Abstract
In the last years, outer membrane vesicles have attracted a lot of attention for the development of vaccines against bacterial pathogens. Extracellular vesicles can be obtained in high yields by genetic mutations, resulting in generalized modules for membrane antigens (GMMA). Methods to check the quality, consistency of production, and stability of GMMA vaccines are of fundamental importance. In this context, analytical methods for size distribution determination and verifying the integrity and possible aggregation of GMMA particles are strongly needed. Herein, GMMA particle size distribution has been evaluated by means of three different techniques. Dynamic light scattering (DLS), multiangle light scattering (MALS) coupled with high-performance liquid chromatography–size exclusion chromatography (SEC), and nanoparticle tracking analysis (NTA) have been compared to characterize GMMA from different mutants of Salmonella typhimurium and Salmonella enteritidis strains. We found that the presence of O-antigen chains on GMMA determined higher Z-average diameters by DLS compared to size estimation by MALS and that the hydrodynamic diameter increased with the number of O-antigen chains per GMMA particle. In the case of SEC-MALS, the size of the whole population better reflects the size of the most abundant particles, whereas DLS diameter is more influenced by the presence of larger particles in the sample. SEC-MALS and NTA are preferable to DLS for the analysis of bimodal samples, as they better distinguish populations of different size. MALS coupled to a size exclusion chromatography module also allows checking the purity of GMMA preparations, allowing determination of generally occurring contaminants such as soluble proteins and DNA. NTA permits real-time visualization with simultaneous tracking and counting of individual particles, but it is deeply dependent on the choice of data analysis parameters. All of the three techniques have provided complementary information leading to a more complete characterization of GMMA particles.
Introduction
Outer membrane vesicles (OMV) are small bilayered membrane structures naturally released from the cell surface of the Gram-negative bacteria. Simplicity of production and high immunogenicity of these particles, which mimic those of the external surface of bacteria, have made OMV particularly attractive for the development of vaccines against bacterial pathogens.1 Extracellular vesicles can be obtained in high yields2 by genetic manipulation, resulting in generalized modules for membrane antigens (GMMA).3,4 Further mutations are usually introduced to reduce GMMA toxicity by modifying, for example, the acylation pattern of lipid A.5,6 Recently, a bivalent formulation of Salmonella typhimurium and Salmonella enteritidis GMMA has been proposed as a vaccine candidate against nontyphoidal Salmonella, the leading cause of morbidity and death in sub-Saharan Africa, for which a vaccine is not yet available.7
GMMA are complex systems and an in-depth characterization is needed to assure consistency of production and stability over time. Particle size distribution is among the characteristics of GMMA to be investigated for their full characterization.8,9
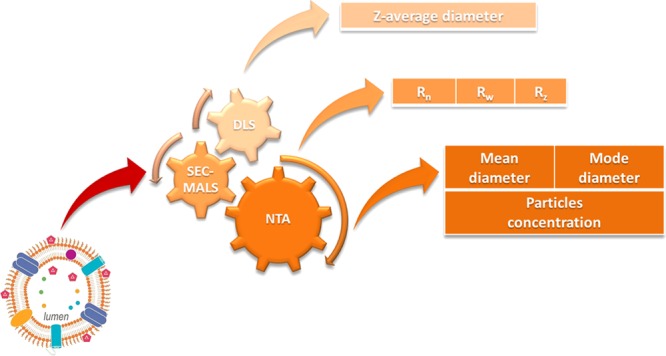
A number of techniques are available for particle size measurement,10−12 including dynamic light scattering (DLS), multiangle light scattering (MALS), and nanoparticle tracking analysis (NTA).
DLS, also known as photon correlation spectroscopy or quasielastic light scattering (QELS), is a popular and routine technique used for the measurement of size distribution of small particles in suspension since 1960s.11,13,14 This technique is used to obtain a mean hydrodynamic diameter (Z-average diameter) and a polydispersity index (PDI), describing the amplitude of the distribution. Hydrodynamic diameter is calculated using the Stokes–Einstein equation, obtaining the diffusion coefficient by measuring intensity fluctuations of scattered light produced by particles as they undergo Brownian motion. MALS represents a static light scattering technique.15,16 Depending on the analysis model, MALS determines, using the angular dependence of the time-averaged scattering intensity, the geometric radius (Rgeo) or the root-mean-square radius, commonly known as the radius of gyration (Rg).
NTA is an alternative light-scattering technique useful for the evaluation of size and number of individual particles,17 ranging from 10 to 2000 nm in size, in liquids.8,18,19 As for the DLS, the particle diameter is calculated using the Stokes–Einstein equation, but by measuring directly the diffusion coefficient of particles moving under Brownian motion, relating the rate of particle motion to particle size.20,21
Here, DLS, HPLC-SEC/MALS, and NTA were applied for determining particle size distribution of GMMA produced by different S. typhimurium- and S. enteritidis-mutated strains. Advantages and limitations of each type of methods are discussed.
Results and Discussion
Size distribution analysis was performed on GMMA from different S. typhimurium- and S. enteritidis-mutated strains. In particular, STm 1418 ΔtolR GMMA and STm 1418 ΔtolR ΔwbaP GMMA were selected to see the differences between OAg-positive (OAg+) and OAg-negative (OAg–) GMMA. SEn 618 ΔtolR ΔmsbB ΔpagP GMMA and STm 2192 ΔtolR ΔpagP ΔmsbB represent the possible candidate vaccines for use in humans, where the deletion of msbB and pagP was used to minimize reactogenicity.
Size Distribution Analysis by DLS
SEn 618 ΔtolR ΔmsbB ΔpagP GMMA exhibited a Z-average diameter of 111.07 nm. STm 2192 ΔtolR ΔpagP ΔmsbB and STm 1418 ΔtolR GMMA were characterized by a Z-average diameter of 103.47 and 91.53 nm, respectively (Table 1, Figure 1A). No differences were found by analyzing the samples at different protein concentrations (in the range 50–200 μg/mL). PDI values were in the range 0.15–0.19, indicating a moderate polydispersity for the GMMA samples analyzed.
Table 1. Z-Average Diameter and Relative PDI of S. enteritidis and S. typhimurium GMMA Samples Analyzed by DLS.
| GMMA | fraction | Z-average diameter (nm) | PDI |
|---|---|---|---|
| SEn 618 ΔtolR ΔmsbB ΔpagP | whole population | 111.07 ± 0.93 | 0.15 |
| HMM | 116.30 ± 0.89 | 0.14 | |
| MMM | 91.28 ± 0.23 | 0.08 | |
| LMM | 84.10 ± 0.62 | 0.16 | |
| STm 2192 ΔtolR ΔpagP ΔmsbB | whole population | 103.47 ± 0.69 | 0.19 |
| HMM | 100.27 ± 0.21 | 0.09 | |
| MMM | 81.69 ± 0.25 | 0.06 | |
| LMM | 77.46 ± 1.15 | 0.16 | |
| STm 1418 ΔtolR | whole population | 91.53 ± 0.46 | 0.18 |
| STm 1418 ΔtolR ΔwbaP | whole population | 57.60 ± 0.53 | 0.26 |
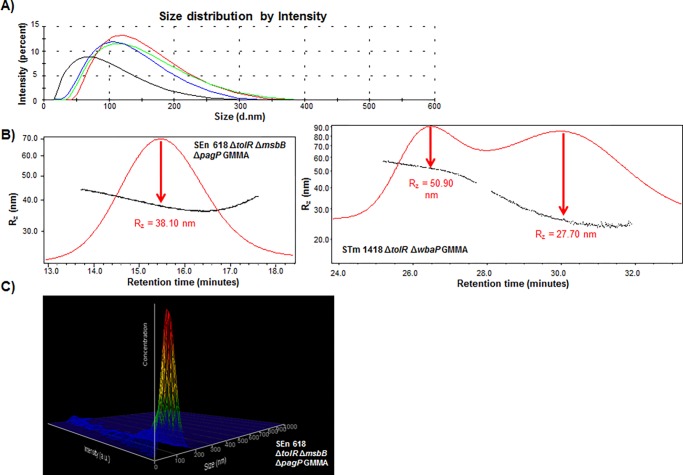
Figure 1.
(A) DLS size distribution of SEn 618 ΔtolR ΔmsbB ΔpagP (red), STm 2192 ΔtolR ΔpagP ΔmsbB (green), STm 1418 ΔtolR (blue), and STm 1418 ΔtolR ΔwbaP GMMA (black). (B) SEC-MALS chromatogram of SEn 618 ΔtolR ΔmsbB ΔpagP and STm 1418 ΔtolR ΔwbaP GMMA, with indication of the Rz value corresponding to the apex of light scattering detection. (C) NTA graph of SEn 618 ΔtolR ΔmsbB ΔpagP GMMA (as an example), in which particle size, particle concentration, and relative intensity are plotted together.
STm 1418 ΔtolR ΔwbaP GMMA were characterized by a smaller size of 57.60 nm and a higher PDI of 0.26 (Figure 1A, Table 1).
For SEn 618 ΔtolR ΔmsbB ΔpagP and STm 2192 ΔtolR ΔpagP ΔmsbB GMMA, three adjacent populations at different molecular mass were separated by HPLC-SEC, and further analyzed by DLS. The Z-average diameter and the PDI values of the collected fractions are summarized in Table 1.
High molecular mass (HMM) fractions, both for S. enteritidis 618 and S. typhimurium 2192 GMMA, showed a particle size distribution similar to that found for unfractionated GMMA. Fractions at the center of the distribution (MMM) and low molecular mass fractions (LMM) showed decreased hydrodynamic diameters with respect to the whole GMMA population.
Size Distribution Analysis by SEC-MALS
All of the GMMA preparations were analyzed by SEC-MALS by applying the “sphere” model. The calculated diameters, averaged with different weights (2 × Rn, 2 × Rw, and 2 × Rz), are reported in Table 2.
Table 2. Diameter Values of S. enteritidis and S. typhimurium GMMA Samples Analyzed by SEC-MALS.
| GMMA | fraction | 2 × Rn (nm) | 2 × Rw (nm) | 2 × Rz (nm) |
|---|---|---|---|---|
| SEn 618 ΔtolR ΔmsbB ΔpagP | whole population | 73.20 ± 0.88 | 74.60 ± 0.90 | 76.20 ± 0.91 |
| HMM | 78.60 ± 4.09 | 79.40 ± 4.05 | 80.20 ± 4.01 | |
| MMM | 67.00 ± 1.01 | 68.40 ± 0.96 | 70.20 ± 0.98 | |
| LMM | 58.00 ± 1.51 | 59.20 ± 1.48 | 61.20 ± 1.53 | |
| STm 2192 ΔtolR ΔpagP ΔmsbB | whole population | 68.20 ± 0.75 | 69.80 ± 0.77 | 72.20 ± 0.79 |
| HMM | 80.00 ± 0.96 | 81.00 ± 0.97 | 82.20 ± 0.99 | |
| MMM | 67.60 ± 0.81 | 68.20 ± 0.82 | 70.00 ± 0.84 | |
| LMM | 58.60 ± 0.94 | 59.80 ± 0.96 | 61.40 ± 0.98 | |
| STm 1418 ΔtolR | whole population | 70.00 ± 0.91 | 71.40 ± 0.86 | 73.80 ± 0.89 |
| STm 1418 ΔtolR ΔwbaP | whole population peak 1 | 98.20 ± 0.59 | 100.00 ± 0.60 | 101.80 ± 0.61 |
| whole population peak 2 | 51.00 ± 1.08 | 52.60 ± 1.00 | 55.40 ± 1.00 |
SEn 618 ΔtolR ΔmsbB ΔpagP, STm 2192 ΔtolR ΔpagP ΔmsbB, and STm 1418 ΔtolR GMMA were characterized by a similar 2 × Rz value (Table 2, Figure 1B).
Unlike DLS, showing one single population, SEC-MALS analysis of STm 1418 ΔtolR ΔwbaP GMMA revealed a bimodal distribution with the maxima at 2 × Rz = 55.40 and 101.80 nm, respectively (Figure 1B).
Samples derived from HPLC-SEC fractionation of SEn 618 ΔtolR ΔmsbB ΔpagP and STm 2192 ΔtolR ΔpagP ΔmsbB GMMA were also analyzed by SEC-MALS. Results relative to the collected fractions are summarized in Table 2.
Size Distribution Analysis by NTA
All of the GMMA samples were analyzed by NTA. Screen gain and camera level (shutter speed and camera gain) were set at values 2 and 16, respectively, and selected based on the visually brightest detection of particles without the occurrence of abundant overscattering events.
After performing a first set of experiments, we verified that the detection threshold needed to be selected by the operator, according to the sample and its dilution, to have consistent results both in terms of size and particles counting.
Similar results, both in terms of size and number of particles per mg of GMMA proteins, were obtained by analyzing SEn 618 ΔtolR ΔmsbB ΔpagP GMMA at different dilutions (Table 3).
Table 3. NTA Analysis of SEn ΔtolR ΔmsbB ΔpagP GMMA Analyzed at Different Dilutions.
| sample dilution | detection threshold | protein concentration (μg/mL) | mean (nm) | mode (nm) | particles (mL) | particles (μg) |
|---|---|---|---|---|---|---|
| 10 000× | 3 | 1.39 × 10–1 | 106.05 | 86.65 | 2.55 × 109 | 1.83 × 1010 |
| 25 000× | 7 | 5.82 × 10–2 | 108.40 | 90.85 | 1.10 × 109 | 1.90 × 1010 |
| 50 000× | 11 | 2.90 × 10–2 | 105.15 | 97.65 | 5.54 × 108 | 1.91 × 1010 |
| 75 000× | 11 | 1.93 × 10–2 | 106.65 | 85.90 | 3.57 × 108 | 1.85 × 1010 |
| 100 000× | 14 | 1.45 × 10–2 | 103.90 | 98.40 | 2.82 × 108 | 1.95 × 1010 |
| average | 106.03 | 91.89 | 1.89 × 1010 |
Similar data were collected for all of the GMMA samples and average results are summarized in Table 4. Standard deviation (SD), D10, D50, and D90 percentile values showed that GMMA samples were quite polydisperse.
Table 4. NTA Results of SEn 618 ΔtolR ΔmsbB ΔpagP, STm 2192 ΔtolR ΔpagP ΔmsbB, STm 1418 ΔtolR, and STm 1418 ΔtolR ΔwbaP GMMA.
| GMMA sample | mean (nm) | mode (nm) | SD | D10 | D50 | D90 | particles (μg) | |
|---|---|---|---|---|---|---|---|---|
| SEn 618 ΔtolR ΔmsbB ΔpagP | whole population | 106.03 | 91.89 | 37.24 | 57.72 | 89.13 | 143.82 | 1.89 × 1010 |
| MMM fraction | 92.88 | 81.38 | 23.15 | 60.63 | 78.05 | 111.83 | 2.24 × 1010 | |
| STm 2192 ΔtolR ΔpagP ΔmsbB | whole population | 102.47 | 78.59 | 38.61 | 60.36 | 79.60 | 140.49 | 1.09 × 1010 |
| MMM fraction | 83.33 | 76.51 | 20.14 | 54.84 | 69.20 | 96.03 | 2.24 × 1010 | |
| STm 1418 ΔtolR | whole population | 95.80 | 85.64 | 31.58 | 56.96 | 79.63 | 120.58 | 1.47 × 1010 |
| MMM fraction | 84.93 | 74.53 | 31.67 | 50.67 | 68.70 | 104.73 | 7.18 × 1010 | |
| STm 1418 ΔtolR ΔwbaP | whole population | 90.13 | 62.63 | 48.23 | 37.17 | 70.07 | 134.33 | 3.32 × 109 |
The analysis of more homogeneous samples was performed by analyzing the MMM fractions from the SEC fractionation of GMMA (Table 4). More similarity was observed between mean and mode hydrodynamic diameter compared to what was found for the unfractionated GMMA. Lower SD and D90 and higher D10 values clearly showed that a more homogeneous sample was obtained.
The mode diameters of the unfractionated samples were similar to the mean values of the fraction corresponding to the central area of the GMMA samples. NTA analysis of STm 1418 ΔtolR ΔwbaP GMMA revealed a high difficulty in proper tracking and enumerating of the particles, probably due to their smaller dimensions and higher polydispersity. Table 4 reports the mean and mode values calculated for the entire population. However, NTA, similar to SEC-MALS, was able to distinguish a major peak with a mode value of 55 nm and a less abundant peak with a mode value of 85 nm.
Comparison of DLS, MALS, and NTA Results
Dimensional analyses performed on SEn 618 ΔtolR ΔmsbB ΔpagP, STm 2192 ΔtolR ΔpagP ΔmsbB, STm 1418 ΔtolR, and STm 1418 ΔtolR ΔwbaP GMMA using the three different methods are summarized in Table 5.
Table 5. Summary of the Analyses Performed by DLS, SEC-MALS, and NTA on SEn 618 ΔtolR ΔmsbB ΔpagP, STm 2192 ΔtolR ΔpagP ΔmsbB, STm 1418 ΔtolR, and STm 1418 ΔtolR ΔwbaP GMMAa.
| |
DLS | SEC-MALS |
NTA |
||||
|---|---|---|---|---|---|---|---|
| GMMA sample | Z-average diameter (nm) | 2 × Rn (nm) | 2 × Rw (nm) | 2 × Rz (nm) | mean diameter (nm) | mode diameter (nm) | |
| SEn 618 ΔtolR ΔmsbB ΔpagP | whole population | 111.07 | 73.20 | 74.60 | 76.20 | 106.03 | 91.89 |
| MMM fraction | 91.28 | 67.00 | 68.40 | 70.20 | 92.88 | 81.38 | |
| STm 2192 ΔtolR ΔpagP ΔmsbB | whole population | 103.47 | 68.20 | 69.80 | 72.20 | 102.47 | 78.59 |
| MMM fraction | 81.69 | 67.60 | 68.60 | 70.00 | 83.33 | 76.51 | |
| STm 1418 ΔtolR | whole population | 91.53 | 70.00 | 71.40 | 73.80 | 95.80 | 85.64 |
| MMM fraction | na | na | na | na | 84.93 | 74.53 | |
| STm 1418 ΔtolR ΔwbaP | whole population | 57.60 | 98.20/51.00 | 100.00/52.60 | 101.80/55.40 | 90.13 | 62.63 |
na: not analyzed.
Analysis of the whole GMMA populations gave NTA mean diameters similar to Z-average diameters by DLS. MMM populations, obtained after the fractionation of GMMA samples, showed similar hydrodynamic diameters, by DLS, and mean diameters, by NTA. Such values were also similar to the NTA mode diameters of the corresponding whole populations.
For all of the OAg+ GMMA, the Z-average size measured by DLS was higher compared to the 2 × Rz value obtained by SEC-MALS. The same behavior was not found for ΔwbaP OAg– GMMA, suggesting that higher values from DLS were not only due to the higher weight of large particles in the average diameter calculation by this method. The difference observed could be related to the presence of the OAg chains displayed on the GMMA surface, which play a role in determining the behavior of GMMA in the solution. OAg+ GMMA were in fact characterized by different Z-average diameters, but similar SEC-MALS diameters of around 70 nm (Table 5).
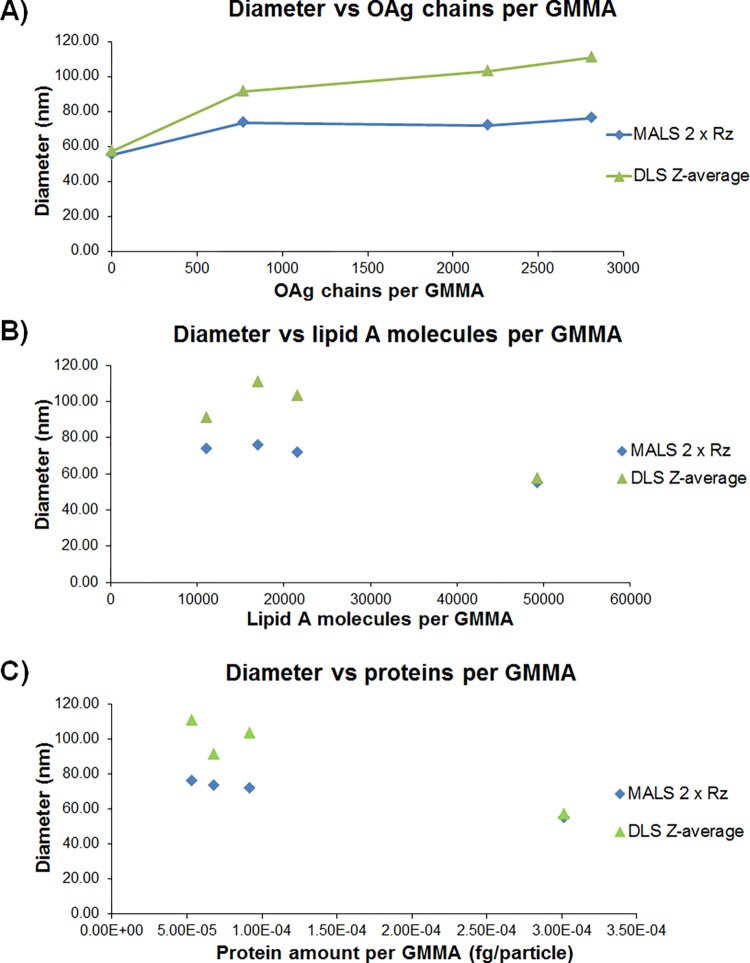
Number of OAg chains per GMMA particle, OAg length, and structural characteristics such as O-acetylation and glucosylation level, as well as the amount of lipid A and its structure, could affect the overall size of GMMA. By looking at these characteristics (Table 6), a correlation between DLS diameters and average number of OAg chains per GMMA particle was found (Figure 2A). The size of OAg chains was similar for all of the OAg+ GMMA tested, and no correlation was found between GMMA size and protein or lipid A content (Figure 2B,C).
Table 6. Characterization of SEn 618 ΔtolR ΔmsbB ΔpagP, STm 2192 ΔtolR ΔpagP ΔmsbB, STm 1418 ΔtolR, and STm 1418 ΔtolR ΔwbaP GMMA Trying To Correlate GMMA Size with Their Main Features.
| GMMA sample | OAg | lipid A structure5 | Z-average (nm) | 2 × Rz (nm) | average μg protein/GMMA | OAg size (kDa)a | average number OAg chains/GMMAb | average number lipid A molecules/GMMAb | ζ-potential (mV) |
|---|---|---|---|---|---|---|---|---|---|
| SEn 618 ΔtolR ΔmsbB ΔpagP | OAg+ | penta | 111.07 | 76.20 | 5.30 × 1011 | 30.00 | 2812 | 16 960 | –3.2 |
| STm 2192 ΔtolR ΔpagP ΔmsbB | OAg+ | penta | 103.47 | 72.20 | 9.19 × 1011 | 34.60 | 2204 | 21 527 | –3.3 |
| STm 1418 ΔtolR | OAg+ | hepta/hexa | 91.353 | 73.80 | 6.78 × 1011 | 32.90 | 768 | 11 035 | –2.7 |
| STm 1418 ΔtolR ΔwbaP | OAg– | hepta/hexa | 57.60 | 55.40 | 3.01 × 1010 | 49 248 | –9.8 |
OAg size was calculated by HPLC-SEC analysis on a TSK gel 3000 PWxl column using dextrans as standards.
Average number of OAg chains per GMMA particle was calculated by sugar quantification by HPAEC-PAD and particles counting by NTA. Average number of lipid A molecules was derived by HPLC-SEC/semicarbazide assay.7
Figure 2.
(A) Correlation between DLS Z-average diameters and MALS 2 × Rz values with the number of OAg chains per GMMA particle. (B) Lack of correlation between DLS Z-average diameters and MALS 2 × Rz values with number of lipid A molecules per GMMA particle. (C) Lack of correlation between DLS Z-average diameters and MALS 2 × Rz values and protein amount per GMMA particle.
Table 6 also reports the ζ-potential values collected for different GMMA. All of them can be considered approximately neutral in phosphate buffered saline (PBS), with similar values for all of the OAg-positive GMMA and a more negative value for the OAg-negative sample. DLS allowed precise and reliable GMMA particle size analysis within few minutes, with a rapid and simple sample preparation and instrument setup. A major drawback of DLS is that it is inherently sensitive to the presence of large particles in the sample used in the analysis,23 as verified by analyzing the unfractionated and fractionated GMMA samples. It is expected that the DLS Z-average size distribution of polydisperse samples is biased by even a small number of large particles because such particles scatter light more efficiently than small ones.24,25 On the contrary, with SEC-MALS, the diameter of the whole population better reflects the size of the most abundant population.
SEC-MALS is also a rapid and robust method for GMMA size characterization. It allows separation and qualitative analysis of generally occurring contaminants, such as free soluble proteins and DNA.
NTA is an alternative light-scattering technology that simultaneously but individually tracks and analyzes the trajectories of GMMA in suspension. NTA can detect small, weakly scattering particles among large, strong-scattering ones that would dominate the size distribution of a particle sample analyzed by DLS. The mean size gives the average size of the whole vesicles population and has a value similar to Z-average diameter by DLS. But mode size characterizes the particle size that appears most often within a given preparation.
NTA allows not only size determination but also counting of the number of particles. However, for NTA analysis, a range of parameters need to be adjusted both for video capture (camera gain and shutter speed) and data elaborations (filter settings, background subtraction, removal of blurring, minimum track length, minimum expected particle size, and detection threshold). This makes standardization of the NTA technique, which is strongly operator dependent,17,23,26−28 difficult to achieve.21 Our study confirmed that detection threshold is one of the parameters that can strongly affect both size and count of particles in NTA analysis.26 In particular, NTA analyses did not show an acceptable reproducibility among experiments performed using scalar-diluted samples at a fixed detection threshold value, both in GMMA size and concentration.
Conclusions
In conclusion, all of the three methods provide size measurements based on absolute analyses of the samples in solution, independent of the calibration standards.
DLS is the most rapid method, but SEC-MALS and NTA are preferable in the case of bimodal samples, allowing better separation of populations at different size. MALS coupled with SEC has also the advantage to detect eventual presence of smaller mass impurities. NTA visualizes and counts single particles also allowing determination of antigens density on GMMA particle. In the specific case of S. typhimurium GMMA, the number of OAg chains on the GMMA surface affects the hydrodynamic radius determined by DLS and mean size by NTA, but not Rgeo by MALS.
To our knowledge, this is one of the first studies describing the use of NTA for OMV characterization29 and comparing the use of different techniques for their size determination. Complete characterization of OMV size is an important research topic in the field of OMV vaccines development because it is essential to check the consistency of production and stability of samples, and to study differences among bacterial strains. All of the techniques tested here provided useful information for a more complete evaluation of the GMMA size.
Experimental Section
Strains for GMMA Production
S. typhimurium isolate SGSC1418 (LT-2 collection, University of Calgary), S. typhimurium 2192 (SGSC2192, SARA collection), and S. enteritidis SA618 (CEESA EASSA II collection of Quotient Bioresearch Limited) were used as parent strains for GMMA production.22 Mutants for GMMA production are described in Table 7 and were obtained as previously described.5tolR deletion increases the GMMA release during bacteria growth, wbaP deletion is associated with the loss of O-antigen (OAg) chains resulting in an OAg-negative strain, whereas ΔpagP and ΔmsbB mutations have an impact on lipid A acylation pattern and are introduced to have a pure penta-acylated lipid A detoxified GMMA.5
Table 7. Mutated Strains Used for GMMA Production and Their Abbreviations.
| strain abbreviation name | strain characteristics | genotype |
|---|---|---|
| STm 1418 ΔtolR | overblebbing, wild type lipid A, OAg positive | S. typhimurium 1418 ΔtolR::aph |
| STm 1418 ΔtolR ΔwbaP | overblebbing, wild type lipid A, OAg negative | S. typhimurium 1418 ΔtolR::aph ΔwbaP::cat |
| STm 2192 ΔtolR ΔpagP ΔmsbB | overblebbing, detoxified lipid A, OAg positive | S. typhimurium 2192 ΔtolR::aph ΔpagP::cat ΔmsbB::tetRA |
| SEn 618 ΔtolR ΔmsbB ΔpagP | overblebbing, detoxified lipid A, OAg positive | S. enteritidis 618 ΔtolR::aph ΔmsbB::tetRA ΔpagP::cat |
GMMA Production
GMMA were produced and purified as previously described.5 After purification, all of the GMMA samples were suspended in phosphate buffered saline (PBS) and then 0.22 μm filtered. GMMA content was estimated as the total protein content by micro Bicinchoninic Acid Protein Assay (BCA). OAg quantification was performed by high-performance anion exchange chromatography with pulsed amperometric detection (HPAEC-PAD) analysis, and OAg size was calculated by HPLC-SEC analysis on a TSK gel 3000 PWxl column using dextrans as standards. Number of lipid A molecules were derived by HPLC-SEC/semicarbazide assay, as previously described.7
Dynamic Light Scattering (DLS)
DLS measurements were performed with a Malvern Zetasizer Nano ZS (Malvern, Herremberg, Germany) equipped with a 633 nm He–Ne laser and operating at an angle of 173°. Scattering light detected at 173° was automatically adjusted by laser attenuation filters. For data analysis, the viscosity and refractive index (RI) of PBS buffer solution (at 25 °C) were used. The software used to collect and analyze the data was the Zetasizer software version 7.11. Temperature was set at 25 °C. Each sample (80 μL) at 50, 125, and 200 μg/mL protein content was characterized in duplicates in single-use polystyrene microcuvette (ZEN0040, Alfatest) with a path length of 10 mm. The hydrodynamic diameter of GMMA was expressed by a Z-average value (general purpose algorithm) of three measurements for each replicate, providing also a PDI of the size values calculated. Size distribution by intensity was preferred to measurements by number or by volume to have more reproducible results and because the RI values of GMMA were not known, respectively.
Size Exclusion Chromatography Coupled with Multiangle Light Scattering (SEC-MALS)
GMMA samples were analyzed by HPLC-SEC with Tosoh TSK gel G6000PW (30 cm × 7.5 mm) + G4000PW (30 cm × 7.5 mm) columns in series equilibrated in PBS (PBS tablets, Medicago) and with in-line UV, fluorescence emission, and MALS detectors. A Wyatt Dawn Heleos II MALS equipped with fused silica cell and a 660 nm laser source were used. A volume of 80 μL of samples with concentrations of 100 μg/mL protein content were injected and eluted with a flow rate of 0.5 mL/min (run time 70 min). All of the dilutions were made in PBS. MALS data were collected using ASTRA 6 software (Wyatt Technology) with “particles” template and analyzed using “Sphere” model. The size of GMMA was expressed by the number average geometric radius Rn, weight average geometric radius Rw, and Z-average geometric radius Rz values.
Nanoparticle Tracking Analysis (NTA)
NS300 Nanosight instrument (Malvern) equipped with a CMOS camera and a 488 nm monochromatic laser beam was used. Data acquisition and processing were performed using NTA software 3.2 build 3.2.16. Automatic settings for the minimum track length, the minimal expected particle size and blur setting were applied. Viscosity settings for water were applied and automatically corrected for the temperature used. Measurements were performed at room temperature ranging from 22 to 25 °C. Particles movement was analyzed by NTA software with camera level at 16, slider shutter at 1300, and slider gain at 512. Different detection threshold values were tested and adjusted for the sample appearance after dilution. For each sample, five replicate videos of 30 s at 25 frames per second were collected, generating five replicate histograms that were averaged. Several dilutions of the samples were analyzed and duplicates were recorded for every diluted sample. GMMA samples were PBS diluted in low-binding Eppendorf tubes, and the dilutions were prepared just before the analysis. Samples were gently mixed and slowly injected in the sample chamber using a 1 mL syringe over 5–10 s. The samples were recorded under controlled flow, using the NanoSight syringe pump (speed 20). Each video was then analyzed to determine the respective mean and mode (particle size that appears most often within a given preparation) GMMA size. In addition to these values, standard deviation (SD) and percentile undersize values (D10, D50, and D90) were collected. SD, D10, D50, and D90 were measure of the spread of particles size distribution within the samples. The concentrations of samples are reported either as particles per mL or particles per frame.
HPLC-SEC
To obtain more homogeneous GMMA samples, a volume of 100 μL with a concentration of 1000 μg/mL protein content was fractionated by HPLC-SEC. Tosoh TSK gel G6000PW (30 cm × 7.5 mm) + G4000PW (30 cm × 7.5 mm) columns in series equilibrated in PBS (PBS tablets, Medicago) were used with in-line UV detector. Samples were eluted with PBS at a flow rate of 0.5 mL/min (run time 70 min). GMMA peaks were fractionated in 1.7 mL low binding Eppendorf tubes monitoring the 280 nm elution profile and collecting fractions at the rate of one tube per minute.
ζ-Potential
ζ-potential measurements were acquired on a Malvern Nano ZS instrument (Zetasizer software ver. 7.11) with the following sample settings: “Protein” as material, “PBS” as dispersant, and “Smoluchowski” as F(ka) selection model. The experiments were performed at 25 °C with 120 s as equilibration time using a disposable folded capillary cell (model DTS 1070) filled with 750 μL of sample. The measurement duration was set as “automatic”, with a minimum and maximum numbers of runs of 10 and 100, respectively, and a measurement number of 3 with a 60 s delay. The attenuation and voltage of measurement as well as the analysis model were set as automatic. The samples were analyzed at 200 μg/mL protein concentration diluted in PBS.
Acknowledgments
We thank Mariaelena Caboni and Aurel Negrea for strains generation (SEn 618 ΔtolR ΔmsbB ΔpagP, STm 2192 ΔtolR ΔpagP ΔmsbB, and STm 1418 ΔtolR) and bacteria growth for GMMA production. We thank Yunshan Goh for generation of the STm 1418 ΔtolR ΔwbaP mutant and Irene Beriotto for the bacteria growth for GMMA production. We thank the GVGH Technical Development Unit for the fermentation, purification, and characterization of GMMA from SEn 618 ΔtolR ΔmsbB ΔpagP, STm 2192 ΔtolR ΔpagP ΔmsbB, Renzo Alfini for characterization of STm 1418 ΔtolR and Roberta Di Benedetto for DLS and SEC-MALS analysis of STm 1418 ΔtolR ΔwbaP GMMA. We thank Amelia Gamini and Sabrina Semeraro for the first analyses of GMMA by NTA at the University of Trieste and their assistance regarding size-analyzing techniques theory and Paola Lo Surdo (GSK Vaccines, Head of High-throughput Vaccines Analytics) for her support for NTA analyses at GSK Siena. We thank Allan Saul for careful revision of the manuscript.
Author Contributions
The manuscript was written through contributions of all of the authors. All of the authors have given approval to the final version of the manuscript.
The authors declare the following competing financial interest(s): Francesca Micoli and Carlo Giannelli are employees of GSK Vaccines Institute for Global Health (GVGH), part of the GSK group of companies. Gianluigi De Benedetto participated in a postgraduate studentship program at GSK Vaccines.
References
- van der Pol L.; Stork M.; van der Ley P. Biotechnol. J. 2015, 10, 1689–1706. 10.1002/biot.201400395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acevedo R.; Fernandez S.; Zayas C.; Acosta A.; Sarmiento M. E.; Ferro V. A.; Rosenqvist E.; Campa C.; Cardoso D.; Garcia L.; Perez J. L. Front. Immunol. 2014, 5, 121 10.3389/fimmu.2014.00121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerke C.; Colucci A. M.; Giannelli C.; Sanzone S.; Vitali C. G.; Sollai L.; Rossi O.; Martin L. B.; Auerbach J.; Di Cioccio V.; Saul A. PLoS One 2015, 10, e0134478 10.1371/journal.pone.0134478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlanda Scorza F.; Colucci A. M.; Maggiore L.; Sanzone S.; Rossi O.; Ferlenghi I.; Pesce I.; Caboni M.; Norais N.; Di Cioccio V.; Saul A.; Gerke C. PLoS One 2012, 7, e35616 10.1371/journal.pone.0035616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi O.; Caboni M.; Negrea A.; Necchi F.; Alfini F.; Micoli F.; Saul A.; MacLennan C. A.; Rondini S.; Gerke C. Clin. Vaccine Immunol. 2016, 23, 304–314. 10.1128/CVI.00023-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi O.; Pesce I.; Giannelli C.; Aprea S.; Caboni M.; Citiulo F.; Valentini S.; Ferlenghi I.; MacLennan C. A.; D’Oro U.; Saul A.; Gerke C. J. Biol. Chem. 2014, 289, 24922–24935. 10.1074/jbc.M114.566570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Benedetto G.; Alfini R.; Cescutti P.; Caboni M.; Lanzilao L.; Necchi F.; Saul A.; MacLennan C. A.; Rondini S.; Micoli F. Vaccine 2017, 35, 419–426. 10.1016/j.vaccine.2016.11.089. [DOI] [PubMed] [Google Scholar]
- Roier S.; Blume T.; Klug L.; Wagner G. E.; Elhenawy W.; Zangger K.; Prassl R.; Reidl J.; Daum G.; Feldman M. F.; Schild S. Int. J. Med. Microbiol. 2015, 305, 298–309. 10.1016/j.ijmm.2014.12.005. [DOI] [PubMed] [Google Scholar]
- Wieser A.; Storz E.; Liegl G.; Peter A.; Pritsch M.; Shock J.; Wai S. N.; Schubert S. Int. J. Med. Microbiol. 2014, 304, 1032–1037. 10.1016/j.ijmm.2014.07.012. [DOI] [PubMed] [Google Scholar]
- Anderson W.; Kozak D.; Coleman V. A.; Jamting A. K.; Trau M. J. Colloid Interface Sci. 2013, 405, 322–330. 10.1016/j.jcis.2013.02.030. [DOI] [PubMed] [Google Scholar]
- van der Pol E.; Coumans F.; Varga Z.; Krumrey M.; Nieuwland R. J. Thromb. Haemostasis 2013, 11, 36–45. 10.1111/jth.12254. [DOI] [PubMed] [Google Scholar]
- Sitar S.; Kejzar A.; Pahovnik D.; Kogej K.; Tusek-Znidaric M.; Lenassi M.; Zagar E. Anal. Chem. 2015, 87, 9225–9233. 10.1021/acs.analchem.5b01636. [DOI] [PubMed] [Google Scholar]
- Pecora R. J. Nanopart. Res. 2000, 2, 123–131. 10.1023/A:1010067107182. [DOI] [Google Scholar]
- Berne B. J.; Pecora R.. Dynamic Light Scattering with Applications to Chemistry, Biology and Physics; Dover Publications, Inc.; Mineola, New York, 2000; pp 1–376. [Google Scholar]
- Vezočnik V.; Rebolj K.; Sitar S.; Ota K.; Tusek-Znidaric M.; Strus J.; Sepcic K.; Pahovnik D.; Macek P.; Zagar E. J. Chromatogr. A 2015, 1418, 185–191. 10.1016/j.chroma.2015.09.048. [DOI] [PubMed] [Google Scholar]
- Wyatt P. J. J. Colloid Interface Sci. 1998, 197, 9–20. 10.1006/jcis.1997.5215. [DOI] [PubMed] [Google Scholar]
- Gardiner C.; Ferreire Y. J.; Dragovic R. A.; Redman C. W.; Sargent I. L. J. Extracell. Vesicles 2013, 2, 19671. 10.3402/jev.v2i0.19671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright M. Methods Mol. Biol. 2012, 906, 511–524. 10.1007/978-1-61779-953-2_41. [DOI] [PubMed] [Google Scholar]
- Saveyn H.; De Baets B.; Thas O.; Hole P.; Smith J.; Van Der Meeren P. Accurate particle size distribution determination by nanoparticle tracking analysis based on 2-D Brownian dynamics simulation. J. Colloid Interface Sci. 2010, 352 (2), 593–600. 10.1016/j.jcis.2010.09.006. [DOI] [PubMed] [Google Scholar]
- Gallego-Urrea J. A.; Tuoriniemi J.; Hasselov M. TrAC, Trends Anal. Chem. 2011, 30, 473–483. 10.1016/j.trac.2011.01.005. [DOI] [Google Scholar]
- Dragovic R. A.; Gardiner C.; Brooks A. S.; Tannetta D. S.; Ferguson D. J.; Hole P.; Carr B.; Redman C. W.; Harris A. L.; Dobson P. J.; Harrison P.; Sargent I. L. Nanomedicine 2011, 7, 780–788. 10.1016/j.nano.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanzilao L.; Stefanetti G.; Saul A.; MacLennan C. A.; Micoli F.; Rondini S. PLoS One 2015, 10, e139847 10.1371/journal.pone.0139847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipe V.; Hawe A.; Jiskoot W. Pharm. Res. 2010, 27, 796–810. 10.1007/s11095-010-0073-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamting Å. K.; Cullen J.; Coleman V. A.; Lawn M.; Herrmann J.; Miles J.; Ford J. Adv. Powder Technol. 2011, 22, 290–293. 10.1016/j.apt.2011.03.009. [DOI] [Google Scholar]
- Hoo C. M.; Starostin N.; West P.; Mecartney M. L. J. Nanopart. Res. 2008, 10, 89–96. 10.1007/s11051-008-9435-7. [DOI] [Google Scholar]
- Maas S. L.; de Vrij J.; van der Vlist E. J.; Geragousian B.; van Bloois L.; Mastrobattista E.; Schiffelers R. M.; Wauben M. H. M.; Broekman M. L. D.; Nolte-’t Hoen E. N. M. J. Controlled Release 2015, 200, 87–96. 10.1016/j.jconrel.2014.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Meeren P.; Kasinos M.; Saveyn H. Methods Mol. Biol. 2010, 906, 525–534. 10.1007/978-1-61779-953-2_42. [DOI] [PubMed] [Google Scholar]
- Hole P.; Sillence K.; Hannell C.; Maguire C. M.; Roesslein M.; Suarez G.; Capracotta S.; Magdolenova Z.; Horev-Azaria L.; Dybowska A.; Cooke L.; Haase A.; Contal S.; Manø S.; Vennermann A.; Sauvain J. J.; Staunton K. C.; Anguissola S.; Luch A.; Dusinska M.; Korenstein R.; Gutleb A. C.; Wiemann M.; Prina-Mello A.; Riediker M.; Wick P. J. Nanopart. Res. 2012, 15, 2101. 10.1007/s11051-013-2101-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerritzen M. J. H.; Martens D. E.; Wijffels R. H.; Stork M. J. Extracell Vesicles 2017, 6, 1333883 10.1080/20013078.2017.1333883. [DOI] [PMC free article] [PubMed] [Google Scholar]