Abstract
Cationic lipid-guided nucleic acid delivery holds great promise in gene therapy and genome-editing applications for treating genetic diseases. However, the major challenge lies in achieving therapeutically relevant efficiencies. Prior findings, including our own, demonstrated that asymmetry in the hydrophobic core of cationic lipids imparted superior transfection efficiencies. To this end, we have developed a lipid nanocarrier system with an asymmetric hydrophobic core (PS-Lips) derived from a mixture of fatty acids of food-grade palmstearin and compared its efficiency with symmetric palmitic acid-based nanocarrier system (P-Lip). PS-Lips exhibited superior transfection efficiencies with both plasmid DNA (pDNA) and mRNA in multiple cultured cells than the control P-Lip. More importantly, PS-Lips exhibited 2-fold superior transfections with linear nucleic acid, green fluorescent protein (GFP) mRNA in hematopoietic cells, when compared with the commercial control lipofectamine RNAiMAX. PS-Lips was also found to be effective in delivering genome-editing tools (CRISPR/Cas9, sgRNA encoded pDNA with a reporter GFP construct) than P-Lip in HEK-293 cells. In the present study, we report that cationic liposomes derivatized from natural food-grade fat palmstearin with a natural hydrophobic core asymmetry are efficient in delivering both linear and circular nucleic acids. In particular, PS-Lips is efficient in delivering mRNA to hematopoietic cells. These findings can be further exploited in the genome-editing approach for treating β-globinopathies.
Introduction
Biological findings in the recent past, including CRISPR/Cas9-mediated genome-editing, have widened the application window of nucleic acid delivery for investigating and manipulating cellular processes.1 Preclinical and clinical studies have demonstrated that the modification of human cells at the genetic level significantly improves their therapeutic potentials.2−4 Vectors play a critical role in the efficient delivery of nucleic acids.5 To date, viral vectors have been well-explored as they are efficient in delivering nucleic acids both in vitro and in vivo. However, the random integration of the virus vector into the host genome, which may interrupt the essential gene expression and cellular processes, limits their therapeutic applications.6 More importantly, the scalability of the vectors is labor intensive and technically demanding; hence, making them into a clinically viable product is a significant challenge.7 Hence, alternate efforts have been directed toward developing nonviral transfection methods.8,9 Effective and safe nucleic acid delivery requires an engineering of synthetic vectors, which are highly biocompatible, nontoxic, and efficient in nature.10,11 Such synthetic lipids are much better than the viral vectors in terms of safety.12 Among the nonviral vectors, cationic lipids hold great promise, owing to their biocompatibility and ease in robust scalability.13 However, lower efficiencies and higher cytotoxicities limit their therapeutic applications.14 To this end, numerous structure–activity investigations have been performed on the molecular architecture for enhancing their efficacy.14−16 Typically, cationic lipids consist of a hydrophilic head group and a hydrophobic domain containing either hydrocarbon chains or steroidal moieties connected with/without a spacer/linker group. The molecular framework of the lipid plays an important role in influencing the transfection properties of the liposomal system. Prior findings, including our own, demonstrated the significance of variation/asymmetry of chain lengths within the hydrophobic region.17−19 Taking cues from these findings, we further demonstrated that the hydrophobic chain asymmetry that originated from food-grade coconut oil enhanced the transfection properties of the liposomal system by greater than 4-fold and was found to be safer, when compared with its symmetric lauric acid (C12) analogue.20 However, greater variation in the chain lengths of the fatty acyl chains from coconut oil, which ranges from C6 to C18:2, makes it difficult to maintain the uniform composition from batch to batch.
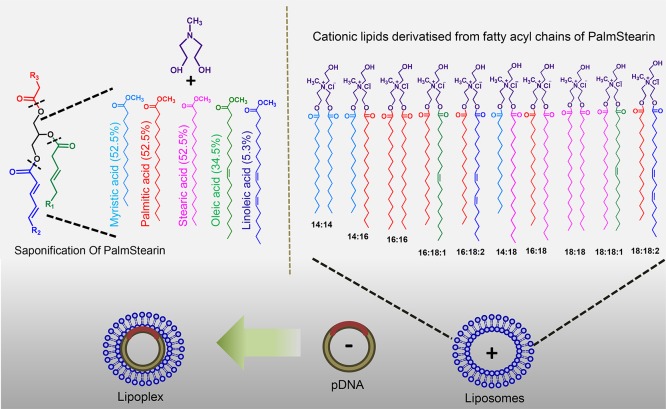
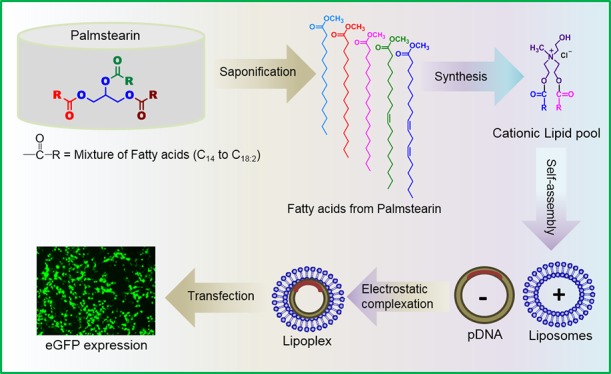
To minimize such a variation, we chose edible fat, palmstearin, which contains minimal variation in fatty acyl chains ranging from myristic acid (C14) to linoleic acid (C18:2) as compared to coconut oil. In the present study, we have developed a cationic lipid nanocarrier system (PS-Lips) with fatty acyl chains from palmstearin and evaluated their nucleic acid delivery efficiencies in vitro using different nucleic acids (Figure 1). Fluorescence resonance energy transfer (FRET), a well-known biophysical technique, was employed to investigate the possible mechanism involved in enhanced transfection efficiencies of the cationic lipid nanocarrier system, PS-Lips. Further, we expand the horizon of the PS-Lips nanocarrier system’s application as a toolset for delivering genome-editing tools.
Figure 1.
Chemical structures of cationic amphiphiles synthesized from palmstearin. Schematic illustration shows the complex formation between PS-Lips and pDNA.
Results and Discussion
Recent advances in nucleic acid delivery and genomic science, including genome-editing with CRISPR/Cas9, have emphasized the significance of developing safer and efficient nonviral nucleic acid delivery reagents.21,22 Among various vectors, cationic lipids are found to be promising in delivering different types of nucleic acids such as plasmid DNA (pDNA), shRNA, siRNA, and microRNA.23,24 Despite the advantages, their application is limited because of lower transfection efficiencies and higher cytotoxicities.25 In our continuous efforts to develop efficient transfection reagents to overcome the limitations, previously, we demonstrated an intriguing strategy of developing cationic lipids from food-grade coconut oil.20 Our prior findings convincingly demonstrated that cationic lipids prepared using a mixture of fatty acids derived from coconut oil impart membrane fusion and enhance gene-delivery efficacies.20 However, the major limitation with cationic lipids derivatized from coconut oil is the reproducibility of the same composition because of a large variation in the fatty acyl chain lengths, which ranges from C6:0 to C18:2 (batch to batch variation). To overcome the limitation, we have developed a cationic lipid pool from palmstearin, which has less variation as compared to coconut oil. Because food-grade palmstearin (triacylglycerols) contains myristoyl (C14) to linoleoyl (C18:2) chain lengths, we envisaged that saponification of palmstearin yields a pool of fatty acyl chains, which can be used as the hydrophobic core for the development of a cationic lipid nanocarrier system containing natural fatty acyl chain asymmetry of palmstearin (Figure 1). We anticipated that such a cationic lipid nanocarrier system would be nontoxic and more efficient in transfection as it is derived from a food-grade source, and such a nanocarrier system mimics the plasma membrane fatty acyl chain composition.
Synthesis
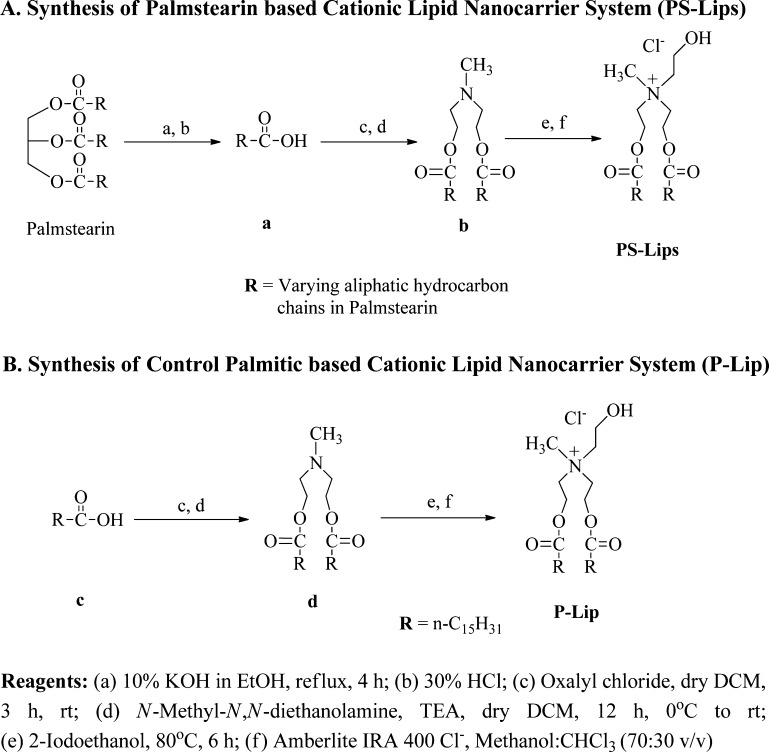
PS-Lips, a cationic lipid nanocarrier system with the hydrophobic core derived from commercially available food-grade palmstearin (fatty acid mixture obtained by the saponification method), was synthesized in three steps, as shown in Scheme 1A. Briefly, palmstearin was saponified using aq KOH, followed by acidification with dil HCl to obtain a fatty acid mixture. Next, the fatty acid mixture extracted from palmstearin was converted to corresponding fatty acyl chlorides using oxalyl chloride and reacted with N-methyl-diethanolamine to give tertiary amine compound, b (Scheme 1A). Finally, intermediate b was reacted with 2-iodo ethanol to yield quaternary iodide salt of PS-Lips, followed by chloride ion exchange using Amberlite IRA 400 Cl– to yield the target compound, PS-Lips (Scheme 1A). As a control lipid, we have synthesized P-Lip (Scheme 1B), which contains only palmitic chains in the hydrophobic core, following the same synthetic protocol used for the preparation of PS-Lips.
Scheme 1. Synthesis of (A) Palmstearin-Based Cationic Lipid Nanocarrier System (PS-Lips) and (B) Control Palmitic-Based Cationic Lipid Nanocarrier System (P-Lip).
Fatty Acid Methyl Ester (FAME) Quantification
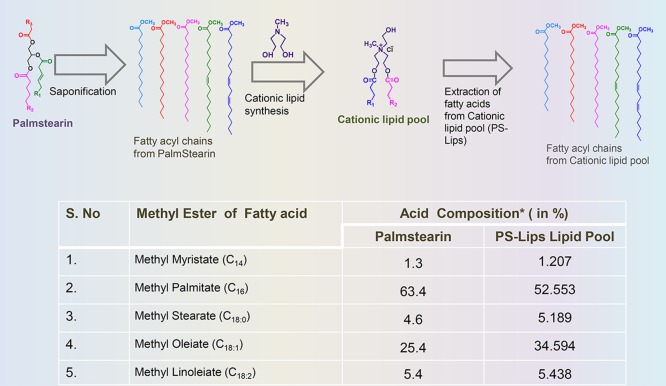
A portion from the mixture of fatty acids extracted from palmstearin following saponification was converted to corresponding fatty acyl methyl esters for FAME quantification using gas chromatography–mass spectrometry (GC–MS). Next, the fatty acids from the cationic lipid nanocarrier system, PS-Lips, were extracted by usual saponification, converted to their fatty acyl methyl esters, and subjected to FAME quantification. The compositions of both palmstearin and PS-Lips derived fatty acids were analyzed (Figure 2). The results showed that palmstearin contains 63% of C16 and 25% of C18:1, whereas PS-Lips had 53% of C16 and 35% of C18:1. Other fatty acids such as C12, C14, C18, and C18:2 were less than 6% (Figure 2). First, we confirmed that the fatty acid compositions of palmstearin did not get grossly affected upon developing into a cationic lipid nanocarrier system, PS-Lips (Figure 1 and Scheme 1A). The percentage of fatty acid methyl esters obtained from the cationic lipid nanocarrier system, PS-Lips, matched <90% with the original fatty acid esters of palmstearin in GC analysis (Figure 2). In both, the palmitate group (C16 ∼55%) and the oleate group (C18:1 ∼35%) were found to be present as the major aliphatic chains with a contribution of >85% (Figure 2).
Figure 2.
Illustration of fatty acid isolation from palmstearin and PS-Lips (above) and the fatty acid composition data of the starting material, palmstearin, and the cationic lipid nanocarrier system, PS-Lips, using GC analysis of fatty acid methyl esters from palmstearin and PS-Lips (below).
Size and Zeta Potential Measurements
Next, we characterized the physicochemical properties such as particle size, surface potentials, and DNA-binding properties of the liposomes PS-Lips and control P-Lip. The size of liposomes obtained by the dynamic light scattering (DLS) study of PS-Lips and P-Lip was found to be 175.3 and 206.6 nm, respectively, whereas the potentials were in the range of 2.6–2.8 meV for both liposomes (Table S1, Supporting Information). Lipoplexes of both PS-Lips and P-Lip showed sizes between 250 and 670 nm across the lipid/DNA charge ratio of 0.5:1 to 8:1 (Table S2, Supporting Information). Similar to the size data, the zeta potentials did not show any significant variation for lipoplexes of PS-Lips and P-Lip. The potentials of PS-Lips lipoplexes were found to be slightly higher (+27.7 to +16.2 meV) across the 8:1 to 2:1 lipid/DNA charge ratio, whereas it was +23.6 to +10.4 meV for P-Lip at the same charge ratios (Table S2, Supporting Information). However, the zeta potentials for lipoplexes of PS-Lips and P-Lip at lower lipid/DNA charge ratios, that is, 1:1 and 0.5:1, exhibited comparable potentials of +2.2 and −6.3 meV (for PS-Lips) and +1.8 and −3.3 meV, respectively (Table S2, Supporting Information). The comparable potentials for PS-Lips and P-Lip could be due to the structural similarity of their cationic lipids, equivalent charges, and hydrodynamic diameters of their lipoplexes.
DNA-Binding Study
DNA-binding studies with liposomes of PS-Lips and P-Lip in complexation with the pDNA (pCMV-SPORT-β-gal expressing β-galactosidase enzyme) were performed using the conventional agarose gel retardation assay (Figure S10, Supporting Information). Both PS-Lips and P-Lips showed high DNA binding across the lipid/DNA charge ratio of 8:1–2:1, whereas moderate DNA binding was observed at 1:1 and 0.5:1 lipid/DNA charge ratios (Figure S10, Supporting Information). Physicochemical studies including DLS and gel retardation studies revealed that the nanocarriers had better DNA-binding efficiency from 2:1 lipid/DNA charge ratios.
In Vitro Transfection Biology
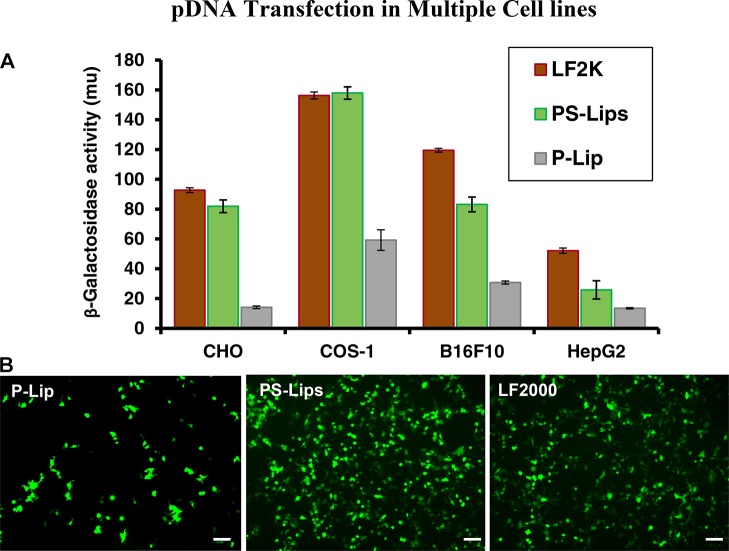
In vitro gene-delivery efficacies of PS-Lips and P-Lip were evaluated in multiple cultured mammalian cells including CHO (Chinese hamster ovary cells), COS-1 (SV40 transformed African green monkey kidney cells), B16F10 (murine melanoma cells), and HepG2 (human hepato carcinoma cells) using pCMV-SPORT-β-gal plasmid DNA as the reporter gene encoding the enzyme β-galactosidase, as described previously.26 Liposomes of PS-Lips exhibited ∼2–5 folds of high transfection as compared to P-Lip in the above listed cell lines at the lipid/DNA charge ratio of 2:1 (Figure 3A). Liposomes of PS-Lips showed comparable gene-transfer efficiencies with Lipofectamine 2000 in CHO and COS-1 cells (Figure 3A).
Figure 3.
Transfection and cellular enhanced green fluorescent protein (eGFP) expression studies with PS-Lips and P-Lip. Comparative in vitro gene-delivery efficiencies of cationic lipid nanocarrier systems, PS-Lips and P-Lip in complexation with pCMV-SPORT-β-gal plasmid DNA using the reporter gene assay in multiple cultured cell lines at a 2:1 lipid/DNA charge ratio (A). Microscopy images for cellular eGFP expression in human embryonic kidney (HEK-293) cells (B). Lipoplexes of PS-Lips and P-Lip with pDNA encoding GFP were transfected in HEK-293 cells at a 2:1 lipid/DNA charge ratio, and images were obtained using an inverted fluorescence microscope. Lipofectamine 2000 was used as the positive control for transfections.
Next, we evaluated the cellular uptake and expression with PS-Lips and P-Lip using eGFP pDNA (plasmid DNA encoding green fluorescence protein) in representative HEK-293 (human embryonic kidney cells) at a 2:1 lipid/DNA charge ratio. Fluorescence microscope images revealed that cells receiving lipoplexes of PS-Lips and pDNA (eGFP) showed higher fluorescence as compared to lipoplexes of P-Lip and pDNA (Figure 3B). Further, the cellular eGFP expression in HEK 293 cells with PS-Lips was found to be on par with Lipofectamine 2000 (Figure 3B). In line with in vitro findings from multiple cultured cells including CHO, COS-1, B16F10, and HepG2, liposomes prepared from PS-Lips exhibited an efficient cellular expression in HEK 293 than their symmetric dipalmitoyl counterpart, P-Lip. Data obtained from transfection studies and the cellular expression suggest that PS-Lips shows an enhanced gene-delivery activity and its efficiency is comparable with commercial Lipofectamine 2000 (Figure 3).
mRNA Transfections
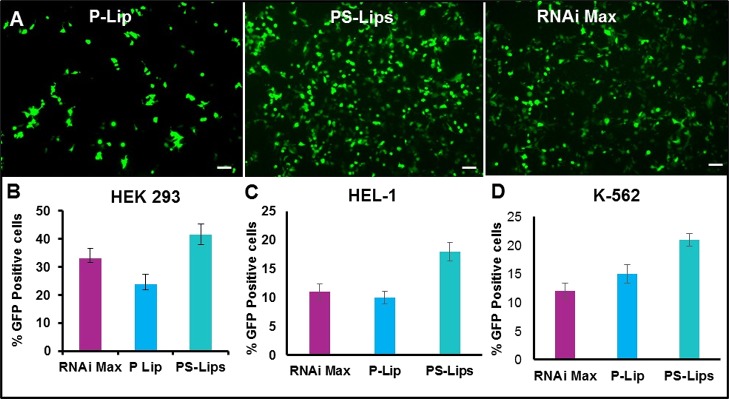
After the successful demonstration of pDNA transfections using the PS-Lips nanocarrier system, we wanted to explore whether this nanocarrier system could deliver linear nucleic acids such as mRNA. In this regard, we evaluated the transfection efficiencies of both cationic nanocarriers (PS-Lips and P-Lip) in the adherent cell line, HEK-293, and in suspension cells, K562 (chronic myelogenous leukemia cell line) and HEL-1 (human erythroleukemia cell line), using GFP encoding mRNA (Figure 4A–D). Surprisingly, liposomes of PS-Lips exhibited ∼2-fold high transfection as compared to P-Lip in HEK-293 at the lipid/mRNA charge ratio of 8:1 (Figure 4A,B) and exhibited comparable transfections with the commercial control RNAiMAX. Interestingly, mRNA transfections with the PS-lips nanocarrier system exhibited 2-fold superior transfection in both K562 and HEL-1 cells than the P-Lip nanocarrier system and commercial control RNAiMAX (Figure 4C,D). PS-Lips nanocarriers exhibited higher transfection efficiencies both with circular pDNA and linear mRNA at different lipid/base charge ratios. For circular DNA, maximum transfection efficiencies were observed at a 2:1 charge ratio, whereas for linear mRNA, it was 8:1. The higher charge ratios for mRNA compared to pDNA is possibly because of the difference in the net ion charge on the nucleic acids. Circular nucleic acids are compacted with a large amount of their counterions, yielding a lower effective negative charge compared to the neutral ones.27 Hence, linear nucleic acids require a higher charge ratio compared to circular nucleic acids.
Figure 4.
mRNA transfection studies with PS-Lips and P-Lip. Microscopic images of in vitro mRNA delivery with cationic lipid nanocarrier systems, PS-Lips and P-Lip in HEK 293 (A). Comparative in vitro mRNA transfection efficiencies of PS-Lips and P-Lip in the adherent cell line, HEK 293 (B), and suspension cell lines, HEL-1 (C) and K-562 (D). Lipofectamine RNAiMAX was used as the positive control for mRNA transfections.
CRISPR/Cas9 pDNA Transfection
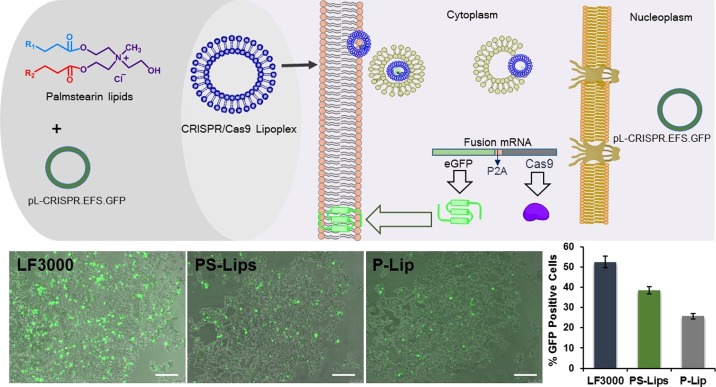
Recent findings demonstrated that lipid-mediated toolsets for delivering CRISPR/Cas9-encoded pDNA were found to be effective.28−30 Transfection results obtained from pDNA and mRNA nucleic acid delivery using PS-Lips and P-Lip compelled us to evaluate their efficacies in delivering CRISPR tools. To explore the ability of PS-Lips in delivering the genome-editing system, we evaluated the efficiency with the pL-CRISPR.EFS.GFP plasmid construct (11.7 kb), which expresses both Cas9 and eGFP proteins driven through the EFS promoter as a single transcript.31 The amount of Cas9 protein expression is directly correlated with the eGFP expression as a single transcript producing both Cas9 and eGFP proteins aided by the self-cleaving P2A peptides, which works through ribosomal skipping of peptide bond formation at the terminal proline residue in the P2A peptide resulting in the production of individual polypeptides. PS-Lips delivered the CRSIPR plasmid 1.5-fold more effectively than P-Lip (Figure 5). However, the efficacy of PS-Lips was found to be less when compared with the commercial control Lipofectamine 3000 (Figure 5). Because the plasmid size is large (11.7 kb), the condensation of pDNA would have been not adequate enough to effectively transfect into the cell, resulting in a lesser transfection efficiency for PS-Lips over Lipofectamine 3000. Further studies directed toward the combined use of PS-Lips with DNA-condensing agents for delivering larger size plasmids such as CRISPR plasmids are warranted. However, to the best of our knowledge, the present study is the first report on developing a cationic toolset designed with the asymmetric hydrophobic chain lengths of food-grade vegetable fat, palmstearin, for delivering CRISPR tools.
Figure 5.
Delivery of all-in-one CRISPR plasmid. Lipoplexes of PS-Lips and P-Lip with pL-CRISPR.EFS.GFP were transfected in HEK-293 cells at a 2:1 lipid to DNA charge ratio; images were obtained using an inverted fluorescence microscope, and quantification data were obtained by flow cytometric analysis.
In vitro transfection studies including pDNA, mRNA, and CRISPR constructs suggested that PS-Lips nanocarriers have better transfection efficiencies compared to the symmetric P-Lip. PS-Lips nanocarriers exhibited unique characteristics that could deliver a wide range of nucleic acids from 1 to 11 kb and linear, circular constructs. Next, we evaluated the cytotoxicities of the nanocarrier systems to confirm whether it had any role in the differential transfection profiles of P-Lip over PS-Lips.
Cell Viability
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT)-based cell viability assay32 was performed in representative CHO cells using the lipoplexes (lipid/DNA complexes) of PS-Lips and P-Lip for 24 h. Cell viability data revealed a remarkably noncytotoxic nature (>90% cell viability, Figure S11, Supporting Information) for both the lipids across the lipid/DNA charge ratios 8:1–0.5:1, which are used in our transfection experiments (Figures 3 and 4) except for P-Lip at a 8:1 lipid/DNA charge ratio (which showed ∼70% cell viability). Thus, these cell viability results summarized in Figure S11, Supporting Information rule out the possibility of any inherent cellular cytotoxicities playing a role behind the relatively compromised/poor transfection efficiencies of P-Lip and PS-Lips. These results also indicate that PS-Lips is safer to use even above the 4:1 lipid/DNA charge ratio.
To understand the superior transfection properties of PS-Lips over P-Lip, first, we evaluated the cellular uptake of the rhodamine-PE labelled lipoplexes and also measured the membrane fusion using FRET.
Cellular Uptake Study
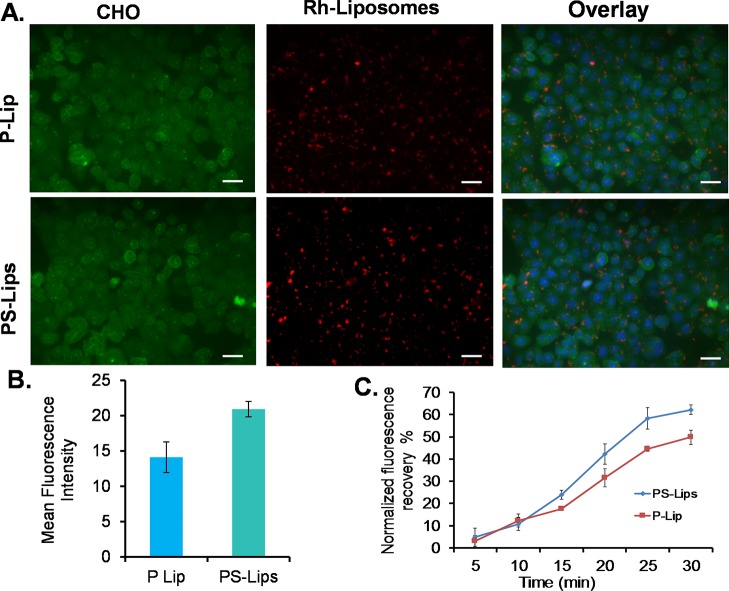
Liposomes of rhodamine-PE labeled PS-Lips and P-Lip in complexation with pDNA (pCMV-SPORT-β-gal) were used to check the cellular uptake of the lipids in representative CHO cells at a 2:1 lipid/DNA charge ratio. The in vitro uptake studies revealed that the lipoplexes of PS-Lips were found to be high when compared to its control dipalmitoyl analogue, P-Lip (Figure 6A,B).
Figure 6.
Cellular uptake study and biomembrane fusogenicity measurements of PS-Lips and P-Lip. Epifluorescence microscopic images of stable plasma membrane eGFP-expressed CHO cells transfected with rhodamine-PE labeled lipoplexes of PS-Lips and P-Lip at a lipid/DNA charge ratio of 2:1 (A) and their quantification graph (B). Biomembrane fusogenicities of liposomes prepared from cationic lipids, PS-Lips and P-Lip (FRET-based assay) (C). Liposome–biomembrane fusion was induced by the addition of PS-Lips and P-Lip liposomes with the dual fluorophore (Rho-PE and NBD-PE)-labeled membrane mimicking DOPC/DOPE/DOPS/Chol liposomal formulation. The values shown are representative of three independent measurements.
The superior cellular uptake of lipoplexes of PS-Lips than P-Lip is corroborated with the transfection studies and also supports the fact that possible high transfection efficacies could be originating from the high membrane fusogenic nature of the cationic lipid nanocarrier system from palmstearin in PS-Lips. To ascertain the fusogenic nature of PS-Lips over P-Lip, we performed FRET studies.
Biomembrane Fusogenicities
To gain mechanistic insight into the enhanced transfection efficiencies of PS-Lips lipid nanocarrier system over the control dipalmitoyl analogue P-Lip, we assessed their relative biomembrane fusogenicities with a dual fluorophore NBD-PE (as a donor) and Rho-PE (as an acceptor)-labelled membrane model, DOPC/DOPE/DOPS/Chol (dioleyol-phosphatidyl choline/dioleyol-phosphatidyl ethanolamine/dioleyol phosphatidylserine/cholesterol at 45:20:20:15, w/w) using the FRET assay.33 The normalized fluorescence recovery at 590 nm for the liposomes of PS-Lips and the control lipid P-Lip was 55 and 45%, respectively (Figure 6C), suggesting higher membrane fusogenicity of the asymmetric cationic lipid nanocarrier system, PS-Lips, than their symmetric lipid analogue, P-Lip. These findings support the notion that enhanced transfection efficiencies of the cationic lipid nanocarrier system might have originated because of their higher plasma membrane fusogenicity (Figure 6C). Efficiency of cationic lipid-mediated transfection critically depends on the lipoplexes entry into the cells (endocytosis), release of pDNA from the endosomes to escape lysosomal degradation, and entry into the nucleus for gene expression. Hydrophobic chain asymmetry imparts higher membrane fusogenicity with both plasma and endosomal membranes. Thus, it is a critical parameter for delivering nucleic acids into the cytoplasm.
Endocytosis Blockers Study
Towards probing the cellular entry of lipoplexes, we pretreated cells with sucrose, a clathrin-mediated endocytosis inhibitor reduced the methyl-β-cyclodextrin (M-β-CD), a cholesterol depleting agent,34 nystatin, an inhibitor of caveolae-mediated endocytosis,35 and dynasore, a small molecule dynamin II inhibitor36 before transfections. Both clathrin and caveolae inhibitors had less effect on transfections with PS-Lips compared to P-Lip. These findings suggested that membrane fusion was driving PS-Lips from endocytosis to direct delivery of pDNA into cytoplasm (Figure 7). Membrane fusion of the lipids is an efficient transport of biomolecules than the endocytosis process.37,38 Liposomal systems were developed to mimic membrane fusion events.39 In line with the prior findings, our results confirmed that the superior transfection activity of PS-Lips over P-Lips is due to the intracellular delivery of lipoplex through membrane fusion and is less endocytosis dependent.
Figure 7.
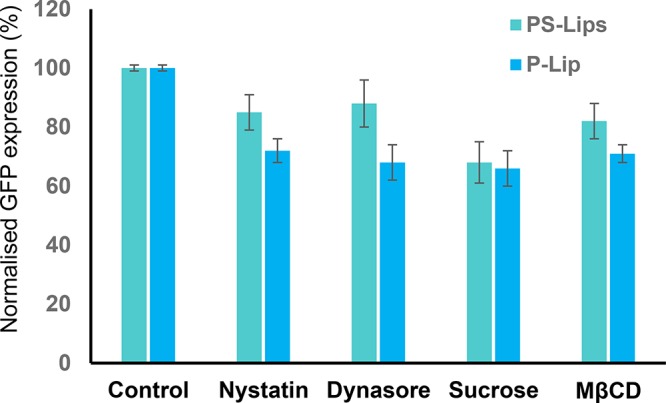
Normalized eGFP expression by endocytosis inhibitors. HEK-293 cells were pretreated with sucrose (450 mM), M-β-CD (2 mM), dynasore (80 μM), or nystatin (100 nM) for 1 h before exposure to the 2:1 lipid/DNA charge ratio of PS-Lips and P-Lip lipoplexes.
Conclusions
In this study, we demonstrated that the cationic lipid nanocarrier system, PS-Lips, prepared using the fatty acid mixture derivatized from food-grade palmstearin exhibited enhanced transfection efficiencies in both adherent and suspension cells than its control dipalmitoyl lipid analogue, P-Lip, which contributes more than 50% of the hydrophobic core of the PS-Lips lipid nanocarrier system. Mechanistic insight into the improved activity of the PS-Lips lipid nanocarrier system than the control symmetric analogue, P-Lip, using FRET and cellular uptake studies revealed that the higher transfection property possibly results from the fusogenic nature of the cationic lipid nanocarrier system (PS-Lips) liposomes. We further demonstrated that the liposomes from the cationic lipid nanocarrier system (PS-Lips) effectively delivered genome-editing tools containing CRISPR/Cas9 encoded pDNA and has an enhanced genome-editing efficiency than their control dipalmitoyl counterpart (P-Lip). In summary, our findings collectively demonstrate that the cationic lipid nanocarrier system (PS-Lips) prepared using the fatty acid mixture from food-grade oils such as palmstearin could be more effective, economic, and safer for nucleic acid delivery and genome-editing applications.
Materials and Methods
General Procedures and Reagents
Proton nuclear magnetic resonance (1H NMR) spectra and electrospray ionization (ESI)-mass spectral data were recorded on an AV 300 MHz NMR spectrometer and an LCQ ion trap mass spectrometer (Thermo Finnigan, SanJose, CA, USA) equipped with an ESI source or a Micromass Quattro LC triple quadrupole mass spectrometer, respectively. Column chromatography was performed with 60–120 mesh silica gel (Acme Synthetic Chemicals, India). All reagents used for synthesis were purchased from Sigma-Aldrich, St. Louis, USA unless otherwise stated. NP-40, antibiotics, and agarose were procured from HiMedia, India. Cell lines (CHO, COS-1, and HEK-293) were procured from the National Centre for Cell Sciences, Pune, India. Suspension cells (Hel-1 and K-562) were obtained from ATCC. Cells were cultured in an incubator in Dulbecco’s modified Eagle’s medium (DMEM) with 10% fetal bovine serum (FBS) at 37 °C in a humidified atmosphere containing 5% CO2/95% air. GlutaMAX and Lipofectamine RNAiMAX transfection reagents were obtained from Thermo Scientific. CleanCap eGFP mRNA was purchased from TriLink BioTechnologies.
Syntheses
The cationic lipids, PS-Lips and P-Lip, were synthesized, as shown in Scheme 1A,B. Optimized synthesis protocols and purification conditions for lipids are described below. Structures of PS-Lips and P-Lip and their synthetic intermediates were confirmed by 1H NMR and ESI-MS. Analysis of the chemical characterizations of the lipids, PS-Lips and P-Lip, as well as those of their synthetic intermediates shown in Scheme 1A,B are described. The purity of both cationic lipids, PS-Lips and P-Lip, was confirmed by reverse-phase analytical high-performance liquid chromatography (HPLC) in two different mobile phases (100% methanol and methanol/water 99:1 v/v), and purity was found to be >95%.
Synthesis of Palmstearin-Based Cationic Lipid Pool (PS-Lips, Scheme 1A)
Steps a and b: Isolation of Palmstearin Fatty Acids (Intermediate “a”, Scheme 1A)
First, fatty acids were isolated from the vegetable fat, palmstearin, following our previously established protocol.20 Briefly, 10 g of palmstearin (Scheme 1A) was saponificated with 10% KOH in ethanol (25 mL) in a 250 mL round-bottom flask under refluxing conditions for 4 h, followed by cooling to room temperature. The volume of the mixture was reduced to half and washed with water and hexane (2 × 25 mL each). The aqueous layer containing potassium salt of fatty acids was collected and acidified with aqueous HCL (30%). The fatty acid mixture of palmstearin was extracted using ethyl acetate (4 × 25 mL) followed by brine solution. The organic layer containing the palmstearin fatty acid mixture was concentrated to yield palmstearin fatty acid mixture as a white solid with intermediate “a” (Scheme 1A, 9.3 g, Rf = 0.4, 10:90 v/v ethyl acetate/hexane).
Step c: Preparation of Palmstearin Fatty Acyl Chlorides (Scheme 1A)
The isolated fatty acid mixture from palmstearin (10 g, 37.7 mmol, intermediate “a”, Scheme 1A) was dissolved in a minimum amount of dry dichloromethane (DCM, 20 mL) with a catalytic amount of dry dimethylformamide, and oxalyl chloride (3.88 mL, 45.2 mmol) was added slowly drop by drop at room temperature with continuous stirring for 3 h. The reaction was stopped after confirming the complete conversion of fatty acids to their corresponding acid chlorides with thin-layer chromatography. The excess oxalyl chloride was removed by downward distillation at 40–45 °C. The residue upon vacuum drying afforded fatty acyl chlorides from palmstearin (10.3 g, 96% yield, Rf = 0.7, 5:95 methanol: chloroform, v/v).
Step d: Synthesis of Palmstearin-Derived N-Methyl-N,N-diethanolamine Difatty Acyl Esters (intermediate “b”, Scheme 1A)
N-Methyl-N,N-diethanolamine (1 g, 8.4 mmol) was added to the fatty acyl chlorides (5.71 g, 20.1 mmol) that were dissolved in 20 mL of dry DCM, with stirring at room temperature followed by the slow addition of triethylamine (2.8 mL, 20.1 mmol) at 0 °C and continued stirring for 12 h at room temperature. After completion of the reaction, first, the mixture was diluted with 20 mL of DCM and washed multiple times with 5% aqueous HCl to remove triethylamine. Next, the organic layer was washed with 5% aqueous sodium bicarbonate solution (3 × 20 mL) followed by brine solution (2 × 20 mL). The combined organic layers were collected, dried over anhydrous sodium sulphate, and concentrated. The residue was subjected to column chromatographic purification using silica gel (60–120 mesh size) with 2% methanol/chloroform, v/v, as the eluent and afforded the intermediate tertiary amine compound N-methyl-N,N-diethanolamine fatty acyl esters of palmstearin (intermediate “b”, Scheme 1A) as a brownish liquid (4.2 g, 82% yield, Rf = 0.8, 5:95 methanol/chloroform, v/v), which solidified upon storage at 4 °C.
1H NMR of Palmstearin-Derived N-Methyl-N,N-diethanolamine Difatty Acyl Esters (300 MHz, CDCl3)
δ/ppm = 0.9 [t, 6H, CH3–CH2–(CH2)11–15−], 1.2–1.4 [br s, 32–52H, −(CH2)9–13−], 1.6 [t, 4H, CH3–(CH2)10–14–CH2– CH2], 2.2 [t, 4H, CH3–(CH2)10–14– CH2– CH2], 2.4 [s, 3H, N–CH3], 2.7 [t, 4H, N–CH2–CH2–O−], 4.15 [t, 4H, −CH2–CH2–O−], 5.3 [t, 2H, CH = CH] ESI-MS m/z: possible combinations (calculated) 539.49 (for C14:C14), 567.52 (for C14:C16), 591.52 (for C14:C18:2), 593.54 (for C14:C18:1), 595.55 (for C16:C16), 595.55 (for C14:C18), 619.55 (for C16:C18:2) 621.57 (for C16:C18:1), 623.59 (for C16:C18), 643.55 (for C18:2:C18:2), 647.59 (for C18:1:C18:1), 647.59 (for C18:C18:2), 649.60 (for C18:C18:1), 651.62 (for C18:C18); found, 540 (M + 1 for C14:C14), 569 (M + 1 for C14:C16), 597 (M + 1 for C16:C16) or (M + 1 for C14:C18), 619 (M+ for C16:C18:2), 621 (M+ for C16:C18:1), 623 (M+ for C16:C18), 648 (M+ for C18:1:C18:1) or (M+ for C18:C18:2), 650 (M+ for C18:C18:1), 652 (M+ for C18:C18).
Step e: Synthesis of Target Palmstearin-Based Cationic Lipid Pool (PS-Lips, Scheme 1A)
Finally, N-methyl-N,N-diethanolamine fatty acyl esters of palmstearin (1.0 g, 1.6 mmol) (intermediate “b”, Scheme 1A) were subjected to quarternization with 2.0 mL of 2-iodoethanol (excess) under stirring at 80 °C for 6 h. After completion of the reaction, the mixture was diluted with DCM (20 mL) and washed with water (3 × 25 mL) followed by brine solution (2 × 20 mL). The combined organic layers were collected, dried over anhydrous sodium sulfate, and concentrated. The crude residue was subjected to column chromatographic purification (using 60–120 mesh silica gel) with 4% methanol/CHCl3, v/v, as the eluent and afforded the iodide salt of PS-Lips as a yellowish solid. The counter iodide ion of PS-Lips was exchanged with the chloride ion on Amberlite IRA 400 chloride resin in the chloroform/methanol solvent mixture (30:70 v/v) and afforded a white solid of target palmstearin-based cationic lipid pool (PS-Lips, Scheme 1A, 0.54 g, 48% yield, Rf = 0.2, 5:95 methanol/chloroform, v/v).
1H NMR of Target Palmstearin-Based Cationic Lipid Pool (300 MHz, CDCl3)
δ/ppm = 0.9 [t, 6H, CH3–CH2– C13H26−], 1.2–1.4 [m, 48H, −(CH2)12−], 1.6 [t, 4H, CH3–(CH2)12–CH2–CH2], 2.4 [t, 4H, CH3–(CH2)12– CH2– CH2], 3.3 [s, 3H, N–CH3], 3.7 [t, 6H, N–CH2–CH2–O], 3.9 [t, 6H, N–CH2–CH2–O], 4.0 [t, 2H N–CH2–CH2–OH], 4.6 [t, 4H, −CO–O–CH2−] ESI-MS m/z: possible combinations (calculated) 584.53 (for C14:C14), 612.56 (for C14:C16), 636.56 (for C14:C18:2), 638.57 (for C14:C18:1), 640.59 (for C14:C18), 640.59 (for C16:C16), 664.59 (for C16:C18:2), 666.60 (for C16:C18:1), 668.62 (for C16:C18), 688.59 (for C18-2:C18:2), 692.62 (for C18:1:C18:1), 692.62 (for C18:C18:2), 694.63 (for C18:C18:1), 696.65 (for C18:C18); found, 585 (M + 1 for C14:C14), 612 (M+ for C14:C16), 640 (M+ for C14:C18) or (M+ for C16:C16), 664 (M+ for C16:C18:2), 666 (M+ for C16:C18:1), 668 (M+ for C16:C18), 692 (M+ for C18:C18:2), 694 (M+ for C18:C18:1), 696 (M+ for C18:C18).
Synthesis of Control Palmitic-Based Cationic Lipid (P-Lip, Scheme 1B)
Step c: Preparation of Palmitoyl Chloride (Scheme 1B)
Palmitoyl chloride was prepared using the same procedure as described above for palmstearin fatty acyl chlorides (step c: preparation of palmstearin fatty acyl chlorides, Scheme 1A) with 5 g of palmitic acid (starting material “c”, 19.5 mmol) and 2 mL of oxalyl chloride (23.4 mmol) to prepare its corresponding acid chloride. Palmitoyl chloride (5.1 g) was obtained with 94% yield (Rf = 0.7, 5:95 methanol/chloroform, v/v).
Step d: Synthesis of N,N-Di[O-hexadecanoyl-2-hydroxyethyl]-N-methyl Amine (Intermediate “d”, Scheme 1B)
Tertiary intermediate was prepared by following the same procedure as described above for palmstearin fatty acyl esters (step d: synthesis of palmstearin-derived N-methyl-N,N-diethanolamine difatty acyl esters, intermediate “b”, Scheme 1A) using 6.4 g of palmitoyl chloride (23.5 mmol, Scheme 1B) and 1 g of N-methyl-N,N-diethanolamine (8.4 mmol) with the addition of 3.27 mL of triethylamine (23.5 mmol). The intermediate tertiary amine compound N,N-di[O-hexadecanoyl-2-hydroxyethyl]-N-methyl amine was a yellowish liquid (4.2 g, 84% yield, Rf = 0.8, 5:95 methanol/chloroform, v/v).
1H NMR of N,N-Di[O-hexadecanoyl-2-hydroxyethyl]-N-methyl Amine (300 MHz, CDCl3)
δ/ppm = 0.9 [t, 6H, CH3–CH2– C13H26−], 1.2–1.4 [m, 48H, −(CH2)12−], 1.6 [t, 4H, CH3–(CH2)12– CH2– CH2], 2.3 [t, 4H, CH3–(CH2)12– CH2– CH2], 2.4 [s, 3H, N–CH3], 2.7 [t, 4H, N–CH2–CH2–O−], 4.2 [t, 4H, N–CH2–CH2–O−] ESI-MS m/z: calcd 595.986 (for C37H73NO4); found, 597 (M + H).
Steps e and f: Synthesis of Target N,N-Di[O-hexadecanoyl-2-hydroxyethyl]-N-hydroxy Ethyl-N-methylammonium Chloride (P-Lip, Scheme 1B)
Target symmetric cationic palmitic lipid, P-Lip, was prepared using the same procedures as described above for the palmstearin target cationic lipid pool, PS-Lips (step e: synthesis of target palmstearin-based cationic lipid pool, PS-Lips, Scheme 1A). For this, 0.5 g of N,N-di[O-hexadecanoyl-2-hydroxyethyl]-N-methyl amine (intermediate “d”, 0.83 mmol) was reacted with 0.6 mL of 2-iodoethanol. The pure target N,N-di[O-hexadecanoyl-2-hydroxyethyl]-N-hydroxy ethyl-N-methyl ammonium chloride was a white solid (P-Lip, Scheme 1B, 0.25 g, 46% yield, Rf = 0.2, 5:95 methanol/chloroform, v/v).
1H NMR of N,N-Di[O-hexadecanoyl-2-hydroxyethyl]-N-hydroxyethyl-N-methyl Ammonium Chloride (300 MHz, CDCl3)
δ/ppm = 0.9 [t, 6H, CH3–CH2– C13H26−], 1.20–1.40 [m, 48H, −(CH2)12−]; 1.5 [t, 4H, CH3–(CH2)12– CH2– CH2], 2.3 [t, 4H, CH3–(CH2)12– CH2– CH2], 3.3 [s, 3H, N–CH3], 3.9 [t, 6H, N–CH2–CH2–O], 3.9 [t, 2H N–CH2–CH2–OH], 4.3 [t, 4H, −CO–O–CH2−] ESI-MS m/z: calcd 641 (for C39H78NO5); found, 641 (M+).
Measurement of Fatty Acid Composition by GC
The fatty acid compositions of the starting material palmstearin and PS-Lips were measured by GC following our previously reported procedure.20 Briefly, 10 mg of palmstearin and PS-Lips in two separate 25 mL round-bottom flasks was dissolved with 5% H2SO4 in methanol solution and refluxed for 4 h. After 4 h, the contents of the reaction mixture were diluted with 5 mL of water and extracted with ethyl acetate. The collected organic layer was washed with brine solution and evaporated to dryness. The fatty acid methyl ester analysis was performed on a gas chromatograph (Agilent 6890 series) equipped with a flame ionization detector and a capillary column DB-23 (30 m × 0.25 mm i.d. × 0.5 μm film thickness).
Preparation of Liposomes and pDNA
Liposomes (0.5 mM) were prepared with 1:1 mol ratios of each lipid and cholesterol using a standard protocol, as reported previously.20,26,40,41 Briefly, PS-Lips/P-Lip and cholesterol in 1:1 mol ratios were made into a thin lipid film and hydrated with 1 mL of diethylpyrocarbonate-treated water overnight. A brief vortex of 2–3 min at room temperature was followed by sonication using a Branson 450 Sonifier at 100% duty cycle and 25 W output power to produce small unilamellar vesicles. The plasmids were amplified in DH5α-strain of Escherichia coli, isolated by alkaline lysis procedure, and finally purified by PEG-8000 precipitation, as described previously.26 The purity of the plasmid was confirmed by the A260/A280 ratio (around 1.9) and 1% agarose gel electrophoresis.
Zeta Potential (ξ) and Global Size Measurements
Neat liposomes of PS-Lips and P-Lip and their corresponding lipoplexes across the lipid/DNA charge ratio of 8:1–0.5:1 were subjected to global size and surface charge measurements using the dynamic laser light scattering method on a Zetasizer 3000HSA (Malvern UK). The measurements for liposomes were carried in deionized water (sample refractive index = 1.59 and viscosity = 0.89). For lipoplexes, the liposomes of PS-Lips and P-Lip were complexed with pDNA in serum-free DMEM and diluted in the same media for size and zeta potential measurements. The system was validated by using the 200 + 5 nm polystyrene polymer (Duke Scientific Corps. Palo Alto, CA). The diameters of particles (liposomes and lipoplexes) were calculated by using the automatic mode. Each size measurement was performed at least 10 times in triplicate (with the zero field correction), and data were tabulated as the average of 3 measurements. Zeta potentials of each sample was measured at least 10 times and tabulated as the average values using the Smoluchowski approximation method for calculation of average values.
DNA-Binding Assay
PS-Lips and P-Lip liposomes’ DNA-binding ability was assessed with the gel retardation assay (1% agarose gel prestained with ethidium bromide) across varying lipid/DNA charge ratios of 8:1 to 0.5:1, as previously described.11 In brief, pCMV-SPORT-β-gal (0.3 μg) and the liposomes of PS-Lips and P-Lip were made to complex in a total volume of 30 μL in N-(2-hydroxyethyl)piperazine-N′-ethanesulfonic acid buffer (pH 7.4) and incubated at room temperature for 20–25 min. The samples were loaded into the wells after mixing with 4 μL of 6× loading buffer [prepared using 0.25% bromophenol blue in 40% (w/v) sucrose with sterile H2O]. A power of 80 V was used for 45 min in running the gel, and DNA bands were visualized in a Bio-Rad Gel Doc XR + imaging system (Bio-Rad, Hercules, CA, USA).
pDNA Transfection Biology
Cells were seeded at a density of 15 000 (for COS-1, CHO, B16F10, and HepG2) per well in a 96-well plate 12–18 h before the transfection experiments. Lipoplexes were prepared using pDNA (0.3 μg) with the liposomes of PS-Lips and P-Lip at a 0.5:1 to 8:1 lipid/DNA charge ratio (0.45–7.2 nmol) in serum-free media to a final volume of 100 μL for 30 min. The prepared lipoplexes were added to the cells and incubated for 3–4 h. After incubation period, the media containing the lipoplexes were replaced with complete media. After 36–48 h of incubation, the reporter gene activity was estimated using our previously reported method.26 Briefly, after the incubation time, the cells were washed with 1× phosphate-buffered saline (PBS) to remove the media and lysed using lysis buffer of 50 μL per well (prepared with 0.25 M Tris-HCl at pH 8.0, added with 0.5% NP40). The reporter gene activity (β-galactosidase activity) was estimated with the addition of substrate solution of 50 μL per well [prepared by dissolving 1.33 mg of ortho-nitrophenyl-β-galactoside (ONPG), 0.2 M sodium phosphate (pH 7.3), and 2 mM MgCl2 in 1 mL volume] into a 96-well plate containing the lysate. Absorbance was measured at 405 nm using a microplate reader, and the transfection efficiency was calculated (as β-galactosidase units) using a calibration curve (obtained with pure β-galactosidase enzyme). The transfection experiments were performed in triplicate, and the transfection efficacies of PS-Lips, P-Lip, and Lipofectamine 2000 are represented as the average of triplicate experiments.
mRNA Transfection Studies
Cells were seeded at a density of 25 000/well (for HEK-293) and 40 000/well (HEL-1 and K-562 in a 48-well plate at least 12 h before the transfection experiments). mRNA plexes were prepared using 0.25 μg of GFP encoding mRNA with varying amounts of liposomes of PS-Lips and P-Lip (1.8–7.2 nmol at a 2:1 to 8:1 lipid/mRNA charge ratio) in serum-free DMEM to a final volume of 100 μL for 30 min. Commercially available RNAi MAX transfection reagent was used for transfection as per manufacture protocol. The complexes were then added to the cells and incubated for 3–4 h. After incubation period, the media containing the lipoplexes were replaced with complete media. GFP expression imaging was done using a fluorescence microscope (Leica DMI 6000B) and quantified in FACS (BD Biosciences Calibur).
Cellular Uptake Studies
Cellular uptake studies of lipoplexes were performed in CHO cell lines, stably expressing GPI-anchored GFP (a gift from Prof. Satyajit Mayor, NCBS, India) using the established protocols.26 In brief, rhodamine-PE labelled liposomes, 0.5 mM liposomes (PS-Lips and P-Lip) were prepared in amber glass vials, as described in the above section of “Preparation of Liposomes” with an additional step of adding 0.005 mM rhodamine-PE (i.e., 1% with respect to the total formulation content, Avanti Polar Lipids, USA). GPI-anchored GFP CHO cells (10 000) were seeded in a 96-well plate (Corning Inc., Corning, NY) in 100 μL of DMEM medium 12–18 h before the transfection experiment. Rhodamine-PE labelled liposomes of PS-Lips and P-Lip were complexed with pCMV-SPORT-β-gal (0.3 μg/well) at a lipid/DNA charge ratio of 2:1 (total volume of 100 μL DMEM) for 15–20 min. The complexes were then added to the cells and incubated for 3–4 h. After 4 h of incubation, the nuclei were stained with 4′,6-diamidino-2-phenylindole, and the cells were washed with PBS (2100 μL) and fixed with 3.8% paraformaldehyde in PBS at room temperature for 10 min. The red fluorescent cells were detected under an inverted fluorescence microscope (Nikon, Japan).
Plasmid Expression Studies
For GFP expression studies, 12–18 h before transfection, 4 × 104 HEK-293 cells were seeded in 24-well plates (Corning Inc., NY). Liposomes of PS-Lips and P-Lip were made to complex with eGFP-N1-encoding pDNA or pL-CRISPR.EFS.GFP (0.9 μg/well) at a 2:1 lipid/DNA charge ratio in serum-free DMEM (total volume made up to 100 μL) for 15–20 min. The final volumes of the complexes were made to 400 μL with DMEM and added to the cells. After 4 h of incubation, the lipoplexes were removed, and the cells were supplemented with complete medium. Post 36 h of transfections, the cells were washed with PBS (100 μL) and fixed with 3.8% paraformaldehyde in PBS at room temperature for 10 min. eGFP-N1 expression was observed under an inverted fluorescence microscope (Nikon, Japan).
Cell Viability Assay
MTT-based colorimetric reduction assay was performed to evaluate the cell viabilities for lipoplexes prepared using liposomes of PS-Lips and P-Lip and pDNA (pCMV-SPORT-β-gal) in CHO cells at varying lipid/DNA charge ratios (8:1–0.5:1), as reported previously.20 The conditions for the MTT assay were maintained same with respect to the cell number per well and the lipid/DNA charge ratio as used for transfection experiments described in “pDNA Transfection Biology” section. Briefly, following the addition of lipoplexes for 4 h, the media were replaced with complete media and incubated for another 24 h. After 24 h of incubation, MTT (5 mg/mL prepared in PBS) was added to the cells and incubated for 4 h at 37 °C. After incubation, the supernatant was removed carefully. The purple formazan crystals were dissolved in dimethylsulfoxide (DMSO)/methanol (1:1 v/v). The absorbance was measured using a microplate reader. The results were expressed as percent viability = [A540 (treated cells) – background/A540 (untreated cells) – background] × 100.
FRET Assay
The membrane fusogenicities of PS-Lips and P-Lip liposomes were measured with a dual fluorophore labelled membrane model, as described previously.20,33 In brief, the model biomembrane lipid composition containing DOPC/DOPE/DOPS/Chol (45:20:20:15, w/w ratio, the total lipid concentration used was 0.5 mM) was labelled with 0.005 mM NBD-PE (acts as donor) and N-Rho-PE (acts as an acceptor) lipids (i.e., 1% with respect to the total biomembrane mimicking formulation content, Avanti Polar Lipids, USA). Fusogenicity experiment was performed by adding a labelled model biomembrane liposomal formulation and PS-Lips and P-Lip liposomes in a black 96-well plate; they were mixed well, and readings were taken in a FLx800 Microplate Fluorescence Reader (BioTek Instruments Inc., U.K.) at room temperature. Starting from 0 to 30 min, for every 5 min, fluorescence intensities were recorded by exciting the sample at 485 nm and emitting at 595 nm. The fluorescent intensity of each time interval (Ft) was normalized with the initial intensity (Fo) and infinity intensity (Fα) of the model membrane by fitting in the equation [Fo – Ft/Fo – Fα] × 100. Infinity fusion (100%) was determined by the emission of Rho-PE fluorescent intensity after the complete dissociation of the labeled biomembrane liposomal formulation with 1% Triton X-100.
Endocytosis Blocking
HEK-293 cells (6 × 104) were seeded in a 24 well plate 22–24 h before the experiment. Prior to the addition of the lipoplexes, the respective cells were pretreated with plain DMEM containing sucrose (450 nM), methyl-β-cyclodextrin (2 mM), dynasore (80 μM), and nystatin (80 μM) individually for 30 min. The medium was then removed and replaced with DMEM with 10% FBS and 1% penicillin–streptomycin. The lipoplexes were then added to the cells, and the cells were incubated at 37 °C and 5% CO2. The reporter gene activity was quantified after 48 h using flow cytometric analysis.
Statistical Analysis
Experimental data reported in each graph are represented as the mean ± SD of obtained values from each experiment performed at least three times. Experimental data from each group were compared with other groups within the graph using student t test. p < 0.05 was considered as significant.
Acknowledgments
H.R.R. thanks the CSIR, India for Doctoral Research Fellowship. S.M. thanks SERB-Fast track grant for young scientists (SB/FT/CS-198/2013), Department of Science and Technology (DST). pL-CRISPR.EFS.GFP was a kind gift from Benjamin Ebert (Addgene plasmid # 57818). CHO cells stably expressing EGFP-GPI (Ai) was a kind gift from Prof. Satyajit Mayor, NCBS, Bengaluru. We thank Dr. Alok Srivastava and the CSCR core facility for supporting the research.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsomega.7b00935.
Spectral characterizations (1HNMR and ESI-MS) for PS-Lips, P-Lip, and their synthetic intermediates, reverse-phase HPLC purity data for both cationic amphiphiles (PS-Lips and P-Lip), global size and surface potential data of individual liposomes and their lipoplexes (PDF)
Author Present Address
⊥ BioSatva Technologies, Golnaka, Hyderabad 500013, India (C.V.).
Author Contributions
∥ Equally contributing authors.
The authors declare no competing financial interest.
Supplementary Material
References
- Naldini L. Gene therapy returns to centre stage. Nature 2015, 526, 351–360. 10.1038/nature15818. [DOI] [PubMed] [Google Scholar]
- Choi S. A.; Yun J.-W.; Joo K. M.; Lee J. Y.; Kwak P. A.; Lee Y. E.; You J.-R.; Kwon E.; Kim W. H.; Wang K.-C.; Phi J.-H.; Kang B.-C.; Kim S.-K. Preclinical Biosafety Evaluation of Genetically Modified Human Adipose Tissue-Derived Mesenchymal Stem Cells for Clinical Applications to Brainstem Glioma. Stem Cells Dev. 2016, 25, 897–908. 10.1089/scd.2015.0324. [DOI] [PubMed] [Google Scholar]
- Sage E. K.; Thakrar R. M.; Janes S. M. Genetically modified mesenchymal stromal cells in cancer therapy. Cytotherapy 2016, 18, 1435–1445. 10.1016/j.jcyt.2016.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpe M.; Mount N. Genetically modified T cells in cancer therapy: opportunities and challenges. Dis. Models & Mech. 2015, 8, 337–350. 10.1242/dmm.018036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H.; Kanasty R. L.; Eltoukhy A. A.; Vegas A. J.; Dorkin J. R.; Anderson D. G. Non-viral vectors for gene-based therapy. Nat. Rev. Genet. 2014, 15, 541–555. 10.1038/nrg3763. [DOI] [PubMed] [Google Scholar]
- Thomas C. E.; Ehrhardt A.; Kay M. A. Progress and problems with the use of viral vectors for gene therapy. Nat. Rev. Genet. 2003, 4, 346–358. 10.1038/nrg1066. [DOI] [PubMed] [Google Scholar]
- Segura M. M.; Kamen A. A.; Garnier A. Overview of current scalable methods for purification of viral vectors. Methods Mol. Biol. 2011, 737, 89–116. 10.1007/978-1-61779-095-9_4. [DOI] [PubMed] [Google Scholar]
- Nayerossadat N.; Ali P. A.; Maedeh T. Viral and nonviral delivery systems for gene delivery. Adv. Biomed. Res. 2012, 1, 27. 10.4103/2277-9175.98152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S.-D.; Huang L. Gene therapy progress and prospects: non-viral gene therapy by systemic delivery. Gene Ther. 2006, 13, 1313–1319. 10.1038/sj.gt.3302838. [DOI] [PubMed] [Google Scholar]
- Mintzer M. A.; Simanek E. E. Nonviral vectors for gene delivery. Chem. Rev. 2009, 109, 259–302. 10.1021/cr800409e. [DOI] [PubMed] [Google Scholar]
- Dharmalingam P.; Marrapu B.; Voshavar C.; Nadella R.; Rangasami V. K.; Shaji R. V.; Abbas S.; Prasad R. B. N.; Kaki S. S.; Marepally S. An anti-oxidant, α-lipoic acid conjugated oleoyl-sn-phosphatidylcholineas a helper lipid in cationic liposomal formulations. Colloids Surf., B 2017, 152, 133–142. 10.1016/j.colsurfb.2017.01.013. [DOI] [PubMed] [Google Scholar]
- Hardee C.; Arévalo-Soliz L.; Hornstein B.; Zechiedrich L. Advances in Non-Viral DNA Vectors for Gene Therapy. Genes 2017, 8, 65. 10.3390/genes8020065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattni B. S.; Chupin V. V.; Torchilin V. P. New Developments in Liposomal Drug Delivery. Chem. Rev. 2015, 115, 10938–10966. 10.1021/acs.chemrev.5b00046. [DOI] [PubMed] [Google Scholar]
- Srinivas R.; Samanta S.; Chaudhuri A. Cationic amphiphiles: promising carriers of genetic materials in gene therapy. Chem. Soc. Rev. 2009, 38, 3326–3338. 10.1039/b813869a. [DOI] [PubMed] [Google Scholar]
- Leuschner F.; Dutta P.; Gorbatov R.; Novobrantseva T. I.; Donahoe J. S.; Courties G.; Lee K. M.; Kim J. I.; Markmann J. F.; Marinelli B.; Panizzi P.; Lee W. W.; Iwamoto Y.; Milstein S.; Epstein-Barash H.; Cantley W.; Wong J.; Cortez-Retamozo V.; Newton A.; Love K.; Libby P.; Pittet M. J.; Swirski F. K.; Koteliansky V.; Langer R.; Weissleder R.; Anderson D. G.; Nahrendorf M. Therapeutic siRNA silencing in inflammatory monocytes in mice. Nat. Biotechnol. 2011, 29, 1005–1010. 10.1038/nbt.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhi D.; Zhang S.; Cui S.; Zhao Y.; Wang Y.; Zhao D. The headgroup evolution of cationic lipids for gene delivery. Bioconjugate Chem. 2013, 24, 487–519. 10.1021/bc300381s. [DOI] [PubMed] [Google Scholar]
- Koynova R.; Tenchov B.; Wang L.; Macdonald R. C. Hydrophobic moiety of cationic lipids strongly modulates their transfection activity. Mol. Pharm. 2009, 6, 951–958. 10.1021/mp8002573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. S.; Moon J.; Kim K. S.; Choi M. M.; Lee J. E.; Heo Y.; Cho D. H.; Jang D. O.; Park Y. S. Gene-transferring efficiencies of novel diamino cationic lipids with varied hydrocarbon chains. Bioconjugate Chem. 2004, 15, 1095–1101. 10.1021/bc049934t. [DOI] [PubMed] [Google Scholar]
- Nantz M. H.; Dicus C. W.; Hilliard B.; Yellayi S.; Zou S.; Hecker J. G. The benefit of hydrophobic domain asymmetry on the efficacy of transfection as measured by in vivo imaging. Mol. Pharm. 2010, 7, 786–794. 10.1021/mp900298f. [DOI] [PubMed] [Google Scholar]
- Chandrashekhar V.; Srujan M.; Prabhakar R.; Reddy R. C.; Sreedhar B.; Rentam K. K. R.; Kanjilal S.; Chaudhuri A. Cationic amphiphiles with fatty acyl chain asymmetry of coconut oil deliver genes selectively to mouse lung. Bioconjugate Chem. 2011, 22, 497–509. 10.1021/bc100537r. [DOI] [PubMed] [Google Scholar]
- Miller J. B.; Zhang S.; Kos P.; Xiong H.; Zhou K.; Perelman S. S.; Zhu H.; Siegwart D. J. Non-Viral CRISPR/Cas Gene Editing In Vitro and In Vivo Enabled by Synthetic Nanoparticle Co-Delivery of Cas9 mRNA and sgRNA. Angew. Chem., Int. Ed. 2017, 56, 1059–1063. 10.1002/anie.201610209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W.; Ji W.; Hall J. M.; Hu Q.; Wang C.; Beisel C. L.; Gu Z. Self-assembled DNA nanoclews for the efficient delivery of CRISPR-Cas9 for genome editing. Angew. Chem., Int. Ed. 2015, 54, 12029–12033. 10.1002/anie.201506030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen J.; Szoka F. C. Nucleic acid delivery: the missing pieces of the puzzle?. Acc. Chem. Res. 2012, 45, 1153–1162. 10.1021/ar3000162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen T. M.; Cullis P. R. Liposomal drug delivery systems: from concept to clinical applications. Adv. Drug Delivery Rev. 2013, 65, 36–48. 10.1016/j.addr.2012.09.037. [DOI] [PubMed] [Google Scholar]
- Karmali P. P.; Chaudhuri A. Cationic liposomes as non-viral carriers of gene medicines: resolved issues, open questions, and future promises. Med. Res. Rev. 2007, 27, 696–722. 10.1002/med.20090. [DOI] [PubMed] [Google Scholar]
- Srujan M.; Chandrashekhar V.; Reddy R. C.; Prabhakar R.; Sreedhar B.; Chaudhuri A. The influence of the structural orientation of amide linkers on the serum compatibility and lung transfection properties of cationic amphiphiles. Biomaterials 2011, 32, 5231–5240. 10.1016/j.biomaterials.2011.03.059. [DOI] [PubMed] [Google Scholar]
- Muñoz-Úbeda M.; Misra S. K.; Barrán-Berdón A. L.; Aicart-Ramos C.; Sierra M. B.; Biswas J.; Kondaiah P.; Junquera E.; Bhattacharya S.; Aicart E. Why is less cationic lipid required to prepare lipoplexes from plasmid DNA than linear DNA in gene therapy?. J. Am. Chem. Soc. 2011, 133, 18014–18017. 10.1021/ja204693f. [DOI] [PubMed] [Google Scholar]
- Wang M.; Zuris J. A.; Meng F.; Rees H.; Sun S.; Deng P.; Han Y.; Gao X.; Pouli D.; Wu Q.; Georgakoudi I.; Liu D. R.; Xu Q. Efficient delivery of genome-editing proteins using bioreducible lipid nanoparticles. Proc. Natl. Acad. Sci. U.S.A. 2016, 113, 2868–2873. 10.1073/pnas.1520244113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X.; Liang X.; Xie H.; Kumar S.; Ravinder N.; Potter J.; de Mollerat du Jeu X.; Chesnut J. D. Improved delivery of Cas9 protein/gRNA complexes using lipofectamine CRISPRMAX. Biotechnol. Lett. 2016, 38, 919–929. 10.1007/s10529-016-2064-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishna S.; Dad A.-B. K.; Beloor J.; Gopalappa R.; Lee S.-K.; Kim H. Gene disruption by cell-penetrating peptide-mediated delivery of Cas9 protein and guide RNA. Genome Res. 2014, 24, 1020–1027. 10.1101/gr.171264.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckl D.; Kowalczyk M. S.; Yudovich D.; Belizaire R.; Puram R. V.; McConkey M. E.; Thielke A.; Aster J. C.; Regev A.; Ebert B. L. Generation of mouse models of myeloid malignancy with combinatorial genetic lesions using CRISPR-Cas9 genome editing. Nat. Biotechnol. 2014, 32, 941–946. 10.1038/nbt.2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukthavaram R.; Marepally S.; Venkata M. Y.; Vegi G. N.; Sistla R.; Chaudhuri A. Cationic glycolipids with cyclic and open galactose head groups for the selective targeting of genes to mouse liver. Biomaterials 2009, 30, 2369–2384. 10.1016/j.biomaterials.2008.12.074. [DOI] [PubMed] [Google Scholar]
- Rajesh M.; Sen J.; Srujan M.; Mukherjee K.; Sreedhar B.; Chaudhuri A. Dramatic influence of the orientation of linker between hydrophilic and hydrophobic lipid moiety in liposomal gene delivery. J. Am. Chem. Soc. 2007, 129, 11408–11420. 10.1021/ja0704683. [DOI] [PubMed] [Google Scholar]
- Ilangumaran S.; Hoessli D. C. Effects of cholesterol depletion by cyclodextrin on the sphingolipid microdomains of the plasma membrane. Biochem. J. 1998, 335, 433–440. 10.1042/bj3350433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayor S.; Parton R. G.; Donaldson J. G. Clathrin-independent pathways of endocytosis. Cold Spring Harbor Perspect. Biol. 2014, 6, a016758. 10.1101/cshperspect.a016758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchhausen T.; Macia E.; Pelish H. E. Use of dynasore, the small molecule inhibitor of dynamin, in the regulation of endocytosis. Methods Enzymol. 2008, 438, 77–93. 10.1016/s0076-6879(07)38006-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y. A.; Scheller R. H. SNARE-mediated membrane fusion. Nat. Rev. Mol. Cell Biol. 2001, 2, 98–106. 10.1038/35052017. [DOI] [PubMed] [Google Scholar]
- Chernomordik L. V.; Kozlov M. M. Mechanics of membrane fusion. Nat. Struct. Mol. Biol. 2008, 15, 675–683. 10.1038/nsmb.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma M.; Bong D. Controlled fusion of synthetic lipid membrane vesicles. Acc. Chem. Res. 2013, 46, 2988–2997. 10.1021/ar400065m. [DOI] [PubMed] [Google Scholar]
- Karmali P. P.; Kumar V. V.; Chaudhuri A. Design, syntheses and in vitro gene delivery efficacies of novel mono-, di- and trilysinated cationic lipids: a structure–activity investigation. J. Med. Chem. 2004, 47, 2123–2132. 10.1021/jm030541+. [DOI] [PubMed] [Google Scholar]
- Karmali P. P.; Majeti B. K.; Sreedhar B.; Chaudhuri A. In vitro gene transfer efficacies and serum compatibility profiles of novel mono-, di-, and tri-histidinylated cationic transfection lipids: a structure–activity investigation. Bioconjugate Chem. 2006, 17, 159–171. 10.1021/bc050194d. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.