Abstract
Overexpression of c-MYC oncogene is associated with cancer pathology. Expression of c-MYC is regulated by the G-quadruplex structure formed in the G-rich segment of nuclease hypersensitive element (NHE III1), that is, “Pu27”, which is localized in the promoter region. Ligand-induced stabilization of the Pu27 structure has been identified as a novel target for cancer therapeutics. Here, we have explored the library of synthetic compounds against the predefined binding site of Pu27. Three compounds were selected based on the docking analyses; they were further scrutinized using all atom molecular dynamics simulations in an explicit water model. Simulated trajectories were scrutinized for conformational stability and ligand binding free energy estimation; essential dynamic behavior was determined using principal component analysis. One of the molecules, “TPP (1-(3-(4-(1,2,3-thiadiazol-4-yl)phenoxy)-2-hydroxypropyl)-4-carbamoylpiperidinium)”, with the best results was considered for further evaluation. The theoretical observations are supported well by biophysical analysis using circular dichroism, isothermal titration calorimetry, and high-resolution NMR spectroscopy indicating association of TPP with Pu27. The in vitro studies were then translated into c-MYC overexpression in the T47D breast cancer cell line. Biological evaluation through the MTT assay, flow cytometric assay, RT-PCR, and reporter luciferase assay suggests that TPP downregulates the expression of c-MYC oncogene by arresting its promoter region. In silico and in vitro observations cumulatively suggest that the novel skeleton of TPP could be a potential anticancer agent by stabilizing the G-quadruplex formed in the Pu27 and consequently downregulating the expression of c-MYC oncogene. Derivation of new molecules on its skeleton may confer anticancer therapeutics for the next generation.
Introduction
Health problems due to cancer are increasing worldwide; according to global demographic characteristics, more than 20 million new cancer cases per year are expected by 2025.1 Limitations in earlier chemotherapeutic cancer treatments such as severe side effects, toxicity, and the emergence of resistance have been overcome by incorporating specificity toward cancer targets. Targeted therapy has proven to be an efficient alternative whereby molecular targets responsible for addictions, dependencies, and vulnerabilities of cancer cells are identified and treated with either an immunotherapy approach or with small molecular chemical entities.1−4 Some of the successes in targeted therapy are as follows: inhibition of EGFR by gefitinib and erlotinib, inhibition of c-kit and PDGFR by imatinib, inhibition of VEGFR by sorafenib, inhibition of protein kinase ALK by crizotinib, inhibition of BRAF by vemurafenib, and so forth.1,4 Recently, Cifuentes et al. studied the interaction of the TTP–DMβCD inclusion complex in targeting ctDNA.5 One of the unattained cancer targets is c-MYC oncogene;6 multiple mutations and overexpression of c-MYC are observed in carcinomas of the breast, colon, and cervix, and in osteosarcomas, glioblastomas, and myeloid leukemias.7c-MYC plays a key role in regulating the cell cycle; it assists in cell proliferation; however, in collaboration with n-MYC, it inhibits the differentiation of cells, thus maintaining the pluripotency and self-renewal of cells and causing malignancy.7−12 Thus, inhibition of c-MYC has been considered as an approach for anticancer therapy. c-MYC can be targeted in three ways; (i) inhibition of c-MYC–MAX dimerization, (ii) inhibition of binding of the dimer with E-box, and (iii) transcriptional regulation of c-MYC by arresting the G-quadruplex formed in its promoter region7 and inhibition of binding of the respective transcription factors with Pu27.13 Here, we have adopted the third mode for downregulation of c-MYC expression, that is, stabilization of the respective quadruplex using small molecules. The mode of arresting the G-quadruplex structure via small molecules has been investigated broadly over the few last decades,14−16 opening a new arena in modern day therapeutic approaches.
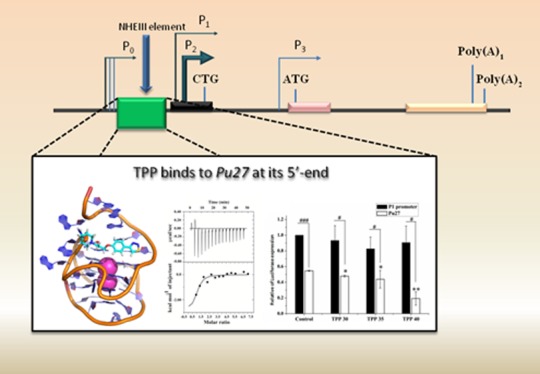
“Nuclease hypersensitivity element” (NHE III1) consists of a 27-bp long purine-rich noncoding sequence named Pu27, which is located at the upstream site of the P1 promoter of c-MYC. G-quadruplex formation in the guanine-rich strand of Pu27 results in silencing of the c-MYC oncogene, whereas in the active state it adopts a duplex secondary structure, thus NHE III1 regulates the transcription of c-MYC through the structural transitions of Pu27.17−22 Natural compounds and some synthetic chemical compounds have been studied for their stabilization effect on the G-quadruplex structure of Pu27 and the further inhibition of expression of the c-MYC oncogene.23−30 Quarfloxin is the only quadruplex binding agent that has entered phase II clinical trials for its anticancer activity;31 it was also found to inhibit c-MYC expression by binding to Pu27.32 Other chemical entities have been found to be transcriptional inhibitors of c-MYC through quadruplex interaction such as the perylene derivative PIPER (N,N′-bis(2-(1-piperidino)ethyl)-3,4,9,10-perylenetetracarboxylic acid diimide),33 the cationic porphyrine derivative TmPyP4,34,35 acridine derivatives,36 quindoline derivatives,27,37 the FDA-approved drug, methylene blue,38 and so forth. The presence of two single-nucleotide loops and one two-nucleotide loop in one of the quadruplex conformers of Pu27 provides the stability to the overall secondary structure in simulated conditions thus allowing the utilization of an in silico approach in studying the Pu27–ligand interaction.39−41 Nasiri et al. screened a fragment library against the promoter region of c-MYC by means of in silico methods and further validated the results using biophysical techniques and biological assays; they proposed some small fragments such as the amino–indole motif and amino–benzodioxan skeleton binding in the groove region as a starting point for further drug design.39 Similarly, Kang et al. determined hit molecules against the promoter region of c-MYC oncogene based on virtual screening, molecular docking, and a molecular dynamics study, and further analyzed their interaction properties using biophysical and biological approaches.23 Also, Rocca et al. determined novel natural alkaloid derivatives with a dual binding property toward h-telo and c-MYC promoter quadruplex by means of virtual screening, molecular docking, and molecular dynamics simulations.28 Shan et al. utilized a similar in silico approach in screening molecules that could disrupt the binding of NM23H2 with Pu27 resulting in the downregulation of c-MYC expression.13 Lee et al. indentified the natural product Fonsecin B as a c-MYC stabilizing agent via a high-throughput virtual screening method.42 In our previous study, we followed the in silico approach for screening the natural drug database with chelerythrine-like molecules and determined the pharmacophoric features essential for stabilizing the quadruplex formed in Pu27.43
In the current study, a library of small compounds has been screened based on their druglikeness using a virtual screening method and further screened for their probable binding capacity toward the G-quadruplex formed in Pu27 using molecular docking. The top three best fitting molecules are further studied for their binding characteristics and stabilizing effect on Pu27 using all atom molecular dynamics simulations in an explicit solvent model. On the basis of the structural stability, essential dynamics properties, and binding free energy estimations, one compound, TPP (1-(3-(4-(1,2,3-thiadiazol-4-yl)phenoxy)-2-hydroxypropyl)-4-carbamoylpiperidinium), with promising results is taken for further scrutiny. Biophysical techniques such as circular dichroism (CD) and isothermal titration calorimetry (ITC) confirm the binding of TPP with Pu27. NMR spectroscopy offers the atomistic details of the binding pattern of TPP, further validating the simulation results. The postulation that the screened compound with a binding phenomenon toward Pu27 can downregulate the expression of c-MYC is established by further ex vivo assessment of c-MYC overexpression in the T47D breast cancer cell line. The flow cytometric assay confirms the killing effect of TPP over cancer cells, reverse transcription (RT)-PCR and the reporter luciferase assay imply that TPP arrests the quadruplex structure and reduces the expression of c-MYC oncogene. Thus, here we have combined the stepwise protocol of computer aided drug design with in vitro and ex vivo approaches to obtain a novel lead molecule that can act as a c-MYC inhibitor. Further optimization of TPP may offer a promising anticancer agent.
Materials and Methods
Virtual Screening and Molecular Docking
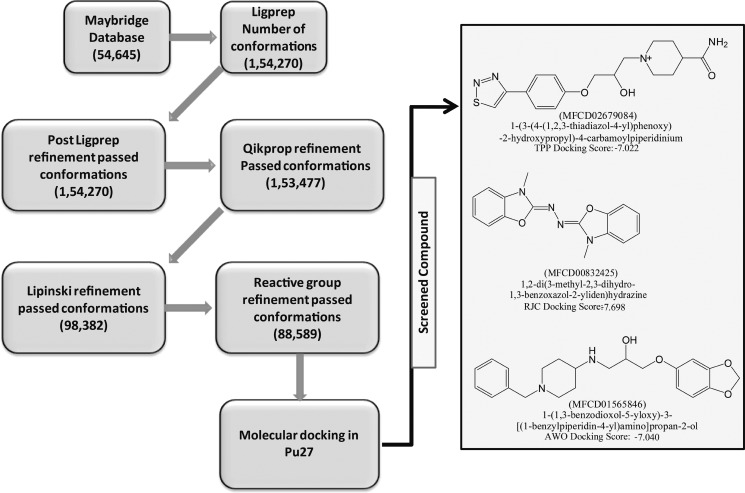
Virtual screening workflow available in Maestro44 was utilized for screening 54 645 chemical compounds obtained from the Maybridge database. The stepwise screening of chemical compounds is as shown in Figure 1. After stepwise refinements, 88 589 conformations were used for further molecular docking.
Figure 1.
Virtual screening protocol followed in stepwise filtering of chemical compounds is given in the flow chart format. Selected compounds are represented in the two-dimensional (2D) format. Respective docking scores, IUPAC naming, and Maybridge database IDs are given below the structures.
The initial model structure of the G-quadruplex formed in Pu27 was kept the same as that reported in the previous study.43 It was built over the template of the NMR solution structure (PDB-ID 2A5P)25 using Maestro45 and further details are as documented previously.43 On the basis of the information provided in the literature,23,25,28,43 the 5′ end of Pu27 was taken as the ligand binding site. A grid with an inner box (10 Å) and an outer box (30 Å) was built at the 5′ end of the quadruplex structure. Screening of compounds was done with three-step molecular docking using Grid based Ligand Docking with energetics (GLIDE);46 (i) all compounds were docked using HTVS protocol and the top 10% of the compounds were screened, (ii) compounds screened through HTVS were re-docked using SP protocol and the top 10% of the compounds were screened, (iii) compounds screened through SP were re-docked using XP protocol. Three compounds with a favorable docking score were taken as a lead for further study. Their IUPAC names, database codes, and respective 2D structures are shown in Figure 1. In further discussion, they are named TPP, RJC, and AWO.
Molecular Dynamics Simulations
Four molecular systems termed unbound-Pu27, Pu27–TPP complex, Pu27–RJC complex, and Pu27–AWO complex were considered for simulation analysis. All atom molecular dynamics simulations in explicit water were conducted using the simulation program AMBER11.47 Two K+ ions were explicitly placed into the central region of the quartet channels using the xleap module of AMBER11. Force field parameters for ligand molecules were generated in the Antechamber48 module of AMBER with AM1-BCC charges. Ions were parameterized using a General Amber Force Field (GAFF)49 and the quadruplex segment was parameterized using the ff99SB50 and parmbsc051 force fields of AMBER11. Sodium ions were incorporated to neutralize the charges, and the systems were submerged within an 8 Å TIP3PBOX water model;52 further simulation stages were the same as that in a prior report.53 Minimization was conducted in two steps using a combination of the steepest descent and conjugate gradient method; initially, water molecules and ions were minimized constraining the biomolecule, and afterwards, the whole system was minimized. Systems were gradually heated to 300 K within 50 ps, and macroscopic factors were equilibrated for 1 ns prior to production. A final production run of 50 ns was conducted in NPT ensemble at 300 K temperature and 1 atm pressure and a calculation was performed for each 2 fs step size. Temperature and pressure was regulated using a Langevin thermostat-barostat, and the SHAKE algorithm was applied for bond length corrections.54−56 Long range electrostatic interactions above the cut off distance of 12 Å were optimized with the Particle Mesh Ewald method (0.1 nm grid space of FFT grid).57 A trajectory was formed with the atomic coordinates saved at each 10 ps time step. Trajectories were further analyzed using the CPPTRAJ module of AMBER, VMD, PyMol, and Chimera tools.58−61
The binding free energy of each ligand was estimated over the entire simulation run by extracting frames at each 500 ps. Both the MMPBSA and MMGBSA approaches were used, as MMPBSA can provide energies close to experimental observations and MMGBSA offers comparative values thus allowing ranking of multiple ligands.62−67 Estimations were based on the following mathematical functions
where ΔGbinding is the binding free energy and GDNA+ligand complex, GDNA, and Gligand represent the free energies of the respective states.
The free energy of each state was calculated as follows
where EMM is the molecular mechanical energy, GPB/GB is the polar contribution toward solvation energy, Poisson–Boltzmann (PB) or Generalized Born (GB) method used for calculations, GSA are the contributions from nonpolar terms toward solvation energy, Eele is the electrostatic energy, EvdW is the van der Waals energy, Eint is the internal energy (bond, angle, and torsional angle energy) SASA is the solvent accessible surface area, γ is the surface tension proportionality constant (0.0072 kcal/mol/Å2), and b is the free energy of nonpolar solvation for a point solute (0 kcal/mol).
In this tool, SASA is computed by molsurf using linear combinations of pairwise overlaps (LCPO), the solute atoms were taken as a probe sphere of 1.4 Å. Binding free energy calculations were averaged over 100 frames taken at the interval of 500 ps over the production run of 50 ns.
Biophysical Techniques and Materials
TPP was purchased from Thermo Fisher Scientific India. Potassium phosphate monobasic, potassium phosphate dibasic, and potassium chloride were purchased from Sigma-Aldrich. The commercially synthesized oligonucleotides, Pu27 and Pu24 were purchased from from Eurofins Genomics India Pvt Ltd.
Pu27-5′-TGGGGAGGGTGGGGAGGGTGGGGAAGG-3′
Pu24-5′-TGAGGGTGGGGAGGGTGGGGAAGG-3′
10 mM phosphate buffer containing 100 mM KCl at pH 7.0 was prepared in millipore water. It was autoclaved and filtered prior to use. Pu27 and Pu24 were dissolved in the same buffer, annealed at 90 °C, cooled to room temperature, and stored at 4 °C for further use. The molecule TPP was dissolved in 100% dimethyl sulfoxide (DMSO), the DMSO concentration was, however, kept below 1% in all experiments.
CD
The CD spectra of Pu27 and the Pu27–TPP complex were monitored using a Jasco 815 spectrometer. Pu27 (20 μM) was dissolved in 10 mM phosphate buffer containing 100 mM KCl at pH 7.0. CD titrations of Pu27 with increasing concentration of TPP were carried out at 25 °C using a cuvette of 0.1 cm path length. To attain equilibration and ensure complete complex formation, each spectrum was recorded after 5 min of addition of TPP. Each spectrum was scanned from 220 to 310 nm with a scan speed of 100 nm/min. The data interval was set at 1 nm with an average of 3 scans. All spectra were base line corrected. To check the stabilization effect of TPP over Pu27, CD melting experiments were carried out. Pu27 (20 μM) was annealed separately in 10 mM phosphate buffer containing 100 mM KCl at pH 7.0 and water (K+ < 1.5 mM) to find out the Tm of unbound-Pu27. To conduct the melting of the Pu27–TPP complex both in phosphate buffer containing 100 mM KCl and in water, TPP was mixed with Pu27 in a 3:1 molar ratio. The thermal melting was recorded from 20 to 95 °C for the Pu27–TPP complex in phosphate buffer and from 20 to 90 °C for the Pu27–TPP complex in water. The temperature interval was 5 °C with a ramp rate of 2.5 °C min–1. The sample was equilibrated for 3.0 min at each temperature before data acquisition.
NMR Spectroscopy
A Bruker AVANCE III 500 MHz NMR spectrometer equipped with a 5 mm SMART probe was used to monitor the NMR spectra at 298 K. Acquisition and data processing were done using TopspinTMv3.1 software. The samples were dissolved in 10 mM phosphate buffer containing 100 mM KCl and 10 % D2O. Trimethylsilylpropanoic acid (TSP) was used as an internal standard (0.0 ppm). An array of one-dimensional proton spectra and proton-decoupled one-dimensional 31P spectra were monitored with increasing concentration of TPP until the peak started broadening employing the standard Bruker pulse programs “zgesgp” and “zgpg30”, respectively. A total of 512 scans were used with a spectral width of 20 ppm. To calculate the rate of exchange of imino protons (Kex) of Pu24, two selective inversion recovery experiments were performed, one for water and the other for imino protons, with inversion recovery delays ranging from 0 to 3.5 s and 0 to 0.0125 s to compute the spin lattice rates, R1W and R1i, respectively. The water-selective magnetization experiment was performed to calculate Kex using the following equation
where I0 and It are the peak intensities of the imino proton at zero and t delay/mixing time, respectively. The detail of the pulse sequence used to commute Kex was described in detail in our previously published paper.68
ITC
An iTC200 Microcalorimeter was used to conduct ITC experiments at room temperature. TPP and Pu27 were degassed ahead of titration to make sure that no bubble formation occurred. Pu27 (700 μM) was injected at an interval of 180 s into a fixed volume of TPP (20 μM). A blank experiment was performed by injecting the same concentration of Pu27 into the buffer solution without TPP to subtract the heat of dilution when Pu27 interacts with the buffer. The isotherm was analyzed with the help of in-built Origin 7.0 software. The data points obtained were best fitted into a “one-site” binding model. The thermodynamic parameters such as change in enthalpy (ΔH), change in entropy (ΔS), and Gibbs free energy (ΔG) were calculated using the following equation.
Cell Culture and Treatment
Human breast ductal carcinoma cell line (T47D) and normal kidney epithelial cell line (NKE) were cultured separately in complete Dulbecco’s modified Eagle’s medium (DMEM) (Himedia; AL007G) and RPMI-1640 (Himedia; AL120A) media, respectively, supplemented with 10% (v/v) fetal bovine serum, 2 mM l-glutamine, 50 μg/mL gentamycin, 1% Pen-Strep, and 2.5 μg/mL Amphotericin B in a fully humidified CO2 incubator (ESCO cell culture CO2 Incubator, Model no. CCL-1708-8-UV) at 37 °C and 5% CO2.
MTT Assay
T47D and NKE cells were subcultured in a 96-well microtiter plate at a density of 104 cells/well in 100 μL of respective culture media and incubated in an ESCO cell culture CO2 incubator at 37 °C and 5% CO2 for 24 h to ensure cell attachment. Cells were treated with an increasing concentration gradient of TPP (1, 5, 10, 20, 30, 40, 50 μM) for 24 h. After treatment, 10 μL of a 5 mg/mL solution in phenol-free media of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) tetrazolium substrate was added to each well and incubated for 4 h at 37 °C in the CO2 incubator. The resulting violet formazan precipitate was solubilized in 100 μL of DMSO. Absorbance for each sample was recorded at 570 nm in the 96-well microtiter plate reader (Polar Star Optima, BMG Labtech) with a reference wavelength of 650 nm. All of the assays were performed in six replicates, and the DMSO-treated set was considered as a negative control to nullify the cytotoxic effects of DMSO (as TPP was solubilized in DMSO). The percentage cell viability was calculated as the percentage of MTT absorption
Flow Cytometry
T47D and NKE cells were individually seeded into 6-well microtiter plates up to 70% confluency, at a density of 1 × 106 cells per well in 2 mL of respective culture media and allowed time for adherence. Cells were treated with an increasing concentration (1–150 μM) of TPP. After 24 h of treatment, cells were trypsinized and washed twice with 1× PBS at 3000 rpm for 5 min at room temperature. The cell pellet was subjected to the flow cytometric assays with a BD Pharmigen Annexin V-FITC Apoptosis detection Kit (Catalog no. 556570). In brief, the cell pellet is re-suspended in 100 μL of 1× binding buffer followed by incubation with 5 μL of Annexin V-FITC and 2 μL of PI under dark conditions for 30 min and 15 min, respectively, at 4 °C. Finally, cell suspensions were diluted with 400 μL of 1× binding buffer immediately prior to the analysis in the BD FACS Verse Flow cytometer. Measurement of individual sets was acquired at 1000 events with medium flow rate and carried out in triplicate. DMSO-treated and 60 μM Etoposide (known to induce apoptosis in cells)-treated cells69 were considered as negative and positive controls, respectively. The gate was set based on the SSC (side scatter) versus FSC (forward scatter) dot plot, and the resulting quadrants of the treated sets reflect the fluorescence properties of the gated population only (Figures S15 and S16).
RT-PCR
T47D cells were subcultured into 6-well microtiter plates up to 70% confluency, at a density of 1 × 106 cells per well in 2 mL of DMEM media and allowed time for adherence. Cells were treated with different concentrations (30, 35, and 40 μM) of TPP. After 24 h of treatment, cells were scraped and washed twice with 1× PBS at 3000 rpm for 5 min at room temperature. Total RNA was isolated from both untreated and TPP treated T47D cells using the TRIzol method (Invitrogen, Carlsbad, CA) as per manufacturer’s instructions. 2 μg of total RNA was processed for cDNA synthesis and reverse transcribed using a Super MuLV RT Kit (Biobharati Life Sciences Pvt. Ltd.). PCR reactions were optimized to 94 °C for 5 min, 45 amplification cycles at 94 °C for 30 s, the appropriate annealing temperature for 45 s, 72 °C for 45 s, and a final extension of 7 min at 72 °C followed by storage at 4 °C. The final products were electrophoresed on 1.5% agarose gels and visualized by ethidium bromide staining along with a 100 bp DNA ladder (Thermo Scientific). Reactions were carried out in triplicate, and the housekeeping gene GAPDH was used as an internal control to normalize the variability in expression levels. Semiquantitative densitometric analyses were performed to determine the relative mRNA expression levels. PCR primers were designed using Primer-BLAST, NCBI, and analyzed in OligoAnalyser 3.1-IDT. The forward and reverse primer sequences and corresponding annealing temperatures are listed in Table 1.
Table 1. Forward and Reverse Primer Sequences and Corresponding Annealing Temperatures.
| gene | sequence (5′−3′) | annealing temperature (°C) |
|---|---|---|
| c-MYC | F-TCTCCACACATCAGCACAAC (20 bp) | 51.8 |
| R-TCCGTTTTAGCTCGTTCCTC (20 bp) | 51.8 | |
| GAPDH | F-GATGCTGGCGCTGAGTACGTCGTG (24 bp) | 62.5 |
| R-AGTGATGGCATGGACTGTGGTCATGAG (24 bp) | 61.3 |
Reporter Luciferase Assay
Customized NHE III1 (−) and NHE III1 (+) P1–P2 promoter sequences cloned into a pGL4.72 vector at the KpnI and HindIII restriction sites and fused with a downstream hRlucCP reporter gene were purchased from Biobharati Life Sciences Pvt. Ltd. T47D cells were seeded into 24-well plates at 70–80% confluency and transfected with 5 μg of reporter plasmid by Lipofectamine 2000 reagent (Invitrogen) using the manufacturer’s protocol. After 30 h of transfection, cells were treated with TPP (25, 35, and 40 μM). Luciferase assays were performed after 24 h of treatment using the Promega Luciferase Assay System (E1500) and Multimode Microplate Reader (Thermo Fisher Scientific) as per the manufacturer’s protocols. Transfection of each construct was performed in triplicate. Readings were recorded for each experiment and triplicates were averaged.
Results and Discussion
Analysis of Molecular Docking
As seen in Table S1, the selected ligand molecules, through virtual screening and molecular docking, fulfill the criteria for druglikeness. All possess a negative docking score (better than chelerythrine43), and respective docked conformations are shown in Figure S1. TPP forms polar interactions with A15 and CH−π interactions with A6 and G14. RJC forms polar interactions with A15 and G11 and π–π stacking interactions with G14. AWO forms polar interactions with A15, G11, and G9, and π–π stacking interactions with G11 and T10. Thus, the 5′ end bases are providing the binding site for ligand molecules. These results are in correlation with our previous findings;43 also, Kang et al. suggested a similar binding pattern for small molecules interacting with Pu27.23 We further explored the binding pattern of selected ligands using all atom molecular dynamics simulations to overcome the limitations of docking in considering the flexibility of receptor and ligand molecules. The results of this are discussed further.
Root-Mean-Square Displacement (RMSD) Analysis
RMSD analysis illustrates the deviation of the structure at a particular time point from that of the reference frame. Here, we considered the coordinates of the structure after the equilibration stage as the reference frame, and RMSD is estimated over the entire 50 ns simulation run using the formula
where N is the total number of atoms under consideration and δ is the distance between two positions of N pairs of equivalent atoms. Higher deviation in the RMSD pattern indicates the flexible nature of a respective structure and if the RMSD is converging to a certain steady value, this implies the convergence of the structure to a stable conformation.
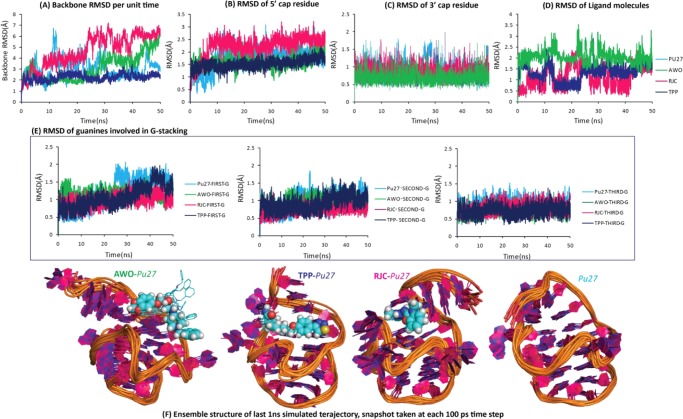
Backbone RMSD represents the integrity of an overall secondary structure; as seen in Figure 2A, the Pu27–TPP complex possesses the lowest backbone RMSD (∼2.0 Å), which is steady over the entire simulation run. Unbound-Pu27 and the Pu27–AWO complex show unsteady RMSD, which fluctuates around ∼3.0 Å. The Pu27–RJC complex has the highest RMSD value, which fluctuates around ∼4.7 Å. Thus, the backbone RMSD estimation suggests that the secondary structure of Pu27 attains the most stable conformation when it is in the bound form with the TPP molecule. 5′ end capping bases (G5, A6, A15) provide the binding surface for ligands; in all of the bound and unbound states, these bases are rigid (Figure 2B) with a steady pattern in RMSD (<∼3.0 Å). In the Pu27–TPP complex, 5′ end capping bases are stabilized to a greater extent, as reflected in the small RMSD value (∼1.5 Å); also, in unbound-Pu27 and the Pu27–AWO complex, these bases are stabilized with RMSD within the range of ∼1.5 to ∼2.0 Å, whereas, in the presence of RJC, the active site bases are fluctuating far from the initial conformation (RMSD ∼ 2.2 Å). RMSD observations imply that TPP is effective in stabilizing the binding site. The characteristic feature of the Pu27 structure, the “GAAG cap” at the 3′ end (G23, A24, A25, G26) (Figure 2C), and the core region of stacked guanines are found to be rigid in all of the systems (Figure 2E) with lower RMSD values (∼1.5 Å). As seen in the RMSD of the ligand molecule (Figure 2D), AWO is flexible as RMSD is fluctuating around ∼2.0 Å; however, RJC and TPP are stabilized at different ranges of RMSD values. TPP is stabilized around ∼1.0 Å for the initial 10 ns and for subsequent 10 ns steps it is stabilized around ∼0.5 Å. During the last 30 ns simulation run TPP has attained a steady RMSD of around ∼1.0 Å. Similar to the RMSD pattern of TPP, the RMSD of RJC is also fluctuating between ∼0.5 and ∼1.0 Å. The flexibility of the ligands is visible in the ensemble structure of the respective system (Figure 2F); TPP and RJC converge to a steady conformation; however, AWO deviates. Thus, among all of the ligand molecules, TPP is found to be effective in enhancing the structural strength of Pu27 over that of its unbound state.
Figure 2.
Segment wise RMSD analysis of all of the states of Pu27 [color key: unbound-Pu27 (sky blue), Pu27–TPP complex (dark blue), Pu27–RJC complex (pink), Pu27–AWO complex (green)]; (A) backbone RMSD of whole structure, (B) RMSD of 5′ cap bases (G5, A6, A15), (C) RMSD of 3′ cap bases (G23, A24, A25, G26), (D) RMSD of ligand molecules, (E) RMSD of guanines involved in core stacking, and (F) superimposed structure of coordinates taken at each 100 ps from last 1 ns trajectory of each system.
B-Factor Analysis
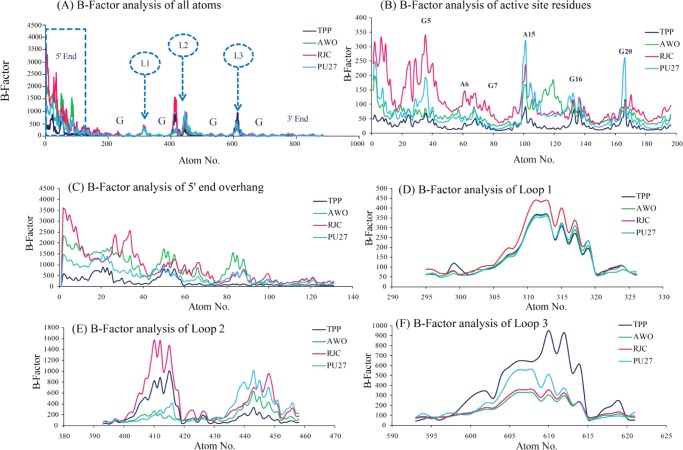
RMSD gives the average deviation of the overall structure from the reference frame at each time point, whereas the B-factor gives the average fluctuation of each atom from the reference frame over the entire simulation run. This means RMSD offers the conformational integrity over the entire structure and B-factor offers the contribution of each atom toward the strength and flexibility of the structure. Segment wise B-factors are determined using the cpptraj58 module of AMBER using the formula
Here, RMSF represents root-mean-square fluctuation.
B-factor analysis (Figure 3A) suggests that the 5′ overhanging nucleotide stretch and loop regions are flexible and the guanines involved in core stacking are rigid; these observations are in correlation with the RMSD data. The B-factor of the bases involved in ligand binding and guanines forming the first G-stack (G5, A6, G7, A15, G16, G20) shows variation in different states (Figure 3B); in the Pu27–TPP complex these bases are stabilized to a greater extent than that of the other states, in the Pu27–RJC complex this region is highly flexible, whereas in unbound-Pu27 and the Pu27–AWO complex these regions are moderately flexible. Both the RMSD and B-Factor data confirm the stabilizing effect of TPP over the ligand binding site. The B-factor of the 5′ overhanging nucleotide stretch (Figure 3C) is found to be lowest in the Pu27–TPP complex, highest in the Pu27–RJC complex, and moderate in both the unbound-Pu27 and Pu27–AWO complex systems. The flexibility of the loop regions increases in the presence of ligand molecules over that of the unbound state. The flexibility of Loop-1 is similar in the Pu27–TPP complex and the Pu27–AWO complex but in the Pu27–RJC complex it is at a higher range (Figure 3D). G13 of Loop-2 is highly flexible (Figure 3E) in the Pu27–RJC complex and in the Pu27–TPP complex whereas G14 of Loop-2 is stabilized in the presence of TPP; visual analysis confirms that TPP forms H–π interactions with G13 thus affecting its flexibility, and G14 is stabilized due to π–π stacking interactions with G13 (refer to the video of simulated trajectory provided in the Supporting Information). Loop-3 (Figure 3F) is highly flexible in the Pu27–TPP complex but stabilized in the presence of RJC and AWO.
Figure 3.
Segment wise B-factor analysis [color key: unbound-Pu27 (sky blue), Pu27–TPP complex (dark blue), Pu27–RJC complex (pink), Pu27–AWO complex (green)], (A) B-factor of all atoms with highlighting key segments, (B) B-factor of ligand binding site residues (G5, A6, G7, A15, G16, G20), (C) B-factor of 5′ end overhanging stretch (T1, G2, G3, G4), (D) B-factor of Loop-1 (T10), (E) B-factor of Loop-2 (G13, G14), and (F) B-factor of Loop-3 (T19).
Hydrogen Bond Analysis
Hydrogen bonds are determined with the distance cut off between donor and acceptor group set to 3 Å and angle cut off set to 20°, “% hydrogen bond occupancy” is calculated as the percentage of time that the hydrogen bond is formed over the entire simulation time using VMD software.59
During the simulation run, TPP forms ∼2 hydrogen bonds with Pu27, whereas both RJC and AWO form only ∼1 hydrogen bond (Figure S2A). AWO, being flexible, does not intercalate with the active site bases and it is highly exposed to the water molecules, as illustrated by water shell analysis (Figure S2F), it forms nearly four hydrogen bonds with water molecules (Figure S2B). However, TPP and RJC are rigidly bound with the active site bases and further stabilized by interacting with water molecules forming ∼3 and ∼1 hydrogen bond interactions, respectively (Figure S2B). As shown in Figure S2G, TPP forms hydrogen bonding interactions with G4, G11, and A15 with high occupancy values and electrostatic interactions with A6, G11, and A15 (Figure S2D). Thus, polar interactions and hydrogen bonding of TPP strengthen the wrapping of 5′ end bases with overhanging stretch. TPP–Pu27 complexation is well supported with solvent molecules. RJC shows poor hydrogen bonding and forms polar interactions with G3, G4, and A15 (Figure S2E); however, unlike the Pu27–TPP complex, wrapping of the overhanging stretch over the 5′ end bases is lacking. AWO forms strong hydrogen bonding interactions with G11 but its overall conformation is not stabilized in the binding site, thus it is excluded from the evaluation of polar interactions. The total number of water molecules in the inner (3.4 Å from ligand) and outer shell (5 Å from ligand) of ligand molecules are as illustrated in Figure S2F. In the Pu27–TPP complex, the total number of water molecules in both the inner and outer shells is in a lower range than that of the other two complex systems. Also, the numbers of total water molecules fluctuate for TPP during the simulation run, unlike the RJC and AWO complex systems that have a steady count of water molecules around the ligands. This implies that binding of TPP is perturbing the structure of the surrounding water molecules, which is absent in RJC and AWO. Binding of TPP and RJC is majorly through interactions with the 5′ end bases; a similar interaction pattern has been reported in analysis of alkaloid derivatives28 and some novel ligands.23
The intramolecular interactions of Pu27 in different states are shown in (Figure S3). In the Pu27–TPP complex and unbound-Pu27, the 5′ end overhanging bases (T1, G2, G3, G4) are capping over the first guanine stack (G7, G11, G16, G20), which is supported by hydrogen bonding interactions among G2–G3 (Pu27–TPP complex), G2–G4 (Pu27–TPP complex), G4–A15 (Pu27–TPP complex), and G3–G4 (unbound-Pu27). Such interactions are absent in the AWO and RJC complex systems. A–A-mismatch (A6–A15) is a characteristic feature of the Pu27 structure, which is highly stabilized in the presence of TPP and in the unbound state; however, it is weakened in the Pu27–RJC complex and Pu27–AWO complex. The guanines involved in core stacking are highly stabilized through Hoogsteen hydrogen bonding in all of the systems, in the Pu27–AWO complex these base pairings are weakened to a small extent at the third guanine stack, as reflected in the reduced hydrogen bond occupancy (Figure S3). Hydrogen bond analyses clearly indicate that binding of TPP is strengthening the secondary structure of quadruplex formed in Pu27 by means of enhancing occupancy of base pairing and other hydrogen bonding interactions.
Water Density Map Analysis and Ion Density Grid Analysis
Density maps of water molecules and ions surrounding the quadruplex were generated using the grid function of CPPTRAJ.58 A grid box of “100 Å × 100 Å × 100 Å” was generated with a mesh size of 0.5 Å. The occupancy of the oxygen atom of a water molecule and sodium ion at each grid point was determined over the average coordinates of Pu27 recorded for the last 10 ns simulation run. The occupancy of water or ion molecule represents the population of the respective atom (oxygen in the case of water and Na+ in the case of ion) that retains the same distance and angle (as present in the reference frame) with that of the corresponding atoms of the quadruplex during the trajectory period (10 ns) under consideration. Though the “simulation time” term is not included in the function of density calculation, it has been indirectly taken into consideration as the population reflects the total number of times the particular condition is satisfied. A denser contour represents the localization of a respective molecule at the associated grid point for a longer duration and vice versa. The generated XPLOR density maps were visualized in Chimera 1.9.61 The approach of correlating water density and ion density with the structural properties of the G-quadruplex structure has been applied in our previous studies.43,53,70
A denser water structure around the macromolecule suggests rigidity of the respective region, whereas a thin water structure is generally observed around the flexible regions. As seen in Figure S4A, the water structure is dense in all of the states of Pu27 reflecting the strength of the overall secondary conformation. Pu27 in complex with the ligand attains a higher water density than that of the unbound state thus indicating a stabilizing effect of the ligands over the G-quadruplex structure. In the Pu27–TPP complex and Pu27–RJC complex, water molecules are densely located in the groove region thus supporting the secondary structure. Further, the flexible nature of the bases present in the loop region is reflected by the deficiency of hydration. In the Pu27–AWO complex, the water structure is unsystematic, loop bases are hydrated thus indicating their rigid nature, which is in correlation with the B-factor analysis.
In the simulation of the quadruplex, the repulsive forces generated due to negative charges over the backbone phosphate atoms and partial negative charges over the carbonyl oxygen atoms are counterbalanced by the explicitly incorporated counterions. As illustrated in Figure S4B, core regions in all of the states of Pu27 are stabilized by channeled K+ ions. In unbound-Pu27, the loop bases, A–A mismatch region, and GAG triad at the 3′ end are supported by counterions. The GAG triad is surrounded by ions in all of the systems, but the rest of the ion structure varies. In the Pu27–TPP complex, ions are centered over the 5′ end, the overhanging stretch at the 5′ end is surrounded by ions and scattered interactions of ions are found in the backbone regions and some in the loop regions. In the Pu27–RJC complex, the A–A mismatch region is in contact with ions, and the other regions show ion interactions to a small extent. Similar to the water structure, the ion structure in the Pu27–AWO complex is different from the other states as Loop-1, the GAG triad, and external portion of the ligand molecule are majorly surrounded by ion molecules.
Principal Component Analysis and Conformational Behavior
The essential dynamics responsible for the overall conformational behavior of the quadruplex was determined using principal component analysis (PCA). PCA assists in extracting the set of coordinate frames that reflects the dynamic pattern of the macromolecule occurring in the simulation run.71−76 All of the states of Pu27 were evaluated using the PCASuite package.77 Each trajectory of 50 ns length was divided into 5 subsections (10 ns each), and they were processed using the PCAzip module for phosphodiester backbone atoms. Noise due to oscillations and tumbling motions was eliminated by a two step alignment; first, all of the frames were superimposed with the initial frame and, accordingly, the average coordinates were computed. Further alignment was performed over the average coordinates based on the Gaussian RMSD algorithm.78 The coordinates of each atom were taken as a random variable to form a covariance matrix, which was further diagonalized to obtain eigenvectors generated with 90% of the variance. The first two eigenvectors were selected based on the cumulative percent contribution for further analysis as they covered a minimum of 40% variance (Figure S5). The porcupine plots representing the essential dynamics were built in PyMOL.60 Lindemann’s coefficient was calculated for each base and averaged over 5 sets, the calculation was based on the formula72,79
where N is the number of atoms, a′ is the empirical constant most probable nonbonded near-neighbor distance and ΔRi2 is the fluctuation of the atom, i.
In unbound-Pu27 (Figure S6), the capping arrangement of the 5′ overhanging stretch is assisted by guanines of Loop-2; however, the dynamicity of the respective bases is in a higher range in the whole simulation period. As found in a previous study,43 the stacking arrangement within T1, G13, and G14 is essential for stabilization of the cap, and this is absent in the current trajectory, which leads to the dynamic behavior of the 5′ overhanging stretch. In the Pu27–TPP complex (Figure S7), the overall dynamicity of the quadruplex and ligand molecule is reduced with simulation time. TPP strengthens the capping of the 5′ overhanging stretch; polar interactions among G2–G3, G2–G4, G4–TPP, and TPP–A15, and H–π stacking interactions in G2/TPP/G14 are found in cap formation. In the RJC and AWO complexes (Figures S8 and S9), the structure is highly dynamic, especially in the loop regions and at the 5′ end. Ligand molecules (RJC and AWO) also show high flexibility.
Zhou et al. and Jamroz et al. applied the Lindemann’s coefficient as a measure of the solid and liquid behavior of protein molecules.72,79 Small values of ΔL (<0.15) indicate a solid nature and higher values (>0.15) indicate a liquid nature of respective bases. In previous studies, we utilized the same concept to determine the dynamic properties of G-quadruplex.43 Here, we compared all of the states of Pu27 and ΔL of the backbone atoms (Figure S10B) suggest that the secondary structure is rigid in all of the systems; however, the 5′ overhanging stretch is highly flexible and stabilized to some extent in the presence of TPP. Here, the backbone atoms are exposed to solvent molecules and thus posses ΔL higher than 0.15; however, the side chain atoms that are buried in the core region have lower ΔL values.79 Side chain atoms of the loop regions and 5′ overhanging stretch are most flexible (Figure S10A). In the presence of TPP, the flexibility of the 5′ overhanging bases is reduced as they are arrested in capped conformation.
Binding Free Energy Estimation
As can be seen in Table 2, the binding of all three ligands is energetically favored as the binding free energy estimated using the MMPBSA and MMGBSA methods is in the negative range. Both electrostatic energies (Eele) and van der Waal’s energies (EvdW) are contributing favorably in the binding of all three ligands; the same was found to be evident in the docked conformations as well as in the simulated conformations. However, the electrostatic energy components (Eele) are counterbalanced by the polar solvation energy components (EPB), and the overall electrostatic interactions impart repulsive forces. Thus, van der Waal’s energies EvdW are found to be deterministic in the binding free energies of the respective ligands. Comparative analysis suggests that TPP and RJC bindings are more favored compared to that of AWO. The energy components leading to binding of TPP are in close correlation with those of chelerythrine (naturally occurring plant alkaloid with anticancer activity) and 12-methoxy chelerythrine, which are reported in a previous study.43 Thus, we can state that, like chelerythrine, TPP can also arrest the G-quadruplex structure formed in Pu27 and it can inhibit the expression of c-MYC oncogene.
Table 2. MMPB(GB)SA Calculations of Binding Free Energy Components of Ligand TPP, RJC, and AWO When Bound to Pu27.
| energy (kcal/mol)a | TPP | RJC | AWO |
|---|---|---|---|
| EvdWb | –36.82 (0.59) | –30.56 (0.44) | –22.84 (0.60) |
| Eelec | –634.58 (3.4) | –1230.41 (3.88) | –1100.84 (5.27) |
| EPBd | 652.70 (3.08) | 1234.24 (3.99) | 1107.36 (5.49) |
| EGBe | 649.30 (3.24) | 1236.06 (3.93) | 1109.49 (5.47) |
| Esurff | –3.66 (0.05) | –2.36 (0.03) | –2.61 (0.05) |
| Ecavityg | –2.62 (0.04) | –2.06 (0.03) | –1.81 (0.05) |
| ΔGgash | –671.40 (3.71) | –1260.98 (4.12) | –1123.68 (5.56) |
| ΔGSolvPBi | 650.08 (2.99) | 1232.18 (3.96) | 1105.55 (5.45) |
| ΔGSolvGB | 645.64 (3.20) | 1233.70 (3.92) | 1106.88 (5.43) |
| ΔGBindingEnergyPBj | –21.32 (1.11) | –28.79 (0.47) | –18.13 (0.59) |
| ΔGBindingEnergyGB | –25.76 (0.71) | –27.28 (0.47) | –16.80 (0.56) |
The energies given are approximate values as predicted by the software and not measured experimentally.
EvdW: nonbonded van der Waals energy.
Eele: nonbonded electrostatic energy.
EPB: polar solvation energy (PB).
EGB: polar solvation energy (GB).
Esurf: nonpolar solvation energy (GB).
Ecavity: nonpolar solvation energy (PB).
ΔGgas: EvdW + Eele + internal energy.
ΔGsolv: polar solvation energy + nonpolar solvation energy.
ΔGbinding energy: binding free energy (ΔGgas + ΔGsolv).
In summary, analysis of the simulated systems suggests that binding of TPP with Pu27 is energetically favorable and well supported by hydrogen bond interactions as well as polar interactions, which impart stabilization over the secondary structure of quadruplex. TPP shows druglike properties, and further, it possesses pharmacophoric features such as a central positive charge (piperidinium motif), moderately flexible skeleton, aromatic ring to form π–π interactions, and polarity imparted by the thiadiazole motif, which assist in the binding of TPP with Pu27. TPP was explored further using an experimental approach to evaluate the proposition that TPP can interact with Pu27 and that it may lead to downregulation of c-MYC expression.
CD
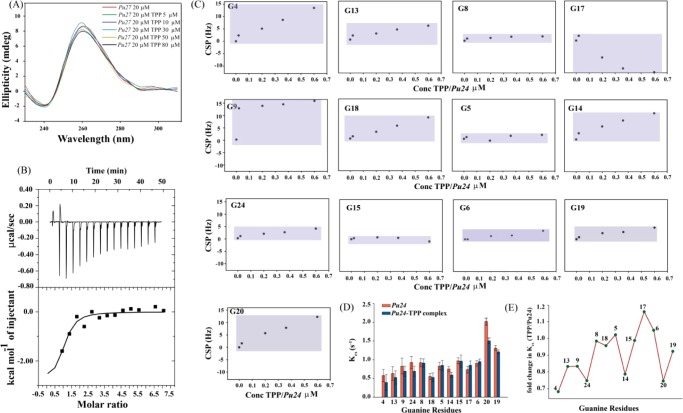
CD spectroscopy provides significant information regarding various topologies, the effect of metal ions, and the thermal stability of G-quadruplex structures. Parallel G-quadruplex structures display a positive peak near 260 nm and a negative peak around 240 nm. CD spectra with a positive peak around 290 nm and a negative minima near 265 nm represent antiparallel topology. A hybrid G-quadruplex structure exhibits a positive peak near 290 nm and a positive shoulder around 270 nm.80,81 The CD spectra of Pu27 exhibits a positive peak at 261 nm with a negative minima at 240 nm, which is indicative of a parallel G-quadruplex structure (Figure 4A). There occurs a slight decrease in elipticity in the CD spectra of Pu27 when titrated with increasing concentrations of TPP. However, the overall topology of Pu27 remains the same after addition of TPP (Figure 4A). This slight alteration in elipticity may arise due to changes in the local environment upon complex formation, such as the development of new interactions between the receptor (Pu27) and ligand (TPP) molecules, and/or changes in the dynamicity of the loop bases. To explore whether TPP imparts any structural stabilization upon binding with Pu27, we carried out thermal melting experiments. The thermal melting profiles of Pu27 and the Pu27–TPP complex reveal that TPP, upon interacting with Pu27, imparts moderate stabilization (ΔTm = ∼3 °C) (Figure S11A,B). Pu27 itself forms a very stable G-quadruplex in the presence of K+ ions (Tm = 77.8 °C), which might be the reason behind the minute increase in Tm upon addition of TPP. To avoid the influence of ions and to establish the major stabilizing effect by TPP on Pu27, melting of the complex was carried out in water (K+ < 1.5 mM). We found a rise in Tm of ∼5 °C in the Pu27–TPP complex compared to that of the unbound-Pu27 in water (Figure S11C). Thus, TPP, when added in a 1:3 ratio (Pu27/TPP), stabilizes the secondary structure of Pu27.
Figure 4.
(A) CD titrations of Pu27 with increasing concentration of TPP. (B) ITC profile generated from the titration of Pu27 with TPP. The upper panel represents the isothermal plot of the Pu27–TPP complex formation, whereas the bottom panel displays the integrated binding isotherm generated from the integration of peak area as a function of molar ratio. The solid line represents the best fit data using the “one-site binding model”. (C) Chemical shift perturbations of imino regions of Pu24 when titrated with increasing concentration of TPP. (D) Rate of hydrogen exchange of imino protons (Kex) of unbound-Pu24 and Pu24–TPP complex. (E) Fold change in Kex (TPP/Pu24).
ITC
The ITC experiment was conducted to determine the thermodynamic parameters behind the complexation of TPP with Pu27. It delivers information concerning the molecular forces, energetics of binding, and most importantly the number of binding sites of the macromolecule (here Pu27) engaged in complex formation with the ligand (here TPP). The thermodynamic parameters for the association of Pu27 with TPP obtained through ITC are listed in Table 3. The thermogram for the interaction is shown in (Figure 4B), portraying a one-site binding event. The Gibbs free energy change is negative (ΔG = −7.5 kcal mol–1) inferring the binding interaction of TPP with Pu27 is energetically favorable. The negative free energy change is attributed to a negative enthalpy change and a positive change in entropy. Stacking interactions of the planar molecule onto the bases of G-quadruplex and electrostatic interactions are majorly responsible for the negative enthalpy. The positive change in entropy of the Pu27–TPP complex may arise from two factors: (i) the entropy of the system may increase if binding of TPP results in release of the solvent molecules or (ii) due to conformational changes in Pu27 upon TPP binding. The negative enthalpy factor and positive entropy factor correlate well with those of the molecular simulation data. The presence of electrostatic interactions and H–π interactions, as obtained from hydrogen bond analysis, may contribute favorably toward the negative enthalpy change of the Pu27–TPP complex. B-factor analysis reveals that the flexibility of Loop-2 and Loop-3 of Pu27 increases upon binding of TPP, thus increasing the conformational entropy of the system (Figure 3E,F). The second factor behind the entropy term may be attributed to the perturbation of the water structure upon binding of TPP with Pu27, as observed in the hydrogen bond analysis (Figure S2B) and water shell analysis (Figure S2F). The binding affinity (Ka) for the Pu27–TPP complex is 3.25 × 105 M–1. The binding affinity obtained is comparable to previously reported literature.14 Small molecules like sanguinarine,82 chelerythrine,26 diarylethynyl amides,83 arene ruthenium(II) complexes,84 aryl indenopyrimidine derivatives,85 and so forth, have similar binding affinity toward c-MYC G-quadruplex structures. The molecule TPP possesses a low Kd (high Ka) value but causes a modest increase in ΔTm upon binding with Pu27. This implies that although the ligand has a moderate effect over the stabilization of the secondary structure, it has significant binding affinity. Such ligands can interfere with the expression of c-MYC, as previously reported by Kang et al.23 Waller et al. suggested that such ligands may be better suited for interfering with protein–quadruplex recognition thus disrupting the assembly of the respective transcription factors.86 The 1:1 binding stoichiometry of TPP with Pu27 found here is in accordance with the earlier reported interaction patterns of small molecules with G-quadruplex DNA. Recently, Bončina et al. studied the thermodynamics of various 1:1 complexes of small molecules interacting with human telomeric G-quadruplexes.87 The Ru(II) complex acts as a groove binder to the c-myc G4 DNA forming 1:1 complex.84 The thermodynamic feasibility (negative value of ΔG) of Pu27–TPP complex formation is also reflected in the negative MMPBSA and MMGBSA values depicted in Table 2.
Table 3. Thermodynamic Parameters Derived from ITC Experiment of Pu27–TPP Complex.
| parameters | Pu27–TPP complex |
|---|---|
| Ka (M–1) | 3.52 × 105 |
| binding stoichiometry (n) | 0.949 |
| ΔH (kcal mol–1) | –2.9 |
| TΔS (kcal mol–1) | 4.6 |
| ΔG (kcal mol–1) | –7.5 |
NMR Titrations
NMR titrations were performed to explicate the interactions of TPP with Pu24 at the atomic level. As Pu27 forms multiple conformations in K+ solution, a truncated 24 mer sequence was used as it gives well-resolved spectra between 10 and 12 ppm.25 The numbering of the Pu24 sequence was done according to the previously reported NMR spectra; to maintain the unambiguity in NMR assigned residue numbers and residue numbers used in simulation, we put the respective numbering of nucleotide bases corresponding to Pu27 in parenthesis. Although the first three bases from the 5′-terminal are absent in the truncated version of Pu27, the bases involved in ligand binding are intact. With increasing concentration of TPP (0.6:1.0, TPP/Pu24), an upfield chemical shift is observed for most of the imino protons but substantial line broadening of most of the imino protons is absent (Figure S12). G17 (G20-Pu27) displays a strong downfield chemical shift (−12.75 Hz) (Figure 4C). The chemical shift perturbation of G4 (G7-Pu27) imino proton is strong (13.47 Hz) but the peak is not broadened upon addition of TPP (Figures 4C and S12). Imino protons of G17 (G20-Pu27) and G9 (G12-Pu27) show a strong chemical shift perturbation along with broadening of the respective peaks (Figures 4C and S12). Moderate perturbations in chemical shift are observed for G13 (G16-Pu27), G18 (G21-Pu27), and G14 (G17-Pu27) without any line broadening (Figures 4C and S12). The maximum perturbations in chemical shift are observed for the 5′ end guanine tetrad (Pu24-G4-G8-G13-G17, Pu27-G7-G11-G16-G20), followed by the middle tetrad (Pu24-G5-G9-G14-G18, Pu27-G8-G12-G17-G21) (Figure 4C). The 3′ end guanine tetrad (Pu24-G6-G15-G19-G24, Pu27-G9-G18-G22-G27) is least perturbed compared to the 5′ end guanine tetrad (Figure 4C). This pattern of perturbation implies that TPP is interacting with Pu24 without forming multiple conformations and the 5′ end guanine tetrad is probably offering a binding site. This is also depicted in the molecular dynamics simulations. B-factor analysis reveals that G4 (G7-Pu27), G13 (G16-Pu27), and G17 (G20-Pu27) of Pu24 are highly stabilized upon association with TPP (Figure 3B). The high chemical shift perturbation of G9 (G12-Pu27) may arise due to flexibility of the Loop-2 and π–π interactions between its two consecutive adjacent bases, G10 (G13-Pu27) and G11 (G14-Pu27), refer to video (tpp-pu27.avi). Interestingly, G20 (G23-Pu27) of the GAG triad, which remains intact over the entire simulation run, shows high chemical shift perturbation of its imino proton in NMR titrations. In visual observations of the simulated trajectory, we found that A24 (A21-Pu24) forms steady π–π stacking interactions with G23 (G20-Pu24) in the Pu27–TPP complex, whereas in unbound-Pu27 A24 (A21-Pu24) is flexible. This enforced structural rigidity over the GAG triad in the presence of TPP might be the reason behind the observed high chemical shift perturbation of G20 (G23-Pu27). An array of one-dimensional 31P titrations of Pu24 with TPP was performed to verify the association of TPP with the phosphate backbone of Pu24 (Figure S13); a similar approach was followed in previous reports.88 Substantial line broadening with a drop in intensity in one-dimensional 31P spectra of Pu24 was observed upon addition of TPP in the molar ratio 0.6:1 (TPP/Pu24), suggesting interaction of TPP with the phosphate backbone of Pu24. These results are in correlation with the interactions observed in simulations where TPP is forming polar interactions with the phosphate backbone of A15 (A12-Pu24) of Pu27 (Figure S2).
Rate of Hydrogen Exchange (Kex)
To understand the base pair stability, as per previous applications,68,89 we calculated the rate of hydrogen exchange of the imino protons (Kex) of the unbound-Pu24 and Pu24–TPP complex. Figure 4D depicts the Kex value of the unbound-Pu24 and Pu24–TPP complex; the rate of exchange of most of the imino protons decreases when TPP is added to Pu24. The calculated fold change (Figure 4E) reveals that the imino proton of G4 (G7-Pu27) is mostly affected, followed by G24 (G27-Pu27), G14 (G17-Pu27), and G20 (G23-Pu27). The G13 (G16-Pu27), G9 (G12-Pu27), and G17 (G20-Pu27) guanine protons show a moderate exchange rate, whereas the rest of the imino protons are least affected upon addition of TPP. The decrease in Kex value of the Pu24–TPP complex compared to that of the unbound-Pu24 shows that the dynamicity of Pu24 is decreasing after complex formation with TPP. Comparative analysis of the Kex value for the individual G-quartet reveals that the 5′ end is mostly affected, followed by the second quartet (Figure 4D,E) and the 3′ end quartet is least affected (Figure 4D,E). It has been previously reported by Lee et al. that regions that are protected from solvent display the slowest exchange rate compared to those of solvent exposed sites.90 Both the rate of exchange and one-dimensional 1H titration correlate well with the molecular dynamics simulations, where the 5′ end tetrad guanines are stabilized when bound with TPP and the 5′ end tetrad guanines are buried by the 5′ end overhanging bases thus reducing solvent exposure to a certain extent. ITC data also reveal that the entropy of the system increases after association of TPP with Pu24; as an increase in entropy may also be attributed to perturbation of solvent molecules associated with Pu24, thus the perturbed solvent structure might be the factor behind the decrease in rate of hydrogen exchange in the Pu24–TPP complex.
The increased compactness of Pu27 after its association with TPP, the 1:1 binding stoichiometry, and the 5′-end of Pu27 being the preferred region of binding are in good agreement with molecular simulation data. Thus, the biophysical techniques implemented in this study support the observations as obtained from molecular dynamics simulations. After analyzing the interaction profile of TPP with Pu27 through simulated conditions and various biophysical experiments, the molecule was proposed for ex vivo study to investigate its potential in arresting intracellular c-MYC expression.
MTT Assay
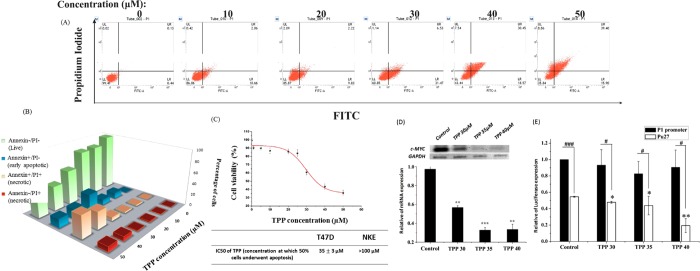
The T47D cell line was taken as an in cellulo model to investigate the effect of TPP on the expression of c-MYC, as earlier studies reported that the T47D cell line displays elevated levels of endogenous c-MYC.91 An MTT assay was carried out in T47D (human breast ductal carcinoma cell line) and NKE (normal kidney epithelial cell line) cells to examine the cytotoxic effect and anticancer property of the molecule TPP. The results depict that TPP significantly and selectively reduces cell viability in T47D cells (19.45% live cells at 50 μM), whereas the NKE cells remain mostly unaffected (74.36% live cells at 50 μM) (Figure S14). This observation underlies the selective anticancer activity of TPP.
Flow Cytometry
In the MTT assay, the depletion of cell viability does not dictate if the cancer cells undergo apoptosis or necrosis. Therefore, we performed flow cytometry-based Annexin V-FITC and propidium iodide (PI) staining experiments for the evaluation of apoptosis and cell viability in T47D and NKE cells treated with TPP at increasing concentrations for 24 h.
The percentages of live, early and late apoptotic, and necrotic or dead populations were quantitatively determined. Annexin V-FITC tags those cells that are en route to early apoptotic events exposing the phosphatidylserine residues to the outer leaflet of the lipid bilayer. PI binds to the nucleic acid and stains the cells that have lost their membrane integrity. In brief, the cells tagged with Annexin V-FITC/PI are classified as follows: dead or necrotic cells (the upper left quadrant; Annexin–/PI+ and the upper right quadrant; Annexin+/PI+), early apoptotic cells (the lower right quadrant; Annexin+/PI–), and live cells (the lower left quadrant; Annexin–/PI–).92,93
The results demonstrate that TPP selectively induces apoptosis in cancer cells, whereas the normal NKE cells were appreciably unaffected (Figures 5A and S17). An escalation in the concentration above 20 μM culminates in a sharp decline in the cell viability with a concomitant increase in the apoptotic population (early apoptosis + late apoptosis) in the T47D cell line. A gradual hike in the early apoptotic and necrotic population is pronounced in the range of 20–50 μM TPP (early apoptotic population ∼10, ∼31, ∼18, ∼16%, necrotic population ∼4, ∼8, ∼38, ∼48% at 20, 30, 40, and 50 μM, respectively) (Figure 5B). The IC50 value (concentration at which 50% of cells are alive) was calculated to be around 35 μM in cancer cells whilst in normal cells around 99% of cells remain alive in the concentration range of 25–85 μM (Figure 5C). However, 50% cell death is not achieved at 150 μM concentration of TPP in NKE cells even unattained at 150 μM concentration of TPP in normal cells. Above 100 μM, a gradual increase in the necrotic population is evident in the case of normal cells (∼6, ∼10, ∼6, ∼16, and ∼28% at 100, 120, 130, 140, and 150 μM, respectively), which suggests that TPP selectively promotes apoptosis in T47D cancer cells (higher c-MYC levels) over normal cells. Therefore, TPP could be a promising lead to show a safe therapeutic index and exert anticancer activity.
Figure 5.
TPP exhibits anticancer properties and abrogates c-MYC transcription in T47D cancer cells. (A) TPP selectively induces apoptosis in cancer cells: FACS dot plots showing apoptosis induction in T47D breast cancer cell line upon treatment with TPP at increasing concentrations up to 50 μM. A DMSO negative control and Etoposide-treated positive control are considered (data included in Figure S11). Normal kidney epithelial cells were exemplified from apoptosis upon TPP treatment up to 100 μM (data included in Figure S12). (B) Percentages of live, early apoptotic, and necrotic cell population are represented in the bar plot. (C) IC50 value of TPP in apoptosis induction in T47D cells was determined to be 35 ± 3 μM. (D) Depleted c-MYC expression profiles at mRNA level are quantified by semiquantitative RT-PCR in TPP treated T47D breast cancer cell line. RT-PCR is performed with specific primers for c-MYCon total RNA isolated from T47D breast cancer cell line under the following conditions: control, 30, 35, and 40 μM TPP treated. In the RT-PCR analysis, the target genes are normalized to the average of internal control gene GAPDH and represented in arbitrary units. (E) Luciferase assay is performed with reporter plasmids containing the P1–P2 promoter with or without Pu27 motif of c-MYC gene in untreated T47D cells and in T47D cells treated with TPP for 24 h. Error bars in bar plot (D, E) represent mean ± s.d. from three independent experiments in triplicate. Asterisks (*) indicated statistical significance as determined from Student’s t-test (* indicates P < 0.05, ** indicates P < 0.01, *** indicates P < 0.001). In the RT-PCR bar plot, asterisks (*) denote the statistical difference in mRNA profiles of c-MYC in TPP treated cells in comparison with the control. In the Luciferase assay, asterisks (*) denote a significant difference compared with the values for untreated cells transfected with the reporter plasmid containing the Pu27 motif. # denotes a significant difference in promoter activity compared with values for cells transfected with the reporter plasmid containing P1–P2 promoter (# P < 0.05, ### P < 0.001).
RT-PCR Study
After examining its anticancer property, we proceeded to study the effect of TPP in c-MYC transcription in T47D cells. We reasoned that if the molecule binds to the G-quadruplex structure developed in the upstream promoter region of the human c-MYC gene, the endogenous expression of c-MYC mRNA will be negatively regulated.94 In this study, T47D cells were treated with TPP in a concentration-dependent manner and the total RNA, extracted from the treated cells, was processed to poly-(A) specific reverse Transcription (RT) to produce cDNA. This cDNA served as the template for specific PCR amplification of the c-MYC gene and the expression level of GAPDH gene was used to normalize the data obtained from different samples. The semiquantitative densitometric analyses demonstrate a notable downregulation in c-MYC expression profiles at the mRNA level with increasing concentrations of TPP. 30 μM TPP treatment renders significant repression (58.28% relative to the control) in c-MYC mRNA expression. An increase in TPP concentration up to 35 μM results in almost a 3-fold repression (66.36% relative to the control) of the intracellular abundance of c-MYC mRNA (Figure 5E). However, further increments in the dose up to 40 μM do not alter the mRNA expression (65.58% relative to the control) compared to that of the 35 μM TPP treated set, which indicates that 35 μM exerts a saturating impact for the abrogation of c-MYC transcription.
Reporter Luciferase Assay
As c-MYC lies at the nexus of innumerable crosstalks and its expression is highly orchestrated by a number of signaling cues, we could not comment from the RT-PCR experiment on whether TPP interferes with the quadruplex in NHE III1 element to repress c-MYC transcription. Therefore, to shed light on TPP’s underlying mechanism of transcription abrogation, we attempted the reporter luciferase assay to verify the potential of TPP in targeting the quadruplex forming motif in the NHE III1 element.24 We transfected the T47D cells with two luciferase vectors: (a) one having the major promoters (P1–P2) only and devoid of the quadruplex forming motif in the NHE III1 (Pu27) and (b) the other comprising of both NHE III1 (Pu27) and P1–P2 promoters. Then, the promoter activities, in the presence of both the vectors, were monitored under different concentrations of TPP. We found that the promoter activity is akin to the control set in the absence of the Pu27 region in the upstream promoter (P1–P2) of c-MYC. This suggests that TPP does not promote any significant alteration in the promoter activity in the absence of the quadruplex forming motif. But, transfection of NHE III1 (+) P1–P2 luciferase vector translates into significant depletion of c-MYC promoter activity with increasing concentrations of TPP. At 40 μM TPP, almost 60% repression in promoter activity is accomplished compared to that of the untreated one (Figure 5F). Therefore, the integration of RT-PCR and reporter assays gives an insight that TPP has no function in c-MYC transcription regulation in the absence of NHE III1, but in the presence of the quadruplex forming motifs it interacts with the quadruplex structure, which in turn probably inhibits the recruitment of RNA polymerase or other transcription factors in the proximal promoter region, rendering transcription repression.
Conclusions
Overexpression of c-MYC is observed in different cancerous conditions; the quadruplex forming motif in NHE III1 found in the vicinity of P1 promoter of c-MYC oncogene significantly represses its transcription and therefore provides a promising therapeutic target in anticancer treatment. Here, we have implemented small molecule induced stabilization of G-quadruplex to interfere with the expression of c-MYC.
A pool of small molecules was screened based on their druglike properties using a virtual screening method and further target specific molecules were mined out using molecular docking. Atomistic binding characterization was investigated theoretically through all atom molecular dynamics simulations with the top three molecules, named “TPP, RJC, and AWO”. Structural analysis of the simulated trajectories inferred the superiority of a small molecule “TPP” over the other two ligands studied (AWO and RJC). The biophysical observations correlate well with the simulation results inferring 1:1 binding stoichiometry with a Kd value in the sub-micro-molar range and ΔG in the negative range. High-resolution NMR spectroscopy confirmed the atomistic details found in simulation inferring the 5′ end guanine stack as a binding site and stabilization of the overall secondary structure with a rigid conformation of TPP. MTT assay and flow cytometry analysis signify the selective cytotoxicity and anti-proliferative effect of TPP on the T47D cancer cells over that of the normal NKE cells. RT-PCR confirms the downregulation c-MYC expression in T47D cancer cells upon treatment with TPP at micromolar concentration. The reporter luciferase assay further validates the proposed mechanism of action of TPP, that is, TPP is arresting the quadruplex forming motif Pu27 and further interfering with the transcription process.
The combined effort of rational drug design with in silico, biophysical, and biological approaches has offered a promising lead molecule “TPP”, which can be explored further as a new generation anticancer agent.
Acknowledgments
S.C. is thankful to Department of Science and technology for Ramanujan Fellowship (SR/S2/RNJ-78/2010 (G), J.B. is thankful to DBT for the fellowship from the project (BT/PR6627/GBD/27/440/2012). S.M. is thankful to UGC Government of India, and P.S. is thankful to CSIR Government of India. We are thankful to Prof. Gaurisankar Sa for providing us with the Cell culture facility at the Division of Molecular Medicine, Bose Institute. Thanks to Mr Souvik Roy, DBT IPLS, CU for helping in the ITC experiment.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsomega.6b00531.
Binding conformation of ligands at the 5′ end of Pu27; ADME properties predicted by QikProp and Docking Score selected molecules after virtual screening; hydrogen bond analysis; schematic representation of topological arrangement of Pu27 and intramolecular hydrogen bonding interactions in quadruplex region; water grid density map and Ion grid of Pu27 in different complexation state; “cumulative % contribution of each eigenvector” derived from PCA analysis; porcupine plots of unbound-Pu27; porcupine plots of the Pu27–TPP complex; porcupine plots of the Pu27–RJC complex; porcupine plots of Pu27–AWO complex; Lindemann’s coefficient of unbound-Pu27, Pu27–TPP complex, Pu27–RJC complex, and Pu27–AWO complex; CD Melting experiments of Pu27 and Pu27–TPP complex; one-dimensional proton NMR spectroscopy Pu24 and Pu24–TPP complex; change in one-dimensional 31P NMR spectra of Pu24 and Pu24–TPP complex; MTT assay in T47D and NKE cell line; positive and negative controls of flow cytometric analysis; gate of flow cytometry; and flow cytometric analysis in NKE cell line upon TPP treatment pu27.avi: trajectory of unbound-Pu27, tpp-pu27.avi: trajectory of Pu27-TPP complex, rjc-pu27.avi: trajectory of Pu27-RJC complex,AWO-PU27.avi: trajectory of Pu27-AWO complex (PDF)
Author Contributions
† J.B. and S.M. have contributed equally.
The authors declare no competing financial interest.
Supplementary Material
References
- Zugazagoitia J.; Guedes C.; Ponce S.; Ferrer I.; Molina-Pinelo S.; Paz-Ares L. Current challenges in cancer treatment. Clin. Ther. 2016, 38, 1551–1566. 10.1016/j.clinthera.2016.03.026. [DOI] [PubMed] [Google Scholar]
- Sawyers C. Targeted cancer therapy. Nature 2004, 432, 294–297. 10.1038/nature03095. [DOI] [PubMed] [Google Scholar]
- Neves H.; Kwok H. F. Recent advances in the field of anti-cancer immunotherapy. BBA Clin. 2015, 3, 280–288. 10.1016/j.bbacli.2015.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoelder S.; Clarke P. A.; Workman P. Discovery of small molecule cancer drugs: Successes, challenges and opportunities. Mol. Oncol. 2012, 6, 155–176. 10.1016/j.molonc.2012.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cifuentes T.; Cayupi J.; Celis-Barros C.; Zapata-Torres G.; Ballesteros R.; Ballesteros-Garrido R.; Abarca B.; Jullian C. Spectroscopic studies of the interaction of 3-(2-thienyl)-[1,2,3]triazolo[1,5-a]pyridine with 2,6-dimethyl-β-cyclodextrin and ctDNA. Org. Biomol. Chem. 2016, 14, 9760–9767. 10.1039/C6OB01573E. [DOI] [PubMed] [Google Scholar]
- Verdine G. L.; Walensky L. D. The challenge of drugging undruggable targets in cancer: lessons learned from targeting BCL-2 family members. Clin. Cancer Res. 2007, 13, 7264–7270. 10.1158/1078-0432.CCR-07-2184. [DOI] [PubMed] [Google Scholar]
- Chen B.-J.; Wu Y.-L.; Tanaka Y.; Zhang W. Small molecules targeting c-Myc oncogene: promising anti-cancer therapeutics. Int. J. Biol. Sci. 2014, 10, 1084–96. 10.7150/ijbs.10190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer C. A.; Groudine M. Control of c-myc regulation in normal and neoplastic cells. Adv. Cancer Res. 1991, 56, 1–48. 10.1016/S0065-230X(08)60476-5. [DOI] [PubMed] [Google Scholar]
- Miller D. M.; Thomas S. D.; Islam A.; Muench D.; Sedoris K. c-Myc and cancer metabolism. Clin. Cancer Res. 2012, 18, 5546–5553. 10.1158/1078-0432.CCR-12-0977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelengaris S.; Rudolph B.; Littlewood T. Action of Myc in vivo proliferation and apoptosis. Curr. Opin. Genet. Dev. 2000, 10, 100–105. 10.1016/S0959-437X(99)00046-5. [DOI] [PubMed] [Google Scholar]
- Marcu K. B.; Bossone S. A.; Patel A. J. Myc function and regulation. Annu. Rev. Biochem. 1992, 61, 809–858. 10.1146/annurev.bi.61.070192.004113. [DOI] [PubMed] [Google Scholar]
- Facchini L. M.; Penn L. Z. The molecular role of Myc in growth and transformation: recent discoveries lead to new insights. FASEB J. 1998, 12, 633–651. [PubMed] [Google Scholar]
- Shan C.; Lin J.; Hou J.-Q.; Liu H.-Y.; Chen S.-B.; Chen A.-C.; Ou T.-M.; Tan J.-H.; Li D.; Gu L.-Q.; Huang Z.-S. Chemical intervention of the NM23-H2 transcriptional programme on c-MYC via a novel small molecule. Nucleic Acids Res. 2015, 43, 6677–6691. 10.1093/nar/gkv641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagano B.; Mattia C. A.; Giancola C. Applications of isothermal titration calorimetry in biophysical studies of G-quadruplexes. Int. J. Mol. Sci. 2009, 10, 2935–57. 10.3390/ijms10072935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salgado G. F.; Cazenave C.; Kerkour A.; Mergny J.-L. G-quadruplex DNA and ligand interaction in living cells using NMR spectroscopy. Chem. Sci. 2015, 6, 3314–3320. 10.1039/C4SC03853C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H. Z.; Ma V. P.; Leung K. H.; Chan D. S.; Yang H.; Cheng Z.; Leung C. H.; Ma D. L. A label-free G-quadruplex-based switch-on fluorescence assay for the selective detection of ATP. Analyst 2012, 137, 1538–40. 10.1039/c2an15999f. [DOI] [PubMed] [Google Scholar]
- Cooney M.; Czernuszewicz G.; Postel E. H.; Flint S. J.; Hogan M. E. Site-specific oligonucleotide binding represses transcription of the human c-myc gene in vitro. Science 1988, 241, 456. 10.1126/science.3293213. [DOI] [PubMed] [Google Scholar]
- Simonsson T.; Pribylova M.; Vorlickova M. A nuclease hypersensitive element in the human c-myc promoter adopts several distinct i-tetraplex structures. Biochem. Biophys. Res. Commun. 2000, 278, 158–166. 10.1006/bbrc.2000.3783. [DOI] [PubMed] [Google Scholar]
- Simonsson T.; Kubista M.; Pecinka P. DNA tetraplex formation in the control region of c-myc. Nucleic Acids Res. 1998, 26, 1167–1172. 10.1093/nar/26.5.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomonaga T.; Levens D. Activating transcription from single stranded DNA. Proc. Natl. Acad. Sci. U.S.A. 1996, 93, 5830–5835. 10.1073/pnas.93.12.5830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelotti E. F.; Tomonaga T.; Krutzsch H.; Levens D. Cellular nucleic acid binding protein regulates the CT element of the human c-myc protooncogene. J. Biol. Chem. 1995, 270, 9494–9499. 10.1074/jbc.270.16.9494. [DOI] [PubMed] [Google Scholar]
- Postel E. H.; Berberich S. J.; Rooney J. W.; Kaetzel D. M. Human NM23/nucleoside diphosphate kinase regulates gene expression through DNA binding to nuclease-hypersensitive transcriptional elements. J. Bioenerg. Biomembr. 2000, 32, 277–284. 10.1023/A:1005541114029. [DOI] [PubMed] [Google Scholar]
- Kang H.-J.; Park H.-J. In silico identification of novel ligands for G-quadruplex in the c-MYC promoter. J. Comput.-Aided Mol. Des. 2015, 29, 339–348. 10.1007/s10822-014-9826-z. [DOI] [PubMed] [Google Scholar]
- Siddiqui-Jain A.; Grand C. L.; Bearss D. J.; Hurley L. H. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl. Acad. Sci. U.S.A. 2002, 99, 11593–11598. 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan A. T.; Kuryavyi V.; Gaw H. Y.; Patel D. J. Small-molecule interaction with a five-guanine-tract G-quadruplex structure from the human MYC promoter. Nat. Chem. Biol. 2005, 1, 167–173. 10.1038/nchembio723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S.; Dasgupta D. Quadruplex forming promoter region of c-myc oncogene as a potential target for a telomerase inhibitory plant alkaloid, chelerythrine. Biochem. Biophys. Res. Commun. 2015, 459, 75–80. 10.1016/j.bbrc.2015.02.072. [DOI] [PubMed] [Google Scholar]
- Ou T.-M.; Lu Y.-J.; Zhang C.; Huang Z.-S.; Wang X.-D.; Tan J.-H.; Chen Y.; Ma D.-L.; Wong K.-Y.; Tang J. C.-O.; et al. Stabilization of G-quadruplex DNA and down-regulation of oncogene c-myc by quindoline derivatives. J. Med. Chem. 2007, 50, 1465–1474. 10.1021/jm0610088. [DOI] [PubMed] [Google Scholar]
- Rocca R.; Moraca F.; Costa G.; Alcaro S.; Distinto S.; Maccioni E.; Ortuso F.; Artese A.; Parrotta L. Structure-Based Virtual Screening of Novel Natural Alkaloid Derivatives as Potential Binders of h-telo and c-myc DNA G-Quadruplex Conformations. Molecules 2014, 20, 206–223. 10.3390/molecules20010206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji X.; Sun H.; Zhou H.; Xiang J.; Tang Y.; Zhao C. The interaction of telomeric DNA and C-myc22 G-quadruplex with 11 natural alkaloids. Nucleic Acid Ther. 2012, 22, 127–136. 10.1089/nat.2012.0342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown R. V.; Danford F. L.; Gokhale V.; Hurley L. H.; Brooks T. A. Demonstration that drug-targeted down-regulation of MYC in non-Hodgkins lymphoma is directly mediated through the promoter G-quadruplex. J. Biol. Chem. 2011, 286, 41018–41027. 10.1074/jbc.M111.274720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drygin D.; Siddiqui-Jain A.; O’Brien S.; Schwaebe M.; Lin A.; Bliesath J.; Ho C.; Proffitt C.; Trent K.; Whitten J.; Lim J.; Von Hoff D.; Andres K.; Rice W. Anticancer activity of CX-3543: a direct inhibitor of rRNA biogenesis. Cancer Res. 2009, 69, 7653–7661. 10.1158/0008-5472.CAN-09-1304. [DOI] [PubMed] [Google Scholar]
- Bidzinska J.; Graziella C.-R.; Zaffaroni N.; Marco F. G-Quadruplex Structure in the Human Genome as novel Therapeutic Targets. Molecules 2013, 18, 12368–12395. 10.3390/molecules181012368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangan A.; Fedoroff O. Y.; Hurley L. H. Induction of duplex to G-quadruplex transition in the c-myc promoter region by a small molecule. J. Biol. Chem. 2001, 276, 4640–4646. 10.1074/jbc.M005962200. [DOI] [PubMed] [Google Scholar]
- Seenisamy J.; Rezler E.; Powell T.; Tye D.; Gokhale V.; Sharma Joshi C.; Siddiqui-Jain A.; Hurley L. The Dynamic Character of the G-Quadruplex Element in the c-MYC Promoter and Modification by TMPyP4. J. Am. Chem. Soc. 2004, 126, 8702–8709. 10.1021/ja040022b. [DOI] [PubMed] [Google Scholar]
- Freyer M. W.; Buscaglia R.; Kaplan K.; Cashman D.; Hurley L.; Lewis E. Biophysical Studies of the c-MYC NHE III1 Promoter: Model Quadruplex Interactions with a Cationic Porphyrin. Biophys. J. 2007, 92, 2007–2015. 10.1529/biophysj.106.097246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao S.-R.; Zhou C.-X.; Wu W.-B.; Ou T.-M.; Tan J.-H.; Li D.; Gu L.-Q.; Huang Z.-S. 12-N-Methylated 5, 6-dihydrobenzo [c] acridine derivatives: A new class of highly selective ligands for c-myc G-quadruplex DNA. Eur. J. Med. Chem. 2012, 53, 52–63. 10.1016/j.ejmech.2012.03.034. [DOI] [PubMed] [Google Scholar]
- Ou T.-M.; Lin J.; Lu Y.-J.; Hou J.-Q.; Tan J.-H.; Chen S.-H.; Li Z.; Li Y.-P.; Li D.; Gu L.-Q.; Huang Z.-S. Inhibition of cell proliferation by quindoline derivative (SYUIQ-05) through its preferential interaction with c-myc promoter G-quadruplex. J. Med. Chem. 2011, 54, 5671–5679. 10.1021/jm200062u. [DOI] [PubMed] [Google Scholar]
- Chan D. S.; Yang H.; Kwan M. H.; Cheng Z.; Lee P.; Bai L. P.; Jiang Z. H.; Wong C. Y.; Fong W. F.; Leung C. H.; Ma D. L. Structure-based optimization of FDA-approved drug methylene blue as a c-myc G-quadruplex DNA stabilizer. Biochimie 2011, 93, 1055–64. 10.1016/j.biochi.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Nasiri H. R.; Bell N.; McLuckie K.; Husby J.; Abell C.; Neidle S.; Balasubramanian S. Targeting a c-MYC G-quadruplex DNA with a fragment library. Chem. Commun. 2014, 50, 1704–1707. 10.1039/c3cc48390h. [DOI] [PubMed] [Google Scholar]
- Huppert J.; Balasubramanian S. Prevelence of quadruplexes in the human genome. Nucleic Acids Res. 2005, 33, 2908–2916. 10.1093/nar/gki609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd A. K.; Johnston M.; Neidle S. Highly prevalent putative quadruplex sequence motifs in human DNA. Nucleic Acid Res. 2005, 33, 2901–2907. 10.1093/nar/gki553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H. M.; Chan D. S.; Yang F.; Lam H. Y.; Yan S. C.; Che C. M.; Ma D. L.; Leung C. H. Identification of natural product fonsecin B as a stabilizing ligand of c-myc G-quadruplex DNA by high-throughput virtual screening. Chem. Commun. 2010, 46, 4680–2. 10.1039/b926359d. [DOI] [PubMed] [Google Scholar]
- Bhat J.; Chatterjee S. Skeleton selectivity in complexation of chelerythrine and chelerythrine-like natural plant alkaloids with the G-quadruplex formed at the promoter of c-MYC oncogene: in silico exploration. RSC Adv. 2016, 6, 36667–36680. 10.1039/C6RA04671A. [DOI] [Google Scholar]
- a Schrödinger Suite 2013-3 Virtual Screening Workflow; Glide, version 6.1, Schrödinger, LLC: New York, NY, 2013;; b LigPrep, version 2.8, Schrödinger, LLC: New York, NY, 2013;; c QikProp, version 3.5, Schrödinger, LLC: New York, NY, 2013.
- Schrödinger Release 2013-3: Maestro, version 9.6; Schrödinger, LLC: New York, NY, 2013.
- Small-Molecule Drug Discovery Suite 2013-3: Glide, version 6.1; Schrödinger, LLC: New York, NY, 2013.
- Case D. A.; Darden T. A.; Cheatham T. E. III; Simmerling C. L.; Wang J.; Duke R. E.; Luo R.; Walker R. C.; Zhang W.; Merz K. M.. AMBER 11; University of California: San Francisco, CA, 2010. [Google Scholar]
- Wang J.; Wang W.; Kollman P. A.; Case D. A. Automatic atom type and bond type perception in molecular mechanical calculations. J. Mol. Graphics Modell. 2006, 25, 247–260. 10.1016/j.jmgm.2005.12.005. [DOI] [PubMed] [Google Scholar]
- Wang J.; Wolf R. M.; Caldwell J. W.; Kollman P. A.; Case D. A. Development and testing of a general amber force field. J. Comput. Chem. 2004, 25, 1157–1174. 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- Hornak V.; Abel R.; Okur A.; Strockbine B.; Roitberg A.; Simmerling C. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins: Struct., Funct., Bioinf. 2006, 65, 712–725. 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez A.; Marchán I.; Svozil D.; Sponer J.; Cheatham T. E.; Laughton C. A.; Orozco M. Refinement of the AMBER force field for nucleic acids: improving the description of α/Î3 conformers. Biophys. J. 2007, 92, 3817–3829. 10.1529/biophysj.106.097782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen W. L.; Chandrasekhar J.; Madura J. D.; Impey R. W.; Klein M. L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. 10.1063/1.445869. [DOI] [Google Scholar]
- Kaulage M.; Maji B.; Bhat J.; Iwasaki Y.; Chatterjee S.; Bhattacharya S.; Muniyappa K. Discovery and Structural Characterization of G-quadruplex DNA in Human Acetyl-CoA Carboxylase Gene Promoters: Its Role in Transcriptional Regulation and as a Therapeutic Target for Human Disease. J. Med. Chem. 2016, 59, 5035. 10.1021/acs.jmedchem.6b00453. [DOI] [PubMed] [Google Scholar]
- Loncharich R. J.; Brooks B. R.; Pastor R. W. Langevin dynamics of peptides: The frictional dependence of isomerization rates of N-acetylalanyl-N′-methylamide. Biopolymers 1992, 32, 523–535. 10.1002/bip.360320508. [DOI] [PubMed] [Google Scholar]
- Izaguirre J. A.; Catarello D. P.; Wozniak J. M.; Skeel R. D. Langevin stabilization of molecular dynamics. J. Chem. Phys. 2001, 114, 2090–2098. 10.1063/1.1332996. [DOI] [Google Scholar]
- Kräutler V.; van Gunsteren W. F.; Hünenberger P. H. A fast SHAKE algorithm to solve distance constraint equations for small molecules in molecular dynamics simulations. J. Comput. Chem. 2001, 22, 501–508. . [DOI] [Google Scholar]
- Darden T.; York D.; Pedersen L. Particle mesh Ewald: An N.log (N) method for Ewald sums in large systems. J. Chem. Phys. 1993, 98, 10089–10092. 10.1063/1.464397. [DOI] [Google Scholar]
- Roe D. R.; Cheatham T. E. III PTRAJ and CPPTRAJ: software for processing and analysis of molecular dynamics trajectory data. J. Chem. Theory Comput. 2013, 9, 3084–3095. 10.1021/ct400341p. [DOI] [PubMed] [Google Scholar]
- Humphrey W.; Dalke A.; Schulten K. VMD: visual molecular dynamics. J. Mol. Graphics 1996, 14, 33–38. 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- DeLano W. L.The PyMOL Molecular Graphics System; Schrödinger, LLC, 2002.
- Pettersen E. F.; Goddard T. D.; Huang C. C.; Couch G. S.; Greenblatt D. M.; Meng E. C.; Ferrin T. E. UCSF Chimera a visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Hou T.; Wang J.; Li Y.; Wang W. Assessing the performance of the molecular mechanics/Poisson Boltzmann surface area and molecular mechanics/generalized Born surface area methods. II. The accuracy of ranking poses generated from docking. J. Comput. Chem. 2011, 32, 866–877. 10.1002/jcc.21666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinde R. N.; Sobhia M. E. Binding and discerning interactions of PTP1B allosteric inhibitors: novel insights from molecular dynamics simulations. J. Mol. Graphics Modell. 2013, 45, 98–110. 10.1016/j.jmgm.2013.08.001. [DOI] [PubMed] [Google Scholar]
- Cheatham T. E. III; Srinivasan J.; Case D. A.; Kollman P. A. Molecular dynamics and continuum solvent studies of the stability of polyG-polyC and polyA-polyT DNA duplexes in solution. J. Biomol. Struct. Dyn. 1998, 16, 265–280. 10.1080/07391102.1998.10508245. [DOI] [PubMed] [Google Scholar]
- Kuhn B.; Kollman P. A. Binding of a diverse set of ligands to avidin and streptavidin: an accurate quantitative prediction of their relative affinities by a combination of molecular mechanics and continuum solvent models. J. Med. Chem. 2000, 43, 3786–3791. 10.1021/jm000241h. [DOI] [PubMed] [Google Scholar]
- Honig B.; Nicholls A. Classical electrostatics in biology and chemistry. Science 1995, 268, 1144–1149. 10.1126/science.7761829. [DOI] [PubMed] [Google Scholar]
- Onufriev A.; Bashford D.; Case D. A. Exploring protein native states and large†scale conformational changes with a modified generalized born model. Proteins: Struct., Funct., Bioinf. 2004, 55, 383–394. 10.1002/prot.20033. [DOI] [PubMed] [Google Scholar]
- Ghosh A.; Kar R. K.; Krishnamoorthy J.; Chatterjee S.; Bhunia A. Double GC: GC Mismatch in dsDNA Enhances Local Dynamics Retaining the DNA Footprint: A High†Resolution NMR Study. ChemMedChem 2014, 9, 2059–2064. 10.1002/cmdc.201402238. [DOI] [PubMed] [Google Scholar]
- Jamil S.; Lam I.; Majd M.; Tsai S. H.; Duronio V. Etoposide induces cell death via mitochondrial-dependent actions of p53. Cancer Cell Int. 2015, 15, 79 10.1186/s12935-015-0231-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondal S.; Bhat J.; Jana J.; Mukherjee M.; Chatterjee S. Reverse Watson-Crick G-G base pair in G-quadruplex formation. Mol. Biosyst. 2016, 12, 18. 10.1039/c5mb00611b. [DOI] [PubMed] [Google Scholar]
- Haider S.; Parkinson G. N.; Neidle S. Molecular dynamics and principal components analysis of human telomeric quadruplex multimers. Biophys. J. 2008, 95, 296–311. 10.1529/biophysj.107.120501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamroz M.; Orozco M.; Kolinski A.; Kmiecik S. Consistent view of protein fluctuations from all-atom molecular dynamics and coarse-grained dynamics with knowledge-based force-field. J. Chem. Theory Comput. 2013, 9, 119–125. 10.1021/ct300854w. [DOI] [PubMed] [Google Scholar]
- Amadei A.; Linssen A.; Berendsen H. J. C. Essential dynamics of proteins. Proteins: Struct., Funct., Bioinf. 1993, 17, 412–425. 10.1002/prot.340170408. [DOI] [PubMed] [Google Scholar]
- Amadei A.; Linssen A. B. M.; De Groot B. L.; Van Aalten D. M. F.; Berendsen H. J. C. An efficient method for sampling the essential subspace of proteins. J. Biomol. Struct. Dyn. 1996, 13, 615–625. 10.1080/07391102.1996.10508874. [DOI] [PubMed] [Google Scholar]
- van Aalten D. M. F.; Findlay J. B. C.; Amadei A.; Berendsen H. J. C. Essential dynamics of the cellular retinol-binding protein evidence for ligand-induced conformational changes. Protein Eng., Des. Sel. 1995, 8, 1129–1135. 10.1093/protein/8.11.1129. [DOI] [PubMed] [Google Scholar]
- Yamaguchi H.; Van Aalten D. M. F.; Pinak M.; Furukawa A.; Osman R. Essential dynamics of DNA containing a cis. syn cyclobutane thymine dimer lesion. Nucleic Acids Res. 1998, 26, 1939–1946. 10.1093/nar/26.8.1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer T.; Ferrer-Costa C.; Perez A.; Rueda M.; Bidon-Chanal A.; Luque F. J.; Laughton C. A.; Orozco M. Essential dynamics: a tool for efficient trajectory compression and management. J. Chem. Theory Comput. 2006, 2, 251–258. 10.1021/ct050285b. [DOI] [PubMed] [Google Scholar]
- Damm K. L.; Carlson H. A. Gaussian-weighted RMSD superposition of proteins: a structural comparison for flexible proteins and predicted protein structures. Biophys. J. 2006, 90, 4558–4573. 10.1529/biophysj.105.066654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y.; Vitkup D.; Karplus M. Native proteins are surface-molten solids: application of the Lindemann criterion for the solid versus liquid state. J. Mol. Biol. 1999, 285, 1371–1375. 10.1006/jmbi.1998.2374. [DOI] [PubMed] [Google Scholar]
- Vorlíčková M.; Kejnovská I.; Sagi J.; Renčiuk D.; Bednářová K.; Motlová J.; Kypr J. Circular dichroism and guanine quadruplexes. Methods 2012, 57, 64–75. 10.1016/j.ymeth.2012.03.011. [DOI] [PubMed] [Google Scholar]
- Ambrus A.; Chen D.; Dai J.; Bialis T.; Jones R. A.; Yang D. Human telomeric sequence forms a hybrid-type intramolecular G-quadruplex structure with mixed parallel/antiparallel strands in potassium solution. Nucleic Acids Res. 2006, 34, 2723–2735. 10.1093/nar/gkl348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S.; Pradhan S. K.; Kar A.; Chowdhury S.; Dasgupta D. Molecular basis of recognition of quadruplexes human telomere and c-myc promoter by the putative anticancer agent sanguinarine. Biochim. Biophys. Acta, Gen. Subj. 2013, 1830, 4189–4201. 10.1016/j.bbagen.2013.03.027. [DOI] [PubMed] [Google Scholar]
- Dash J.; Shirude P. S.; Hsu S.-T. D.; Balasubramanian S. Diarylethynyl amides that recognize the parallel conformation of genomic promoter DNA G-quadruplexes. J. Am. Chem. Soc. 2008, 130, 15950–15956. 10.1021/ja8046552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q.; Chen T.; Zhang Z.; Liao S.; Wu X.; Wu J.; Mei W.; Chen Y.; Wu W.; Zeng L.; Zheng W. Microwave-assisted synthesis of arene ruthenium (ii) complexes [(η 6-RC 6 H 5) Ru (m-MOPIP) Cl] Cl (R = —H and–CH 3) as groove binder to c-myc G4 DNA. Dalton Trans. 2014, 43, 9216–9225. 10.1039/c3dt53635a. [DOI] [PubMed] [Google Scholar]
- Diveshkumar K. V.; Sakrikar S.; Harikrishna S.; Dhamodharan V.; Pradeepkumar P. I. Targeting Promoter G-Quadruplex DNAs by Indenopyrimidine-Based Ligands. ChemMedChem 2014, 9, 2754–2765. 10.1002/cmdc.201402394. [DOI] [PubMed] [Google Scholar]
- Waller Z. A. E.; Shirude P. S.; Rodriguez Rl; Balasubramanian S. Triarylpyridines: a versatile small molecule scaffold for G-quadruplex recognition. Chem. Commun. 2008, 12, 1467–1469. 10.1039/b718854d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonc̆ina M.; Podlipnik Č.; Piantanida I.; Eilmes J.; Teulade-Ficho M.-P.; Vesnaver G.; Lah J. Thermodynamic fingerprints of ligand binding to human telomeric G-quadruplexes. Nucleic Acids Res. 2015, 43, 10376–10386. 10.1093/nar/gkv1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S.; Jana J.; Kar R. K.; Chatterjee S.; Dasgupta D. Plant Alkaloid Chelerythrine Induced Aggregation of Human Telomere Sequence—A Unique Mode of Association between a Small Molecule and a Quadruplex. Biochemistry 2015, 54, 974–986. 10.1021/bi501117x. [DOI] [PubMed] [Google Scholar]
- Mondal S.; Jana J.; Sengupta P.; Jana S.; Chatterjee S. Myricetin arrests human telomeric G-quadruplex structure: a new mechanistic approach as an anticancer agent. Mol. Biosyst. 2016, 12, 2506. 10.1039/C6MB00218H. [DOI] [PubMed] [Google Scholar]
- Lee J.-H.; Jucker F.; Pardi A. Imino proton exchange rates imply an induced-fit binding mechanism for the VEGF165-targeting aptamer, Macugen. FEBS Lett. 2008, 582, 1835–1839. 10.1016/j.febslet.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H.; Radisky D. C.; Yang D.; Xu R.; Radisky E. S.; Bissell M. J.; Bishop J. M. MYC suppresses cancer metastasis by direct transcriptional silencing of [alpha] v and [beta] 3 integrin subunits. Nat. Cell Biol. 2012, 14, 567–574. 10.1038/ncb2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S.; Zhao Q.; Xiang H.; Liu M.; Zhang Q.; Xue W.; Song B.; Yang S. Antiproliferative activity and apoptosis-inducing mechanism of constituents from Toona sinensis on human cancer cells. Cancer Cell Int. 2013, 13, 12 10.1186/1475-2867-13-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings B. S.; Wills L. P.; Schnellmann R. G. Measurement of cell death in Mammalian cells. Current Protocols Pharmacol. 2012, 12.8, 24. 10.1002/0471141755.ph1208s56. [DOI] [PubMed] [Google Scholar]
- Chen W.-J.; Zhou C.-X.; Yao P.-F.; Wang X.-X.; Tan J.-H.; Li D.; Ou T.-M.; Gu L.-Q.; Huang Z.-S. Disubstituted 1, 8-dipyrazolcarbazole derivatives as a new type of C-myc G-quadruplex binding ligands. Bioorg. Med. Chem. 2012, 20, 2829–2836. 10.1016/j.bmc.2012.03.031. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.