Abstract
Herein, we introduce a new facile method of luminescent gold nanocluster (Au NC) synthesis on the surface of bacteria for detection, counting, and strain differentiation. The limit of detection was 740 ± 14 colony-forming unit (CFU)/mL for the Gram-negative and was 634 ± 16 CFU/mL for the Gram-positive bacteria. Brief treatment with lysozyme could differentiate the Gram strains based on their luminescence intensities. The current method could also detect bacterial contaminants from water sources and kanamycin-resistant strains rapidly. This quick synthesis of Au NCs on a bacterial template attributes an easy and rapid method for enumeration and detection of bacterial contaminants and kanamycin-resistant strains.
1. Introduction
Rapid and easy detection of numerous drug-resistant bacteria is a burgeoning field of research in the recent time. New methods and tools based on the advanced biochemical techniques, such as PCR,1−4 mass spectroscopy,5 immunological assays/microarrays,1,6 and enzyme assays,7 have been adopted for bacterial detection. However, along with the time constraints and cost, these methods employ prolonged steps for sample preparation. Detection of antibiotic-resistant bacteria using optical methods,8 standard disk diffusion assays, and E-test at various antibiotic concentrations on strips requires more than 24 h.9
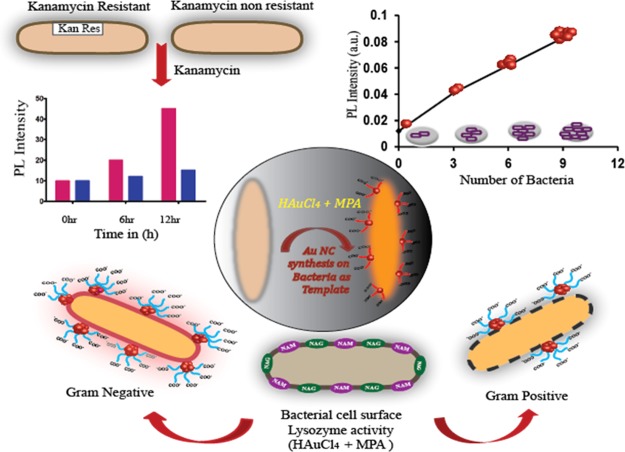
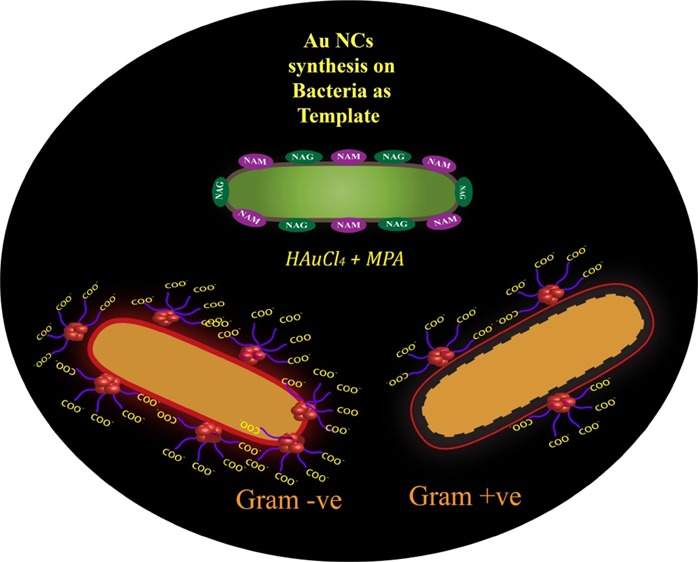
Nanotechnology-based solutions have gained much attention owing to unique physiochemical properties because of small size over the conventional methods for bacterial detection.10 Many nanoparticle (NP)-based detection methods require functional conjugation of suitable probes,11,12 ligands,13−15 antibodies,16 and aptamers.17 Recent progress in this field has led to the evolution of a “lab on a chip” for multiplex analysis with heightened sensitivity. Among this diversity of methods, the luminescence-based methods are fascinating because of rapid response time, easy operation, and sensitivity.18−20 However, organic fluorophores employed in the routine exercises have several limitations, which decline their widespread applications. Further, all NP-based detections involve two-way systems—first, the synthesis of desired functionalized NPs and then the detection, and thus, the overall process is time-consuming (12–14 h for bacterial growth only and separately for synthesis and detection). In fact, the optical determination of antibiotic-resistant strains involves extended procedure of synthesis and purification of fluorophores.8 However, metal nanoclusters (NCs) have been reported to overcome the inherent limitations of the conventional organic dyes.21 In the size domain of (<2 nm), the NPs do not support surface plasmon resonance, which is the common phenomenon associated with the metal NPs; however, they exhibit bright luminescence because of discretization of the continuous electronic states of the metal. The synthesis of the metal NCs offers huge challenge in their stabilization because of their ultrasmall size, which often results in agglomerations of the NCs or formation of plasmonic NPs. Macromolecules provide an essential template for the stabilization of the NCs. A great repertoire of studies has employed bio-macromolecules, such as proteins, small molecules, and DNA as the template for the synthesis of the NCs that are useful for bioimaging and sensing applications.21−24 A recent report has demonstrated the synthesis of the gold NCs (Au NCs) inside cancer cells for the use of bioimaging.25 Synthesis of Au NCs was reported using Escherichia coli enoyl-acyl-carrier protein reductase, an enzyme commonly known as FabI.26 Other reports of bacterial detection using Au NCs showed a long synthesis and detection procedure.27,28 Hence, the use of entire bacteria as the template for the synthesis of NCs could be explored to utilize luminescence of the NCs for bacterial detection. Herein, we report a one-way procedure of bacterial detection by synthesizing highly luminescent Au NCs on bacterial cells by slightly modifying the reaction concentrations of mercaptopropionic acid (MPA) and gold chloride from already established protocol of our previous work.29 For the present work, the reaction mixture was exposed to (50 °C) heating for 2 min, which is less than pasteurization. Interestingly, the luminescence intensity of the Au NCs changes with the number of bacteria, offering a quick method to enumerate the number of bacteria present in the samples. The method is very versatile, where both Gram-positive and Gram-negative bacteria were used to synthesize Au NCs, and at the same time, this method can be employed for the detection of bacterial contamination in various water sources (samples) based on its luminescence. Further, with increase in the number of antibiotic-resistant bacteria, the present method can also be employed to detect antibiotic-resistant strains in short time where distinctive difference can be observed in 6 h. Therefore, this current investigation emphasizes on the development of a simple, fast detection method for “point of care” diagnosis and prognosis, based on the luminescence property of the Au NCs on the bacterial surface. The schematic representation of Au NC-synthesized bacteria and their role in enumeration and differentiation between Gram-positive and Gram-negative bacteria and kanamycin-resistant strains has been explicated in Scheme 1.
Scheme 1. Schematic Representation of Au NCs Synthesized on Bacteria and Their Role in Enumeration and Differentiation between Gram-Positive and Gram-Negative Bacteria and Kanamycin-Resistant Strains.
2. Results and Discussion
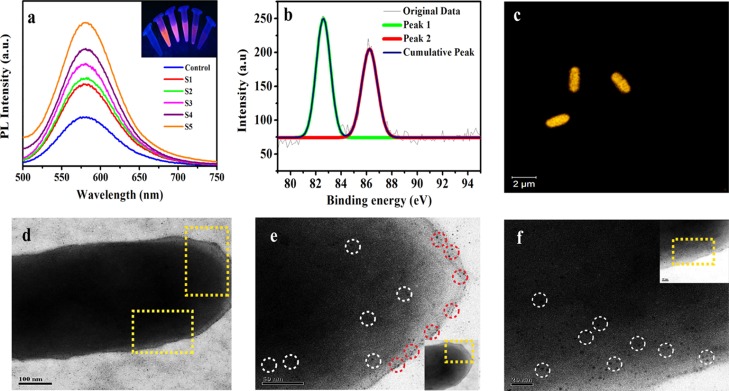
Herein, Au NCs were synthesized using bacteria as their template by slightly modifying our previous work.29 Overnight-grown bacteria were collected by centrifugation and then redispersed in water at different dilutions. To this bacterial solution, 0.01 M MPA and 10 mM gold solution (HAuCl4) were added, followed by heating at 50 °C for 2 min. The formation of the Au NCs was confirmed by the fluorescence spectroscopy by observing the luminescence peak at 580 nm after the formation of Au NCs (Figure S1a, Supporting Information); however, the plasmon peak around 520 nm, which is a typical signature of the Au NPs, was not seen in UV–vis spectroscopy (Figure S1b, Supporting Information). The control bacteria, that is, without Au NC synthesis, have no peak in the red region, whereas after synthesis of the Au NCs, strong luminescence was observed at 580 nm when excited at 320 nm (Figure S1a, Supporting Information). Interestingly, the luminescence intensity gradually increases with bacterial numbers as depicted in Figure 1a, and the same phenomenon could be visualized in the inset of Figure 1a. This experiment was performed on two Gram-positive (Bacillus cereus MTCC 1305, Enterococcus faecalis MTCC 439) and two Gram-negative strains (E. coli MTCC 433, Pseudomonas aeruginosa MTCC 2488), where luminescence increased with the number of bacteria (Figure S2). The luminescence of Au NCs originates from ligand to metal charge transfer where MPA and gold salts form −S–Au complexes. Also, it may be speculated that attachment of ligand-stabilized Au NCs on the bacterial cell wall leads to the restricted motion of the ligands. This might have led to lowering of nonradiative transitions and energy loss, thus increasing the luminescence quantum yield. This observation is consistent with the literature reports of aggregation-induced emission enhancement and other such enhancements of photoluminescence (PL).30,31 Here, MPA served as the surface passivizing agent for the Au NCs and was responsible for the optical and colloidal stability of Au NCs by forming −S–Au bonds. Whereas bacterial cell wall served as the template for the synthesis of Au NCs, possibly interacting with the carboxyl groups (−COOH) of MPA and ultimately producing stable luminescent Au NCs. Hence, the stability of Au NCs was imparted through interactions with the surface proteins of the bacterial cell wall. To confirm the formation of Au NCs, X-ray photoelectron spectroscopy (XPS) was performed, which exhibited peaks at 82.62 and 86.27 eV corresponding to Au (4f5/2, 4f7/2) (Figure 1b). At the same time, matrix-assisted laser desorption/ionization-time of flight (MALDI-TOF)-based mass spectrometric measurements (using sinapinic acid as the matrix) analysis of the as-synthesized sample revealed suggestive information about the formation of Au19 and Au22 atomic clusters (Figure S3a–d, Supporting Information). The peaks (m/z) were obtained at 5807, 5788, 5385, 5382, and 6332, which were assigned to 19, 19, 19, 19, and 22 atoms of gold, respectively. However, stabilizing ligands were also considered for the calculation of masses, which were calculated to be [Au19 + MPA19 + 3Na – 3H], [Au19 + MPA18 + 7 Na – 8H], [Au19 + MPA15 + 3Na – 4H], [Au19MPA15 + 3Na – 3H], and [Au22 + MPA19], respectively. Whereas no such peak was observed in control bacteria (Figure S4, Supporting Information). Formation of Au NCs on the bacterial surface was also observed under confocal laser microscopy, where the bright yellow orange luminescence was observed on the surface of bacteria (Figure 1c) on exciting it with 405 nm. The bright-field image of the same is shown in Figure S5, Supporting Information. The microscopic images clearly stated that Au NCs were synthesized only on bacteria, as a result of which bacteria as a whole showed luminescence (Figure S6, Supporting Information). Transmission electron microscopy (TEM) image clearly depicted the presence of Au NCs on the bacterial surface (Figure 1d–f). For TEM analysis, the as-synthesized Au NCs on bacterial sample was drop-cast into the TEM grid after centrifuging at 8000 rpm for 5 min to remove the unreacted materials and redispersed into deionized water. The Au NCs were prominent on the magnified image of the highlighted bacterial surface (Figure 1e,f), while the surface of the control bacteria remained clear as illustrated in Figure S7, Supporting Information. The magnified TEM image of Au NCs synthesized on bacteria revealed that the average particle size was found to be less than 2 nm. The energy-dispersive X-ray (EDX) spectrum was also recorded during TEM analysis, which confirmed the presence of the metal Au(0) (Figure S8, Supporting Information) on Au NCs synthesized on a bacterium. However, the EDX spectrum of the control bacterium does not reveal the same (Figure S9, Supporting Information). Further, to enlighten the role of bacteria for Au NC synthesis, few control experiments were performed. Au NCs synthesized without bacteria were not stable for longer time. To check the stability of Au NCs without bacteria, time-dependent florescence measurements were carried out by probing the luminescence of control Au NCs (without bacteria) and Au NCs synthesized on bacteria. It was found that the luminescence of the control significantly declined within 60 min, whereas in 60 min there was a slight reduction in the luminescence of Au NCs synthesized on bacteria (Figures S10 and S11, Supporting Information). At the same time, the luminescence of Au NCs synthesized on bacteria was much higher as compared to that of the control (Figure S12, Supporting Information).
Figure 1.
(a) Plot of luminescence intensity vs wavelength in the presence of increasing number of bacteria in the medium of synthesis. Control: Au NCs synthesized (with HAuCl4 and MPA) in the absence of bacteria. S1, S2, S2, S4, and S5 correspond to the bacterial number in the medium 0.74 × 103, 0.74 × 104, 0.74 × 105, 0.74 × 106, and 1.48 × 106, respectively. (b) XPS spectrum of Au NCs synthesized on bacteria. (c) Confocal laser scanning microscopy (CLSM) image of Au NCs synthesized on a bacterium. TEM images of Au NCs synthesized on a bacterium. (d) Au NC-synthesized bacterium; the portion to be magnified was marked in yellow. (e,f) Magnified image of the same sample clearly showing the formation of the Au NCs on the surface; some of the Au NCs are highlighted by white and red circles.
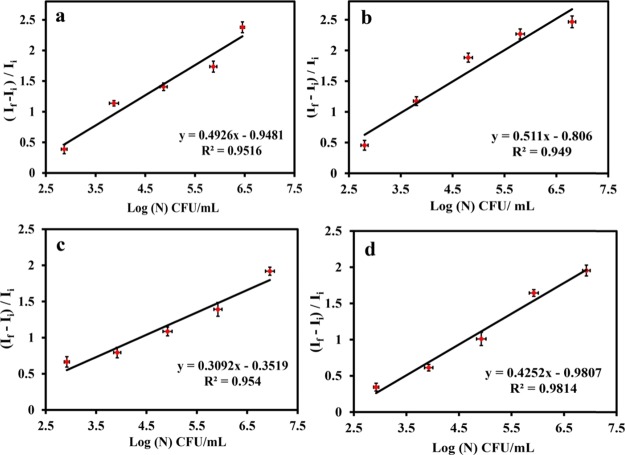
Importantly, the emission intensity of Au NCs was found to be gradually augmented with increasing number of both Gram-positive and Gram-negative bacteria. This offers plausibility to count the number of bacteria present in the test sample. For this purpose, the bacterial strains were grown in the culture media for 12 h. The cells were collected by centrifugation at 6000 rpm for 5 min, and the pellets were resuspended in water. The Au NCs were synthesized using the same reaction condition by varying the numbers of bacteria and were prepared by serial dilutions (102, 103, 104, 105, and 106) of the original stock of the bacteria. It was observed that by keeping the concentration of the gold salt and MPA fixed, the emission intensity of the Au NCs increased with the number of the respective bacterial strains while maintaining an identical reaction condition. Because the emission intensity of the Au NCs varied based on the number of bacteria present in the test sample, it was used to correlate with the number of bacteria. The increased luminescence intensity was normalized with respect to the initial value (the luminescence intensity of control, i.e., MPA and HAuCl4 only) and plotted with log of colony-forming unit (CFU)/mL of bacteria (where If = final emission intensity and Ii = initial emission intensity), and a linear relationship was observed. The experiments were carried out on two Gram-positive bacteria (B. cereus MTCC 1305 and E. faecalis MTCC 439) and two Gram-negative bacteria (E. coli MTCC 433 and P. aeruginosa MTCC 2488) (Figure 2a–d). Interestingly, the results revealed that the slope values were different for Gram-positive and Gram-negative bacteria. The linearity of the plot observed herein signified that the emission of Au NCs synthesized on the surface of bacteria was directly dependent on the log number of CFU/mL for respective strains of Gram-positive and Gram-negative bacteria. The difference in the slope could possibly be due to the variation in the cell wall compositions of the Gram-positive and Gram-negative bacteria, where it might have acted as scaffolds for Au NC synthesis. To find out the exact number of bacteria, a standard plate count method was followed, and the values were obtained after multiplication with the corresponding dilution factors. For this, the same amount of bacteria (by following the serial dilution method) used for Au NC synthesis was grown in the agar plate to find out the exact number of bacteria (CFU/mL). The average limit of detection was found to be 634 ± 16 and 740 ± 14 CFU/mL for Gram-positive and Gram-negative bacteria, respectively. The quantum yield of Au NCs synthesized on bacteria was found to be 1.6% (Figure S13, Supporting Information). Thus, the interaction between the clusters and the functional groups on the bacterial surface was evident to correlate the luminescence intensity of bacteria and finally validates the direct relation of PL enhancement with the number of bacteria used for Au NC synthesis. Further, the restriction of the motion of the stabilizing ligand, upon binding with the cell wall, might have prevented the loss of energy into nonradiative pathways, thus enhancing the luminescence quantum yield.
Figure 2.
Relative increase in the luminescence intensity with the log number of bacteria (CFU/mL): (a) Bacillus cerius MTCC 1305, (b) E. faecalis MTCC 439, (c) E. coli MTCC 433, and (d) P. aeruginosa MTCC 2488. Here, If = final emission intensity and Ii = initial emission intensity. Data are represented by taking average ± standard deviation (SD) of three individual experiments.
The outer layer of the cell walls of both Gram-positive and Gram-negative bacteria contains peptidoglycan, though the thickness is significantly higher in Gram-positive than in Gram-negative bacteria. In Gram-positive bacteria it has been reported that the phosphoryl group present in the linkage unit along with two sugars of teichoic acid is covalently linked with the sugars of the peptidoglycan layer, which are the active cation binding sites. In contrast, Gram-negative bacteria have an extra outer membrane along with the peptidoglycan layer, which contains phospholipids, lipoproteins, lipopolysaccharides, and proteins, where phosphoryl groups are present in phospholipids in the same fashion as teichoic acid. The negative charge of the cell wall could play a major role in metal adsorption, as reported by the molecular stimulation techniques.32
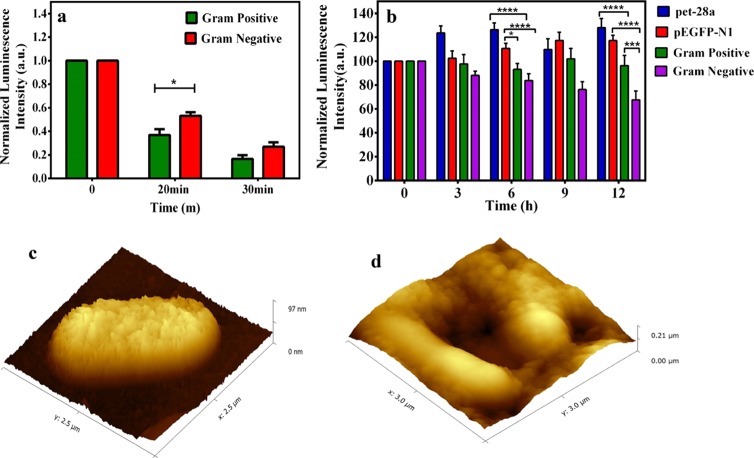
Additionally, the intrinsic properties of the bacterial cell wall composition were investigated to distinguish between the Gram-positive and the Gram-negative strains. It is evident from TEM images that the primary outer layer, that is, peptidoglycan of the cell wall, served as the scaffold for the luminescent Au NCs, which can be tuned for bacterial strain selectivity. Thus the role of the cell wall in stabilization of Au NCs was explored for both Gram -positive and Gram- negative bacteria. For this the respective bacterial strains were treated with lysozyme (1 mg/mL) for various time periods (0, 20 and 30 min). It is to be noted here that lysozyme used here is only to degrade the cell wall of bacteria,33 which had been removed from the medium following treatment. Thereafter, the cells were collected, washed with phosphate-buffered saline, and used as templates for Au NC synthesis maintaining the same reaction condition and precursors (i.e., HAuCl4 and MPA). The results revealed that the emission intensity of Au NCs synthesized after lysozyme treatment in the case of Gram-positive bacteria was low as compared to that of the lysozyme-treated Gram-negative bacteria at both the time points, indicating the possibility of the role of the bacterial cell wall as a scaffold for Au NCs (Figure 3a). It is well-known that both Gram-positive and Gram-negative bacteria have peptidoglycan as cell wall components; however, its thickness is different in Gram-positive than in Gram-negative bacteria. Gram-positive bacteria have a thick peptidoglycan layer, which is known to be sensitive toward lysozyme.33 Whereas Gram-negative bacteria have two layered wall structures with a thinner peptidoglycan layer, which is covered by the outer membrane comprising phospholipids, lipoproteins, lipopolysaccharides, and proteins and is thus prevented or less affected by the lysozyme action. Hence, on lysozyme treatment, the cell wall of the Gram-positive bacteria degraded fast, and therefore, the luminescence of the product Au NCs was less in comparison to that of Au NCs formed in the presence of the lysozyme-treated Gram-negative bacteria. Thus, the method offered the scope of distinguishing Gram-positive and Gram-negative bacteria based on the luminescence of Au NCs. Here, the control stands for Au NC synthesis on Gram-positive and Gram-negative bacteria without treatment with lysozyme, which has been converted to 100% for comparison with the treated groups (lysozyme treatment at two time points), respectively.
Figure 3.
(a) Luminescence intensity of Au NCs synthesized after lysozyme treatment on both Gram-positive and Gram-negative bacteria at different time intervals (0, 20, and 30 min). (b) Normalized luminescence intensity of Au NCs synthesized on different strains of bacteria. pet-28a and pEGFP-N1 are antibiotic (kanamycin)-resistant strain and Gram-positive (B. cereus MTCC 1305) and Gram-negative (E. coli MTCC 433) are wild strains. A plot was generated by average ± SD of three individual experiments. The analysis of variance test revealed the statistical significance, which is represented by “*” (p < 0.05), “***” (p < 0.001), and “****” (p < 0.0001). (c) AFM image of control bacteria and (d) AFM image of Au NC-synthesized bacteria.
At the same time, the current method was employed to test the presence of bacteria (bacterial contaminants) in water for its practicability. Water from three different sources, namely, laboratory wastewater, river water, and pond water, along with a control group (i.e., Milli-Q grade water), were collected. To detect bacteria in water samples, Au NCs were synthesized as mentioned earlier using water from different sources. The luminescence peak was observed with all the water samples except in the control group (Figure S14, Supporting Information). Subsequently, the presence of bacteria was confirmed by the agar plate colony count method, where no colonies were observed with Milli-Q water and the highest numbers of colonies were found in the laboratory wastewater, as the number of bacteria was the highest in that case. Thus, the probe for bacterial detection, that is, the luminescence property of Au NCs synthesized on bacteria, is directly proportional to the number of bacteria in the system. Further, the luminescence of Au NCs on bacteria was explored to find out the antibiotic-resistant strains as well. To circumvent the threat of antibiotic-resistant bacteria via early detection, the current method was used to find out the antibiotic-resistant strain based on the luminescence intensity of Au NCs synthesized on bacteria. It is to be noted here that mostly antibiotic disks were used to find out antibiotic- resistant strains which take 10–12 h to form colonies. For this application, pet-28a (DH5-α) and pEGFP-N1 (DH5-α) which are kanamycin antibiotic-resistant strains and Gram-positive B. cereus MTCC 1305 and Gram-negative E. coli MTCC 433 bacteria nonkanamycin-resistant strains were chosen. First, the respective strains of bacteria (5 × 106) with kanamycin (10 μL of 50 mg/mL) were incubated and then at different time points (0, 3, 6, 9, and 12 h) bacteria were collected by centrifugation, followed by Au NC synthesis. Remarkably, after 6 h, significant difference in the emission of Au NCs was observed in the case of kanamycin-resistant strains (pet-28a and pEGFP-N1) in comparison to that of the nonkanamycin-resistant strains (Figure 3b). This is mainly because kanamycin inhibits the growth of nonresistant bacteria (Gram-positive B. cereus MTCC 1305 and Gram-negative E. coli MTCC 433), whereas resistant strains grow with time. As already mentioned before, the linearity of the plots indicated that the emission of Au NCs synthesized directly depended on the log number of CFU/mL of bacteria. Hence, the current method can be employed to find out kanamycin-resistant strains within 6 h based on Au NCs synthesized on the bacterial surface where emission of Au NCs is directly proportional to the number of bacteria. Hence, the overall time for detection has been drastically reduced as Au NC synthesis was possible at all the respective time points while growing the bacteria. Therefore, the luminescence of Au NCs was useful for the detection of bacteria in a very short time, while normal bacterial detection needs at least 12 h for their growth followed by different procedures of detection. At the same time, the cellular cytotoxicity of the Au NCs synthesized on bacteria was also checked following incubation with HEK-293 cells for 24 h and was found to be nontoxic (Figure S15, Supporting Information). The atomic force microscopy (AFM) analysis was also performed to monitor the structural changes on the surface of bacteria (Gram-negative E. coli MTCC 433). It has been found that the bacterial surface was rough and indented after the synthesis of Au NCs, as compared to the control bacteria (Figure 3c,d). The surface roughness and indentation of the bacteria after Au NC synthesis were quantified using Gwyddion software analysis (Table S1, Supporting Information).
3. Conclusions
In summary, we have developed a new method of synthesis of Au NCs using bacteria as the template. The luminescence property of the as-synthesized Au NCs was probed for bacterial detection and counting. This method was applied to analyze bacterial contamination in water from various sources. Furthermore, the method distinguished between kanamycin-resistant bacterial strains using luminescence of Au NCs on the bacteria. The major advantage of this method is to detect bacteria within short time as direct synthesis on bacteria can be used for detection with opposed to available routine detection techniques. Thus, the current method is a new, easy, rapid, nontoxic, and low-cost approach for detection and enumeration of Gram-positive and Gram-negative kanamycin-resistant strains and bacterial contamination in water sources.
Acknowledgments
The authors greatly acknowledge the financial support from the Department of Electronics and Information Technology, Government of India [no. 5(9)/2012-NANO (Vol. II)] and the Department of Biotechnology, India, for the DBT Programme Support (BT/PR13560/COE/34/44/2015). The authors also acknowledge the instrumental support of the Centre for Nanotechnology and the Central Instruments Facility at Indian Institute of Technology, Guwahati. The authors are thankful to Nilanjan Mandal, Srestha Basu, and Anushree Dutta for their help.
Glossary
Abbreviations
- LOD
limit of detection
- CFU/mL
colony-forming unit
- Au NCs
gold nanoclusters
- MPA
mercaptopropionic acid
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsomega.8b00504.
Materials and methods, synthesis procedures, characterization method details, luminescence and UV–vis spectra, MALDI-TOF spectrum of control as well as Au NC-synthesized bacterium, CLSM and TEM image of control samples, quantum yield details, luminescence of bacteria from different water samples, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay, and surface roughness and indentation statistics of control bacteria and bacteria after Au NC synthesis (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Chung H. J.; Castro C. M.; Im H.; Lee H.; Weissleder R. A Magneto-DNA Nanoparticle System for Rapid Detection and Phenotyping of Bacteria. Nat. Nanotechnol. 2013, 8, 369–375. 10.1038/nnano.2013.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adzitey F.; Huda N.; Ali G. R. R. Molecular Techniques for Detecting and Typing of Bacteria, Advantages and Application to Foodborne Pathogens Isolated from Ducks. 3 Biotech 2013, 3, 97–107. 10.1007/s13205-012-0074-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble R. T. A Review of Technologies for Rapid Detection of Bacteria in Recreational Waters. J. Water Health 2005, 3, 381–392. 10.2166/wh.2005.051. [DOI] [PubMed] [Google Scholar]
- Gopinath S. C. B.; Tang T.-H.; Chen Y.; Citartan M.; Lakshmipriya T. Bacterial Detection: From Microscope to Smartphone. Biosens. Bioelectron. 2014, 60, 332–342. 10.1016/j.bios.2014.04.014. [DOI] [PubMed] [Google Scholar]
- Anhalt J. P.; Fenselau C. Identification of Bacteria Using Mass Spectrometry. Anal. Chem. 1975, 47, 219–225. 10.1021/ac60352a007. [DOI] [Google Scholar]
- Delehanty J. B.; Ligler F. S. A Microarray Immunoassay for Simultaneous Detection of Proteins and Bacteria. Anal. Chem. 2002, 74, 5681–5687. 10.1021/ac025631l. [DOI] [PubMed] [Google Scholar]
- Magliulo M.; Simoni P.; Guardigli M.; Michelini E.; Luciani M.; Lelli R.; Roda A. A Rapid Multiplexed Chemiluminescent Immunoassay for the Detection of Escherichia Coli O157:H7, Yersinia Enterocolitica, Salmonellatyphimurium, and Listeria Monocytogenes Pathogen Bacteria. J. Agric. Food Chem. 2007, 55, 4933–4939. 10.1021/jf063600b. [DOI] [PubMed] [Google Scholar]
- Khan S.; Sallum U. W.; Zheng X.; Nau G. J.; Hasan T. Rapid Optical Determination of β-Lactamase and Antibiotic Activity. BMC Microbiol. 2014, 14, 84. 10.1186/1471-2180-14-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macia M. D.; Borrell N.; Perez J. L.; Oliver A. Detection and Susceptibility Testing of Hypermutable Pseudomonas Aeruginosa Strains with the Etest and Disk Diffusion. Antimicrob. Agents Chemother. 2004, 48, 2665–2672. 10.1128/aac.48.7.2665-2672.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray P. C.; Khan S. A.; Singh A. K.; Senapati D.; Fan Z. Nanomaterials for Targeted Detection and Photothermal Killing of Bacteria. Chem. Soc. Rev. 2012, 41, 3193. 10.1039/c2cs15340h. [DOI] [PubMed] [Google Scholar]
- Disney M. D.; Zheng J.; Swager T. M.; Seeberger P. H. Detection of Bacteria with Carbohydrate-Functionalized Fluorescent Polymers. J. Am. Chem. Soc. 2004, 126, 13343–13346. 10.1021/ja047936i. [DOI] [PubMed] [Google Scholar]
- Chen W.; Li Q.; Zheng W.; Hu F.; Zhang G.; Wang Z.; Zhang D.; Jiang X. Identification of Bacteria in Water by a Fluorescent Array. Angew. Chem., Int. Ed. 2014, 53, 13734–13739. 10.1002/anie.201407606. [DOI] [PubMed] [Google Scholar]
- Phillips R. L.; Miranda O. R.; You C.-C.; Rotello V. M.; Bunz U. H. F. Rapid and Efficient Identification of Bacteria Using Gold-Nanoparticle–Poly(para-Phenyleneethynylene) Constructs. Angew. Chem., Int. Ed. 2008, 47, 2590–2594. 10.1002/anie.200703369. [DOI] [PubMed] [Google Scholar]
- Miranda O. R.; Li X.; Garcia-Gonzalez L.; Zhu Z.-J.; Yan B.; Bunz U. H. F.; Rotello V. M. Colorimetric Bacteria Sensing Using a Supramolecular Enzyme–Nanoparticle Biosensor. J. Am. Chem. Soc. 2011, 133, 9650–9653. 10.1021/ja2021729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden S. C.; Zhao G.; Saha K.; Phillips R. L.; Li X.; Miranda O. R.; Rotello V. M.; El-Sayed M. A.; Schmidt-Krey I.; Bunz U. H. F. Aggregation and Interaction of Cationic Nanoparticles on Bacterial Surfaces. J. Am. Chem. Soc. 2012, 134, 6920–6923. 10.1021/ja301167y. [DOI] [PubMed] [Google Scholar]
- Zhao X.; Hilliard L. R.; Mechery S. J.; Wang Y.; Bagwe R. P.; Jin S.; Tan W. A Rapid Bioassay for Single Bacterial Cell Quantitation Using Bioconjugated Nanoparticles. Proc. Natl. Acad. Sci. U.S.A. 2004, 101, 15027–15032. 10.1073/pnas.0404806101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H.; Wang J.; Liu H.; Li Z.; Jiang F.; Wang F.-B.; Yuan Q. Rapid and Selective Detection of Pathogenic Bacteria in Bloodstream Infections with Aptamer-Based Recognition. ACS Appl. Mater. Interfaces 2016, 8, 19371–19378. 10.1021/acsami.6b06671. [DOI] [PubMed] [Google Scholar]
- Tseng Y.-T.; Chang H.-T.; Chen C.-T.; Chen C.-H.; Huang C.-C. Preparation of Highly Luminescent Mannose–gold Nanodots for Detection and Inhibition of Growth of Escherichia Coli. Biosens. Bioelectron. 2011, 27, 95–100. 10.1016/j.bios.2011.06.021. [DOI] [PubMed] [Google Scholar]
- Sahoo A. K.; Sharma S.; Chattopadhyay A.; Ghosh S. S. Quick and Simple Estimation of Bacteria Using a Fluorescent Paracetamol dimer–Au Nanoparticle Composite. Nanoscale 2012, 4, 1688. 10.1039/c2nr11837h. [DOI] [PubMed] [Google Scholar]
- Wu S.; Duan N.; Shi Z.; Fang C.; Wang Z. Simultaneous Aptasensor for Multiplex Pathogenic Bacteria Detection Based on MulticolorUpconversion Nanoparticles Labels. Anal. Chem. 2014, 86, 3100–3107. 10.1021/ac404205c. [DOI] [PubMed] [Google Scholar]
- Wang H.-H.; Lin C.-A. J.; Lee C.-H.; Lin Y.-C.; Tseng Y.-M.; Hsieh C.-L.; Chen C.-H.; Tsai C.-H.; Hsieh C.-T.; Shen J.-L.; Chan W.-H.; Chang W. H.; Yeh H.-I. Fluorescent Gold Nanoclusters as a Biocompatible Marker for In Vitro and In Vivo Tracking of Endothelial Cells. ACS Nano 2011, 5, 4337–4344. 10.1021/nn102752a. [DOI] [PubMed] [Google Scholar]
- Couleaud P.; Adan-Bermudez S.; Aires A.; Mejías S. H.; Sot B.; Somoza A.; Cortajarena A. L. Designed Modular Proteins as Scaffolds To Stabilize Fluorescent Nanoclusters. Biomacromolecules 2015, 16, 3836–3844. 10.1021/acs.biomac.5b01147. [DOI] [PubMed] [Google Scholar]
- Yang J.; Xia N.; Wang X.; Liu X.; Xu A.; Wu Z.; Luo Z. One-Pot One-Cluster Synthesis of Fluorescent and Bio-Compatible Ag14 Nanoclusters for Cancer Cell Imaging. Nanoscale 2015, 7, 18464–18470. 10.1039/c5nr06421j. [DOI] [PubMed] [Google Scholar]
- Yuan Z.; Chen Y.-C.; Li H.-W.; Chang H.-T. Fluorescent Silver Nanoclusters Stabilized by DNA Scaffolds. Chem. Commun. 2014, 50, 9800. 10.1039/c4cc02981j. [DOI] [PubMed] [Google Scholar]
- Wang J.; Zhang G.; Li Q.; Jiang H.; Liu C.; Amatore C.; Wang X. In Vivo Self-Bio-Imaging of Tumors through in Situ Biosynthesized Fluorescent Gold Nanoclusters. Sci. Rep. 2013, 3, 1157. 10.1038/srep01157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding H.; Li H.; Liu P.; Hiltunen J. K.; Wu Y.; Chen Z.; Shen J. Templated in-Situ Synthesis of Gold Nanoclusters Conjugated to Drug Target Bacterial Enoyl-ACP Reductase, and Their Application to the Detection of Mercury Ions Using a Test Stripe. Microchim. Acta 2014, 181, 1029–1034. 10.1007/s00604-014-1201-1. [DOI] [Google Scholar]
- Chan P.-H.; Ghosh B.; Lai H.-Z.; Peng H.-L.; Mong K. K. T.; Chen Y.-C. Photoluminescent Gold Nanoclusters as Sensing Probes for Uropathogenic Escherichia Coli. PLoS One 2013, 8, e58064 10.1371/journal.pone.0058064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherji R.; Samanta A.; Illathvalappil R.; Chowdhury S.; Prabhune A.; Devi R. N. Selective Imaging of Quorum Sensing Receptors in Bacteria Using Fluorescent Au Nanocluster Probes Surface Functionalized with Signal Molecules. ACS Appl. Mater. Interfaces 2013, 5, 13076–13081. 10.1021/am404093m. [DOI] [PubMed] [Google Scholar]
- Goswami U.; Basu S.; Paul A.; Ghosh S. S.; Chattopadhyay A. White Light Emission from Gold Nanoclusters Embedded Bacteria. J. Mater. Chem. C 2017, 5, 12360–12364. 10.1039/c7tc04554a. [DOI] [Google Scholar]
- Ren Y.; Lam J. W. Y.; Dong Y.; Tang B. Z.; Wong K. S. Enhanced Emission Efficiency and Excited State Lifetime due to Restricted Intramolecular Motion in Silole Aggregates. J. Phys. Chem. B 2005, 109, 1135–1140. 10.1021/jp046659z. [DOI] [PubMed] [Google Scholar]
- Leung N. L. C.; Xie N.; Yuan W.; Liu Y.; Wu Q.; Peng Q.; Miao Q.; Lam J. W. Y.; Tang B. Z. Restriction of Intramolecular Motions: The General Mechanism behind Aggregation-Induced Emission. Chem.—Eur. J. 2014, 20, 15349–15353. 10.1002/chem.201403811. [DOI] [PubMed] [Google Scholar]
- Johnson K. J.; Cygan R. T.; Fein J. B. Molecular Simulations of Metal Adsorption to Bacterial Surfaces. Geochim. Cosmochim. Acta 2006, 70, 5075–5088. 10.1016/j.gca.2006.07.028. [DOI] [Google Scholar]
- Salazar O.; Asenjo J. A. Enzymatic Lysis of Microbial Cells. Biotechnol. Lett. 2007, 29, 985–994. 10.1007/s10529-007-9345-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.