Abstract
Single-molecule force spectroscopy greatly benefits from site-specific surface immobilization and specific probing with a functionalized cantilever. Here, we describe a streamlined approach to such experiments by covalently attaching mechanically stable receptors onto proteins of interest (POI) to improve pickup efficiency and specificity. This platform provides improved throughput, allows precise control over the pulling geometry, and allows for multiple constructs to be probed with the same ligand-modified cantilever. We employ two orthogonal enzymatic ligation reactions [sortase and phosphopantetheinyl transferase (Sfp)] to covalently immobilize POI to a pegylated surface and to subsequently ligate the POI to a mechanically stable dockerin domain at the protein’s C-terminus for use as a high-strength pulling handle. Our configuration permits expression and folding of the POI to proceed independently from the mechanically stable receptor used for specific probing and requires only two short terminal peptide sequences (i.e., ybbR-tag and sortase C-tag). We applied this system successfully to proteins expressed using in vitro transcription and translation reactions without a protein purification step and to purified proteins expressed in Escherichia coli.
Introduction
In recent years, the field of single-molecule force spectroscopy (SMFS) has implemented many developments in bioconjugation to improve upon the classical approach of nonspecific pulling experiments by moving to specific, often covalent surface functionalization.1,2 Traditionally, polyproteins are recombinantly expressed as fusion constructs framed by several repeats of marker domains of known unfolding patterns (often Ig-like domains) and nonspecifically deposited onto a surface.3 A bare cantilever tip is then indented into the surface in an attempt to pickup and stretch single polyprotein chains on opposing ends by nonspecific adhesion. In case the number of domain unfoldings in the recorded data trace exceeds the number of domains on each side of the proteins of interest (POI), an N- to C-terminal stretching of the POI can be concluded.
In contrast to the nonspecific attachment, site-specific anchoring and probing approaches offer many advantages. They allow for homogeneous surface preparation as the immobilization geometry is defined; the usage of spacer molecules such as polyethyleneglycol (PEG) diminishes possible surface interaction effects. Drawbacks of unspecific probing—such as low-pickup efficiencies or the requirement of recombinant expression of large polyproteins—have been addressed by utilizing the receptor–ligand pairs as pulling handles to provide a specific interaction by which force can be applied to the POI. Systems such as StrepII-tag-Strep-Tactin,4 streptavidin–biotin,5,6 GCN4-peptide–antibody,7 and cohesin–dockerin domains8−10 are only a few of the interactions that have been employed for this purpose.
These pulling handles classically are genetically appended to the POI and expressed as fusion proteins. The fusion proteins are then covalently immobilized through one end of the POI and probed by ligand-functionalized cantilever-tips that recognize the respective receptor on the other end. A wide range of forces are accessible by utilizing short tags such as the StrepII-tag (116 pN at a loading rate of 4 nN s–1 if the tag is C-terminal and 46 pN at 4 nN s–1 if the tag is N-terminal4) and biotin (257 pN5), as well as that with larger handles such as the interaction between type-3 dockerin and cohesin E from Ruminococcus flavefaciens, reaching up to 700 pN at 100 nN s–1.10 These high-force interactions allow characterization of very stable proteins such as the unfolding of several Titin-Ig domains in series.
However, recombinant expression of a fusion between a (possibly large) POI and a large handle-protein (e.g., 29 kDa for CohE) can be cumbersome. The resulting fusion proteins might be insoluble or the correct folding of the POI might be affected by the presence of the fusion domains during translation and folding. Here, we utilize two orthogonal enzymatic ligation reactions to achieve sortase and phosphopantetheinyl transferase (Sfp)-mediated covalent surface attachment and post-translational modification of several POIs with dockerin handles by sortase-mediated11,12 ligation. This allows the expression of only the protein domain of interest without risking to affect proper folding. The very robust interaction of type-3 dockerin and cohesin from R. flavefacienswas already shown to be functional over repeated measurements of about 24 h,10 which is an important requirement for multiplexing atomic force microscopy (AFM) experiments.
Furthermore, we combined this technique with in vitro expression of the POI in a cell-free system. Because smaller proteins are, in general, expressed with higher yields,13 the reduced size of the protein construct to be expressed is beneficial. This allows for a fast and easy workflow from plasmid DNA to covalently immobilized proteins containing mechanostable handles without the need for bulk expression. We anticipate that our approach will aid in highly parallel mechanical screening of mutant proteins, which benefits from the in vitro expression, obsoleting the need for protein purifications and benefitting from the enhancements in force spectroscopy throughput and robustness.
Materials and Methods
Experimental Design
We selected Titin-Ig domains14 and superfolder green fluorescent protein (sfGFP)15 as the exemplary POIs for this study, as they are well-documented in the literature, and enable comparison with established methods. The POIs were cloned with a ybbR-peptide tag16 at their N-terminus and a sortase A recognition sequence17 LPETGG at their C-terminus. For force-spectroscopy handles, we used GGG-dockerin,10 which was recombinantly expressed in Escherichia coli, purified, and ligated to the C-terminus of the POI using ligation with sortase A. While preliminary experiments were carried out with wild-type sortase A, an evolved mutant18 was ultimately used because of its superior performance. On the cantilever side, CohE-CBM-ybbR was used and immobilized at the ybbR-tag via Sfp-catalyzed ligation.16
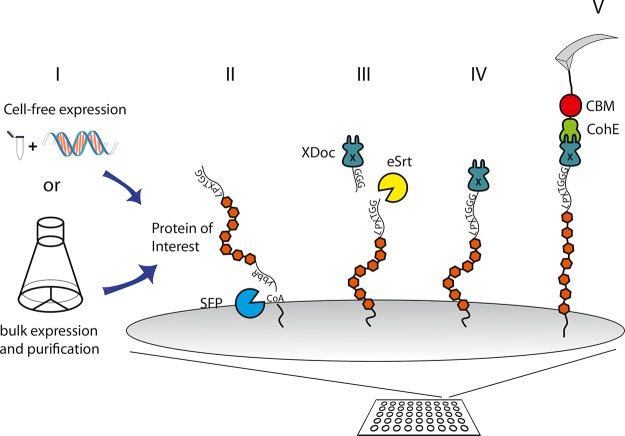
The two specific enzymatic recognition sites located on the termini of the POI ensure that only fully expressed proteins are measured in SMFS-experiments. Figure 1 shows a schematic overview of the experiment.
Figure 1.
Schematic of the experimental setup. (I) POIs were either expressed in bulk or synthesized using a cell-free expression mix. POIs contained a ybbR-tag at the N-terminus and a sortase LPETGG tag at the C-terminus. (II) Surface-bearing PEG-coenzyme A is covalently modified with POIs via Sfp-catalyzed ligation. (III) Next, GGG-Doc is ligated to the POI at the C-terminal end using the LPETGG sortase-tag for use as a force spectroscopy pulling handle. (IV,V) Unfolding experiments are conducted by approaching and retracting a CohE-CBM-functionalized cantilever.
Cloning
Modified pET28a plasmids encoding for ybbR-His-XylanaseT6(T129C) (Geobacillus stearothermophilus)-Doc3 (R. flavefaciens), ybbR-His-sfGFP-DocI (Clostridium thermocellum), and Titin-Ig domains (repeats 27 to 32, repeat 34, human) were used as templates for polymerase chain reaction (PCR) with subsequent reconstitution by Gibson19 assembly. The previously reported18 d59 sortase(P94R/D160N/D165A/K190E/K196T) mutant was created by introducing the mutations via overlap extension PCR followed by ligating the linearized plasmid using Kinase–Ligase–DpnI (KLD) enzyme mix and KLD reaction buffer from the Q5 site-directed mutagenesis kit (New England Biolabs, MA, USA). The chemically competent E. coli DH5-α cells were transformed [Life Technologies GmbH, Frankfurt, Germany; 30 min on ice, 30 s heat shock at 42 °C followed by 37 °C for 1 h in a super optimal broth with catabolite repression medium] and plated on kanamycin-supplemented agar plates. For amino acid sequences, see the Supporting Information.
Protein Expression and Purification
All proteins were expressed in NiCo21(DE3)RIPL cells, which were cultivated in ZYM-5052 autoinduction media20 supplemented with kanamycin and chloramphenicol. After pelleting, the cells were lysed by sonication and then centrifuged at 4 °C, 39 000 rcf for 60 min. The supernatant was filtered to 0.22 μm and applied to Ni-NTA columns (HisTrap FF, GE Healthcare Europe GmbH, Freiburg, Germany). After washing with 6 column volumes of a buffer containing 25 mM Tris, pH 8.4, 300 mM NaCl, 20 mM imidazole, and 0.5 vol % Triton X-100, the bound fraction was eluted with an elution buffer containing 25 mM Tris, pH 8.4, 300 mM NaCl, and 300 mM imidazole.
All protein solutions were concentrated using Amicon centrifugal filter units (10k MWCO, Merck KGaA, Darmstadt, Germany), followed by buffer exchange to Ca-TBS buffer (25 mM Tris, pH 7.2, 75 mM NaCl, and 1 mM CaCl2) using polyacrylamide spin desalting columns. Proteins were stored at −80 °C with glycerol added to 10% (v/v). For cell-free expression, 25 μM reactions of PURExpress In Vitro Protein Synthesis Kit (New England Biolabs, Ipswich, Massachusetts) were incubated for 2 h at 37 °C, containing 300 ng plasmid DNA coding for the POIs.
In case of MGGG-His-Doc, the N-terminal methionine cleavage in E. coli was sufficient for the preparation of GGG-His-Doc, so that no additional protease digestion was necessary.
Surface Preparation
Surfaces and cantilevers for force spectroscopy were silanized using (3-aminopropyl)-dimethyl-ethoxysilane (APDMES, ABCR GmbH, Karlsruhe, Germany) and PEGylated with α-maleimide-hexanoic-ω-NHS PEG (NHS-PEG5000-Mal, Rapp Polymere, Tübingen, Germany) dissolved to 25 mM in 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid buffer (HEPES), 50 mM, pH 7.5 to provide suitable conditions for NHS coupling. Then, the PEGylated surfaces and cantilevers were coupled to coenzyme A (CoA, 1 mM) in sodium phosphate buffer, pH 7.2.
Silicon nitride cantilevers (BioLever mini BL-AC40TS-C2, Olympus, Tokyo, Japan) were used as force probes. Silicone masks with a grid of 1 mm-diameter holes (CultureWell Reusable Gaskets, Grace Bio-Labs, Bend, OR, USA), were applied to the CoA-functionalized glass slides to create separated incubation wells. Each purified POI was diluted to 50 μM in Ca-TBS that was supplemented with 20 mM MgCl2 and Sfp enzyme was added to 10 μM. The reaction mixtures were added to the single incubation wells in the mask, enabling covalent immobilization via Sfp-catalyzed ligation of CoA and the ybbR tags.
For cell-free expression of the POIs, the cell-free expression reaction mix (PURExpress, New England Biolabs, MA, USA) was prepared to contain 100 ng of plasmid DNA. The expression mix was incubated at 37 °C for 2 h, then supplemented with Sfp enzyme to 10 μM and directly applied to the micowells without further purification. Sfp ligation reactions were performed for 2 h at room temperature. After subsequent rinsing with Ca-TBS, the wells were incubated with 100 μM GGG-Doc protein and 10 μM sortase A for 1 h. After rinsing with Ca-TBS, the silicon mask was removed, providing an array of covalently linked proteins that were modified with the dockerin handle at one end.
The sortase-catalyzed ligation reactions for Figure 2 contained 10 μM ybbR-Titin-LPETGG, 10 μM GGG-Doc, and 10 μM of either wild-type d59 sortase or the evolved pentamutant.18 The ligation reactions were incubated for 1 h at 37 °C.
Figure 2.
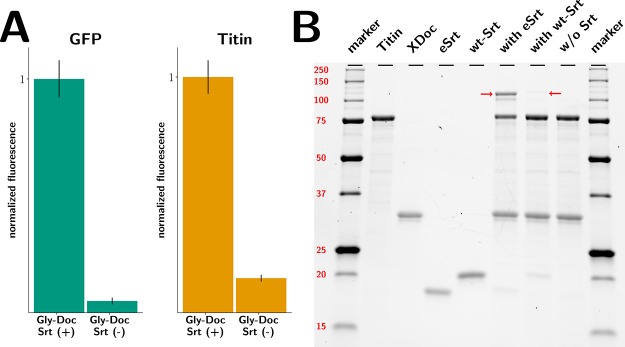
(A) Averaged fluorescence intensities of a CohE-CBM-ybbR-CoA647-labeled surface functionalized with ybbR-Titin-Ig-LPETGG and ybbR-sfGFP-LPETGG. Each protein was immobilized at two separate spots that were then incubated with either GGG-dockerin and sortase or GGG-dockerin but not with sortase. To test for successful ligation of dockerins, CohE-CBM-ybbR-CoA647 was allowed to bind for 10 min at 300 nM, then rinsed and imaged immediately afterward. Fluorescent intensities of each construct were normalized to the intensity of the sortase-positive spot. (B) SDS-PAGE demonstrating the ligation of GGG-dockerin to ybbR-Titin-LPETGG with wild-type sortase A (wt-Srt), pentamutant sortase A (eSrt), or no sortase as negative control. The red arrows are indicating the ligation products.
For surface functionalization tests, CohE-CBM-ybbR was labeled with CoA647 (New England Biolabs, MA, USA) in a reaction containing 25 μM CoA647, 10 μM CohE-CBM-ybbR, and 2 μM Sfp in Ca-TBS supplemented with 20 mM MgCl2. The labeling reaction was incubated for 4 h at 37 °C. Free dye and Sfp enzyme were removed via preparative gel filtration with Ca-TBS as the running buffer through a Yarra 3 μm SEC-3000 (Phenomenex, Torrance, California, USA) column. Appropriate fractions [evaluated via sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE)] were pooled, diluted to 3 μM, supplemented with glycerol to 10% (v/v), and stored at −80 °C.
Labeled surfaces were imaged using ChemiDoc MP (Bio-Rad, Hercules, California, USA), with 625(30) nm/695(55) nm emission/excitation filters. The exposure time was 30 s; for background subtraction, a blank and clean cover slip was imaged with the same settings and an average background signal was subtracted from the measured average intensities. Intensities were quantified via Image Lab 5.2 (Bio-Rad, Hercules, California, USA) volume tool.
Single-Molecule Force Spectroscopy
All data were obtained using Ca-TBS. Measurements were taken with custom-built instruments (driven by PI-731 piezo actuators, Physik Instrumente, Germany) in conjunction with MFP-3D AFM controllers (Asylum Research, Santa Barbara, USA). Upon approaching the sample surface with the cantilever tip, the complex between cohesin/dockerin (C/D) was formed, and the cantilever was retracted from the surface at a constant velocity of 800 nm s–1 while recoding the distance and cantilever deflection at a sampling rate of 12 500 Hz. After each force–extension curve was recorded, the sample was moved laterally by 100 nm to probe a different molecule. For data analysis, force–distance curves were transformed into contour length space using a freely rotating chain model with quantum mechanical corrections for peptide backbone stretching21 and then sorted by contour length increments.22 Loading rates prior to domain unfolding or complex dissociation were extracted by applying a linear fit to the last 3 nm before the respective event and then used in fitting the rupture-force histograms with the Bell–Evans model.23
Results and Discussion
To test for successful surface functionalization, we incubated surfaces that had been prepared as described in the Materials and Methods section with fluorescently labeled cohesin. Figure 2A confirms that if sortase is ommited, no dockerin functionalization is achieved, whereas if sortase was present to perform the ligation reaction, binding of CoE-CBM-ybbR-CoA647 is observed. Figure 2B demonstrates successful ligation of GGG-dockerin to ybbR-Titin-LPETGG and illustrates the superior performance of the evolved sortase mutant d95/P94R/D160N/D165A/K190E/K196T18 in comparison with wild-type sortase A.
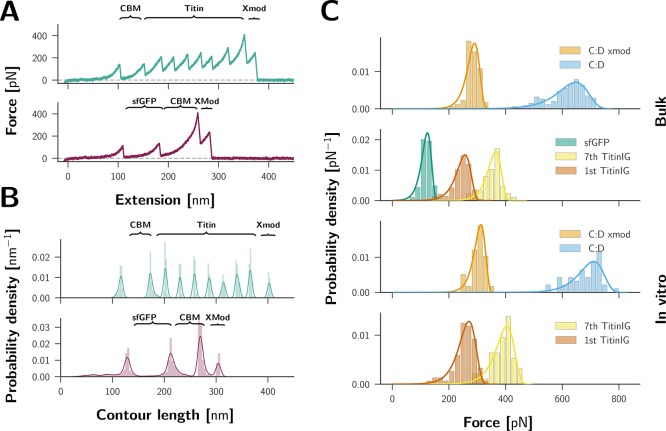
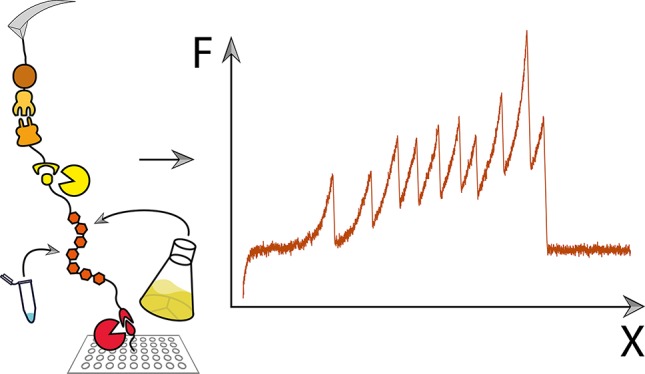
Typical single-molecule force–distance unfolding patterns for the sortase-incubated spots are shown in Figure 3A. They exhibit the unbinding pattern of CohE–Doc dissociation as characterized in previous publications,10 where dissociation can occur with or without unfolding of the dockerin subdomain called x-module. The resulting force–distance curves were transformed into contour length space and then sorted by comparing the observed unfolding increments (3B). Only curves exhibiting the 56 nm increment corresponding to a full unfolding of the CBM-domain were classified to be the result of probing a CohE–Doc complex. Furthermore, the curves were sorted to exhibit no more than one increment corresponding to the unfolding of sfGFP 79 nm and no more than seven increments corresponding to Titin-Ig unfolding 28 nM. These increments result from the added free contour length of the peptide chain upon unfolding the folded protein domains and match the previously reported values.14,15,24 For these traces, unfolding forces of the domains of interest were histogrammed with a bin width of 20 pN (Figure 3C).
Figure 3.
SMFS on Ctta-dockerin-labeled 7× Titin-Ig and sfGFP. (A) Force distance traces showing complete unfolding of the POI (Titin-Ig unfolding is shown in the upper trace, sfGFP in the lower trace). (B) Transformation of traces from (A) into contour-length space. (C) Force histograms of complex dissociation events and unfolding events of the POI: the upper two panels contain data from the bulk-expressed proteins and the lower two panels contain data from in vitro-expressed proteins. C/D complex dissociation can occur with [as in both sample traces shown in (A)] or without prior unfolding of the x-module, which is a subdomain of the dockerin, resulting in two populations of the dissociation forces. Each population was fitted with the Bell–Evans model.23
Despite its narrow tip apex, each cantilever is typically
functionalized
with multiple cohesin-anchors; hence, multiple receptor–ligand
complexes can form if dockerin-decorated surface is densely populated.
Therefore, we went for a rather sparse surface functionalization which
can be tuned by the incubation times of Sfp and sortase-catalyzed
ligation reactions and/or the substrate concentrations. Alternatively,
cantilevers with blunter tips could be used when more interactions
are desired. The achieved surface densities were in a suitable range
for SMFS, sparse enough to avoid multiple interactions but dense enough
to acquire good statistics. Probing attempts (1.24%) resulted in single
molecule unfolding traces satisfying the outlined criteria. In total,
142 Titin-Ig and 92 sfGFP single molecule traces were obtained within
11 h of measurement with a single cantilever (spring constant: 0.093
N m–1). If sortase had been omitted, no traces showing
unfolding of CBM and one of the POI were recorded. For probing of
in vitro-expressed Titin-Ig, 0.33% of attempts were successful, yielding
72 Titin-Ig unfoldings in 9 h of measurement, which was also probed
with a single cantilever (spring constant: 0.097 N m–1). Figure 3C shows
force histograms for unfolding events of sfGFP, the last of seven
Titin-Ig domain to unfold and the complex dissociation itself. This
was carried out for bulk-expressed and purified sfGFP and Titin-Ig,
as well as for Titin-Ig expressed in the cell-free system. Complex
dissociation events cluster into two populations that are characteristic
of Doc/Coh unbinding.10 The most probable
forces at which the POI unfold are 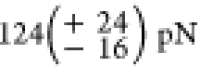 for sfGFP,
for sfGFP, 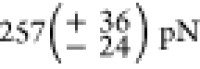 for the first, and
for the first, and 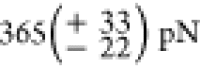 for the last Titin-Ig domain to
unfold
(
for the last Titin-Ig domain to
unfold
(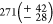 and
and 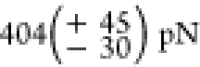 for Titin-Ig expressed in the
cell-free
system), the asymmetrical full widths at half maximum of the distributions
are given in brackets. The most probable forces were determined by
fitting each histogram of unfolding forces with the Bell–Evans
model.23
for Titin-Ig expressed in the
cell-free
system), the asymmetrical full widths at half maximum of the distributions
are given in brackets. The most probable forces were determined by
fitting each histogram of unfolding forces with the Bell–Evans
model.23
The differences between the most probable unfolding forces observed for the POI expressed in the cell-free system and the bulk-expressed proteins are within tolerance of errors resulting from cantilever calibration.25
This method can be easily applied to any recombinantly expressed protein by adding the terminal peptide tags necessary for covalent surface attachment and post-translational sortase-mediated ligation. Owing to the terminal location of these tags, only nondigested and fully expressed proteins are probed. This is especially advantageous for cell-free expression systems, where the small quantity of expressed protein often makes the usually necessary affinity purification cumbersome.
Conclusions
We developed a method that enables acquisition of SMFS datasets of specifically probed and covalently immobilized single molecules. By post-translationally modifying the POI with the high-force interactions of the Coh/Doc receptor–ligand system via sortase ligation, we can probe even resilient proteins such as Titin-Ig domains with high specificity and throughput, improving on the nonspecific polyprotein method and eliminating the requirement of expressing the POI as large fusion constructs with handle domains. The modular system of post-translational attachment of the mechanostable pulling handle allowed us to probe different proteins with the same cantilever. We also applied this approach to proteins expressed in cell-free systems without further purification while still selecting for only fully expressed proteins owing to the specificity provided by the high-affinity pulling handle.
Acknowledgments
This work was supported by the EU 7th Framework Programme NMP4-SL-2013-604530 (CellulosomePlus) and the Society in Science—the Branco Weiss Fellowship from ETH Zürich. The authors thank M. A. Jobst for the instrument control software, L. F. Milles for providing the force curve analysis software, and Thomas Nicolaus as well as Angelika Kardinal for laboratory assistance.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsomega.7b00478.
Amino acid sequences (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Ott W.; Jobst M. A.; Schoeler C.; Gaub H. E.; Nash M. A. Single-molecule force spectroscopy on polyproteins and receptor–ligand complexes: The current toolbox. J. Struct. Biol. 2017, 197, 3–12. 10.1016/j.jsb.2016.02.011. [DOI] [PubMed] [Google Scholar]
- Alsteens D.; Gaub H. E.; Newton R.; Pfreundschuh M.; Gerber C.; Müller D. J. Atomic force microscopy-based characterization and design of biointerfaces. Nat. Rev. Mater. 2017, 2, 17008. 10.1038/natrevmats.2017.8. [DOI] [Google Scholar]
- Li L.; Huang H. H.-L.; Badilla C. L.; Fernandez J. M. Mechanical unfolding intermediates observed by single-molecule force spectroscopy in a fibronectin type III module. J. Mol. Biol. 2005, 345, 817–826. 10.1016/j.jmb.2004.11.021. [DOI] [PubMed] [Google Scholar]
- Baumann F.; Bauer M. S.; Milles L. F.; Alexandrovich A.; Gaub H. E.; Pippig D. A. Monovalent Strep-Tactin for strong and site-specific tethering in nanospectroscopy. Nat. Nanotechnol. 2016, 11, 89–94. 10.1038/nnano.2015.231. [DOI] [PubMed] [Google Scholar]
- Moy V. T.; Florin E.-L.; Gaub H. E. Intermolecular forces and energies between ligands and receptors. Science 1994, 266, 257–259. 10.1126/science.7939660. [DOI] [PubMed] [Google Scholar]
- Edwards D. T.; Faulk J. K.; Sanders A. W.; Bull M. S.; Walder R.; LeBlanc M.-A.; Sousa M. C.; Perkins T. T. Optimizing 1-μs-Resolution Single-Molecule Force Spectroscopy on a Commercial Atomic Force Microscope. Nano Lett. 2015, 15, 7091–7098. 10.1021/acs.nanolett.5b03166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morfill J.; Neumann J.; Blank K.; Steinbach U.; Puchner E. M.; Gottschalk K.-E.; Gaub H. E. Force-Based Analysis of Multidimensional Energy Landscapes: Application of Dynamic Force Spectroscopy and Steered Molecular Dynamics Simulations to an Antibody Fragment–Peptide Complex. J. Mol. Biol. 2008, 381, 1253–1266. 10.1016/j.jmb.2008.06.065. [DOI] [PubMed] [Google Scholar]
- Stahl S. W.; Nash M. A.; Fried D. B.; Slutzki M.; Barak Y.; Bayer E. A.; Gaub H. E. Single-molecule dissection of the high-affinity cohesin-dockerin complex. Proc. Natl. Acad. Sci. U.S.A. 2012, 109, 20431–20436. 10.1073/pnas.1211929109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otten M.; Ott W.; Jobst M. A.; Milles L. F.; Verdorfer T.; Pippig D. A.; Nash M. A.; Gaub H. E. From genes to protein mechanics on a chip. Nat. Methods 2014, 11, 1127–1130. 10.1038/nmeth.3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoeler C.; Malinowska K. H.; Bernardi R. C.; Milles L. F.; Jobst M. A.; Durner E.; Ott W.; Fried D. B.; Bayer E. A.; Schulten K.; Gaub H. E.; Nash M. A. Ultrastable cellulosome-adhesion complex tightens under load. Nat. Commun. 2014, 5, 5635. 10.1038/ncomms6635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazmanian S. K.; Liu G.; Ton-That H.; Schneewind O. Staphylococcus aureus sortase, an enzyme that anchors surface proteins to the cell wall. Science 1999, 285, 760–763. 10.1126/science.285.5428.760. [DOI] [PubMed] [Google Scholar]
- Guimaraes C. P.; Witte M. D.; Theile C. S.; Bozkurt G.; Kundrat L.; Blom A. E. M.; Ploegh H. L. Site-specific C-terminal and internal loop labeling of proteins using sortase-mediated reactions. Nat. Protoc. 2013, 8, 1787–1799. 10.1038/nprot.2013.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagoski D.; Polinkovsky M. E.; Mureev S.; Kunert A.; Johnston W.; Gambin Y.; Alexandrov K. Performance benchmarking of four cell-free protein expression systems. Biotechnol. Bioeng. 2016, 113, 292–300. 10.1002/bit.25814. [DOI] [PubMed] [Google Scholar]
- Rief M.; Gautel M.; Oesterhelt F.; Fernandez J. M.; Gaub H. E. Reversible unfolding of individual titin immunoglobulin domains by AFM. Science 1997, 276, 1109–1112. 10.1126/science.276.5315.1109. [DOI] [PubMed] [Google Scholar]
- Kufer S. K.; Dietz H.; Albrecht C.; Blank K.; Kardinal A.; Rief M.; Gaub H. E. Covalent immobilization of recombinant fusion proteins with hAGT for single molecule force spectroscopy. Eur. Biophys. J. 2005, 35, 72–78. 10.1007/s00249-005-0010-1. [DOI] [PubMed] [Google Scholar]
- Yin J.; Straight P. D.; McLoughlin S. M.; Zhou Z.; Lin A. J.; Golan D. E.; Kelleher N. L.; Kolter R.; Walsh C. T. Genetically encoded short peptide tag for versatile protein labeling by Sfp phosphopantetheinyl transferase. Proc. Natl. Acad. Sci. U.S.A. 2005, 102, 15815–15820. 10.1073/pnas.0507705102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popp M. W.-L.; Antos J. M.; Ploegh H. L.. Site-Specific Protein Labeling via Sortase-Mediated Transpeptidation. In Current Protocols in Protein Science; John Wiley & Sons, Inc., 2009; pp 15.3.1–15.3.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen I.; Dorr B. M.; Liu D. R. A general strategy for the evolution of bond-forming enzymes using yeast display. Proc. Natl. Acad. Sci. U.S.A. 2011, 108, 11399–11404. 10.1073/pnas.1101046108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson D. G.; Young L.; Chuang R.-Y.; Venter J. C.; Hutchison C. A.; Smith H. O. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 2009, 6, 343–345. 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- Studier F. W. Protein production by auto-induction in high-density shaking cultures. Protein Expression Purif. 2005, 41, 207–234. 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- Livadaru L.; Netz R. R.; Kreuzer H. J. Stretching Response of Discrete Semiflexible Polymers. Macromolecules 2003, 36, 3732–3744. 10.1021/ma020751g. [DOI] [Google Scholar]
- Puchner E. M.; Franzen G.; Gautel M.; Gaub H. E. Comparing proteins by their unfolding pattern. Biophys. J. 2008, 95, 426–434. 10.1529/biophysj.108.129999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans E.; Ritchie K. Dynamic strength of molecular adhesion bonds. Biophys. J. 1997, 72, 1541–1555. 10.1016/s0006-3495(97)78802-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietz H.; Rief M. Exploring the energy landscape of GFP by single-molecule mechanical experiments. Proc. Natl. Acad. Sci. U.S.A. 2004, 101, 16192–16197. 10.1073/pnas.0404549101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson C. T.; Smith D. A.; Roberts C. J. Calibration of silicon atomic force microscope cantilevers. Nanotechnology 2005, 16, 234–238. 10.1088/0957-4484/16/2/009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.