-CTA at 70 °C in DMF for 5 h.
| polymer | [M]/[CTA]/[AIBN] | conv.d (%) | Mn,GPCe (g/mol) | Đe | Mn,NMRf (g/mol) | Mn,theog (g/mol) |
|---|---|---|---|---|---|---|
| P(Boc-Ala-HEMA)14a | 25/1/0.1 | 50 | 3900 | 1.10 | 4700 | 4200 |
| P(Boc-Leu-HEMA)15a | 25/1/0.1 | 64 | 4800 | 1.18 | 5500 | 5900 |
| P(Boc-Phe-HEMA)10a | 25/1/0.1 | 60 | 4300 | 1.10 | 4200 | 6000 |
| P(Boc-Ala-HEMA)14-b-PPEGMA60b | 50/1/0.1 | 87 | 20 500 | 1.12 | 22 700 | 17 800 |
| P(Boc-Ala-HEMA)14-b-PMMA37c | 50/1/0.1 | 77 | 8200 | 1.14 | 8400 | 8500 |
[M]/[CTA]/[AIBN] = [Boc-AA-HEMA]/[CDP]/[AIBN], Boc-AA-HEMA = Boc-Ala-HEMA or Boc-Leu-HEMA or Boc-Phe-HEMA.
[M]/[CTA]/[AIBN] = [PEGMA]/[P(Boc-Ala-HEMA)-macro-CTA]/[AIBN].
[M]/[CTA]/[AIBN] = [MMA]/[P(Boc-Ala-HEMA)-macro-CTA]/[AIBN].
Calculated gravimetrically.
Measured by GPC.
Determined by 1H NMR study.
Mn,theo = (([monomer]/[CTA] × average molecular weight (MW) of monomer × Conv.) + (MW of CTA)).
Next, PEGMA and MMA were polymerized using P(Boc-Ala-HEMA)-macro-CTA at a constant ratio of [monomer (M)]/[CTA]/[AIBN] = 50/1/0.1 in DMF at 70 °C for 5 h to synthesize the P(Boc-Ala-HEMA)-b-PPEGMA and P(Boc-Ala-HEMA)-b-PMMA block copolymers, respectively. These block copolymers were characterized by 1H NMR spectroscopy in CDCl3 (Figures S5 and S6). Comparison of the integration areas from the terminal group at 2.4–2.6 ppm and side-chain —OCH3 protons in the PPEGMA block at 3.38 ppm allowed calculation of DPn for the PPEGMA segment (Figure S5A). Similarly, the DPn of the PMMA block in the P(Boc-Ala-HEMA)-b-PMMA block copolymer was determined by comparing the integration areas at 2.4–2.6 ppm and side-chain —OCH3 protons at 3.6 ppm from the MMA units (Figure S6A). The DPn of each block is denoted by the subscripts after each block abbreviation; for example, P(Boc-Ala-HEMA)14-b-PPEGMA60 represents the block copolymer, which consists of a P(Boc-Ala-HEMA) block of DPn = 14 and PPEGMA block of DPn = 60. The Mn,NMR values of the block copolymers were also determined by NMR chain-end analysis (Table 1) using the following equation: Mn,NMR = [(DPn,PEGMA/MMA × MPEGMA/MMA) + molecular weight of P(Boc-Ala-HEMA) macro-CTA], where DPn and M are the number-average degree of polymerization of the PPEGMA/PMMA segment and molecular weight of the PEGMA/MMA monomer, respectively. Unimodal GPC RI traces of the block copolymers were shifted toward higher molecular weight (lower elution volume) with respect to P(Boc-Ala-HEMA)14 (Figure S1). The Mn,GPC, Đ, and Mn,theo values of all block copolymers are summarized in Table 1.
Incorporation of cationic charge into our amino acid-based polymers is an essential requirement for good antimicrobial activity via bacterial negatively charged cell wall disruption through electrostatic interaction.43,44 To instill cationic charges into the polymers, deprotection of the side-chain Boc groups from the Boc-protected homopolymers and block copolymers was achieved by trifluoroacetic acid (TFA) at room temperature (Scheme 1). Successful deprotection was proven by the disappearance of Boc proton signals at around 1.44 ppm in the 1H NMR spectrum (Figures S2–S6). The —NH2 signal was lost in the 1H NMR spectrum because these protons are exchangeable with the surrounding deuterated solvent (D2O). After deprotection, the homopolymers (P(AA-HEMA)) and block copolymers (P(Ala-HEMA)-b-PPEGMA/PMMA) were soluble in aqueous media as the —NH2 group of the side chain becomes —NH3+ in an acid medium. The aqueous solubility test was performed for all of the five polymers after deprotection (Scheme S1), wherein each of the polymer solution concentrations was 10 g/L (Table S1). Thus, cationic charge was introduced into our polymers, and this was already proved by our group through the measurement of zeta potential.45,46
Antibacterial Activity against E. coli
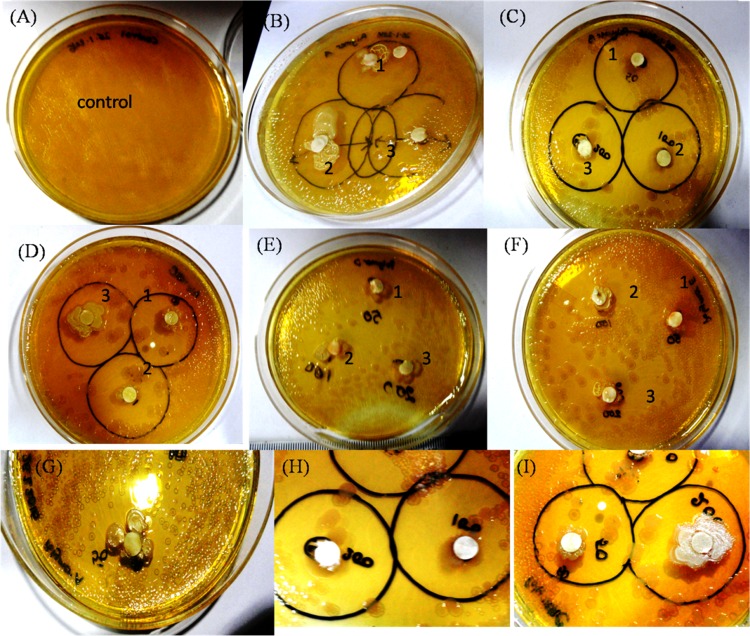
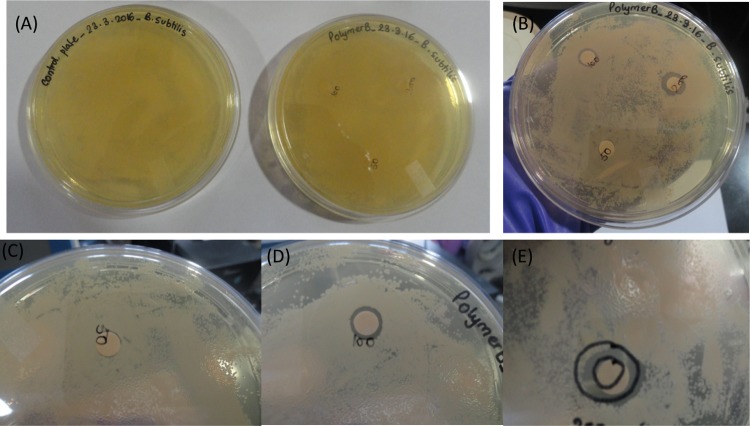
Cationic amphiphilic copolymers have long attracted significant attention from the scientific community due to their capability to control bacterial growth in solution and on surfaces by a mechanism involving the disruption of bacterial cytoplasmic membranes.47 Alkyl quaternary ammonium groups have been widely used as cationic groups, and are likely responsible for polymer binding to bacteria and membrane disruption imitating the mechanism of AMPs.48 Polymers containing cationic pendent primary ammonium groups on the side chain exhibiting higher antimicrobial efficacy have been reported,16,36 as primary ammonium group bearing polymers can extensively imitate the amphiphilic properties and cationic functionalities of AMPs.49,50 Our amino acid-based polymers also contain cationic primary amine groups at their side chain, hence we expected to observe antimicrobial activity with these cationic polymers. Also, they are biocompatible and noncytoxic, which has been previously reported by our group.51 Thus, the bacterial growth inhibitory properties of our side-chain amino acid-based cationic polymers was proven by the zone of inhibition method. Three homopolymers and two block copolymers were tested against a Gram-negative bacterium (E. coli) at volumes of 50, 100, and 200 μL from an initial stock of 10 mg/mL (Figure S7). One schematic representation of a petriplate is shown in Figure S8 for further clarification about the inhibition zone. A clear zone of inhibition was observed for the three homopolymers, which indicates a strong bacterial growth inhibitory effect. Such antibacterial properties do not appear to be prominent when both the block copolymers were tested after 12 h incubation (Figures 1 and S7, where the circled portion of the figure indicates the zone of inhibition). This could be due to the different hydrophobicity and less positive charges in the block copolymer systems. In addition to electrostatic interactions, the effect of the hydrophobicity of a polymer on the antimicrobial activity is well reported.52,53 Many research groups have already proposed the insertion of hydrophobic substituents into the bacterial cell membrane that could cause leakage of the cytoplasm causing cell death.54,55 Hence, in addition to electrostatic interactions, more hydrophobicity could result in better antimicrobial activity. Conversely, introduction of a small mol % of hydrophilic PEGMA into poly(vinylpyridine) was reported to improve the antimicrobial efficacy due to enhancement of the surface wettability of the hydrophobic copolymer.56 In our case, the polymers were already soluble in aqueous media, hence PEGMA could not affect the water solubility and surface wettability. Addition of a sufficiently large hydrophilic PPEGMA60 block to the P(Ala-HEMA)14 segment may have caused the decrease in hydrophobicity leading to the lower cell penetration efficiency of P(Ala-HEMA)14-b-PPEGMA60 compared to that of the P(Ala-HEMA)14 homopolymer, leading to a lower antimicrobial efficiency of the resulting block copolymer. However, despite the hydrophobicity of the PMMA block, the resulting block copolymer P(Ala-HEMA)14-b-PMMA37 showed a lower antibacterial efficiency compared to that of the pure homopolymer. This could be due to the amphiphilic balance of antimicrobial polymers, which is an important parameter that controls antimicrobial activities.57,58 It is most likely that P(Ala-HEMA)14-b-PMMA37 formed a micelle,42 and as a result, the hydrophobic PMMA segments became unavailable for interaction with the lipid membranes of the bacteria. Thus, the aggregation of P(Ala-HEMA)14-b-PMMA37 in solution prevented its antimicrobial efficiency.59
Figure 1.
Zone of inhibition (circled portion) against E. coli treatment: (A) control (without polymer), (B) treated with P(Ala-HEMA)14, (C) P(Leu-HEMA)15, (D) P(Phe-HEMA)10, (E) P(Ala-HEMA)14-b-PPEGMA60, and (F) P(Ala-HEMA)14-b-PMMA37 at (1) 50 μL, (2) 100 μL, and (3) 200 μL from 10 mg/mL stock solution and zoomed view of zone of inhibition treatment with (G) P(Ala-HEMA)14, (H) P(Leu-HEMA)15, and (I) P(Phe-HEMA)10. Bacterial growth is not inhibited in control disk in the absence of polymer, but when the disk was loaded with polymer, the inhibition zone was prominent for three homopolymers and expanded with increasing concentration of the polymer solution, on the contrary, no clear inhibitory effect of the two block copolymers was observed. Each experiment was run in duplicate.
Table 2 provides the quantitative data of the area of zone of inhibition for the three homopolymers. The hydrophobic effect of the side-chain −R group of the amino acid-based polymer on bacterial growth inhibition is observed here. P(Ala-HEMA)14, P(Leu-HEMA)15, and P(Phe-HEMA)10 have methyl, isopropyl, and benzyl as their −R group, respectively. Isopropyl and benzyl are more hydrophobic compared to the methyl group resulting in greater cell wall penetration ability. Hence, a greater area of zone of inhibition for P(Leu-HEMA)15 and P(Phe-HEMA)10 is observed compared to that of P(Ala-HEMA)14 (Table 2).
Table 2. Quantitative Values of Zone of Inhibition against E. coli XL10.
| polymer | volume (μL) | radius of zone of inhibition (R1) (cm) | area of zone of inhibition (cm2)a |
|---|---|---|---|
| P(Ala-HEMA)14 | 50 | 1.3 | 5.18 |
| 100 | 1.3 | 5.18 | |
| 200 | 1.3 | 5.18 | |
| P(Leu-HEMA)15 | 50 | 2.0 | 12.43 |
| 100 | 2.0 | 12.43 | |
| 200 | 2.0 | 12.43 | |
| P(Phe-HEMA)10 | 50 | 1.3 | 5.18 |
| 100 | 1.5 | 6.94 | |
| 200 | 2.0 | 12.43 |
Zone of inhibition = π(R12 – r2), r = radius of sterilized filter paper = 0.2 cm, area πr2 = π(0.2)2 = 0.13 cm2.
Additional evidence of the bacterial growth inhibitory efficacy of our polymer was obtained by performing the Minimum inhibitory concentration (MIC) experiment with P(Leu-HEMA)15 on E. coli cells. The MIC value was determined as 60 μg/mL, which is quite impressive compared to that of some other reported cationic antimicrobial polymers that exhibit high MIC values.16,36 Hence, we can comment that our polymer is much more efficient as an antimicrobial agent compared to several reported cationic antibacterial polymers.
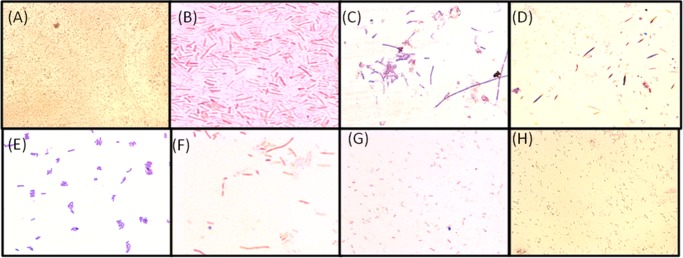
In the next stage, Gram staining was performed only with the homopolymer-treated bacterial cell which gave a prominent zone of inhibition compared to that of the two block copolymers. Gram staining was performed with cells from the close vicinity of the homopolymer-treated region and from the periphery of the agar plate, which showed the effect of polymer treatment on the bacterial cell morphology (Figure 2). The polymer-treated cells were found to be clustered to each other in comparison to the normal bacterial cell, though the individual cell morphology was very difficult to interpret based on optical microscopy images. This observation is most prominent in the case of the P(Leu-HEMA)15-treated bacterial cells from within the zone of inhibition (Figure 2). Upon polymer treatment, retention of crystal violet (CV) within the bacterial cells is another interesting observation (Figure 2). The general mechanism of Gram staining allows for positively charged CV molecules to passively disperse into the cell and electrostatically bind to available anionic surfaces. Introduction of a mordant (typically a solution of iodine and potassium iodide) allows it to react with cationic CV, producing a CV–mordant precipitate. Cells were washed with an alcohol to remove the primary stain (i.e., decolorization) followed by counter staining (typically using the red dye, safranin O). However, during antimicrobial polymer treatment, the cytoplasm leaked out into the periplasmic space due to cell membrane disruption and interacted with the primary stain to some extent leading to retention of the color.
Figure 2.
Optical microscope images of E. coli cells following Gram staining: (A) control (40× resolution), (B) control (100× resolution), (C) treated with P(Ala-HEMA)14 within the zone of inhibition (100× resolution), (D) treated with P(Ala-HEMA)14 away from the zone of inhibition (100× resolution), (E) treated with P(Leu-HEMA)15 within the zone of inhibition (100× resolution), (F) treated with P(Leu-HEMA)15 away from the zone of inhibition (100× resolution), (G) treated with P(Phe-HEMA)10 within the zone of inhibition (100× resolution), and (H) treated with P(Phe-HEMA)10 away from the zone of inhibition (100× resolution). Polymer-treated bacterial cells appear to be stacked and CV color is retained in the vicinity of the zone of inhibition and the effect becomes less prominent with increasing distance from the inhibitory zone.
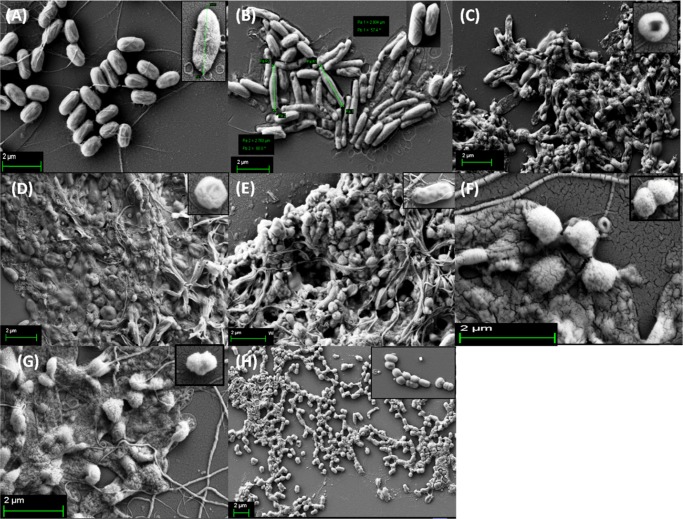
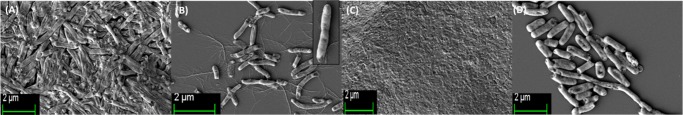
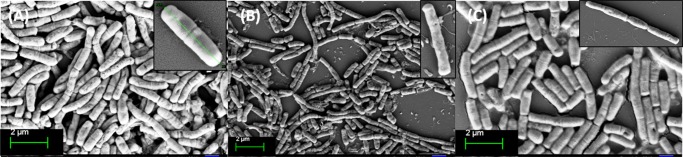
Individual cell morphology was observed by FESEM (Figure 3). Imaging was performed with the three homopolymer-treated bacterial cells from the near and far areas of the zone of inhibition. A morphological change of the bacteria after incubation with poly(ionic liquid) membranes has been reported, where aggregation of lipid vesicles and collapsed cell walls on the membrane surface were the crucial observations.55 Bacterial cellular morphology in the presence of complex natural products with antibacterial activity, such as honey, has been investigated, wherein the cell morphology was analyzed during lag- and log-phase growth; and cell shape transformation (length or width), cell lysis (breakage of cells or leakage of cytoplasm indicating cell envelope or growth abnormalities), and detection of chromosomal DNA abnormalities by DAPI staining were the crucial observations.60 Bacterial cell morphology was also investigated upon treatment with Tween20, heparin, and disodium tetraborate.61 Further, the morphological investigation of E. coli cells after the destructive extraction of phospholipids from the peptidoglycan layer by graphene nanosheets through transmission electron microscopy has been reported.62 Our observation was exclusively different, as the polymer-treated cells appeared to be spherical compared to the rod shape of the control cells. In addition to the peptidoglycan wall, the actin-like MreB protein and several membrane proteins interacting with MreB are essential for the production and preservation of the rod shape morphology of bacteria, and MreB has an extended-filament architecture whose localization, in turn, may affect the shape of the cell wall, causing the rod to spherical transformation.63 The morphological switching is prominent in close vicinity of the polymer-treated region. The effect becomes less prominent at regions further away from the zone of inhibition, but the switching characteristics were still observed to some extent. The smooth cell membrane of untreated bacteria was preserved whereas the presence of a corrugated cell surface and debris of polymer-treated cells suggests that polymers show antibacterial activity through a membrane disruption mechanism (Figure 3E,G). The E. coli cell consists of an OM and IM, which are separated by a cross-linked porous peptidoglycan layer. The surface of E. coli is negatively charged, and mainly consists of lipopolysaccharides and anionic phospholipids of the OM.64 The cationic polymer may first interact with the negatively charged OM disrupting it through electrostatic interactions thereby penetrating the peptidoglycan mesh. It then interacts with the inner cell membrane through electrostatic and hydrophobic interactions. Disruption of the cell membrane results in the leakage of cytoplasm which causes cell death. During this cell disruption process, the morphological switching observed here could be a possibility (Figure 3F–H). The best variation is observed for the P(Leu-HEMA)15 treated cells. This is may be due to the greater cell penetration ability of this polymer compared to that of the others, as the hydrophobicity of the side-chain isopropyl group could play an important role in cell penetration. Another interesting observation was the decrease in bacterial cell size upon polymer treatment (less than 1 μm), indicating E. coli may not be able to grow to the maximum length during treatment (Figure 3D,E,H).
Figure 3.
FESEM images of E. coli cells: (A) control, where the smooth bacterial cell membrane was preserved, (B) treated with P(Ala-HEMA)14 within the zone of inhibition, stacking of cells was observed, (C) treated with P(Ala-HEMA)14 away from the zone of inhibition, presence of corrugated cell surface was found, (D) treated with P(Leu-HEMA)15 within the zone of inhibition, the cells were stacked through leakage of cytoplast and a spherical morphology appeared, (E) treated with P(Leu-HEMA)15 away from the zone of inhibition, spherical cells and cell debris were observed, (F) treated with P(Phe-HEMA)10 within the zone of inhibition, cleavage of bacterial cell was found during treatment, (G) treated with P(Phe-HEMA)10 away from the zone of inhibition, debris of polymer-treated cells appeared, and (H) treated with P(Leu-HEMA)15 at the vicinity of polymer-treated region, bacterial cells appear as spherical.
As an efficient bacterial growth inhibitory property of side-chain amino acid-based homopolymers in solid media has been established by the above experiments, their efficacy in preventing bacterial growth in liquid media was studied next to verify whether these polymers show efficient antimicrobial properties in liquid media. Luria Broth (LB) was used as the liquid media. The experiment was performed with only P(Leu-HEMA)15 treatment as this polymer gave the best morphological switching of the bacterial cells. The OD600 value of the bacterial culture with P(Leu-HEMA)15 was recorded at different time intervals and plotted against time, and showed no exponential enhancement curve (Figure 4). In comparison, exponential cell growth was observed in the flask without the polymer. The absence of the exponential enhancement curve indicates that the polymer prevents the growth of bacterial cells.
Figure 4.
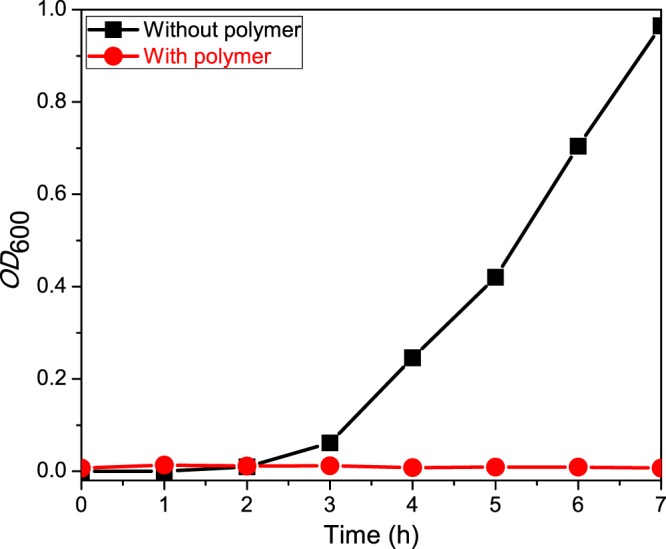
Growth curve of E. coli cells in LB media in the presence and absence of P(Leu-HEMA)15. For the control experiment, where the polymer was absent, exponential cell growth was observed, and this was absent in the presence of the polymer.
Gram staining results (Figure S9) with each fraction of P(Leu-HEMA)15-treated bacterial culture at 1 h time intervals show retention of the primary stain color to some extent. The observation is not so clear because of the smaller cell size. Again, the population of bacteria was found to be much lower compared to that of the control (bacterial culture without polymer) during Gram staining after 12 h incubation (Figure S9), which confirms the bacterial growth inhibitory effect in a liquid medium. The bacterial cell morphology in LB media due to polymer treatment was investigated by FESEM analysis (Figure 5). Distinct morphological switching, as was observed in solid media, is not observed here. However, a sheetlike structure (Figure 5C) and stacking of the cells (Figure 5D) are the crucial observations of this experiment after 7 h incubation, when cell growth was completely prevented, and indicate cell death had occurred.
Figure 5.
FESEM images of E. coli during bacterial growth in LB media: control (without P(Leu-HEMA)15 polymer) images of bacterial cell from (A) congested cell area and (B) discrete cell area, where cell size and morphology were intact; P(Leu-HEMA)15 treated cell images from (C) congested cell area (sheetlike structure) and (D) discrete cell area (stacking of cells) after 7 h incubation.
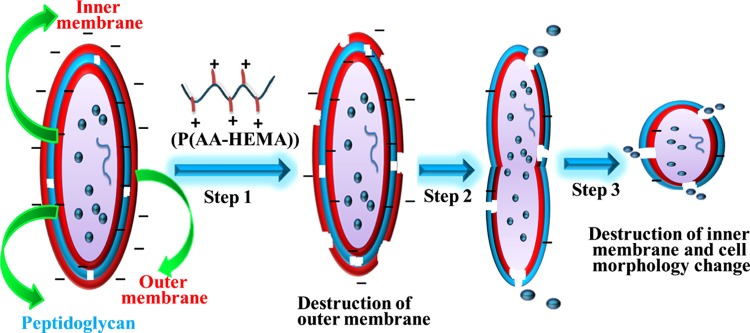
The Gram-negative bacterial cell wall is composed of the OM, intermediate peptidoglycan layer, and IM. The cytoplasmic membrane of E. coli is rich in phosphatidylethanolamine and anionic phosphatidylgylcerol lipids, which are present in a roughly 4:1 ratio.65 Our polymers exhibited bacterial killing efficacy due to the presence of the cationic pendent primary amine groups and hydrophobic −R group (methyl, isopropyl, and benzyl) at their side chains. This is the major structural difference from other non-antimicrobial polymers. The molecular mechanisms of membrane binding and bacterial cell disruption by cationic amphiphilic polymer were reported earlier by flourometric assay.66 The polymer-induced leakage of small dye molecules from liposomes, lipid vesicles synthesized mimicking the phospholipid composition of a bacterial cell, was the reported procedure to quantify the membrane permeability of the polymers.67 On the basis of these earlier reports, a possible mechanism of morphological switching of the Gram-negative bacterial cell is summarized in Figure 6. In step 1, the positively charged polymer destroys the negatively charged OM of the bacterial cell wall through electrostatic interactions. The peptidoglycan layer consists of pores called a peptidiglycan mesh. In step 2, the cationic polymer, especially the hydrophobic group, crosses the peptidoglycan layer through the peptidoglycan mesh and interacts with the IM causing disruption via electrostatic and hydrophobic interactions leading to cytoplasm leakage. The cell wall disruption proceeds through a cleavable intermediate morphological variation (Figure 3F). In step 3, bacterial cell morphology completely switches from a rod shape to a spherical shape during treatment with the antibacterial cationic polymer (Figure 3H).
Figure 6.
Step 1: positively charged polymer disrupts the OM of Gram-negative bacterial cell wall through electrostatic interactions. Step 2: polymer penetrates the intermediate peptidoglycan layer and interacts with the IM through cleavable intermediate morphological variation. Step 3: total morphological switching of bacterial cell from rod shape to spherical shape with destruction of inner cell membrane.
Antibacterial Activity against B. subtilis
In the next stage, the antimicrobial activity of our cationic polymers on a Gram-positive bacterium (B. subtilis) is investigated by the zone of inhibition method. The experiment was performed only with P(Leu-HEMA)15 at three different volumes of 50, 100, and 200 μL from an initial 10 mg/mL stock solution (Figure 7). After 12 h incubation, no inhibition zone was observed on treatment with the 50 μL polymer solutions, whereas a very clear inhibition zone was noticed when treated with 100 and 200 μL of polymer solutions (Figure 7). However, the inhibitory effect is localized and the area of zone of inhibition increases with increasing concentration of P(Leu-HEMA)15 solution (Figure 7 and Table 3). The quantitative values of area of zone of inhibition (Table 3) indicate a lower antibacterial activity of P(Leu-HEMA)15 on B. subtilis compared to that on E. coli at equivalent concentrations of polymer solution treatment. The explanation for this is based on the variation of cell wall structure of the Gram-positive and Gram-negative bacteria. For Gram-negative bacteria, the cell wall is more anionic and hydrophilic compared to that of the Gram-positive one,68 hence leading to stronger electrostatic interactions between the anionic cell wall and cationic P(Leu-HEMA)15.
Figure 7.
Zone of inhibition for B. subtilis treatment with P(Leu-HEMA)15: (A) control (without polymer), (B) after polymer treatment, and zoomed view of zone of inhibition at (C) 50 μL, (D) 100 μL, and (E) 200 μL from 10 mg/mL stock solution. Inhibitory effect is localized and area of zone of inhibition increases with increasing concentration of P(Leu-HEMA)15 solution. Each experiment was run in duplicate.
Table 3. Quantitative Values of Zone of Inhibition for B. subtilis Treatment with P(Leu-HEMA)15a.
| polymer | volume (μL) | radius of zone of inhibition (R1) (cm) | area of zone of inhibition (cm2) |
|---|---|---|---|
| P(Leu-HEMA)15 | 50 | 0.0 | 0.0 |
| 100 | 0.4 | 0.4 | |
| 200 | 0.6 | 1.0 |
Zone of inhibition = π(R12 – r2), r = radius of sterilized filter paper (disk) = 0.2 cm, area πr2 = π(0.2)2 = 0.13 cm2.
The effect of P(Leu-HEMA)15 on cell morphology was analyzed by Gram staining (Figure S10) and FESEM (Figure 8). B. subtilis has a very thick outer cell wall composed of a negatively charged peptidoglycan layer (polysaccharide with amino acid side chains) and inner cytoplasmic membrane. However, during polymer treatment, stacking of cells was observed, although the overall cell morphology and average cell length remain unchanged. The fusion of the cell membrane results in an assemblage of lipid vesicles, hence causing surface collapse in the Gram-positive bacterial cells (B. subtilis).
Figure 8.
FESEM images of B. subtilis cells: (A) control (without polymer treatment), P(Leu-HEMA)15 treated cells (B) near and (C) away from the zone of inhibition. During polymer treatment, stacking of cells was observed, although overall cell morphology and average cell length remain unchanged from control set.
Conclusions
Side-chain amino acid-based cationic polymers with pendant alanine, leucine, and phenylalanine moieties showed efficient antibacterial activity on both Gram-negative (E. coli) and Gram-positive (B. subtilis) bacteria. Considerable switching of bacterial cell morphology from a rod shape to a spherical shape was clearly observed through FESEM analysis during polymer treatment of E. coli cells. The most prominent effect was observed for treatment of E. coli cells with the leucine-based cationic homopolymer, whereas B. subtilis cells did not show any drastic morphological change. Stacking of cells was observed in the cases of E. coli and B. subtilis. During polymeric treatment, sometimes the E. coli cells could not grow to the maximum bacterial length due to the harsh polymeric environment; the effect is most obvious at the vicinity of the polymer-treated region; however it was also observed to some extent far away from the inhibitory zone. With increasing distance from the polymer-treated region on the petriplate, the morphology switching effect is lower, as expected. Such a widespread effect is absent in the case of B. subtilis, although the bacterial growth inhibition zone is more clear compared to that for E. coli at the vicinity of the polymer on the petriplates. Thus, the area of zone of inhibition for E. coli is larger than that for B. subtilis. Therefore, we can conclude that for Gram-negative bacteria the polymer has a more spread-out antibacterial effect through morphological switching, whereas in the case of Gram-positive bacteria, the effect is very clear and localized with indiscrete bacterial cell morphology and cell size.
Materials and Methods
Materials
Boc-l-alanine (Boc-l-Ala-OH, 99%), Boc-l-phenylalanine (Boc-l-Phe-OH, 99%), and TFA (99.5%) were purchased from Sisco Research Laboratories Pvt. Ltd., India. Boc-l-leucine (Boc-l-Leu-OH, 99%), 4-dimethylaminopyridine (99%), anhydrous N,N-dimethylformamide (DMF, 99.9%), dicyclohexylcarbodiimide (99%), and 2-hydroxyethyl methacrylate (HEMA, 97%) were obtained from Sigma-Aldrich. MMA (Sigma-Aldrich, 99%) and PEGMA (molecular weight 300 g/mol, Sigma-Aldrich, 99%) were passed through a basic alumina column prior to polymerization. 2,2′-Azobisisobutyronitrile (AIBN, Sigma, 98%) was recrystallized twice from methanol. CDCl3 (99.8% D) and D2O (99% D) were purchased from Cambridge Isotope Laboratories, Inc., for NMR study. Amino acid-based vinyl monomers,39 Boc-l-alanine methacryloyloxyethyl ester (Boc-Ala-HEMA), Boc-l-leucine methacryloyloxyethyl ester (Boc-Leu-HEMA), Boc-l-phenylalanine methacryloyloxyethyl ester (Boc-Phe-HEMA), and 4-cyano-4-(dodecylsulfanylthiocarbonyl)sulfanylpentanoic acid (CDP)40 as CTA were synthesized by previously reported procedures. The solvents, such as hexanes (mixture of isomers), acetone, dichloromethane (DCM), and so forth, were purified by standard procedures. Agar, tryptone, sodium chloride (NaCl), and yeast extract were obtained from Merck, India. Petriplates were obtained from Tarsons Products Pvt. Ltd., India. Phosphate buffered saline (PBS) tablets were received from Sigma-Aldrich. Milli-Q filtered water was used to prepare solutions and autoclaved before using. The bacterial stains used for experiments were E. coli XL10 (E. coli) and B. subtilis.
Instrumentation
GPC measurements were conducted in tetrahydrofuran at 30 °C with a flow rate of 1.0 mL/min (equipped with a Waters Model 515 HPLC pump, one PolarGel-M guard column, and two PolarGel-M analytical columns (300 × 7.5 mm2)). Detection consisted of a Waters Model 2414 RI detector. Narrow molecular weight poly(methyl methacrylate) (PMMA) standards (Mp values ranging from 1280 to 199 000 g/mol) were used to calibrate the GPC system. NMR spectra were acquired in a Bruker AvanceIII 500 MHz spectrometer at 25 °C. Gram staining images of bacteria were taken using an optical microscope at 40× and 100× resolution before and after polymer treatment. Optical density (OD) measurements of bacterial solutions with and without polymer at 600 nm (OD600) were performed using a Hitachi U2900 spectrometer.
Synthesis of Homopolymers
A typical polymerization procedure is described as follows: Boc-Ala-HEMA (1.0 g, 3.3 mmol), CDP (53.6 mg, 0.13 mmol), AIBN (2.13 mg, 13.0 μmol; 1.06 g stock solution of 4.0 mg AIBN in 2.0 g DMF), and DMF (2.9 g) were sealed in a 20 mL vial equipped with a magnetic stir bar. The vial was purged with dry N2 for 20 min and was placed in a preheated reaction block at 70 °C. The polymerization reaction was quenched by cooling the vial in an ice–water bath and exposing the solution to air after 5 h. The solution was diluted with acetone and precipitated into cold hexanes. The polymer, P(Boc-Ala-HEMA), was reprecipitated four times from acetone/hexanes and dried under vacuum at 40 °C for 6 h. Similarly, Boc-Leu-HEMA and Boc-Phe-HEMA were polymerized to obtain the corresponding polymers P(Boc-Leu-HEMA) and P(Boc-Phe-HEMA), respectively. The purified polymers were isolated as yellowish white powders.
Synthesis of Block Copolymers
A typical block copolymerization procedure is described as follows: PEGMA (0.45 g, 1.50 mmol), P(Boc-Ala-HEMA)-macro-CTA (Mn,GPC = 3900 g/mol, dispersity (Đ) = 1.10, 100 mg, 0.03 mmol), AIBN (0.49 mg, 3.0 μmol; 0.25 g stock solution of 4.0 mg AIBN in 2.0 g DMF), and DMF (1.0 g) were added to a 20 mL polymerization vial equipped with a magnetic bar and purged with dry N2 gas for 15 min. The reaction vial was put in a preheated reaction block at 70 °C for 5 h. The resulting block copolymer, P(Boc-Ala-HEMA)-b-PPEGMA, was purified as mentioned above for the homopolymer. Another block copolymer, P(Boc-Ala-HEMA)-b-PMMA, was synthesized by polymerization of MMA using P(Boc-Ala-HEMA)-macro-CTA following the above-mentioned procedure.
Deprotection of Boc-Protected Polymers
Typically, 2.0 mL of TFA was added to a solution containing 0.3 g of polymer in 1.0 mL of DCM in a 20 mL glass vial. The solution was stirred for 2 h at room temperature, precipitated four times in hexanes from acetone solutions, and finally dried under vacuum at 40 °C for 8 h.
Antibacterial Activity: Zone of Inhibition Method
Constituents of Luria Bertani (LB) agar (1.0 g tryptone, 1.0 g NaCl, and 0.5 g yeast extract in 100 mL de-ionized (DI) water) were weighed and autoclaved. Sterile LB agar plates were prepared and 100 μL of inoculum (either E. coli or B. subtilis) was spread on the surface homogeneously. The plates were allowed to dry for 10 min. UV sterile disks made of filter paper (radius = 0.2 cm, area 0.13 cm2) were soaked in the polymer solution prepared in sterilized distilled water and placed on the agar plates. For each plate, three different volumes were used for study; 50, 100, and 200 μL of each polymer solution from an initial stock 10 mg/mL. A control plate was also prepared without polymer. Duplicate plates were prepared for each polymer. The agar plates were incubated at 37 °C for 12 h. The area of the zone up to which the polymer prevents the bacterial growth was measured by simple mathematical calculation. Photographs were captured using a digital camera.
MIC was determined by an LB microdilution technique. Here, 200 μL of E. coli culture (OD600 = 0.5) and our cationic polymer solution from the initial 10 mg/mL stock were added to the 5 mL of LB media and incubated at 37 °C for 18 h (overnight). A number of experimental sets were arranged whereby the final concentrations of the polymer solutions were 10, 20, 30, 40, 50, 60, 70, 80, 120, 160, 200, 240, 280, 320, and 360 μg/mL. MIC is the lowest concentration of antibacterial agent to prevent the appearance of visible haziness after overnight incubation, that is, bacterial growth is resisted completely.13 The tests were conducted in duplicate.
Gram Staining
Gram staining was performed with cells collected from the zone of inhibition and from the periphery of the plate, that is, far from the zone of inhibition, to check for the effect of polymer on the cell morphology. Gram staining was performed following standard published protocol41 and the slides were observed under a light microscope.
FESEM Analysis
Bacterial cells (E. coli or B. subtilis) were collected from within the zone of inhibition and away from the zone of inhibition from each homopolymer-treated plate and from the control plate (without polymer). The cultures were centrifuged at 5000 rpm for 5 min. The precipitates were washed with DI water two times and then with 1% PBS (pH 7.2). 1 mL of 2.5% glutaraldehyde in PBS was added for 0.5 μL culture in the next stage. The samples were incubated at room temperature for 30 min and then overnight at 4 °C. The pellets were collected by centrifugation and washed with PBS three times. Dehydration of the samples was performed in different ethanol grades (10, 30, 50, 60, 70, 80, 90, and 100% – each volume 200 μL for 10 min). Samples were incubated in 100% ethanol for 1 h. Finally, FESEM samples were prepared as follows: an aliquot of sample solution was drop-casted on a cover slip, dried, and coated with gold/palladium (20:80). Finally, images were recorded using a Carl Zeiss-Sigma instrument.
Antibacterial Activity: LB Media
Polymer antibacterial activity was determined against E. coli cells cultured in LB. Overnight cultures were prepared and used as the starter culture for the growth experiment. Four conical flasks were fixed: one as blank (LB media), one as control (culture), and two experimental set ups (culture + 200 μL of polymer). Cells were cultured at 37 °C at 180 rpm. OD was measured using a U2900 UV-vis spectrometer at 1 h time intervals to plot the bacterial growth curve in the presence and absence of polymer. Fractions were collected for Gram staining and FESEM analysis.
Acknowledgments
Ishita Mukherjee acknowledges the Council of Scientific and Industrial Research (CSIR), Government of India, for her junior research fellowship.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsomega.7b00181.
GPC and NMR characterization plots, zone of inhibition experiment against E. coli treatment, and Gram staining results, as noted in the article (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Zhang Q.; Lambert G.; Liao D.; Kim H.; Robin K.; Tung C.-k.; Pourmand N.; Austin R. H. Acceleration of Emergence of Bacterial Antibiotic Resistance in Connected Microenvironments. Science 2011, 333, 1764–1767. 10.1126/science.1208747. [DOI] [PubMed] [Google Scholar]
- Martínez J. L. Antibiotics and Antibiotic Resistance Genes in Natural Environments. Science 2008, 321, 365–367. 10.1126/science.1159483. [DOI] [PubMed] [Google Scholar]
- Ford T. E.; Colwell R. R.. Global Decline in Microbiological Safety of Water: A Call for Action; American Academy of Microbiology: Washington, DC, 1996; pp 1–40. [Google Scholar]
- Hoque J.; Akkapeddi P.; Yarlagadda V.; Uppu D. S.; Kumar P.; Haldar J. Cleavable Cationic Antibacterial Amphiphiles: Synthesis, Mechanism of Action, and Cytotoxicities. Langmuir 2012, 28, 12225–12234. 10.1021/la302303d. [DOI] [PubMed] [Google Scholar]
- Asensi G. F.; dos Reis E. M. F.; Del Aguila E. M.; dos P. Rodrigues D.; Silva J. T.; Paschoalin V. M. F. Detection of Escherichia coli and Salmonella in Chicken Rinse Carcasses. Br. Food J. 2009, 111, 517–527. 10.1108/00070700910965989. [DOI] [Google Scholar]
- Zhou C.; Wang M.; Zou K.; Chen J.; Zhu Y.; Du J. Antibacterial Polypeptide-Grafted Chitosan-Based Nanocapsules As an “Armed” Carrier of Anticancer and Antiepileptic Drugs. ACS Macro Lett. 2013, 2, 1021–1025. 10.1021/mz400480z. [DOI] [PubMed] [Google Scholar]
- Krizsan A.; Volke D.; Weinert S.; Sträter N.; Knappe D.; Hoffmann R. Insect-Derived Proline-Rich Antimicrobial Peptides Kill Bacteria by Inhibiting Bacterial Protein Translation at the 70 S Ribosome. Angew. Chem., Int. Ed. 2014, 53, 12236–12239. 10.1002/anie.201407145. [DOI] [PubMed] [Google Scholar]
- Poole K. Efflux-mediated Multiresistance in Gram-negative Bacteria. Clin. Microbiol. Infect. 2004, 10, 12–26. 10.1111/j.1469-0691.2004.00763.x. [DOI] [PubMed] [Google Scholar]
- Zhou C.; Wang F.; Chen H.; Li M.; Qiao F.; Liu Z.; Hou Y.; Wu C.; Fan Y.; Liu L.; Wang S.; Wang Y. Selective Antimicrobial Activities and Action Mechanism of Micelles Self-Assembled by Cationic Oligomeric Surfactants. ACS Appl. Mater. Interfaces 2016, 8, 4242–4249. 10.1021/acsami.5b12688. [DOI] [PubMed] [Google Scholar]
- Brender J. R.; McHenry A. J.; Ramamoorthy A. Does cholesterol play a role in the bacterial selectivity of antimicrobial peptides?. Front. Immunol. 2012, 3, 195 10.3389/fimmu.2012.00195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paslay L. C.; Abel B. A.; Brown T. D.; Koul V.; Choudhary V.; McCormick C. L.; Morgan S. E. Antimicrobial Poly(methacrylamide) Derivatives Prepared via Aqueous RAFT Polymerization Exhibit Biocidal Efficiency Dependent upon Cation Structure. Biomacromolecules 2012, 13, 2472–2482. 10.1021/bm3007083. [DOI] [PubMed] [Google Scholar]
- Yeaman M. R.; Yount N. Mechanisms of Antimicrobial Peptide Action and Resistance. Pharmacol. Rev. 2003, 55, 27–55. 10.1124/pr.55.1.2. [DOI] [PubMed] [Google Scholar]
- Michl T. D.; Locock K. E. S.; Stevens N. E.; Hayball J. D.; Vasilev K.; Postma A.; Qu Y.; Traven A.; Haeussler M.; Meagher L.; Griesser H. J. RAFT-Derived Antimicrobial Polymethacrylates: Elucidating the Impact of End-Groups on Activity and Cytotoxicity. Polym. Chem. 2014, 5, 5813–5822. 10.1039/C4PY00652F. [DOI] [Google Scholar]
- Matsuzaki K. Control of Cell Selectivity of Antimicrobial Peptides. Biochim. Biophys. Acta 2009, 1788, 1687–1692. 10.1016/j.bbamem.2008.09.013. [DOI] [PubMed] [Google Scholar]
- Hancock R. E.; Sahl H.-G. Antimicrobial and Host-Defense Peptides as New Anti-Infective Therapeutic Strategies. Nat. Biotechnol. 2006, 24, 1551–1557. 10.1038/nbt1267. [DOI] [PubMed] [Google Scholar]
- Punia A.; Mancuso A.; Banerjee P.; Yang N. Nonhemolytic and Antibacterial Acrylic Copolymers with Hexamethyleneamine and Poly(ethylene glycol) Side Chains. ACS Macro Lett. 2015, 4, 426–430. 10.1021/acsmacrolett.5b00102. [DOI] [PubMed] [Google Scholar]
- Oda Y.; Kanaoka S.; Sato T.; Aoshima S.; Kuroda K. Block Versus Random Amphiphilic Copolymers as Antibacterial Agents. Biomacromolecules 2011, 12, 3581–3591. 10.1021/bm200780r. [DOI] [PubMed] [Google Scholar]
- Engler A. C.; Tan J. P. K.; Ong Z. Y.; Coady D. J.; Ng V. W. L.; Yang Y. Y.; Hedrick J. L. Antimicrobial Polycarbonates: Investigating the Impact of Balancing Charge and Hydrophobicity Using a Same-Centered Polymer Approach. Biomacromolecules 2013, 14, 4331–4339. 10.1021/bm401248t. [DOI] [PubMed] [Google Scholar]
- Kuroda K.; DeGrado W. F. Amphiphilic Polymethacrylate Derivatives as Antimicrobial Agents. J. Am. Chem. Soc. 2005, 127, 4128–4129. 10.1021/ja044205+. [DOI] [PubMed] [Google Scholar]
- Ilker M. F.; Nüsslein K.; Tew G. N.; Coughlin E. B. Tuning the Hemolytic and Antibacterial Activities of Amphiphilic Polynorbornene Derivatives. J. Am. Chem. Soc. 2004, 126, 15870–15875. 10.1021/ja045664d. [DOI] [PubMed] [Google Scholar]
- Mowery B. P.; Lee S. E.; Kissounko D. A.; Epand R. F.; Epand R. M.; Weisblum B.; Stahl S. S.; Gellman S. H. Mimicry of Antimicrobial Host-Defense Peptides by Random Copolymers. J. Am. Chem. Soc. 2007, 129, 15474–15476. 10.1021/ja077288d. [DOI] [PubMed] [Google Scholar]
- Sgolastra F.; Deronde B. M.; Sarapas J. M.; Som A.; Tew G. N. Designing Mimics of Membrane Active Proteins. Acc. Chem. Res. 2013, 46, 2977–2987. 10.1021/ar400066v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler A. C.; Wiradharma N.; Ong Z. Y.; Coady D. J.; Hedrick J. L.; Yang Y. Y. Emerging Trends in Macromolecular Antimicrobials to Fight Multi-Drug-Resistant Infections. Nano Today 2012, 7, 201–222. 10.1016/j.nantod.2012.04.003. [DOI] [Google Scholar]
- Uppu D. S. S. M.; Bhowmik M.; Samaddar S.; Haldar J. Cyclization and Unsaturation Rather than Isomerisation of Side Chains Govern the Selective Antibacterial Activity of Cationic-Amphiphilic Polymers. Chem. Commun. 2016, 52, 4644–4647. 10.1039/C5CC09930G. [DOI] [PubMed] [Google Scholar]
- Kenawy E.-R.; Worley S. D.; Broughton R. The Chemistry and Applications of Antimicrobial Polymers: A State-of-the-Art Review. Biomacromolecules 2007, 8, 1359–1384. 10.1021/bm061150q. [DOI] [PubMed] [Google Scholar]
- Ulubayram K.; Calamak S.; Shahbazi R.; Eroglu I. Nanofibers Based Antibacterial Drug Design, Delivery and Applications. Curr. Pharm. Des. 2015, 21, 1930–1943. 10.2174/1381612821666150302151804. [DOI] [PubMed] [Google Scholar]
- Haldar J.; Kondaiah P.; Bhattacharya S. Synthesis and Antibacterial Properties of Novel Hydrolyzable Cationic Amphiphiles. Incorporation of Multiple Head Groups Leads to Impressive Antibacterial Activity. J. Med. Chem. 2005, 48, 3823–3831. 10.1021/jm049106l. [DOI] [PubMed] [Google Scholar]
- Shirbin S. J.; Lam S. J.; Jun-An Chan N.; Ozmen M. M.; Fu Q.; O’Brien-Simpson N.; Reynolds E. C.; Qiao G. G. Polypeptide-Based Macroporous Cryogels with Inherent Antimicrobial Properties: The Importance of a Macroporous Structure. ACS Macro Lett. 2016, 5, 552–557. 10.1021/acsmacrolett.6b00174. [DOI] [PubMed] [Google Scholar]
- Avery C. W.; Palermo E. F.; McLaughlin A.; Kuroda K.; Chen Z. Investigations of the Interactions between Synthetic Antimicrobial Polymers and Substrate-Supported Lipid Bilayers Using Sum Frequency Generation Vibrational Spectroscopy. Anal. Chem. 2011, 83, 1342–1349. 10.1021/ac1025804. [DOI] [PubMed] [Google Scholar]
- Palermo E. F.; Kuroda K. Structural Determinants of Antimicrobial Activity in Polymers Which Mimic Host Defense Peptides. Appl. Microbiol. Biotechnol. 2010, 87, 1605–1615. 10.1007/s00253-010-2687-z. [DOI] [PubMed] [Google Scholar]
- Gabriel G. J.; Som A.; Madkour A. E.; Eren T.; Tew G. N. Infectious Disease: Connecting Innate Immunity to Biocidal Polymers. Mater. Sci. Eng., R 2007, 57, 28–64. 10.1016/j.mser.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lienkamp K.; Kumar K.; Som A.; Nüsslein K.; Tew G. N. “Doubly Selective” Antimicrobial Polymers: How Do They Differentiate between Bacteria?. Chem.–Eur. J. 2009, 15, 11710–11714. 10.1002/chem.200802558. [DOI] [PubMed] [Google Scholar]
- Kellenberger E.; Ryter A. Cell Wall and Cytoplasmic Membrane of Escherichia coli. J. Biophys. Biochem. Cytol. 1958, 4, 323–326. 10.1083/jcb.4.3.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz-Bonilla A.; Fernández-García M. Polymeric Materials with Antimicrobial Activity. Prog. Polym. Sci. 2012, 37, 281–339. 10.1016/j.progpolymsci.2011.08.005. [DOI] [Google Scholar]
- Yuan H.; Liu Z.; Liu L.; Lv F.; Wang Y.; Wang S. Cationic Conjugated Polymers for Discrimination of Microbial Pathogens. Adv. Mater. 2014, 26, 4333–4338. 10.1002/adma.201400636. [DOI] [PubMed] [Google Scholar]
- Punia A.; He E.; Lee K.; Banerjee P.; Yang N. Cationic Amphiphilic Non-Hemolytic Polyacrylates with Superior Antibacterial Activity. Chem. Commun. 2014, 50, 7071–7074. 10.1039/c4cc01583e. [DOI] [PubMed] [Google Scholar]
- Bauri K.; Roy S. G.; De P. Side-Chain Amino-Acid-Derived Cationic Chiral Polymers by Controlled Radical Polymerization. Macromol. Chem. Phys. 2016, 217, 365–379. 10.1002/macp.201500271. [DOI] [Google Scholar]
- Roy S. G.; De P. pH Responsive Polymers with Amino Acids in the Side Chains and Their Potential Applications. J. Appl. Polym. Sci. 2014, 131, 41084 10.1002/app.41084. [DOI] [Google Scholar]
- Sun H.; Gao C. Facile Synthesis of Multiamino Vinyl Poly(amino acid)s for Promising Bioapplications. Biomacromolecules 2010, 11, 3609–3616. 10.1021/bm101060m. [DOI] [PubMed] [Google Scholar]
- Moad G.; Chong Y. K.; Postma A.; Rizzardo E.; Thang S. H. Advances in RAFT Polymerization: The Synthesis of Polymers with Defined End-Groups. Polymer 2005, 46, 8458–8468. 10.1016/j.polymer.2004.12.061. [DOI] [Google Scholar]
- Wilhelm M. J.; Sheffield J. B.; Sharifian M. G.; Wu Y.; Spahr C.; Gonella G.; Xu B.; Dai H. Gram’s Stain Does Not Cross the Bacterial Cytoplasmic Membrane. ACS Chem. Biol. 2015, 10, 1711–1717. 10.1021/acschembio.5b00042. [DOI] [PubMed] [Google Scholar]
- Kumar S.; Roy S. G.; De P. Cationic Methacrylate Polymers Containing Chiral Amino Acid Moieties: Controlled Synthesis via RAFT Polymerization. Polym. Chem. 2012, 3, 1239–1248. 10.1039/c2py00607c. [DOI] [Google Scholar]
- Ji E.; Parthasarathy A.; Corbitt T. S.; Schanze K. S.; Whitten D. G. Antibacterial Activity of Conjugated Polyelectrolytes with Variable Chain Lengths. Langmuir 2011, 27, 10763–10769. 10.1021/la2018192. [DOI] [PubMed] [Google Scholar]
- Locock K. E. S.; Michl T. D.; Valentin J. D. P.; Vasilev K.; Hayball J. D.; Qu Y.; Traven A.; Griesser H. J.; Meagher L.; Haeussler M. Guanylated Polymethacrylates: A Class of Potent Antimicrobial Polymers with Low Hemolytic Activity. Biomacromolecules 2013, 14, 4021–4031. 10.1021/bm401128r. [DOI] [PubMed] [Google Scholar]
- Bauri K.; Roy S. G.; Pant S.; De P. Controlled Synthesis of Amino Acid-Based pH-Responsive Chiral Polymers and Self-Assembly of Their Block Copolymers. Langmuir 2013, 29, 2764–2774. 10.1021/la304918s. [DOI] [PubMed] [Google Scholar]
- Haldar U.; Nandi M.; Ruidas B.; De P. Controlled Synthesis of Amino-Acid Based Tadpole-Shaped Organic/Inorganic Hybrid Polymers and Their Self-Assembly in Aqueous Media. Eur. Polym. J. 2015, 67, 274–283. 10.1016/j.eurpolymj.2015.04.005. [DOI] [Google Scholar]
- Wu M.; Maier E.; Benz R.; Hancock R. E. W. Mechanism of Interaction of Different Classes of Cationic Antimicrobial Peptides with Planar Bilayers and with the Cytoplasmic Membrane of Escherichia coli. Biochemistry 1999, 38, 7235–7242. 10.1021/bi9826299. [DOI] [PubMed] [Google Scholar]
- Mizutani M.; Palermo E. F.; Thoma L. M.; Satoh K.; Kamigaito M.; Kuroda K. Design and Synthesis of Self-Degradable Antibacterial Polymers by Simultaneous Chain- and Step-Growth Radical Copolymerization. Biomacromolecules 2012, 13, 1554–1563. 10.1021/bm300254s. [DOI] [PubMed] [Google Scholar]
- Tew G. N.; Scott R. W.; Klein M. L.; Degrado W. F. De Novo Design of Antimicrobial Polymers, Foldamers, and Small Molecules: From Discovery to Practical Applications. Acc. Chem. Res. 2010, 43, 30–39. 10.1021/ar900036b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mowery B. P.; Lee S. E.; Kissounko D. A.; Epand R. F.; Epand R. M.; Weisblum B.; Stahl S. S.; Gellman S. H. Mimicry of Antimicrobial Host-Defense Peptides by Random Copolymers. J. Am. Chem. Soc. 2007, 129, 15474–15476. 10.1021/ja077288d. [DOI] [PubMed] [Google Scholar]
- Kumar S.; Acharya R.; Chatterji U.; De P. Side-Chain Amino-Acid-Based pH-Responsive Self-Assembled Block Copolymers for Drug Delivery and Gene Transfer. Langmuir 2013, 29, 15375–15385. 10.1021/la403819g. [DOI] [PubMed] [Google Scholar]
- Sambhy V.; Peterson B. R.; Sen A. Antibacterial and Hemolytic Activities of Pyridinium Polymers as a Function of the Spatial Relationship between the Positive Charge and the Pendant Alkyl Tail. Angew. Chem. 2008, 120, 1270–1274. 10.1002/ange.200702287. [DOI] [PubMed] [Google Scholar]
- Gabriel G. J.; Maegerlein J. A.; Nelson C. F.; Dabkowski J. M.; Eren T.; Nüsslein K.; Tew G. N. Comparison of Facially Amphiphilic versus Segregated Monomers in the Design of Antibacterial Copolymers. Chem.–Eur. J. 2009, 15, 433–439. 10.1002/chem.200801233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trewyn B. G.; Whitman C. M.; Lin V. S. Y. Morphological Control of Room-Temperature Ionic Liquid Templated Mesoporous Silica Nanoparticles for Controlled Release of Antibacterial Agents. Nano Lett. 2004, 4, 2139–2143. 10.1021/nl048774r. [DOI] [Google Scholar]
- Guo J.; Xu Q.; Zheng Z.; Zhou S.; Mao H.; Wang B.; Yan F. Intrinsically Antibacterial Poly(ionic liquid) Membranes: The Synergistic Effect of Anions. ACS Macro Lett. 2015, 4, 1094–1098. 10.1021/acsmacrolett.5b00609. [DOI] [PubMed] [Google Scholar]
- Sellenet P. H.; Allison B.; Applegate B. M.; Youngblood J. P. Synergistic Activity of Hydrophilic Modification in Antibiotic Polymers. Biomacromolecules 2007, 8, 19–23. 10.1021/bm0605513. [DOI] [PubMed] [Google Scholar]
- Mowery B. P.; Lindner A. H.; Weisblum B.; Stahl S. S.; Gellman S. H. Structure–activity Relationships among Random Nylon-3 Copolymers That Mimic Antibacterial Host-Defense Peptides. J. Am. Chem. Soc. 2009, 131, 9735–9745. 10.1021/ja901613g. [DOI] [PubMed] [Google Scholar]
- Palermo E. F.; Kuroda K. Chemical Structure of Cationic Groups in Amphiphilic Polymethacrylates Modulates the Antimicrobial and Hemolytic Activities. Biomacromolecules 2009, 10, 1416–1428. 10.1021/bm900044x. [DOI] [PubMed] [Google Scholar]
- Feder R.; Dagan A.; Mor A. Structure-Activity Relationship Study of Antimicrobial Dermaseptin S4 Showing the Consequences of Peptide Oligomerization on Selective Cytotoxicity. J. Biol. Chem. 2000, 275, 4230–4238. 10.1074/jbc.275.6.4230. [DOI] [PubMed] [Google Scholar]
- Lu J.; Carter D. A.; Turnbull L.; Rosendale D.; Hedderley D.; Stephens J.; Gannabathula S.; Steinhorn G.; Schlothauer R. C.; Whitchurch C. B.; Harry E. J. The Effect of New Zealand Kanuka, Manuka and Clover Honeys on Bacterial Growth Dynamics and Cellular Morphology Varies According to the Species. PLoS One 2013, 8, e55898 10.1371/journal.pone.0055898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camesano T. A.; Natan M. J.; Logan B. E. Observation of Changes in Bacterial Cell Morphology Using Tapping Mode Atomic Force Microscopy. Langmuir 2000, 16, 4563–4572. 10.1021/la990805o. [DOI] [Google Scholar]
- Tu Y.; Lv M.; Xiu P.; Huynh T.; Zhang M.; Castelli M.; Liu Z.; Huang Q.; Fan C.; Fang H.; Zhou R. Destructive Extraction of Phospholipids from Escherichia coli Membranes by Graphene Nanosheets. Nat. Nanotechnol. 2013, 8, 594–601. 10.1038/nnano.2013.125. [DOI] [PubMed] [Google Scholar]
- Reimold C.; Soufo H. J. D.; Dempwolff F.; Graumann P. L. Motion of Variable-Length MreB Filaments at the Bacterial Cell Membrane Influences Cell Morphology. Mol. Biol. Cell 2013, 24, 2340–2349. 10.1091/mbc.E12-10-0728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timofeeva L.; Kleshcheva N. Antimicrobial Polymers: Mechanism of Action, Factors of Activity, and Applications. Appl. Microbiol. Biotechnol. 2011, 89, 475–492. 10.1007/s00253-010-2920-9. [DOI] [PubMed] [Google Scholar]
- Sovadinova I.; Palermo E. F.; Huang R.; Thoma L. M.; Kuroda K. Mechanism of Polymer-Induced Hemolysis: Nanosized Pore Formation and Osmotic Lysis. Biomacromolecules 2011, 12, 260–268. 10.1021/bm1011739. [DOI] [PubMed] [Google Scholar]
- Palermo E. F.; Lee D.-K.; Ramamoorthy A.; Kuroda K. Role of Cationic Group Structure in Membrane Binding and Disruption by Amphiphilic Copolymers. J. Phys. Chem. B 2011, 115, 366–375. 10.1021/jp1083357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palermo E. F.; Sovadinova I.; Kuroda K. Structural Determinants of Antimicrobial Activity and Biocompatibility in Membrane-Disrupting Methacrylamide Random Copolymers. Biomacromolecules 2009, 10, 3098–3107. 10.1021/bm900784x. [DOI] [PubMed] [Google Scholar]
- Li P.; Zhou C.; Rayatpisheh S.; Ye K.; Poon Y. F.; Hammond P. T.; Duan H.; Chan-Park M. B. Cationic Peptidopolysaccharides Show Excellent Broad-Spectrum Antimicrobial Activities and High Selectivity. Adv. Mater. 2012, 24, 4130–4137. 10.1002/adma.201104186. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.